This study used next-generation sequencing to analyze norovirus whole genomes from infected immunocompromised patients. We identified 2 of these patients as linked by the same infection, and analysis of the viral variants showed the direction of transmission between them.
Keywords: norovirus, next-generation sequencing, nosocomial transmission, whole genome
Abstract
Background. Noroviruses are a highly transmissible and major cause of nosocomial gastroenteritis resulting in bed and hospital-ward closures. Where hospital outbreaks are suspected, it is important to determine the routes of spread so that appropriate infection-control procedures can be implemented. To investigate a cluster of norovirus cases occurring in children undergoing bone marrow transplant, we undertook norovirus genome sequencing by next-generation methods. Detailed comparison of sequence data from 2 linked cases enabled us to identify the likely direction of spread.
Methods. Norovirus complementary DNA was amplified by overlapping polymerase chain reaction (PCR) from 13 stool samples from 5 diagnostic real-time PCR–positive patients. The amplicons were sequenced by Roche 454, the genomes assembled by de novo assembly, and the data analyzed phylogenetically.
Results. Phylogenetic analysis indicated that patients were infected by viruses similar to 4 distinct GII.4 subtypes and 2 patients were linked by the same virus. Of the 14 sites at which there were differences between the consensus sequences of the 2 linked viral genomes, 9 had minor variants present within one or the other patient. Further analysis confirmed that minor variants at all 9 sites in patient B were present as the consensus sequence in patient A.
Conclusions. Phylogenetic analysis excluded a common source of infection in this apparent outbreak. Two of 3 patients on the same ward had closely related viruses, raising the possibility of cross-infection despite protective isolation. Analysis of deep sequencing data enabled us to establish the likely direction of nosocomial transmission.
Noroviruses are the leading cause of infectious gastroenteritis in high-income countries, with an estimated 3 million community episodes annually in England and Wales [1]. Within healthcare settings, they account for up to half of all gastroenteritis outbreaks [2–4]. In a previous study, almost half of all norovirus cases in a hospital in the Netherlands were estimated to be hospital acquired [5]. In the absence of therapeutic or preventative options, control of norovirus outbreaks relies on the implementation of infection prevention and control policies including hand washing, environmental cleaning, and exclusion of infected patients. This can result in bed, ward, and institutional closure, with a consequent loss of capacity and significant cost. It has been estimated that 65% of the annual cost of infectious gastroenteritis to the UK National Health Service, approximately £115 million, is attributable to norovirus outbreaks [2]. Classical symptoms of norovirus infection include fever, violent vomiting, and diarrhea. Consequently, environmental contamination is common, and this, together with the low infectious dose [6, 7], high environmental stability [8, 9], and long-term shedding in immunocompromised patients [10], contributes to the ongoing problem of norovirus within healthcare settings.
Norovirus (of the family Caliciviridae) is a single-stranded RNA virus with a genome (7500 nucleotides) typically comprising 3 open reading frames (ORFs). ORF1 encodes a nonstructural polyprotein that includes an RNA-dependent RNA polymerase (RdRP), and ORF2 encodes the major capsid protein VP1. The major capsid protein comprises 2 regions, the shell and protruding domains, with the latter subdivided into the P1 and P2 regions. P2 represents the most exposed domain of the protein and contains histo-blood group antigen (HBGA) binding sites as well as the major antigenic determinants of the capsid [11–13]. ORF3 encodes a protein that is required for viral replication [14]. Noroviruses are classified into 5 genogroups, of which only GI, GII, and GIV infect humans, each of which is further divided into multiple genotypes. Globally, GII.4 is currently the most prevalent genotype, responsible for approximately 60% of norovirus outbreaks [15, 16].
