Abstract
Hepatocellular carcinoma (HCC) affects more than half a million people worldwide and is the third most common cause of cancer deaths. Because mammalian target of rapamycin (mTOR) signaling is up-regulated in 50% of HCCs, we compared the effects of the U.S. Food and Drug Administration–approved mTOR-allosteric inhibitor, RAD001, with a new-generation phosphatidylinositol 3-kinase/mTOR adenosine triphosphate–site competitive inhibitor, BEZ235. Unexpectedly, the two drugs acted synergistically in inhibiting the proliferation of cultured HCC cells. The synergistic effect closely paralleled eukaryotic initiation factor 4E-binding protein 1 (4E-BP1) dephosphorylation, which is implicated in the suppression of tumor cell proliferation. In a mouse model approximating human HCC, the drugs in combination, but not singly, induced a marked regression in tumor burden. However, in the tumor, BEZ235 alone was as effective as the combination in inhibiting 4E-BP1 phosphorylation, which suggests that additional target(s) may also be involved. Microarray analyses revealed a large number of genes that reverted to normal liver tissue expression in mice treated with both drugs, but not either drug alone. These analyses also revealed the down-regulation of autophagy genes in tumors compared to normal liver. Moreover, in HCC patients, altered expression of autophagy genes was associated with poor prognosis. Consistent with these findings, the drug combination had a profound effect on UNC51-like kinase 1 (ULK1) dephosphorylation and autophagy in culture, independent of 4E-BP1, and in parallel induced tumor mitophagy, a tumor suppressor process in liver. These observations have led to an investigator-initiated phase 1B-2 dose escalation trial with RAD001 combined with BEZ235 in patients with HCC and other advanced solid tumors.
INTRODUCTION
Hepatocellular carcinoma (HCC) is the fifth most common cause of cancer and—because of late diagnosis, poor treatment options, and aggressive disease—ranks third in cancer deaths (1). Most patients present with intermediate- or advanced-stage disease, and surgical resection is an option for less than 20% of these patients (2). Although the number of HCC cases in North America is relatively small, it is the most rapidly expanding tumor type (3, 4). Two-thirds of these cases are attributed to chronic alcohol use, exposure to toxic agents, or prolonged hepatitis B or C infection (5); however, the remaining third have been linked to nonalcoholic steatohepatitis, most likely driven by the recent epidemic in obesity.
Currently, sorafenib, a multiprotein kinase inhibitor, shows unprecedented clinical response in HCC patients (6, 7). However, the response is not enduring, underscoring the need for novel therapies. One candidate target that has emerged is the mammalian target of rapamycin (mTOR) signaling pathway, which is hyperactivated in 40 to 50% of HCC cases. Moreover, recent studies have shown that HCC incidence and progression are significantly augmented by a high-fat diet (8), which is known to lead to an increase in circulating branched-chain amino acids (BCAAs) and induction of mTOR signaling independent of phosphatidylinositol 3-kinase (PI3K) signaling (9, 10). On the basis of these observations, rapamycin and two derivatives, everolimus (RAD001) and temsirolimus (CCI-779), are under evaluation in phase 1, 1–2, 2, 2–3, and 3 clinical trials for the treatment of HCC (11).
mTOR can be found in two multiprotein kinase complexes: mTORC1 and mTORC2. Both complexes contain mLST8 and a number of distinct interacting proteins, including raptor and rictor, which define mTORC1 and mTORC2, respectively. Although both complexes respond to hormones and mitogens, only mTORC1 responds to nutrients, including BCAAs, and cellular energy inputs (9). Mitogens initiate mTORC1 signaling by the canonical PI3K/protein kinase B (PKB/Akt) pathway (12, 13). The most studied effectors downstream of mTORC1 are the ribosomal protein S6 kinases (S6K1/2) and the eukaryotic protein synthesis initiation factor 4E-binding proteins (4E-BP1/2). mTORC2 mediates activation of PKB/Akt and serum/glucocorticoidregulated kinase 1. The mTOR complexes are key regulators of multiple cellular processes including translation, growth, proliferation, metabolism, and autophagy (14, 15).
The rapamycins form a complex with the immunophilin FKBP12, which binds to an allosteric site near the kinase domain to inhibit mTOR signaling. Mutation of a single residue in the rapamycin- FKBP12 binding site confers complete resistance (16). Although the rapamycins are used clinically, they potentiate PI3K activation through inhibition of the mTORC1/S6K1 negative feedback loop (17, 18) and incompletely suppress mTORC1 signaling to 4E-BP1 (19). Therefore, we chose an mTOR adenosine triphosphate (ATP)–site competitive inhibitor to test efficacy in the treatment of HCC. We made the unexpected observation that RAD001 and BEZ235 synergized at low doses on mTORC1 and mTORC2, causing tumor regression in mouse models best approximating human HCC (20, 21). Moreover, this effect was associated with a marked increase in autophagy, which correlated with UNC51-like kinase 1 (ULK1) dephosphorylation in cell culture, independent of S6K1 or 4E-BP1.
