Abstract
Interferon cytokine family members shape the immune response to protect the host from both pathologic infections and tumorigenesis. To mediate their physiologic function, interferons evoke a robust and complex signal transduction pathway that leads to the induction of interferon-stimulated genes with both proinflammatory and anti-viral function. Numerous mechanisms exist to tightly regulate the extent and duration of these cellular responses. Among such mechanisms, the post-translational conjugation of ubiquitin polypeptides to protein mediators of interferon signaling has emerged as a crucially important mode of control. In this mini-review we highlight recent advances in our understanding of these ubiquitin-mediated mechanisms, their exploitation by invading viruses and their possible utilization for medical intervention.
Key terms: ubiquitin, interferon, E3 ligase, virus
Interferon signaling and protein ubiquitination
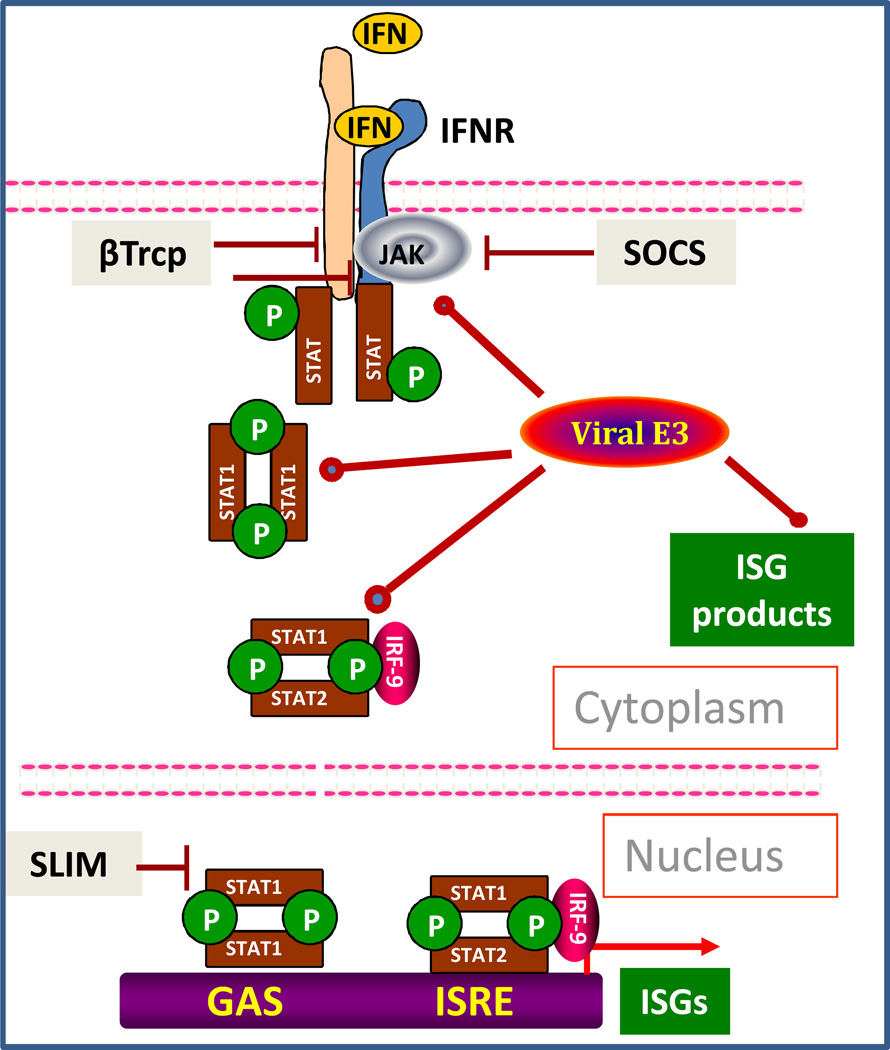
Interferons (IFNs) encompass three cytokine families that play key roles in shaping immune responses that protect the host from pathogenic infection as well as from tumor development (59). Type 1 IFNs (including IFNβ and diverse species of IFNα) are produced by numerous cell types, whereas secretion of Type 2 (IFNγ) and Type 3 IFNs (IFNλ, also termed IL28/29) is largely restricted to immune and epithelial cells, respectively. Each type of IFN interacts with its own cognate receptor (Figure 1). Whereas receptors for Type 1 IFN (consisting of a complex of IFNAR1 and IFNAR2c chains) and Type 2 IFN (a complex of IFNGR1 and IFNGR2) are ubiquitously expressed, not all tissues are responsive to IFNλ (17). Signal transduction downstream of the receptors is similar for Type 1 and Type 3 IFN, and involves activation of JAK1 and TYK2 members of the Janus kinase (JAK) family. JAK activation promotes subsequent tyrosine phosphorylation of signal transduction and activators of transcription (STAT) proteins that form transcriptionally active STAT1 homodimers or STAT1/STAT2/IRF9 complexes (17, 59). Type 2 IFN activates JAK1 and JAK2, ultimately leading to formation of STAT1 homodimers. These transcriptionally active complexes translocate to the nucleus and induce the expression of a diverse family of interferon-stimulated genes (ISGs, Figure 1). Protein products of these genes act in concert to mediate the anti-viral, anti-tumorigenic and immunomodulatory effects of IFNs (17, 59).
Figure 1. IFN signaling and the regulatory role of ubiquitination.
Upon engaging a cognate receptor complex (IFNAR1-IFNAR2 for Type 1 IFN, IFNGR1-IFNGR2 for Type 2 IFN or IL28RA-IL10R2 for Type 3 IFN) IFN activate receptor-associated JAKs. A resulting tyrosine phosphorylation of STAT proteins leads to the formation of transcriptionally active complexes (STAT1 homodimers for all types and additional STAT1-STAT2-IRF9 complexes for Types 1/3 IFN) and ensuing induction of ISGs. Cellular (β-Trcp, SOCS, SLIM) and diverse viral E3 ubiquitin ligases facilitate the ubiquitination of various mediators of IFN signaling pathway to limit the extent and duration of this signaling.