When a cluster of hospital-acquired infections occurs, identification of the sources and routes of nosocomial transmission is required to inform implementation of appropriate infection prevention and control procedures. Here, using next-generation sequencing (Roche 454), we investigate a cluster of norovirus infections that occurred in immunocompromised bone marrow transplant (BMT) patients in a UK hospital in 2010. Unlike Sanger sequencing, which is commonly used to type norovirus infection, next-generation sequencing yields information about the total viral population within an infected individual, and the direction of transmission may be identified by comparing the viral alleles in patients linked by infection.
METHODS
Patients
BMT patients with diarrhea were investigated for infectious and noninfectious causes. All patients were nursed in positive pressure protective isolation cubicles. Beyond routine investigation for bacterial causes of diarrhea, stool samples were examined weekly, by electron microscopy, for the presence of viral particles. Gut biopsy to look for graft-vs-host-disease was performed in cases of unexplained chronic diarrhea. Five BMT patients with congenital immunodeficiency syndromes were identified as norovirus positive following the introduction of a multiplex 1-step diagnostic reverse transcription PCR to target GI [17] and GII [18]. Sampling times, onset of symptoms, and ward location are shown in Figure 1A (patient C was sampled at admission). Information on symptoms among staff and family was collected by the infection control team. Environmental swabbing and PCR for norovirus is carried out when epidemiologic investigations suggest transmission on the ward and evidence is sought to demonstrate a possible environmental reservoir and monitor the cleaning and control processes.
Figure 1.
Analysis of a cluster of 5 norovirus cases in hospitalized patients. A, Timeline of sampling. B, Phylogenetic tree showing the relationships. The tree was rooted with GII.3 (GenBank accession number GU9805851) and nodes supported by a posterior probability ≥0.9 or ≥0.7 are indicated with asterisks and a single dagger, respectively. The gray box highlights patients A and B, who appear linked by the same infection.
Reverse Transcription and PCR
Viral RNA was extracted from 10% (vol/vol) stool suspensions using the Qiagen BioRobot MDx automated workstation. RNA from each stool sample was reverse transcribed using random hexamers and Invitrogen SuperScript III reverse transcriptase.
Norovirus complementary DNA (cDNA) was amplified using 22 published [19] overlapping PCR primer sets optimized to cover the full genome (Supplementary Table 1A and 1B). Each reaction consisted of 0.2 μL high-fidelity KOD polymerase (Merck), 2 μL 10 × KOD buffer, 200 μM deoxyribonucleotide triphosphate, 1.2 μL magnesium sulfate, 1.5 μM of primer pair, and 2 μL cDNA. Following denaturation at 95°C for 2 minutes, each reaction underwent 40 cycles of denaturation at 95°C for 20 seconds, annealing at 50°C for 10 seconds, and extension at 70°C for 40 seconds, followed by a final extension at 70°C for 5 minutes. Amplicons were purified with the Illustra GFX PCR DNA and Gel Band Purification Kit using the manufacturer's instructions. Large (600–1100 bp) and small (300–600 bp) amplicons were pooled separately, with the former sheared mechanically to generate 500-bp fragments. Amplicons from a single sample were tagged with a unique sequence identification to allow multiplex sequencing.
Sequence Assembly and Analysis
Contigs were assembled de novo using Newbler (Roche) and aligned across a reference genome (GenBank accession number GU325839) using MUMmer [20]. Each read-set was realigned against the de novo sequence using SMALT (http://www.sanger.ac.uk/resources/software/smalt; accessed 6 March 2013) to correct for errors, and pileup files were generated using SAMTools [21]. Consensus sequences were called with the QUASR (http://sourceforge.net/projects/quasr/; accessed 24 September 2012) module “pileup Consensus” and a 50% frequency threshold (ie, no ambiguities were included). Full-genome Sanger sequencing of samples B (day 34) and E (day 30) was carried out to confirm the de novo mapping.