RESULTS
RAD001 and BEZ235 synergistically inhibit mTOR signaling in HCC cells
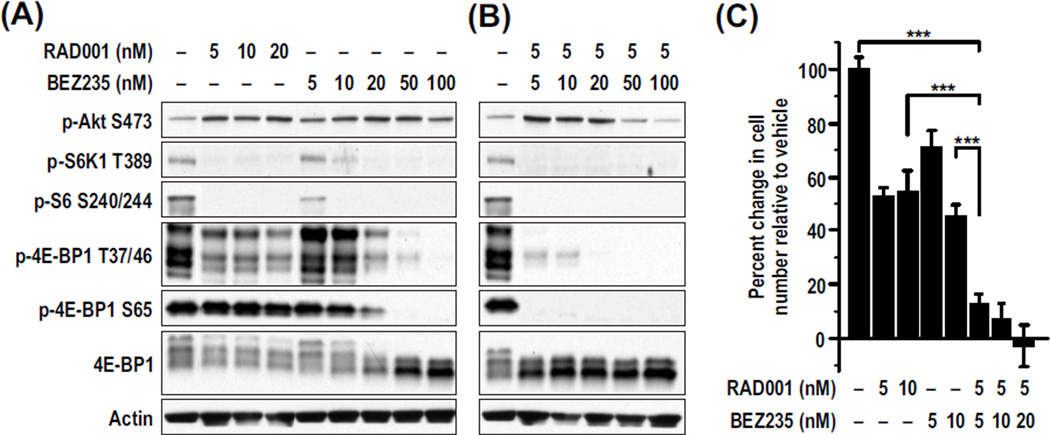
Treatment of the human HCC cell line Huh7 with 5 nM RAD001abolished S6K1 activation, as measured by S6K1 Thr389 (T389) and S6 Ser240/Ser244 (S240/244) phosphorylation (Fig. 1A). This treatment was associated with an approximate threefold increase in PKB/Akt S473 phosphorylation because of suppression of the mTORC1/S6K1 negative feedback loop (18) (Fig. 1A). RAD001 had some effect on 4E-BP1 T37/46 but virtually no effect on S65 phosphorylation (Fig. 1A). BEZ235 treatment also led to inhibition of S6K1 T389 phosphorylation and an approximate threefold potentiation of PKB/Akt S473 phosphorylation, consistent with mTORC1/S6K1 inhibition (Fig. 1A). However, at doses of 100 nM BEZ235, mTORC2 began to be inhibited, as evidenced by PKB/Akt S473 dephosphorylation. Unlike RAD001, BEZ235 caused both S6K1 and 4E-BP1 dephosphorylation (Fig. 1A). These data suggest that low BEZ235 concentrations selectively inhibit mTORC1 and the negative feedback loop, but higher BEZ235 concentrations inhibit both mTORC1 and mTORC2.
Fig. 1.
Huh7 RAD001 and BEZ235 synergistically inhibit mTOR signaling and decrease proliferation in Huh7 cells. Cells were treated with the indicated concentrations of RAD001 and/or BEZ235 for 48 hours. (A and B) Amounts of phosphorylated and total proteins were measured in cell lysates by Western blot analysis. Actin was used as a loading control. (C) Cell proliferation was measured using the CellTiter-Glo luminescent assay. The percent change in cell number from the beginning of the treatment relative to vehicle is plotted. Results are means ± SD (n = 3). ***P < 0.0001.
To test the effect of the two drugs together, we kept the RAD001 concentration at 5 nM and gradually increased the BEZ235 concentration. Unexpectedly, at 5 nM BEZ235, phosphorylation of 4E-BP1 S65 and T37/46 was largely abolished in the presence of RAD001 (Fig. 1B), an effect requiring 50 nM BEZ235 when used alone (Fig. 1A). Moreover, the potentiation of PKB/Akt S473 phosphorylation was blunted at 50 nM BEZ235 in combination with 5 nM RAD001, whereas this was not observed when BEZ235 was used alone at 50 nM (compare Fig. 1, A and B). These findings indicate that the two drugs act synergistically to inhibit both mTORC1 and mTORC2 signaling. Next, we determined whether the effects of drug treatment on cell proliferation more closely followed 4E-BP1 or PKB/Akt dephosphorylation. RAD001 alone at all concentrations tested inhibited cell proliferation by about 50%, whereas BEZ235 caused a dose-dependent inhibition of proliferation, reaching a maximum at 100 nM (Fig. 1C). In combination, proliferation was almost completely abolished at the lowest concentration of each drug, 5 nM (Fig. 1C). Using the Chou-Talalay equation (22), we achieved synergy at 5 nM RAD001 with either 5 or 10 nm BEZ235 (table S1), with inhibition of proliferation closely paralleled by 4E-BP1 dephosphorylation (Fig. 1B). The parallel effects on 4E-BP1 dephosphorylation and cell proliferation are not cell line–dependent, because synergy was also observed in the human HCC Alexander cell line and mouse HCC cell lines derived from either a primary diethylnitrosamine (DEN)–induced tumor (8) or a transgenic E2F1-induced tumor (23), although at different concentrations (fig. S1, A to C). These observations suggest that the effects of RAD001 in combination with BEZ235 more closely follow the inhibition of mTORC1 than mTORC2, on the basis of 4E-BP1 phosphorylation compared to that of PKB/Akt.