Numerous additional mechanisms play a key role in shaping these pathways and limit their extent (59, 81). While mediating host protection, IFNs also exert negative effects on cell growth, proliferation and viability. Therefore, the extent of IFN signaling is tightly regulated at many levels to limit these detrimental effects. Notably, conjugation of ubiquitin (termed ubiquitination) is of paramount importance in restricting the IFN signaling (29). Ubiquitination involves conjugation of ubiquitin, a small polypeptide, to lysine side chains within the substrate. This reaction is catalyzed by a cascade of enzymatic reactions mediated by ubiquitin-activating (E1), ubiquitin-conjugating (E2) and ubiquitin-ligating (E3) enzymes. E3 ubiquitin ligases recognize specific substrates and determine the efficacy of ubiquitination (16). Importantly, the next round of ubiquitination can attack lysines within ubiquitin (e.g., Lys48 or Lys63 or Lys11) resulting in the formation of polyubiquitin chains. Protein ubiquitination is an important post-translational modification that plays a key role in regulating numerous intracellular signaling pathways and biological processes through both proteolytic and non-proteolytic modes of action (11, 16, 36). For example, Lys-48-linked polyubiquitin targets substrate proteins for proteasomal degradation, whereas Lys63-based chains can stimulate endocytosis and lysosomal degradation of membrane proteins or alternatively contributes to activation of stress-activated protein kinases in response to inflammatory cytokines (reviewed in (11, 16, 36)). The importance of protein ubiquitination in regulating cytokine signaling in general and IFN signaling in particular is underscored by the propensity of tumor cells and viruses to hijack this mode of regulation to evade IFN control and interfere with ability of a host to suppress malignant growth and viral replication (reviewed in (20, 82)).
This mini-review aims to highlight the mechanisms by which protein ubiquitination contributes to the regulation of IFN signaling (Figure 1). Because of size limitations we will focus on IFN signaling per se and will not address production of IFN or the relative importance of other ubiquitin-like proteins (such as ISG15 or Sumo). These areas have been extensively covered in a number of outstanding review articles (5, 32, 37, 68, 87). Wherever possible, we will describe how cellular regulatory mechanisms can be subverted by diverse viruses to evade anti-viral control by IFNs. Viruses may express proteins that activate intracellular signaling that stimulate targeting of IFN pathway mediators for ubiquitination by the cellular E3 ligases. In addition, viruses may express proteins that either act alone or combine cellular proteins to function as unique viral/complex E3 ubiquitin ligases. Examples of the latter scenario are listed in Table 1.
Table 1.
Examples of virus-encoded proteins that function as E3 ubiquitin ligases to target the mediators of IFN pathway (alone or in combination with cellular proteins)
Regulation of IFN receptors stability
Protein ubiquitination has emerged as a prominent mechanism that governs downregulation of signaling receptors. In addition to its impact on intracellular proteasomal degradation of receptor precursors, ubiquitination of receptors localized to the plasma membrane often accelerates receptor internalization and/or directs the post-internalization fate of these receptors towards degradation in the lysosomal compartment (reviewed in (29)).
The IFNAR1 chain of the Type 1 IFN receptor is a key signaling mediator (81) that is often used as model signaling receptor for studies on the role of ubiquitination in endocytosis. A Cullin-based E3 ubiquitin ligase facilitates the ubiquitination and degradation of IFNAR1 (40) following its recruitment through interactions with substrate-recognizing F-box proteins, such as β-Trcp (21). Despite being itself unstable (46), β-Trcp can tether IFNAR1 with three other components of the E3 ligase (proteins Skp1, Cullin1 and Roc1/Rbx1) to facilitate polyubiquitination of IFNAR1 predominantly within a cluster of three lysine residues (38, 39). This polyubiquitination directs already internalized IFNAR1 to the lysosomal compartment, thereby promoting its subsequent degradation (38, 40).
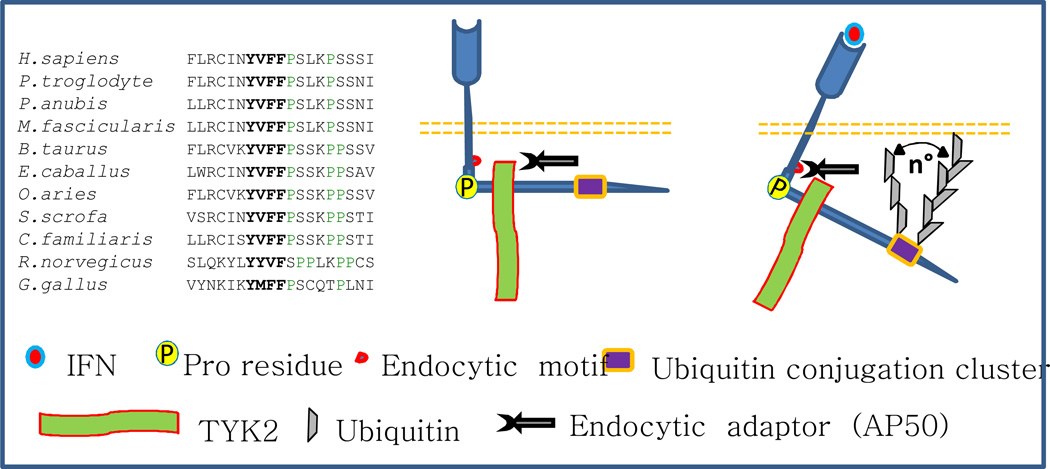
IFNAR1 ubiquitination also robustly stimulates IFNAR1 internalization by exposing its linear endocytic motif so that it can interact with AP50, the endocytic machinery adaptor (Figure 2 and ref. (38)). In the absence of ubiquitination, this motif is masked by associated TYK2, thereby preventing basal endocytosis and degradation of IFNAR1 (25, 41, 64). It is unclear how IFNAR1 ubiquitination unmasks the linear endocytic motif. Given that TYK2 remains associated with IFNAR1 throughout early endocytic events and trafficking stages (56), it is plausible that ubiquitination alters spatial arrangements between the receptor, other participating proteins and the plasma membrane, allowing for functional interactions of the linear motif with endocytic machinery. For example, a highly conserved proline residue following the endocytic motif (e.g. Pro470 in human receptor) likely induces a turn that orients the polypeptide chain of IFNAR1 parallel to the membrane (Figure 2). Subsequent addition of an ubiquitin moiety may alter this orientation, exposing the endocytic motif and enhancing recognition/binding by AP50. Intriguingly, both Lys48- and Lys63-linked polyubiquitin chains contribute significantly to IFNAR1 endocytosis and subsequent degradation (38). Given that these two types of polyubiquitin chains adopt very different conformations (74), interactions between the endocytic motif and endocytic machinery could be augmented and/or stabilized by the presence of both chain types (Figure 2).