Consensus sequences were aligned using MAFFT version 6 [22] and Bayesian phylogenetic trees generated by BEAST version 1.7 [23] under a general-time-reversible model of nucleotide substitution with gamma-shaped rate variation and a proportion of invariable sites (chosen using jModeltest version 0.1.1 [24]). The Markov chain Monte Carlo algorithms were run for 25 000 000 iterations with a thinning of 1000 and checked for convergence (an effective sample size of at least 200 for all parameters). TreeAnnotator version 1.7 (http://beast.bio.ed.ac.uk/TreeAnnotator; accessed 13 May 2013) was used to obtain the tree with the highest clade credibility.
Identification of Transmission
During transmission, only some of the alleles within the donor viral population will establish a new infection in the recipient. Here a minor allele, one that occurs in less than half of the viral genomes, from the donor may by chance become the major allele in the recipient [25]. By comparing allelic data between patients who are linked by the same infection (as shown by a phylogeny), we can trace the direction of transmission in a cluster of linked infections. Variant profiling for each dataset was performed using VarScan version 2.2.11 [26] with the following parameters: basecall quality ≥20, read depth ≥50, reads supporting minor allele ≥2. Variant calls showing directional strand bias ≥0.85 were flagged for manual inspection.
RESULTS
Patient Characteristics
The underlying medical conditions and drug treatment of the patients are shown in Table 1. At the time of sampling, all patients had diarrhea but no vomiting. Patients A, B, D, and E had never previously been nursed on the same ward. Patient A was transferred to ward 1 one week before the onset of symptoms. None of the staff reported symptoms of norovirus infection, although one of patient B's relatives gave a history of having developed mild diarrhea around the same time as patient A.
Table 1.
Patient Characteristics Including Underlying Diagnoses and Drug Treatments
| Characteristic | Patient A | Patient B | Patient C | Patient D | Patient E |
|---|---|---|---|---|---|
| Age | 5 mo | 6 y | 3 mo | 11 y | 5 y |
| Sex | Male | Male | Female | Male | Male |
| Underlying diagnosis | SCID | MUNC-deficient HLH | ADA deficiency | Cartilage hair hypoplasia | IPEX syndrome (FOX P3 mutation) |
| Artificially fed PN | Yes | Yes plus enteral | No | No | Yes |
| GVHD diagnosed on gut biopsy | No | Yes (skin and gut) | NA | No | Yes (gut) |
| Transplant statusa | Post-BMT + 21 d | Post-BMT + 160 d | Pre-BMT –60 d | Post-BMT + 1 d | Post BMT + 210 d |
| Immunosuppressive therapya | MMF, Cyclosporin | Infliximab, methylprednisolone | None | Cyclosporin | Prednisolone, cyclosporin, infliximab |
| Days of diarrhea before first norovirus positive sample | 14 | 160 | 0 | >720 | >365 |
| Duration in hospital before first sample, d | 60 | 200 | 0 | 9 | 350 |
| Inpatient visits prior to current admission | 8 | 27 | 0 | 7 | 2 |
| EM result of stool prior to norovirus positive sample | Negative | Negative | NA | Negative | Negative |
| Clinical outcomes | 100% donor engraftment (immune reconstitution) | Ongoing admissions for pulmonary alveolar proteinosis | 100% donor engraftment (immune reconstitution) | 100% donor engraftment (immune reconstitution) | Died 5 months after sampling from pneumonitis advancing to multiorgan failure |
Abbreviations: ADA, adenosine deaminase; BMT, bone marrow transplant; EM, electron microscopy; GVHD, graft-vs-host-disease; HLH, hemophagocytic lymphohistiocytosis; IPEX, immunodysregulation polyendocrinopathy enteropathy X-linked; MMF, mycophenolate mofetil; MUNC, mammalian uncoordinated; NA, not applicable; PN, parenteral nutrition; SCID, severe combined immunodeficiency .
a At time of first sample.