RAD001 and BEZ235 synergistically inhibit the in vitrokinase activity of mTORC1 and mTORC2
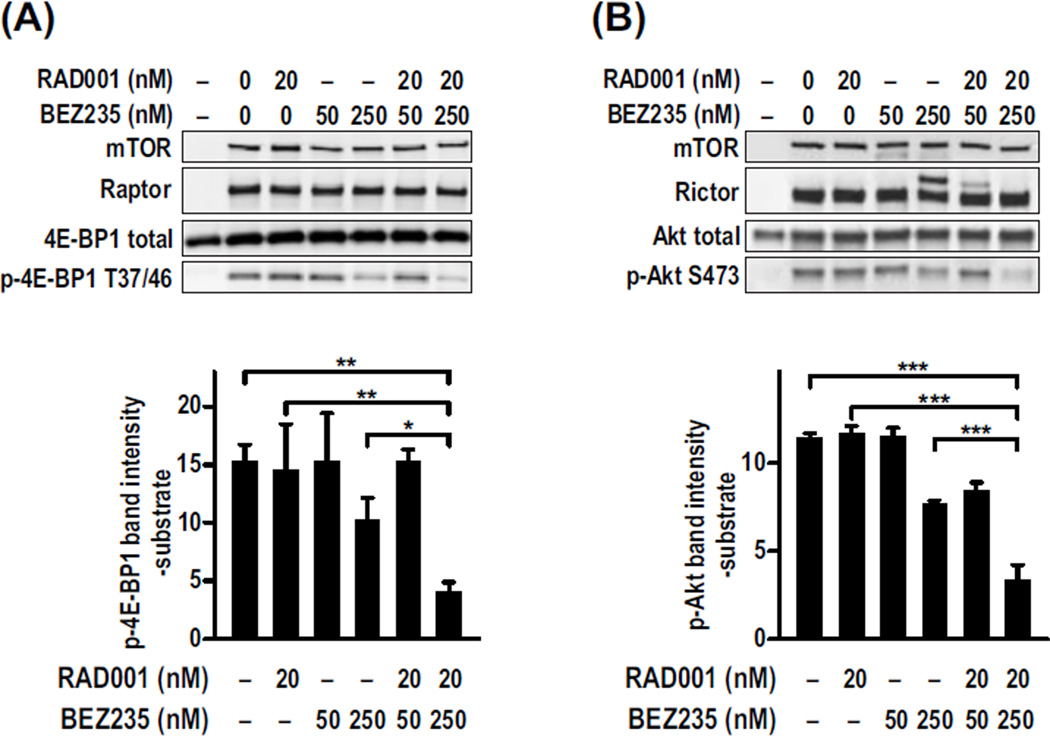
To determine whether the synergistic effects of RAD001 and BEZ235 were elicited at the level of mTOR, we tested the drugs in an mTORC1 in vitro kinase assay, after immunoprecipitation with a raptor antibody and using 4E-BP1 as a substrate (24). The phosphorylation of 4E-BP1 T37/46 was not significantly inhibited by 20 nM RAD001, in contrast to increasing concentrations of BEZ235 from 50 to 250 nM (Fig. 2A). Critically, the combination of 20 nM RAD001 and 250 nM BEZ235 resulted in synergistic inhibition of mTORC1 activity compared to inhibition with the same concentration of either drug alone (Fig. 2A). The ability of RAD001 to sensitize PKB/Akt S473 to BEZ235-induced dephosphorylation in Huh7 cells can be attributed to the loss of the negative feedback loop from mTORC1/S6K1 to PKB/Akt. However, these effects may also result from the binding of RAD001/FKBP12 to mTORC2 (25). We found that BEZ235 inhibited mTORC2 phosphorylation of PKB/Akt in vitro, and this effect was enhanced by RAD001 (Fig. 2B), suggesting that the observed synergy is through inhibition of mTORC2, not through other targets.
Fig. 2.
RAD001 and BEZ235 synergistically inhibit the kinase activity of mTORC1 and mTORC2 in vitro. Endogenous mTORC1 or mTORC2 complexes were immunoprecipitated from HEK293T cell lysates using a raptor or rictor antibody, respectively. (A and B) Kinase activity was assayed for (A) mTORC1 or (B) mTORC2 using 4E-BP1 or Akt as substrates, respectively. Each assay was performed a minimum of three times. A representative Western blot analysis shows the amount of total and phosphorylated proteins in each reaction. The graphs show the means ± SD (n = 3) of the amount of phosphorylated substrate, minus the background level of the substrate alone, in each treatment. *P < 0.05; **P = 0.001; ***P < 0.0001
RAD001 and BEZ235 act synergistically to inhibit HCC progression
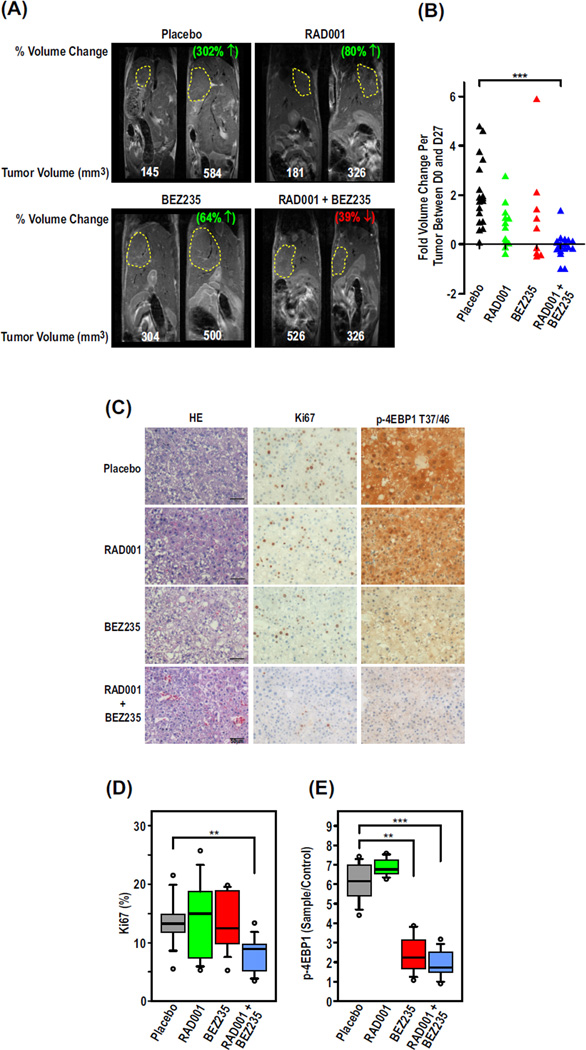
Primary events leading to human HCC are best represented in mouse models initiated by injury, resulting in compensatory proliferation of liver cells (20, 21, 26). To address this issue, we used the DEN-induced HCC model (27), whose gene expression profile corresponds closely to that of human HCC with unfavorable outcome (20, 21). C57BL/6 mice treated with DEN at 2 weeks exhibited tumors (fig. S2A) between 1.05 and 2618 mm3 at ~44 weeks, as measured by magnetic resonance imaging (MRI) (Fig. 3A). Tumor-bearing mice were divided into four treatment arms on the basis of tumor burden (Fig. 3A) and gavaged daily for 28 days with the recommended doses (Novartis Oncology) of RAD001 (10 mg/kg), BEZ235 (30 mg/kg), or a combination of RAD001 (2.5 mg/kg) and BEZ235 (18 mg/kg). Such treatment had no apparent effect on body weight throughout the course of the experiment (fig. S2B). Similarly, treatment with either drug alone or in combination had no adverse effects on body weight of transgenic mice engineered to ectopically express E2F1/c-Myc in the liver (fig. S4A), a mouse model of human HCC, with better prognosis (20).