Figure 2. A hypothetical mechanism for ubiquitination-mediated stimulation of IFNAR1 internalization.
Sequence alignment of membrane-adjacent proximal fragments of the cytoplasmic tail of IFNAR1 is shown on the left. Tyr-based endocytic motif YXXΨ, where Ψ is a hydrophobic residue, is shown in bold letters. Proline residues conferring the IFNAR1 tail turns are marked by green letters. Figure also shows an unstimulated receptor whose endocytic motif does not interact with endocytic machinery (e.g., AP50 protein) due to spatial interference by associated TYK2. Upon ligand binding and ensuing ubiquitination, a putative conformational change can allow the recognition of the endocytic motif and subsequent internalization. We speculate that formation of two different types of polyubiquitin chains (Lys48- versus Lys63-linked) oriented toward plasma membrane at different angles may stabilize the conformation conducive for endocytosis.
A key event in regulating IFNAR1 ubiquitination is its phosphorylation on specific Ser residues following Type 1 IFN engagement. Such phosphorylation promotes recruitment of β-Trcp and the remaining components of the E3 ubiquitin ligase (39), resulting in receptor downregulation that limits further IFN responses (51, 88, 90). Intriguingly, a number of physiological and/or pathological regulators can trigger phosphorylation-induced IFNAR1 ubiquitination and degradation even in the absence of IFN, thereby desensitizing cells to future encounters with ligand. Notably, signaling through the Bcr-Abl oncogene (4), vascular endothelial growth factor (89), additional inflammatory cytokines (30), or ER stress pathway (48) can all have a negative impact IFN signaling by promoting ubiquitination-dependent receptor internalization.
The ER stress-induced mechanism of IFNAR1 downregulation probably accounts (at least in part) for many instances of widely observed downregulation of IFN signaling following overexpression of viral proteins. In addition to triggering endocytosis and lysosomal degradation of mature receptors, these proteins can also accelerate the proteasomal proteolysis of immature non-glycosylated proteins. For example, the EBV-encoded latent membrane proteins, LMP2A and LMP2B, attenuate signaling by Type 1 IFN by stimulating the intracellular turnover of the receptors precursors (69). Among other viruses implicated in accelerating the turnover of IFNAR1 are herpes simpex virus (63), hepatitis C (3, 15, 48, 60) and B (12) viruses, vesicular stomatitis virus (3, 48), and SARS coronavirus (52).
Little is known about the role of ubiquitin-mediated mechanisms in the downregulation of Type 2 and Type 3 IFN receptors. However, some effects of viruses have been suggested. Expression of the abovementioned EBV proteins, LMP2A/B was suggested to interfere with IFNγ signaling via accelerating turnover of Type 2 IFN receptor precursors (69). In addition, specific RING proteins K3 and K5 (also known as MIR1 and MIR2) encoded by Kaposi’s sarcoma-associated herpesvirus (KSHV) stimulate ubiquitination, endocytosis and degradation of IFNGR1, leading to attenuation of cellular responses to Type 2 IFN (45). Future studies will identify and characterize cellular and viral factors that mediate ubiquitination of IFNγ and IFNλ receptors.
Ubiquitination-dependent regulation of JAK
Given that TYK2 and JAK1 might escort IFNAR1 on its trafficking route to the endosomal compartment (56), it is plausible that these (and perhaps other) JAKs may undergo limited lysosomal co-degradation along with their associated receptors. However, it appears that ubiquitination-mediated proteasomal degradation plays a greater role in regulating JAK stability. For example, RANKL-induced JAK1 proteolysis is required to alleviate the suppression of osteoclastogenesis mediated by IFNβ signaling (43). Mechanistically, RANKL induction of suppressors of cytokine signaling (SOCS1/3) proteins, which function as E3 ubiquitin ligase adaptors, may directly facilitate JAK1 ubiquitination within this pathway. In support, JAK2 ubiquitination and degradation depends on SOCS1 (1, 33, 34, 79). In addition, recent reports also suggest that the Notch-induced Asb2 protein serves as a substrate recognition subunit to recruit Cullin-based E3 ubiquitin ligases to JAK2 independently of SOCS participation (54). Finally, viruses have been implicated in stimulating ubiquitination and degradation of JAKs. For example, human metapneumovirus, responsible for most respiratory infections in infants, inhibits type 1 IFN, at least in part, by promoting proteosomal degradation of JAK1 and TYK2 (66).
Mechanisms of STAT ubiquitination: cellular and viral factors
Although IFN-activated STAT1 was one of the original transcription factors found to be ubiquitinated (35), the identity of cellular E3 ubiquitin ligases that target STAT1 and STAT2 for ubiquitination and the mechanisms regulating these events remain poorly understood. A report that β-Trcp-based E3 ubiquitin ligase may be also involved in ubiquitination and degradation of STAT1 phosphorylated by MAPK (70) remains to be confirmed. It has been proposed that ubiquitination of activated STAT1 in the nucleus is mediated by STAT-interacting LIM (SLIM, currently termed PDLIM2) protein (72, 80). Intriguingly, this ubiquitination and subsequent degradation can be stimulated by exposure of macrophages or mammary epithelial tumor cells to a secreted glycoprotein, osteopontin. SLIM/PDLIM2 was reported to function as a bona fide STAT1 E3 ubiquitin ligase that facilitates STAT1 turnover to negatively regulate IFNγ signaling (22, 23, 27). It is suggested that SLIM/PDLIM2 E3 ligase also targets STAT3 (73).
Viruses are notorious for hijacking cellular factors that target STAT proteins for ubiquitination and degradation in order to evade IFN-imposed restrictions. For example, accelerated turnover of STAT1 is triggered by infection with Sendai virus (24), paramixoviruses (76, 77), and mumps virus (62, 78, 85). STAT2 appears to be a target of ubiquitin-mediated proteasomal degradation stimulated by human cytomegalovirus (42, 75), respiratory syncytial virus (19, 66, 71), paramyxoviruses (61, 76, 77) and Dengue virus (2) among others. Use of virus-encoded proteins to enable the recruitment of STAT1/2 to cellular Cullin-based ubiquitin ligases appears to be a common theme underlying viral targeting of STAT1/2 proteins for proteasomal degradation (65). Although the abovementioned PDLIM2 was shown to associate with the NS1 protein of highly pathogenic avian H5N1 influenza A virus (86), the significance of this interaction for STAT1/2 stability remains to be elucidated.