Phylogenetic Analyses
Near-full-length genome sequences (85.6%–99% coverage) were generated from all 13 samples (GenBank accession numbers KC810020–KC810032). The mean read depth was 423–951 reads per base (Supplementary Table 2). The norovirus genotyping tool (http://www.rivm.nl/mpf/norovirus/typingtool; accessed 7 January 2013) indicated that all of the sequences are GII.4: patients A and B showed homology to Minerva_2006b, which replaced the Hunter strains in the United States and Europe in 2006 [27]. Patient C is similar to New_Orleans_2009, which emerged as the dominant strain in the United States (in place of Minerva) [28, 29]. Patient D was infected by the Grimsby_1995 strain, which was the most prevalent GII.4 norovirus strain across Europe and the United States between 1995 and 2002 [30]. Finally, both samples from patient E showed homology to Yerseke_2006a, which, along with Minerva, replaced the Hunter strains in Europe and the United States [27]. Of the 3 patients on the same ward, viruses from A and B grouped together into a single clade (Figure 1B). The viruses sampled from the other patient cluster separately support the genotyping results.
Analyses of Variation
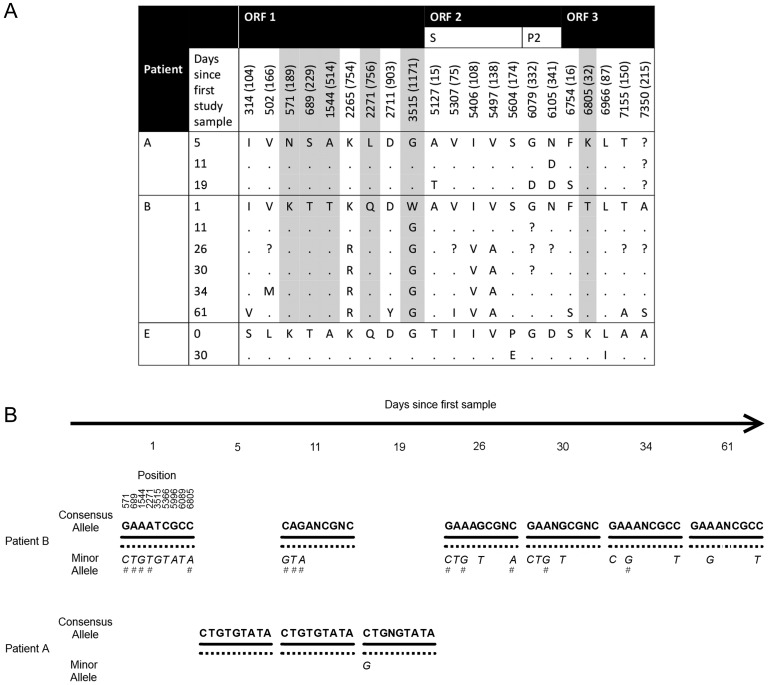
At the consensus level, we identified 1082 sites that were polymorphic across all of the samples (846 were synonymous and 236 were nonsynonymous). These polymorphisms were distributed throughout the genome, although ORF2 showed the highest diversity: 18% of the sites were polymorphic compared with 13% in ORF1 and 16% in ORF3. Of the nonsynonymous polymorphisms in ORF2, 50% occur in the immunogenic P2 region, although there is an excess of synonymous substitutions (67 polymorphic synonymous sites compared to 40). For the patients who were multiply sampled, Figure 2A shows the amino acid variation over time. Temporal changes in the P2 region were only identified for patient A's virus, and none of these coincide with sites that have been documented as hypervariable or antigenically important, or occur in known epitopes [31–37] (although the 341 mutation is next to a site previously linked to an outbreak in 2002 [37]). There were no substitutions in the P2 region for patient B. None of the ORF1 nonsynonymous substitutions in patients A and B occur in previously documented sites. From the longitudinal sampling, we estimated the accumulation rate of amino acid changes in the norovirus genome as 0.47, 0.31, and 0.07 amino acids per day for patients A, B, and E, respectively (0.33, 0.16, and 0.03 amino acids per day for the capsid protein only). The accumulation rates from patients A and B are greater than the rate reported previously in an immunocompromised patient [38], although this may be due to the more frequent sampling in our study. The norovirus substitution rates for patients A, B, and E are estimated as 5.18 × 10−2, 3.10 × 10−2, and 1.93 × 10−2 substitutions per site per year, respectively.