Fig. 3.
The combination of RAD001 and BEZ235 inhibits HCC progression better than the single agents alone. Male C57BL/6 mice (n = 9 to 10) were injected with DEN at 2 weeks. After 44 weeks, mice were treated daily with placebo, RAD001, and/or BEZ235. (A) MRI scans from days 0 and 28 of treatment are shown from a representative mouse from each group. The percentage depicts the change in volume between the two time points. (B) Fold change in tumor volume between days 0 and 28 based on quantification of the MRI scans. ***P < 0.0001, Kruskal-Wallis test followed by Dunn’s multiple comparisons test. (C) Representative 100× magnification images of hematoxylin and eosin (H&E), Ki67, and phospho–4E-BP1 (p-4E-BP1) T37/T46–stained tumor sections from mice from each of the treatment groups. (D and E) Percent proliferating cells (D) or p-4E-BP1 (E) per preneoplastic focus or tumor based on quantification of immunohistochemical analysis of paraffin-embedded tissue sections stained with Ki67 or p-4E-BP1 T37/T46 antibodies, respectively. **P < 0.001; ***P < 0.0001, Mann-Whitney test.
On the basis of MRI analyses, tumor volumes of placebo-treated mice doubled on average within the 28 days of the study, whereas treatment with either RAD001 or BEZ235 alone had a pronounced inhibitory effect on this response (Fig. 3, A and B). More striking, the lower doses of the drug combination induced a marked effect in HCC regression, relative to mice treated with either drug alone at optimal doses (Fig. 3, A and B). This was particularly evident by comparing the ratio of liver weight to body weight (fig. S2C). The reduction in HCC progression in the combined treatment could be accounted for, in part, by the cumulative effect of an increase in apoptosis (fig. S2D) and a decrease in proliferation, as determined by quantitative immunohistochemistry (IHC) of Ki67-stained tumor sections (Fig. 3, C and D). Similar results were obtained for HCCs of E2F1/c-Myc–treated mice (fig. S4, B and C). Unexpectedly, in DEN-induced tumors, unlike cells in culture (Fig. 1), 4E-BP1 T37/46 phosphorylation was inhibited to the same extent by BEZ235 alone as in combination with RAD001 (Fig. 3, C and D, and fig. S3A), as confirmed by Western blot analyses (fig. S3, B and C). Likewise, by Western blot analyses or IHC, dephosphorylation of PKB/Akt S473 induced by BEZ235 alone was as potent as the drug combination (fig. S3, B and D to F), suggesting that in addition to 4E-BP1 and PKB/Akt, other targets are involved in the synergistic response in tumor regression.
RAD001 and BEZ235 cause reversal of gene expression levels in tumors
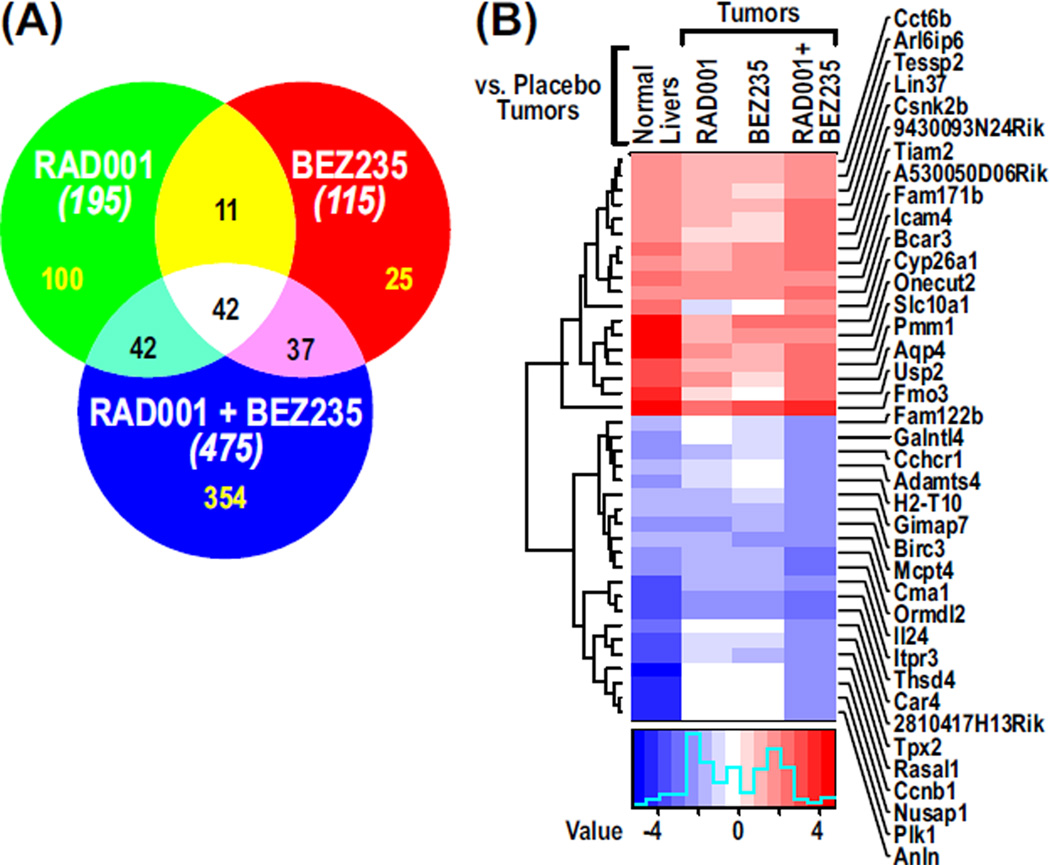
For deeper insights into the effects of differential drug treatments, DEN-induced tumors and normal livers were profiled by gene expression microarrays at the end of the 28-day treatment period. Four comparisons were made: placebo-treated liver versus placebo-treated tumor, and placebo-treated tumor versus each of the drug regimens. Gene expression analysis identified 5665 genes that were significantly altered (false discovery rate <0.25) between placebo-treated livers and