Examples of regulation of ISGs via ubiquitination
IFN stimulates expression of many genes, and their protein products are likely to be regulated by ubiquitination. For example, IFNs induce expression of MHC Class I and Class II molecules that play a paramount role in immunomodulatory effects of IFN. The robust IFN-induced synthesis and assembly of MHC Class I molecules is equally vigorously counteracted by the ER-associated degradation pathway facilitated by ubiquitination facilitated by HRD1 E3 ubiquitin ligase (10).
The K3 and K5 Kaposi’s virus E3 ligases mentioned above are clearly implicated in downregulation and degradation of both MHC Class I (6–8, 28, 44) and MHC Class II molecules (67). This virus has also been shown to stimulate ubiquitination and degradation of IFN-induced tetherin (18, 50, 55), a cell-surface protein whose expression is induced by IFNα. This protein plays a key role in IFN-induced defenses against retroviruses (47) and human cytomegaloviruses (83). Accelerated degradation of both tetherin and another IFN-induced anti-retroviral protein, APOBEC, has also been described in cells exposed to HIV (18, 26, 49, 53).
Ubiquitin-dependent positive regulation of IFN signaling: an uncharted territory
A paramount role for non-proteolytic ubiquitination in promoting signaling induced by inflammatory cytokines such as interleukin-1 or tumor necrosis factor-α has been uncovered in the past decade and a half (11). A specific role for polyubiquitin chains linked either via internal Lys63 conjugation or in a linear manner following induction of stress-activated protein kinases such as p38 kinase and IκB kinases (IKK) is well supported. Traditionally, IFNs are viewed as important components of the inflammatory cytokine milieu (5, 11), however, there is a relative paucity of data regarding the positive effects of protein ubiquitination on IFN signaling. Although IFN can clearly activate p38 kinases (59) and IKK (57), the mechanisms underlying this activation remain largely unclear. While it has been suggested that TYK2 is decorated by a non-proteolytic Lys63-linked polyubiquitin chain (58), the significance of this modification is yet to be understood.
Whereas TRAF2 E3 ubiquitin ligase (implicated in the ubiquitination-dependent activation of signaling elicited by tumor necrosis factor-α (11)) is recruited to Type 1 IFN receptor (84), the role of this recruitment for the downstream signaling needs to be investigated. In addition, non-canonical mechanisms of IKK activation may contribute to activation of these kinases in response to IFN in a ubiquitination-independent manner. Such mechanisms that may involve either the PKR kinase (9, 13, 31) or phosphoinositol-3 kinase-Akt signaling axis (reviewed in (57)) have been described.
Conclusions and medical significance
As seen from the review of abovementioned literature, the importance of protein ubiquitination as a mechanism for controlling the extent of IFN signaling continues to emerge. In addition to cell-autonomous mechanisms, ubiquitination stimulated by viral factors is of tremendous biologic significance. Besides the negative modes of regulation where ubiquitination limits the extent of IFN signaling by proteolytic degradation of pathway mediators via lysosomal or proteasomal routes, it is plausible that positive regulation may also occur.
Further delineation of these mechanisms is expected to yield novel means for pharmaceutical control of IFN pathways. Intriguingly, both activators and inhibitors of this pathway may find their application in medicine. Given that various types of IFN are widely used for treatment of chronic viral infections and cancers (29, 57, 59), interfering with the ubiquitination-dependent mechanisms of negative regulation is expected to improve the efficacy of these therapies. Conversely, future identification of stimuli/effectors that promote ubiquitination of IFN signaling intermediates may pave the road for novel therapies against autoimmune diseases (e.g., systemic lupus erythematosus) whose pathogenesis is fueled by IFN (14). There is a great deal of hope that these research efforts will come to translational fruition within the next decade or two.
Acknowledgements
The author expresses his sincere apologies to all colleagues whose work was not extensively cited due to space limitations. The editorial help from Dr. L.B. King and support from Mari Lowe Center for Comparative Oncology and NIH/NCI grants CA142425 and CA092900 are gratefully acknowledged.
Footnotes
Declaration of Interest
The author reports no declarations of interest.