Figure 2.
A, Amino acid variation in longitudinally sampled patients A, B, and E (amino acid position is shown in brackets). Dots indicate no difference with the site in the first sample (from each patient), and “?” indicates missing data. Amino acid sites highlighted in gray were informative for norovirus transmission between patient A and B. B, Norovirus allele profiles in patients A and B indicate that the minor alleles in B are present in patient A in the majority. The hash marks indicate those positions where the minor allelic frequency is ≥5%. Abbreviation: ORF, open reading frame.
Transmission Between Patients A and B
From the phylogeny, patients A and B's viruses are closely linked (Figure 1B). Not all of the samples from the 2 patients cluster separately—fluctuations in the allele frequencies at 2 sites in patient B (571 and 1544) resulted in the consensus sequence for the second sample clustering more closely with those of patient A's virus (Figure 2A and 2B). The virus from patient B was more heterogeneous with between 0.86% and 1.62% biallelic positions compared to between 0.34% to 0.47% in patient A. At the consensus level, the viruses from the first samples from patients A and B differed at 14 positions, of which we observed a minor allele at 9 sites. Six of these loci were in ORF1 (5 nonsynonymous substitutions), 3 synonymous loci in ORF2, and 1 nonsynonymous in ORF3 (Figure 2B). None of these sites coincide with previously documented sites such as RdRP active sites or antigenic sites. The minor alleles present at all 9 sites in patient B were the consensus (major) sequence in patient A. All 6 sequences from patient B had a minor allele present at 2 or more of the 9 positions.
DISCUSSION
Here, we have used next-generation sequencing of whole genomes to elucidate patterns of norovirus transmissions in a clinical setting. We analyzed samples collected from a cluster of BMT inpatients with symptoms of norovirus infection. The phylogenetic patterns observed in our study show different sources of infection for most of our patients, indicating that several strains are cocirculating among temporally linked symptomatic patients. However, 2 patients, despite being nursed in protective isolation, were infected by closely related viruses, suggesting infection from a common source or transmission. Comparison of the next-generation sequence variant data demonstrated that one patient had been infected by the other. Specifically, viral minor alleles in patient B were present as the only allele in patient A, indicating a direction of transmission from B to A. Moreover, the number of biallelic loci in patient A's samples was much lower than that of patient B, which is consistent with a population bottleneck following viral transmission. Additionally, patient B was symptomatic before A was born, lending circumstantial support. Whereas patient B's symptoms may have partly been due to concomitant gut pathology (Table 1), patient A became symptomatic only following admission to ward 1 (Figure 1A), suggesting that the transmitted virus was virulent. To our knowledge, this is the first report of direct transmission of a virulent virus between 2 immunosuppressed patients.
It is unlikely that transmission between patients A and B occurred through an intermediate host as this may have disrupted the variant signal showing cross-infection (due to viral changes in the intermediate host). In particular, infection in immunocompetent hosts has been associated with changes in the P2 region at sites thought to be associated with immune-driven selection [13, 32, 35]. No nonsynonymous changes were observed in P2 between the first samples from patient A and B. Diagnostic norovirus quantitative PCR carried out by the hospital (data not shown) showed contamination of the communal kitchen used by relatives of patients A and B providing a possible environment for transmission. These findings helped to target infection control resources toward improving hygiene among staff and relatives of children on ward 1.
Increased viral heterogeneity in patients with poor immune function has been postulated to act as a reservoir for new variants, some of which could contribute to further outbreaks [39]. In this study, none of the minor alleles at nonsynonymous loci within patient B and which had been transmitted to patient A occurred in previously documented sites, such as HBGA binding. Nonetheless, transmitted variants may have amino acid differences from the dominant viral strain within a donor host that, for example, alter epitopes, allowing pathogen escape from herd immunity. Interestingly, at the consensus level over the entire genome, there was an excess of synonymous site variation including in the immunogenic P2 region. Thus, despite infecting immunocompromised hosts, these viruses appear to be under purifying natural selection; that is, in the context of a virulent virus, such as the strain infecting patients A and B, nonsynonymous changes may be deleterious.