placebo-treated tumors, whereas 245, 146, and 708 genes were significantly changed in placebo-treated tumors compared to tumors treated with RAD001, BEZ235, and BEZ235 plus RAD001, respectively. Of the genes significantly affected in placebo-treated liver compared to placebo-treated tumor, 195, 115 and 475 genes in tumors treated with RAD001, BEZ235, or RAD001 plus BEZ235, respectively, reverted to roughly baseline expression levels of placebo-treated liver (Fig. 4A and table S2). Assessment of the gene sets using the Fisher’s exact test revealed that a significant number of cancer genes renormalized to placebo-treated liver (P < 2.2 × 10−16) in all three treatment groups. Only 50% of the genes affected by RAD001 were also affected by BEZ235, whereas the combined treatment affected 354 distinct genes, providing confirmation of cooperative interaction between BEZ235 and RAD001 in vivo (Fig. 4A). The ability of the RAD001/BEZ235 combination, compared with either agent alone, to induce reversion to the gene expression phenotype of placebo-treated liver is depicted in the heat map of the data (Fig. 4B). Gene Set Enrichment Analysis (GSEA) identified cell cycle inhibition as one of the major pathways altered by the combination of both drugs, which was not observed in the single-drug treatments (table S3). These data suggest that the interaction of the two drugs in vivo is distinct from either alone.
Fig. 4.
RAD001 and BEZ235 cause reversal of gene expression levels in tumors. (A) Venn diagram showing the numbers of genes reverting to expression levels equivalent to those of normal liver in tumors treated with RAD001 and/or BEZ235. The numbers in parentheses show the total number of genes in each treatment. (B) Heat map showing renormalizing genes selected on the basis of a fourfold change in tumors treated with RAD001 and BEZ235 compared to placebo-treated tumors.
RAD001 and BEZ235 synergize on autophagy
In the pairwise comparative microarray analyses, we noted changes in a number of autophagy genes. Although a few were up-regulated in tumors, almost half were significantly down-regulated (table S4), two of which, Atg5 and Atg7, are tumor suppressors in liver (28, 29). Analysis of existing microarray data sets from HCC patients, having the signature of A (poor prognosis) versus B (good prognosis) (20), showed that the altered expression of autophagy genes is associated with those patients having a poor prognosis, as shown in the Kaplan-Meier plot of these subsets of HCC patients (P < 0.01) (fig. S5). Although BEZ235 and RAD001 stimulate autophagy (30), a role for autophagy in suppressing DEN-induced HCC (Fig. 3) would not be consistent with reports thatautophagy is mediated by 4E-BP1 (31, 32), because BEZ235 alone induces 4E-BP1 dephosphorylation to almost the same extent as that of the drug combination (Fig. 3E).
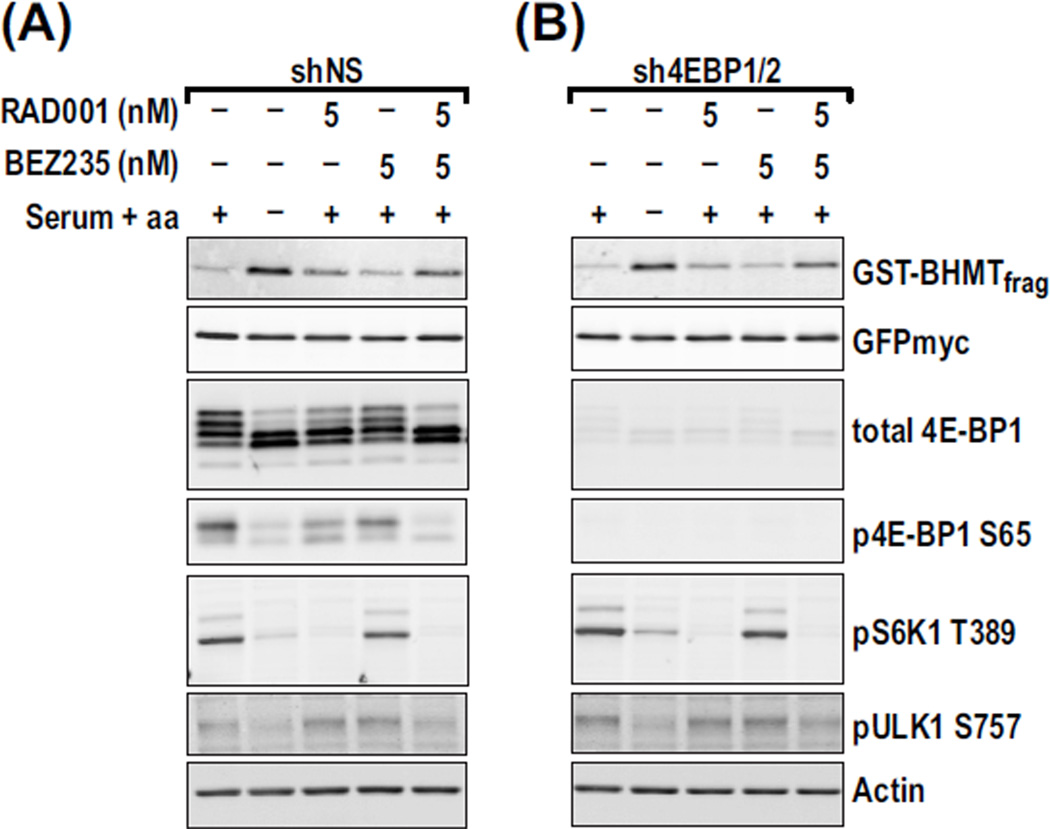