REFERENCES
- 1.Ali S, Nouhi Z, Chughtai N. SHP-2 regulates SOCS-1-mediated Janus kinase-2 ubiquitination/degradation downstream of the prolactin receptor. J Biol Chem. 2003;278:52021–52031. doi: 10.1074/jbc.M306758200. [DOI] [PubMed] [Google Scholar]
- 2.Ashour J, Laurent-Rolle M, Shi PY, Garcia-Sastre A. NS5 of dengue virus mediates STAT2 binding and degradation. J Virol. 2009;83:5408–5418. doi: 10.1128/JVI.02188-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhattacharya S, HuangFu WC, Liu J, Veeranki S, Baker DP, Koumenis C, Diehl JA, Fuchs SY. Inducible priming phosphorylation promotes ligand-independent degradation of the IFNAR1 chain of type I interferon receptor. J Biol Chem. 2010;285:2318–2325. doi: 10.1074/jbc.M109.071498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhattacharya S, Zheng H, Tzimas C, Carroll M, Baker DP, Fuchs SY. Bcr-abl signals to desensitize chronic myeloid leukemia cells to IFNalpha via accelerating the degradation of its receptor. Blood. 2011;118:4179–4187. doi: 10.1182/blood-2010-12-325373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bibeau-Poirier A, Servant MJ. Roles of ubiquitination in pattern-recognition receptors and type I interferon receptor signaling. Cytokine. 2008;43:359–367. doi: 10.1016/j.cyto.2008.07.012. [DOI] [PubMed] [Google Scholar]
- 6.Boname JM, de Lima BD, Lehner PJ, Stevenson PG. Viral degradation of the MHC class I peptide loading complex. Immunity. 2004;20:305–317. doi: 10.1016/s1074-7613(04)00047-0. [DOI] [PubMed] [Google Scholar]
- 7.Boname JM, Lehner PJ. What has the study of the K3 and K5 viral ubiquitin E3 ligases taught us about ubiquitin-mediated receptor regulation? Viruses. 2011;3:118–131. doi: 10.3390/v3020118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boname JM, Thomas M, Stagg HR, Xu P, Peng J, Lehner PJ. Efficient internalization of MHC I requires lysine-11 and lysine-63 mixed linkage polyubiquitin chains. Traffic. 2010;11:210–220. doi: 10.1111/j.1600-0854.2009.01011.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonnet MC, Weil R, Dam E, Hovanessian AG, Meurs EF. PKR stimulates NF-kappaB irrespective of its kinase function by interacting with the IkappaB kinase complex. Mol Cell Biol. 2000;20:4532–4542. doi: 10.1128/mcb.20.13.4532-4542.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burr ML, Cano F, Svobodova S, Boyle LH, Boname JM, Lehner PJ. HRD1 and UBE2J1 target misfolded MHC class I heavy chains for endoplasmic reticulum-associated degradation. Proc Natl Acad Sci U S A. 2011;108:2034–2039. doi: 10.1073/pnas.1016229108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen ZJ. Ubiquitin signalling in the NF-kappaB pathway. Nat Cell Biol. 2005;7:758–765. doi: 10.1038/ncb0805-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cho IR, Oh M, Koh SS, Malilas W, Srisuttee R, Jhun BH, Pallegrini S, Fuchs SY, Chung YH. Hepatitis B virus X protein inhibits extracellular IFN-alpha-mediated signal transduction by downregulation of type I IFN receptor. Int J Mol Med. 2012 doi: 10.3892/ijmm.2012.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chu WM, Ostertag D, Li ZW, Chang L, Chen Y, Hu Y, Williams B, Perrault J, Karin M. JNK2 and IKKbeta are required for activating the innate response to viral infection. Immunity. 1999;11:721–731. doi: 10.1016/s1074-7613(00)80146-6. [DOI] [PubMed] [Google Scholar]
- 14.Crow MK, Kirou KA. Interferon-alpha in systemic lupus erythematosus. Curr Opin Rheumatol. 2004;16:541–547. doi: 10.1097/01.bor.0000135453.70424.1b. [DOI] [PubMed] [Google Scholar]
- 15.Datta S, Hazari S, Chandra PK, Samara M, Poat B, Gunduz F, Wimley WC, Hauser H, Koster M, Lamaze C, Balart LA, Garry RF, Dash S. Mechanism of HCV's resistance to IFN-alpha in cell culture involves expression of functional IFN-alpha receptor 1. Virol J. 2011;8:351. doi: 10.1186/1743-422X-8-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Bie P, Ciechanover A. Ubiquitination of E3 ligases: self-regulation of the ubiquitin system via proteolytic and non-proteolytic mechanisms. Cell Death Differ. 2011;18:1393–1402. doi: 10.1038/cdd.2011.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Donnelly RP, Kotenko SV. Interferon-lambda: a new addition to an old family. J Interferon Cytokine Res. 2010;30:555–564. doi: 10.1089/jir.2010.0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Douglas JL, Gustin JK, Viswanathan K, Mansouri M, Moses AV, Fruh K. The great escape: viral strategies to counter BST-2/tetherin. PLoS Pathog. 2010;6:e1000913. doi: 10.1371/journal.ppat.1000913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elliott J, Lynch OT, Suessmuth Y, Qian P, Boyd CR, Burrows JF, Buick R, Stevenson NJ, Touzelet O, Gadina M, Power UF, Johnston JA. Respiratory syncytial virus NS1 protein degrades STAT2 by using the Elongin-Cullin E3 ligase. J Virol. 2007;81:3428–3436. doi: 10.1128/JVI.02303-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fuchs SY. The role of ubiquitin-proteasome pathway in oncogenic signaling. Cancer Biol Ther. 2002;1:337–341. [PubMed] [Google Scholar]
- 21.Fuchs SY, Spiegelman VS, Kumar KG. The many faces of beta-TrCP E3 ubiquitin ligases: reflections in the magic mirror of cancer. Oncogene. 2004;23:2028–2036. doi: 10.1038/sj.onc.1207389. [DOI] [PubMed] [Google Scholar]
- 22.Gao C, Guo H, Mi Z, Grusby MJ, Kuo PC. Osteopontin induces ubiquitin-dependent degradation of STAT1 in RAW264.7 murine macrophages. J Immunol. 2007;178:1870–1881. doi: 10.4049/jimmunol.178.3.1870. [DOI] [PubMed] [Google Scholar]
- 23.Gao C, Mi Z, Guo H, Kuo PC. Osteopontin regulates ubiquitin-dependent degradation of Stat1 in murine mammary epithelial tumor cells. Neoplasia. 2007;9:699–706. doi: 10.1593/neo.07463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garcin D, Marq JB, Strahle L, le Mercier P, Kolakofsky D. All four Sendai Virus C proteins bind Stat1, but only the larger forms also induce its mono-ubiquitination and degradation. Virology. 2002;295:256–265. doi: 10.1006/viro.2001.1342. [DOI] [PubMed] [Google Scholar]