Norovirus genotyping has generally focused on the capsid gene, which has been sufficient to reconstruct phylogenetic relationships and identify linked infections [5, 40]. Although our data showed the same phylogenetic relationships for capsid and whole genome sequences (Supplementary Figure 1), limiting our sequencing to ORF2 would have excluded 4 of the 9 sites that were informative for the direction of transmission. Longer sequences or whole genomes, which include mutations outside ORF2, also provide important phylogenetic data where outbreaks occur over short time periods or for closely related genomes [19].
In summary, we have shown the utility of next-generation sequencing for investigating clusters of infection and highlighting routes of transmission. Variation across the genome, not just in the capsid region, provides useful information for determining direction of transmission. Whereas in this study we have used 22 preoptimized primer sets, our most recent pipeline utilizes substantially fewer primer pairs, which, when combined with bench-top sequencers, can result in turnaround times of 48 hours. This makes the method tractable for management of nosocomial norovirus infections within a clinically relevant timeframe.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online (http://cid.oxfordjournals.org/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. The authors thank K. Green and S. Sosnovtsev for supplying the primer sequences, and M. Cotton and P. Kellam for sequencing assistance. We thank the MRC Centre for Molecular Medical Virology for infrastructure support and Great Ormond Street Hospital Departments of Medical Microbiology and Infection Control for the samples.
Financial support. This work was supported by a fellowship from Sanofi-Pastuer MSD to J. L.; the National Institute for Health Research (NIHR) and University College London/University College London Hospitals (UC/UCLH) Comprehensive Biomedical Research Centre to S. K.; the Medical Research Council Centre for Molecular Medical Virology to D. P. D.; the Wellcome Trust to I. G. and Y. C.; and the UC/UCLH Comprehensive Biomedical Research Centre to J. B. Finally, I. G. acknowledges the support of the Imperial College Healthcare Trust NIHR Biomedical Research Centre.
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Tam CC, Rodrigues LC, Viviani L, et al. Longitudinal study of infectious intestinal disease in the UK (IID2 study): incidence in the community and presenting to general practice. Gut. 2012;61:69–77. doi: 10.1136/gut.2011.238386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lopman B, Vennema H, Kohli E, et al. Increase in viral gastroenteritis outbreaks in Europe and epidemic spread of new norovirus variant. Lancet. 2004;363:682–8. doi: 10.1016/S0140-6736(04)15641-9. [DOI] [PubMed] [Google Scholar]
- 3.Green KY, Belliot G, Taylor JL, et al. A predominant role for Norwalk-like viruses as agents of epidemic gastroenteritis in Maryland nursing homes for the elderly. J Infect Dis. 2002;185:133–46. doi: 10.1086/338365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Billgren M, Christenson B, Hedlund K-O, Vinjé J. Epidemiology of Norwalk-like human caliciviruses in hospital outbreaks of acute gastroenteritis in the Stockholm area in 1996. J Infect. 2002;44:26–32. doi: 10.1053/jinf.2001.0946. [DOI] [PubMed] [Google Scholar]
- 5.Sukhrie FH, Beersma MFC, Wong A, et al. Using molecular epidemiology to trace transmission of nosocomial norovirus infection. J Clin Microbiol. 2011;49:602–6. doi: 10.1128/JCM.01443-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Illingworth E, Taborn E, Fielding D, Cheesbrough J, Diggle PJ, Orr D. Is closure of entire wards necessary to control norovirus outbreaks in hospital? Comparing the effectiveness of two infection control strategies. J Hosp Infect. 2011;79:32–7. doi: 10.1016/j.jhin.2011.04.024. [DOI] [PubMed] [Google Scholar]