Recent studies argue that mTORC1 can directly suppress autophagy by phosphorylating autophagy-initiating kinase ULK1 at S757 (33). Thus, we asked whether RAD001 and BEZ235 synergize on the autophagic response and ULK1 S757 dephosphorylation and whether these effects were 4E-BP1/2–dependent. To measure autophagy and 4E-BP1/2 dependence, we took advantage of a glutathione S-transferase–tagged betaine homocysteine methyltransferase (GST-BHMT) reporter, whose cleavage represents a cargo-based autophagy end point (34), and human embryonic kidney (HEK) 293 cells stably expressing either a nonsilencing short hairpin RNA (shRNA), (shNS) or an shRNA against 4E-BP1/2 (sh4E-BP1/2) (19). Withdrawal of aminoacids and serum from shNS cells led to dephosphorylation of S6K1, 4E-BP1, and ULK1 (Fig. 5A), with similar results obtained for S6K1 and ULK1 in sh4E-BP1/2 cells (Fig. 5B). However, basal levels of the GST-BHMT fragment were indistinguishable in shNS versus sh4EBP1/2 cells, as was the extent of fragment accumulation caused by serum and amino acid withdrawal (Fig. 5, A and B). Treatment with 5 nM RAD001 induced S6K1 dephosphorylation but had little impact on 4E-BP1 and ULK1 phosphorylation or on the accumulation of the GST-BHMT fragment in shNS cells (Fig. 5A), with equivalent results obtained for S6K1 and ULK1 in sh4E-BP1/2 cells (Fig. 5B). The combination of the two drugs had a synergistic effect on the accumulation of the GST-BHMT fragment and ULK1 S757 dephosphorylation (Fig. 5, A and B) independent of 4E-BP1/2 (Fig. 5B) and S6K1 (Fig. 5, A and B). Thus, the induction of autophagy can occur independently of 4E-BP1/2 and S6K1.
Fig. 5.
The combination of RAD001 and BEZ235 synergistically affects autophagy independently of 4E-BP1. (A and B) HEK293 cells stably expressing shNS (A) or sh4E-BP1/2 (B) were transfected with GST-BHMT and treated 48 hours later with the indicated concentrations of each drug for 6 hours. Cells were maintained in full medium or starved of serum and amino acids for 6 hours as negative and positive controls for autophagy, respectively. The GST-BHMT immunoprecipitates were analyzed by Western blot analysis to measure the indicated proteins in each sample. GFPmyc is a control for plasmid expression level.
RAD001 and BEZ235 induce autophagy in liver tumors
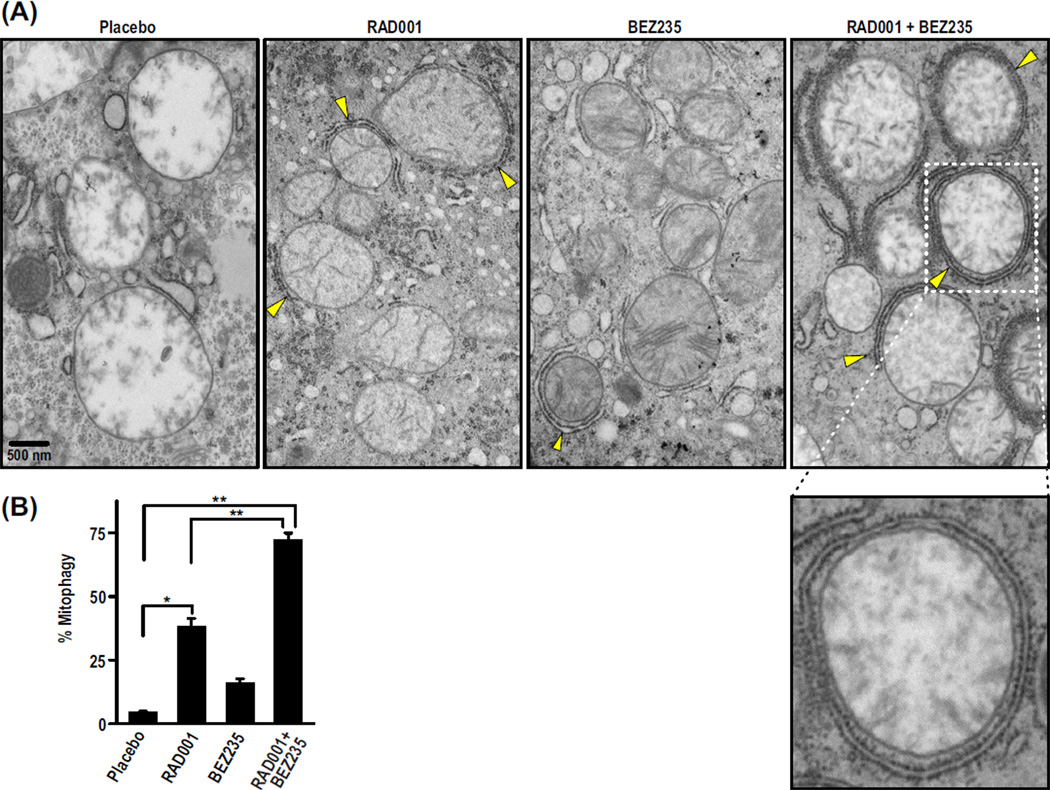
The findings above raised the question of whether autophagy might more closely follow regression of DEN-induced tumors than 4E-BP1 dephosphorylation (Fig. 3). To explore this, we examined liver tumors for autophagosome formation by transmission electron microscopy (TEM). The TEM images revealed double-membrane vesicles indicative of autophagosomes, which were studded with small particles resembling ribosomes (Fig. 6A), consistent with autophagosomes being derived from the endoplasmic reticulum (35, 36). By morphometric analysis, many more autophagosome-like structures were detected in the RAD001/BEZ235 combination compared to tumors treated with placebo or either drug alone (Fig. 6, A and B). Mitochondria were the apparent target (Fig. 6A), consistent with the induction of mitophagy in nutrient-deprived hepatocytes (37). Although tumor regression in HCC tumors treated with a combination of RAD001 and BEZ235 may be due to multiple factors, the data suggest that autophagy, specifically mitophagy, is a major effector.