- 25.Gauzzi MC, Barbieri G, Richter MF, Uze G, Ling L, Fellous M, Pellegrini S. The amino-terminal region of Tyk2 sustains the level of interferon alpha receptor 1, a component of the interferon alpha/beta receptor. Proc Natl Acad Sci U S A. 1997;94:11839–11844. doi: 10.1073/pnas.94.22.11839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goila-Gaur R, Strebel K. HIV-1 Vif, APOBEC, and intrinsic immunity. Retrovirology. 2008;5:51. doi: 10.1186/1742-4690-5-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo H, Mi Z, Bowles DE, Bhattacharya SD, Kuo PC. Osteopontin and protein kinase C regulate PDLIM2 activation and STAT1 ubiquitination in LPS-treated murine macrophages. J Biol Chem. 2010;285:37787–37796. doi: 10.1074/jbc.M110.161869. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Hewitt EW, Duncan L, Mufti D, Baker J, Stevenson PG, Lehner PJ. Ubiquitylation of MHC class I by the K3 viral protein signals internalization and TSG101-dependent degradation. EMBO J. 2002;21:2418–2429. doi: 10.1093/emboj/21.10.2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huangfu WC, Fuchs SY. Ubiquitination-dependent regulation of signaling receptors in cancer. Genes Cancer. 2010;1:725–734. doi: 10.1177/1947601910382901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huangfu WC, Qian J, Liu C, Liu J, Lokshin AE, Baker DP, Rui H, Fuchs SY. Inflammatory signaling compromises cell responses to interferon alpha. Oncogene. 2011 doi: 10.1038/onc.2011.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ishii T, Kwon H, Hiscott J, Mosialos G, Koromilas AE. Activation of the I kappa B alpha kinase (IKK) complex by double-stranded RNA-binding defective 11 and catalytic inactive mutants of the interferon-inducible protein kinase PKR. Oncogene. 2001;20:1900–1912. doi: 10.1038/sj.onc.1204267. [DOI] [PubMed] [Google Scholar]
- 32.Jackson PK. A new RING for SUMO: wrestling transcriptional responses into nuclear bodies with PIAS family E3 SUMO ligases. Genes Dev. 2001;15:3053–3058. doi: 10.1101/gad.955501. [DOI] [PubMed] [Google Scholar]
- 33.Kamizono S, Hanada T, Yasukawa H, Minoguchi S, Kato R, Minoguchi M, Hattori K, Hatakeyama S, Yada M, Morita S, Kitamura T, Kato H, Nakayama K, Yoshimura A. The SOCS box of SOCS-1accelerates ubiquitin-dependent proteolysis of TEL-JAK2. J Biol Chem. 2001;276:12530–12538. doi: 10.1074/jbc.M010074200. [DOI] [PubMed] [Google Scholar]
- 34.Kamura T, Sato S, Haque D, Liu L, Kaelin WG, Jr, Conaway RC, Conaway JW. The Elongin BC complex interacts with the conserved SOCS-box motif present in members of the SOCS, ras, WD-40 repeat, and ankyrin repeat families. Genes Dev. 1998;12:3872–3881. doi: 10.1101/gad.12.24.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim TK, Maniatis T. Regulation of interferon-gamma-activated STAT1 by the ubiquitin-proteasome pathway. Science. 1996;273:1717–1719. doi: 10.1126/science.273.5282.1717. [DOI] [PubMed] [Google Scholar]
- 36.Kirkin V, Dikic I. Ubiquitin networks in cancer. Curr Opin Genet Dev. 2011;21:21–28. doi: 10.1016/j.gde.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 37.Komuro A, Bamming D, Horvath CM. Negative regulation of cytoplasmic RNA-mediated antiviral signaling. Cytokine. 2008;43:350–358. doi: 10.1016/j.cyto.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kumar KG, Barriere H, Carbone CJ, Liu J, Swaminathan G, Xu P, Li Y, Baker DP, Peng J, Lukacs GL, Fuchs SY. Site-specific ubiquitination exposes a linear motif to promote interferon-alpha receptor endocytosis. J Cell Biol. 2007;179:935–950. doi: 10.1083/jcb.200706034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kumar KG, Krolewski JJ, Fuchs SY. Phosphorylation and specific ubiquitin acceptor sites are required for ubiquitination and degradation of the IFNAR1 subunit of type I interferon receptor. J Biol Chem. 2004;279:46614–46620. doi: 10.1074/jbc.M407082200. [DOI] [PubMed] [Google Scholar]
- 40.Kumar KG, Tang W, Ravindranath AK, Clark WA, Croze E, Fuchs SY. SCF(HOS) ubiquitin ligase mediates the ligand-induced down-regulation of the interferon-alpha receptor. EMBO J. 2003;22:5480–5490. doi: 10.1093/emboj/cdg524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kumar KG, Varghese B, Banerjee A, Baker DP, Constantinescu SN, Pellegrini S, Fuchs SY. Basal ubiquitin-independent internalization of interferon alpha receptor is prevented by Tyk2-mediated masking of a linear endocytic motif. J Biol Chem. 2008;283:18566–18572. doi: 10.1074/jbc.M800991200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Le VT, Trilling M, Wilborn M, Hengel H, Zimmermann A. Human cytomegalovirus interferes with signal transducer and activator of transcription (STAT) 2 protein stability and tyrosine phosphorylation. J Gen Virol. 2008;89:2416–2426. doi: 10.1099/vir.0.2008/001669-0. [DOI] [PubMed] [Google Scholar]
- 43.Lee Y, Hyung SW, Jung HJ, Kim HJ, Staerk J, Constantinescu SN, Chang EJ, Lee ZH, Lee SW, Kim HH. The ubiquitin-mediated degradation of Jak1 modulates osteoclastogenesis by limiting interferon-beta-induced inhibitory signaling. Blood. 2008;111:885–893. doi: 10.1182/blood-2007-03-082941. [DOI] [PubMed] [Google Scholar]
- 44.Lehner PJ, Hoer S, Dodd R, Duncan LM. Downregulation of cell surface receptors by the K3 family of viral and cellular ubiquitin E3 ligases. Immunol Rev. 2005;207:112–125. doi: 10.1111/j.0105-2896.2005.00314.x. [DOI] [PubMed] [Google Scholar]
- 45.Li Q, Means R, Lang S, Jung JU. Downregulation of gamma interferon receptor 1 by Kaposi's sarcoma-associated herpesvirus K3 and K5. J Virol. 2007;81:2117–2127. doi: 10.1128/JVI.01961-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Y, Gazdoiu S, Pan ZQ, Fuchs SY. Stability of homologue of Slimb F-box protein is regulated by availability of its substrate. J Biol Chem. 2004;279:11074–11080. doi: 10.1074/jbc.M312301200. [DOI] [PubMed] [Google Scholar]