- 7.Teunis PFM, Moe CL, Liu P, et al. Norwalk virus: how infectious is it? J Med Virol. 2008;80:1468–76. doi: 10.1002/jmv.21237. [DOI] [PubMed] [Google Scholar]
- 8.Richards GP, Watson MA, Meade GK, Hovan GL, Kingsley DH. Resilience of norovirus GII.4 to freezing and thawing: implications for virus infectivity. Food and Environmental Virology. 2012;4:192–7. doi: 10.1007/s12560-012-9089-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim SJ, Si J, Lee JE, Ko G. Temperature and humidity influences on inactivation kinetics of enteric viruses on surfaces. Environ Sci Technol. 2012;46:13303–10. doi: 10.1021/es3032105. [DOI] [PubMed] [Google Scholar]
- 10.Ludwig A, Adams O, Laws H-J, Schroten H, Tenenbaum T. Quantitative detection of norovirus excretion in pediatric patients with cancer and prolonged gastroenteritis and shedding of norovirus. J Med Virol. 2008;80:1461–7. doi: 10.1002/jmv.21217. [DOI] [PubMed] [Google Scholar]
- 11.Cao S, Lou Z, Tan M, et al. Structural basis for the recognition of blood group trisaccharides by norovirus. J Virol. 2007;81:5949–57. doi: 10.1128/JVI.00219-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen R, Neill JD, Estes MK, Prasad BVV. X-ray structure of a native calicivirus: structural insights into antigenic diversity and host specificity. Proc Natl Acad Sci U S A. 2006;103:8048–53. doi: 10.1073/pnas.0600421103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lochridge VP, Jutila KL, Graff JW, Hardy ME. Epitopes in the P2 domain of norovirus VP1 recognized by monoclonal antibodies that block cell interactions. J Gen Virol. 2005;86:2799–806. doi: 10.1099/vir.0.81134-0. [DOI] [PubMed] [Google Scholar]
- 14.Thorne L, Bailey D, Goodfellow I. High-resolution functional profiling of the norovirus genome. J Virol. 2012;86:11441–56. doi: 10.1128/JVI.00439-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siebenga JJ, Vennema H, Zheng D-P, et al. Norovirus illness is a global problem: emergence and spread of norovirus GII.4 variants, 2001–2007. J Infect Dis. 2009;200:802–12. doi: 10.1086/605127. [DOI] [PubMed] [Google Scholar]
- 16.Zheng D-P, Widdowson M-A, Glass RI, Vinjé J. Molecular epidemiology of genogroup II-genotype 4 noroviruses in the United States between 1994 and 2006. J Clin Microbiol. 2010;48:168–77. doi: 10.1128/JCM.01622-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kageyama T, Kojima S, Shinohara M, et al. Broadly reactive and highly sensitive assay for Norwalk-like viruses based on real-time quantitative reverse transcription-PCR. J Clin Microbiol. 2003;41:1548–57. doi: 10.1128/JCM.41.4.1548-1557.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stals A, Baert L, Botteldoorn N, et al. Multiplex real-time RT-PCR for simultaneous detection of GI/GII noroviruses and murine norovirus 1. J Virol Methods. 2009;161:247–53. doi: 10.1016/j.jviromet.2009.06.019. [DOI] [PubMed] [Google Scholar]
- 19.Bok K, Abente EJ, Realpe-Quintero M, et al. Evolutionary dynamics of GII.4 noroviruses over a 34-year period. J Virol. 2009;83:11890–901. doi: 10.1128/JVI.00864-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Delcher AL, Phillippy A, Carlton J, Salzberg SL. Fast algorithms for large-scale genome alignment and comparison. Nucleic Acids Res. 2002;30:2478–83. doi: 10.1093/nar/30.11.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H, Handsaker B, Wysoker A, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katoh K. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–66. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29:1969–73. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Posada D. jModelTest: phylogenetic model averaging. Mol Biol Evol. 2008;25:1253–6. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- 25.Bull RA, Eden J-S, Luciani F, McElroy K, Rawlinson WD, White PA. Contribution of intra- and interhost dynamics to norovirus evolution. J Virol. 2012;86:3219–29. doi: 10.1128/JVI.06712-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koboldt DC, Zhang Q, Larson DE, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–76. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kroneman A, Vennema H, Harris J, et al. Increase in norovirus activity reported in Europe. Euro Surveill. 2006;11:E061214.1. doi: 10.2807/esw.11.50.03093-en. [DOI] [PubMed] [Google Scholar]
- 28.Yen C, Wikswo ME, Lopman BA, Vinje J, Parashar UD, Hall AJ. Impact of an emergent norovirus variant in 2009 on norovirus outbreak activity in the United States. Clin Infect Dis. 2011;53:568–71. doi: 10.1093/cid/cir478. [DOI] [PubMed] [Google Scholar]
- 29.Vega E, Barclay L, Gregoricus N, Williams K, Lee D, Vinjé J. Novel surveillance network for norovirus gastroenteritis outbreaks, United States. Emerg Infect Dis. 2011;17:1389–95. doi: 10.3201/eid1708.101837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mathijs E, Stals A, Baert L, et al. A review of known and hypothetical transmission routes for noroviruses. Food Environ Virol. 2012;4:131–52. doi: 10.1007/s12560-012-9091-z. [DOI] [PubMed] [Google Scholar]
- 31.Siebenga JJ, Vennema H, Renckens B, et al. Epochal evolution of GGII.4 norovirus capsid proteins from 1995 to 2006. J Virol. 2007;81:9932–41. doi: 10.1128/JVI.00674-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lindesmith LC, Donaldson EF, Baric RS. Norovirus GII.4 strain antigenic variation. J Virol. 2011;85:231–42. doi: 10.1128/JVI.01364-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lindesmith LC, Donaldson EF, Lobue AD, et al. Mechanisms of GII.4 norovirus persistence in human populations. PLoS Med. 2008;5:e31. doi: 10.1371/journal.pmed.0050031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tan M, Huang P, Meller J, Zhong W, Farkas T, Jiang X. Mutations within the P2 domain of norovirus capsid affect binding to human histo-blood group antigens: evidence for a binding pocket. J Virol. 2003;77:12562–71. doi: 10.1128/JVI.77.23.12562-12571.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lindesmith LC, Beltramello M, Donaldson EF, et al. Immunogenetic mechanisms driving norovirus GII .4 antigenic variation. PLoS Pathog. 2012;8:1–18. doi: 10.1371/journal.ppat.1002705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Debbink K, Donaldson EF, Lindesmith LC, Baric RS. Genetic mapping of a highly variable norovirus GII.4 blockade epitope: potential role in escape from human herd immunity. J Virol. 2012;86:1214–26. doi: 10.1128/JVI.06189-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Allen DJ, Gray JJ, Gallimore CI, et al. Analysis of amino acid variation in the P2 domain of the GII-4 norovirus VP1 protein reveals putative variant-specific epitopes. PLoS One. 2008;3:1–9. doi: 10.1371/journal.pone.0001485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schorn R, Höhne M, Meerbach A, et al. Chronic norovirus infection after kidney transplantation: molecular evidence for immune-driven viral evolution. Clin Infect Dis. 2010;51:307–14. doi: 10.1086/653939. [DOI] [PubMed] [Google Scholar]
- 39.Bok K, Green KY. Norovirus gastroenteritis in immunocompromised patients. N Engl J Med. 2012;367:2126–32. doi: 10.1056/NEJMra1207742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Verhoef L, Williams KP, Kroneman A, Sobral B, Van Pelt W, Koopmans M. Selection of a phylogenetically informative region of the norovirus genome for outbreak linkage. Virus Genes. 2012;44:8–18. doi: 10.1007/s11262-011-0673-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.