Fig. 6.
Autophagosomes targeting mitochondria increase in numbers in tumors treated with RAD001 and BEZ235 compared to tumors treated with placebo or either drug alone. (A) Representative TEM images from one mouse per treatment show mitophagy (yellow arrows point to forming autophagosomes). (B) Percent mitophagy was quantified by counting the total number of mitochondria partially or fully engulfed with autophagosomes relative to the total number of mitochondria counted per image. Results are means ± SD (n = 8 to 11). *P < 0.05; **P < 0.001.
DISCUSSION
We set out to determine whether BEZ235 would be a more effective inhibitor of HCC progression than RAD001. Unexpectedly, the two in combination are more potent than either agent alone in inhibiting proliferation of HCC cells in culture and tumors in vivo (Figs.1 and 3). Consistent with earlier findings that rapamycin affects substrate specificity, not kinase activity (38), recent studies show that the ability of the rapamycins to inhibit mTORC1 signaling is more pronounced for S6K1 than 4E-BP1 (39, 40). This has led to the suggestion that S6K1, but not 4E-BP1, is excluded from interacting with mTORC1 because of its relative larger size (41, 42). However, S6K1 is about half the size of ULK1, whose phosphorylation is largely unaffected by rapamycin. It is more likely that mTORC1 activity is dependent on the conformation of a ternary complex that includes the kinase, the substrate, and ATP. In contrast, the new mTOR ATP-binding site competitive drugs inhibit both mTOR complexes and block 4E-BP1 phosphorylation to the same extent as S6K1 (39, 40, 43). Downstream of mTORC1, we recently showed that the main effects of these inhibitors on cell proliferation are attributed to activation of the 4E-BPs (19). These findings are consistent with our observations using RAD001 and BEZ235 in HCC cell lines (Fig. 1 and fig. S1); however, inhibition of 4E-BP1 alone is not sufficient to explain the effects on HCC progression, which eventually led us to examine the role of autophagy.
It has been demonstrated that prolonged treatment with rapamycin affects mTORC2, in addition to mTORC1 (44). This response is more difficult to discern, because rapamycin also relieves the negative feedback loop from mTORC1/S6K1 to PKB/Akt (Fig. 1A). A similar effect was observed with BEZ235 at lower concentrations (Fig. 1A), suggesting that mTORC1 may be more readily targeted than mTORC2. As with mTORC1, mTORC2 inhibition by BEZ235 was greatly enhanced by RAD001 (Fig. 1B). Moreover, from in vitro studies, these effects appear to be elicited at the level of mTORC1 and mTORC2 (Fig. 2). Although in HCC the major effects on proliferation appear to be through mTORC1, it is clear in other tumor types, including phosphatase and tensin homolog deleted from chromosome 10 (PTEN)–deficient prostate tumors, that the effects on tumor progression are mTORC2- dependent (45). The therapeutic advantage of combining RAD001 with BEZ235 is that it should be efficacious in either tumor type. Moreover, because of the findings here and those of Nyfeler et al. (41), one would predict that the rapamycins could be used in combination with any mTOR ATP-binding site competitive inhibitor. Combination treatment should decrease the effective dose of either drug, reducing off target effects of the mTOR ATP-binding site competitive inhibitor.
We tested the efficacy of RAD001 and BEZ235 in HCC with the DEN mouse model, which best represents human HCC with unfavorable outcome (21). Gene expression profiling showed that the major classes of genes affected in both mouse and human HCCs with poor prognosis were cell proliferation and antiapoptotic genes (21). We find that DEN induced HCCs treated with RAD001 and BEZ235 have a significant cell cycle inhibition signature. Moreover, the drug combination, unlike either RAD001 or BEZ235 alone, revealed a significant number of genes reverting to roughly baseline expression levels of normal livers, suggesting that the effect of the two drugs together cannot be recapitulated by increasing the dose of either drug alone. Recent data in ovarian cancer cells and non–small cell lung cancer cells in culture and xenografts suggest that c-Myc is a major regulator of the tumor response to rapamycin or RAD001 in combination with a PI3K/mTOR inhibitor (46, 47). However, we found no evidence of significant alterations in genes transcriptionally regulated by c-Myc in placebo- or drug-treated HCC DEN tumors. Our findings suggest that the mechanisms at play may be unique to a syngeneic tumor confronted with an intact cytokine and immune response arising from a natural history in the endogenous stroma or to HCC itself, rather than cultured cell–initiated xenografts in immunocompromised mice.