- 47.Liberatore RA, Bieniasz PD. Tetherin is a key effector of the antiretroviral activity of type I interferon in vitro and in vivo. Proc Natl Acad Sci U S A. 2011;108:18097–18101. doi: 10.1073/pnas.1113694108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu J, HuangFu WC, Kumar KG, Qian J, Casey JP, Hamanaka RB, Grigoriadou C, Aldabe R, Diehl JA, Fuchs SY. Virus-induced unfolded protein response attenuates antiviral defenses via phosphorylation-dependent degradation of the type I interferon receptor. Cell Host Microbe. 2009;5:72–83. doi: 10.1016/j.chom.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mangeat B, Gers-Huber G, Lehmann M, Zufferey M, Luban J, Piguet V. HIV-1 Vpu neutralizes the antiviral factor Tetherin/BST-2 by binding it and directing its beta-TrCP2-dependent degradation. PLoS Pathog. 2009;5:e1000574. doi: 10.1371/journal.ppat.1000574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mansouri M, Viswanathan K, Douglas JL, Hines J, Gustin J, Moses AV, Fruh K. Molecular mechanism of BST2/tetherin downregulation by K5/MIR2 of Kaposi's sarcoma-associated herpesvirus. J Virol. 2009;83:9672–9681. doi: 10.1128/JVI.00597-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marijanovic Z, Ragimbeau J, Kumar KG, Fuchs SY, Pellegrini S. TYK2 activity promotes ligand-induced IFNAR1 proteolysis. Biochem J. 2006;397:31–38. doi: 10.1042/BJ20060272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Minakshi R, Padhan K, Rani M, Khan N, Ahmad F, Jameel S. The SARS Coronavirus 3a protein causes endoplasmic reticulum stress and induces ligand-independent downregulation of the type 1 interferon receptor. PLoS One. 2009;4:e8342. doi: 10.1371/journal.pone.0008342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mitchell RS, Katsura C, Skasko MA, Fitzpatrick K, Lau D, Ruiz A, Stephens EB, Margottin-Goguet F, Benarous R, Guatelli JC. Vpu antagonizes BST-2-mediated restriction of HIV-1 release via beta-TrCP and endo-lysosomal trafficking. PLoS Pathog. 2009;5:e1000450. doi: 10.1371/journal.ppat.1000450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nie L, Zhao Y, Wu W, Yang YZ, Wang HC, Sun XH. Notch-induced Asb2 expression promotes protein ubiquitination by forming non-canonical E3 ligase complexes. Cell Res. 2011;21:754–769. doi: 10.1038/cr.2010.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pardieu C, Vigan R, Wilson SJ, Calvi A, Zang T, Bieniasz P, Kellam P, Towers GJ, Neil SJ. The RING-CH ligase K5 antagonizes restriction of KSHV and HIV-1 particle release by mediating ubiquitin-dependent endosomal degradation of tetherin. PLoS Pathog. 2010;6:e1000843. doi: 10.1371/journal.ppat.1000843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Payelle-Brogard B, Pellegrini S. Biochemical monitoring of the early endocytic traffic of the type I interferon receptor. J Interferon Cytokine Res. 2010;30:89–98. doi: 10.1089/jir.2009.0044. [DOI] [PubMed] [Google Scholar]
- 57.Pfeffer LM. The role of nuclear factor kappaB in the interferon response. J Interferon Cytokine Res. 2011;31:553–559. doi: 10.1089/jir.2011.0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Piganis RA, De Weerd NA, Gould JA, Schindler CW, Mansell A, Nicholson SE, Hertzog PJ. Suppressor of cytokine signaling (SOCS) 1 inhibits type I interferon (IFN) signaling via the interferon alpha receptor (IFNAR1)-associated tyrosine kinase Tyk2. J Biol Chem. 2011;286:33811–33818. doi: 10.1074/jbc.M111.270207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Platanias LC. Mechanisms of type-I-and type-II-interferon-mediated signalling. Nat Rev Immunol. 2005;5:375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- 60.Poat B, Hazari S, Chandra PK, Gunduz F, Balart LA, Alvarez X, Dash S. SH2 modified STAT1 induces HLA-I expression and improves IFN-gamma signaling in IFN-alpha resistantHCV replicon cells. PLoS One. 2010;5 doi: 10.1371/journal.pone.0013117. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 61.Precious B, Childs K, Fitzpatrick-Swallow V, Goodbourn S, Randall RE. Simian virus 5 V protein acts as an adaptor, linking DDB1 to STAT2, to facilitate the ubiquitination of STAT1. J Virol. 2005;79:13434–13441. doi: 10.1128/JVI.79.21.13434-13441.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Puri M, Lemon K, Duprex WP, Rima BK, Horvath CM. A point mutation, E95D, in the mumps virus V protein disengages STAT3 targeting from STAT1 targeting. J Virol. 2009;83:6347–6356. doi: 10.1128/JVI.00596-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Qian J, Zheng H, Huangfu WC, Liu J, Carbone CJ, Leu NA, Baker DP, Fuchs SY. Pathogen recognition receptor signaling accelerates phosphorylation-dependent degradation of IFNAR1. PLoS Pathog. 2011;7:e1002065. doi: 10.1371/journal.ppat.1002065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ragimbeau J, Dondi E, Alcover A, Eid P, Uze G, Pellegrini S. The tyrosine kinase Tyk2 controls IFNAR1 cell surface expression. EMBO J. 2003;22:537–547. doi: 10.1093/emboj/cdg038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ramachandran A, Horvath CM. Paramyxovirus disruption of interferon signal transduction: STATus report. J Interferon Cytokine Res. 2009;29:531–537. doi: 10.1089/jir.2009.0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ren J, Kolli D, Liu T, Xu R, Garofalo RP, Casola A, Bao X. Human metapneumovirus inhibits IFN-beta signaling by downregulating Jak1 and Tyk2 cellular levels. PLoS One. 2011;6:e24496. doi: 10.1371/journal.pone.0024496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schmidt K, Wies E, Neipel F. Kaposi's sarcoma-associated herpesvirus viral interferon regulatory factor 3 inhibits gamma interferon and major histocompatibility complex class II expression. J Virol. 2011;85:4530–4537. doi: 10.1128/JVI.02123-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Seth RB, Sun L, Chen ZJ. Antiviral innate immunity pathways. Cell Res. 2006;16:141–147. doi: 10.1038/sj.cr.7310019. [DOI] [PubMed] [Google Scholar]
- 69.Shah KM, Stewart SE, Wei W, Woodman CB, O'Neil JD, Dawson CW, Young LS. The EBV-encoded latent membrane proteins, LMP2A and LMP2B, limit the actions of interferon by targeting interferon receptors for degradation. Oncogene. 2009;28:3903–3914. doi: 10.1038/onc.2009.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Soond SM, Townsend PA, Barry SP, Knight RA, Latchman DS, Stephanou A. ERK and the F-box protein betaTRCP target STAT1 for degradation. J Biol Chem. 2008;283:16077–16083. doi: 10.1074/jbc.M800384200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 71.Straub CP, Lau WH, Preston FM, Headlam MJ, Gorman JJ, Collins PL, Spann KM. Mutation of the elongin C binding domain of human respiratory syncytial virus non-structural protein 1 (NS1) results in degradation of NS1 and attenuation of the virus. Virol J. 2011;8:252. doi: 10.1186/1743-422X-8-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tanaka T, Soriano MA, Grusby MJ. SLIM is a nuclear ubiquitin E3 ligase that negatively regulates STAT signaling. Immunity. 2005;22:729–736. doi: 10.1016/j.immuni.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 73.Tanaka T, Yamamoto Y, Muromoto R, Ikeda O, Sekine Y, Grusby MJ, Kaisho T, Matsuda T. PDLIM2 Inhibits T Helper 17 Cell Development and Granulomatous Inflammation Through Degradation of STAT3. Sci Signal. 2011;4:ra85. doi: 10.1126/scisignal.2001637. [DOI] [PubMed] [Google Scholar]
- 74.Tenno T, Fujiwara K, Tochio H, Iwai K, Morita EH, Hayashi H, Murata S, Hiroaki H, Sato M, Tanaka K, Shirakawa M. Structural basis for distinct roles of Lys63-and Lys48-linked polyubiquitin chains. Genes Cells. 2004;9:865–875. doi: 10.1111/j.1365-2443.2004.00780.x. [DOI] [PubMed] [Google Scholar]
- 75.Trilling M, Le VT, Fiedler M, Zimmermann A, Bleifuss E, Hengel H. Identification of DNA-damage DNA-binding protein 1 as a conditional essential factor for cytomegalovirus replication in interferon-gamma-stimulated cells. PLoS Pathog. 2011;7:e1002069. doi: 10.1371/journal.ppat.1002069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ulane CM, Horvath CM. Paramyxoviruses SV5 and HPIV2 assemble STAT protein ubiquitin ligase complexes from cellular components. Virology. 2002;304:160–166. doi: 10.1006/viro.2002.1773. [DOI] [PubMed] [Google Scholar]
- 77.Ulane CM, Kentsis A, Cruz CD, Parisien JP, Schneider KL, Horvath CM. Composition and assembly of STAT-targeting ubiquitin ligase complexes: paramyxovirus V protein carboxyl terminus is an oligomerization domain. J Virol. 2005;79:10180–10189. doi: 10.1128/JVI.79.16.10180-10189.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ulane CM, Rodriguez JJ, Parisien JP, Horvath CM. STAT3 ubiquitylation and degradation by mumps virus suppress cytokine and oncogene signaling. J Virol. 2003;77:6385–6393. doi: 10.1128/JVI.77.11.6385-6393.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ungureanu D, Saharinen P, Junttila I, Hilton DJ, Silvennoinen O. Regulation of Jak2 through the ubiquitin-proteasome pathway involves phosphorylation of Jak2 on Y1007 and interaction with SOCS-1. Mol Cell Biol. 2002;22:3316–3326. doi: 10.1128/MCB.22.10.3316-3326.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ungureanu D, Silvennoinen O. SLIM trims STATs: ubiquitin E3 ligases provide insights for specificity in the regulation of cytokine signaling. Sci STKE. 2005;2005:pe49. doi: 10.1126/stke.3042005pe49. [DOI] [PubMed] [Google Scholar]
- 81.Uze G, Schreiber G, Piehler J, Pellegrini S. The receptor of the type I interferon family. Curr Top Microbiol Immunol. 2007;316:71–95. doi: 10.1007/978-3-540-71329-6_5. [DOI] [PubMed] [Google Scholar]
- 82.Viswanathan K, Fruh K, DeFilippis V. Viral hijacking of the host ubiquitin system to evade interferon responses. Curr Opin Microbiol. 2010;13:517–523. doi: 10.1016/j.mib.2010.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Viswanathan K, Smith MS, Malouli D, Mansouri M, Nelson JA, Fruh K. BST2/Tetherin enhances entry of human cytomegalovirus. PLoS Pathog. 2011;7:e1002332. doi: 10.1371/journal.ppat.1002332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yang CH, Murti A, Pfeffer SR, Fan M, Du Z, Pfeffer LM. The role of TRAF2 binding to the type I interferon receptor in alternative NF kappaB activation and antiviral response. J Biol Chem. 2008;283:14309–14316. doi: 10.1074/jbc.M708895200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yokosawa N, Yokota S, Kubota T, Fujii N. C-terminal region of STAT-1alpha is not necessary for its ubiquitination and degradation caused by mumps virus V protein. J Virol. 2002;76:12683–12690. doi: 10.1128/JVI.76.24.12683-12690.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yu J, Li X, Wang Y, Li B, Li H, Li Y, Zhou W, Zhang C, Rao Z, Bartlam M, Cao Y. PDlim2 selectively interacts with the PDZ binding motif of highly pathogenic avian H5N1 influenza A virus NS1. PLoS One. 2011;6:e19511. doi: 10.1371/journal.pone.0019511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang D, Zhang DE. Interferon-stimulated gene 15 and the protein ISGylation system. J Interferon Cytokine Res. 2011;31:119–130. doi: 10.1089/jir.2010.0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zheng H, Qian J, Baker DP, Fuchs SY. Tyrosine phosphorylation of protein kinase D2 mediates ligand-inducible elimination of the Type 1 interferon receptor. J Biol Chem. 2011;286:35733–35741. doi: 10.1074/jbc.M111.263608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zheng H, Qian J, Carbone CJ, Leu NA, Baker DP, Fuchs SY. Vascular endothelial growth factor-induced elimination of the type 1 interferon receptor is required for efficient angiogenesis. Blood. 2011;118:4003–4006. doi: 10.1182/blood-2011-06-359745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zheng H, Qian J, Varghese B, Baker DP, Fuchs S. Ligand-stimulated downregulation of the alpha interferon receptor: role of protein kinase D2. Mol Cell Biol. 2011;31:710–720. doi: 10.1128/MCB.01154-10. [DOI] [PMC free article] [PubMed] [Google Scholar]