It has been known for some time that inhibition of mTOR signaling in hepatocytes is associated with the activation of autophagy (48). Moreover, recent studies describe the spontaneous induction of liver adenomas in mice with a mosaic deletion of Atg5 or a liver specific deletion of Atg7 (28). However, in other systems, autophagy supports tumor persistence by maintaining cells under nutrient deprived conditions, thus acting as a survival factor (49). In our hands, RAD001 and BEZ235 synergize at the level of autophagy as shown by accumulation of the GST-BHMT fragment (Fig. 5). The increase in autophagy is independent of 4E-BP1 and correlates with the dephosphorylation of ULK1 at S757, an mTORC1 phosphorylation site (33). These findings suggest that activation of autophagy, in a 4E-BP1/2/eIF-4G– independent manner, may be implicated in HCC regression observed in tumors treated with combined RAD001/BEZ235 (Fig. 3). With the exception of Atg3, we did not observe significant changes in the gene expression of autophagy genes in tumors treated with the combination of RAD001 and BEZ235, compared to vehicle-treated tumors. The significant autophagic response to mTOR inhibitors in the absence of major transcriptional changes suggests that transcriptional reprogramming of autophagy genes was not required for the mitophagy response. At this point, it will be important to determine the extent to which the activation of mitophagy in DEN-induced HCCs contributes to tumor regression.
RAD001 has been approved by the U.S. Food and Drug Administration for renal clear cell carcinoma, TSC-associated subependymal giant cell astrocytoma, and neuroendocrine tumors. However, in all cases, RAD001 delayed tumor progression, but there were no complete responses. It may be that such effects are due to partial suppression of mTORC1 signaling by rapamycin derivatives, or resistant mechanisms may develop over time. The combination of an ATP-binding site competitive mTOR inhibitor with a rapamycin derivative may prove more effective in inhibiting additional targets of mTORC1. Our hypothesis is that synergy may arise as a function of the ATP-competitive inhibitors having enhanced access to the active site of the kinase. To our knowledge, there are no other examples where two inhibitors act synergistically on the same target; thus, these studies provide a strategy to increase the specificity of ATP-competitive inhibitors. On the basis of our in vivo data, we have begun an investigator-initiated phase 1B-2 dose escalation study of BEZ235 in combination with RAD001 in patients with HCC or other solid tumors. Since rapamycin and its derivatives have already been approved clinically, their combination with a PI3K/mTOR ATP-competitive inhibitor, such as BEZ235, would be a rapid strategy to test the efficacy of this class of drugs in cancer and to fast-track their approval.
MATERIALS AND METHODS
Cell Culture and Proliferation assay
HEK293T cells purchased from the American Type Culture Collection and Huh7 cells from Riken Cell Bank were maintained in Dulbecco’s modified Eagle’s medium (Hyclone) containing 10% fetal bovine serum (Hyclone). Cell proliferation assays were performed with the CellTiter-Glo Luminescent Cell Viability assay (Promega).
Animal Studies
C57BL/6 mice were obtained from the Jackson Laboratory. At 2 weeks, male mice were injected intraperitoneally with DEN (50 mg/kg) (Sigma). At ~44 weeks, the animals (n = 9 to 10) were gavaged twice daily for 28 days with the specified doses of RAD001 and/or BEZ235. The mice were starved for 3 hours before sacrifice and received their last dose of RAD001 at the time of starvation and/or BEZ235 1 hour before sacrifice. Liver and tumor tissues were immediately snap-frozen in liquid nitrogen for protein and RNA extraction or fixed in 10% formalin for IHC analysis. Generation of the E2F1/c-Myc mice was previously described (23). All in vivo studies were approved by the Institutional Animal Care and Use Committee and performed following the ethical guidelines of the University of Cincinnati and Cincinnati Children’s Hospital Medical Center.
Statistical Analyses
Data are presented as means ± SD unless otherwise specified. All the statistical analyses were performed with GraphPad Prism 5. Unless otherwise specified in the figure legend, statistical significance was determined with a one-way analysis of variance (ANOVA) followed by a Bonferroni multiple group comparison test, with P values of <0.05, 0.001, and 0.0001 considered to be significant and represented with *, **, and ***, respectively. Synergy was calculated with the ComboSyn Synergy software (version 3.0.1) on the basis of the Chou-Talalay method (22).
Immunoblot analysis
Refer to Supplementary Materials.
In vitro mTOR kinase assays
Refer to Supplementary Materials.
Immunohistochemistry
Refer to Supplementary Materials.
BHMT assay
Refer to Supplementary Materials.
Magnetic resonance imaging
Refer to Supplementary Materials.
Microarray analyses
Refer to Supplementary Materials.
Electron microscopy
Refer to Supplementary Materials.
Supplementary Materials
www.sciencetranslationalmedicine.org/cgi/content/full/4/139/139ra84/DC1
Materials and Methods
Fig. S1. RAD001 and BEZ235 synergistically inhibit mTOR signaling in both human and mouse HCC cell lines.
Fig. S2. RAD001 in combination with BEZ235 inhibits HCC progression better than the single agents alone in DEN-injected mice.
Fig. S3. RAD001 and BEZ235 inhibit mTOR signaling in livers and tumors of DEN-injected mice.
Fig. S4. RAD001 and BEZ235 treatment decreases HCC proliferation in E2F1/c-Myc mice.
Fig. S5. Kaplan-Meier plot grouped by gene expression of autophagy genes can predict overall survival of HCC patients.
Table S1. Chou and Talalay calculation of synergy in RAD001- and/or BEZ235-treated Huh7 cells.
Table S2. Renormalizing genes in RAD001- and/or BEZ235-treated tumors.
Table S3. Pairwise comparisons of differentially expressed pathways by GSEA in placebo-, RAD001-, and/or BEZ235-treated tumors.
Table S4. Pairwise comparisons of autophagy-related genes between treatment groups.
Supplementary Material
Acknowledgments
We thank V. Zimmerman, M. Bruns, A. Karr, and J. Park for help with in vivo experiments. We are grateful to D. Lindquist and S. Dunn for discussions in planning and executing MRI imaging. We thank A. Selvaraj, Z. Tsuchihashi, and A. Huang for critical discussions and G. Doerman for preparation of figures. We are grateful to Novartis for supplying RAD001 and NVP-BEZ235. Funding: Supported by National Cancer Institute grants U01CA120475 and U01CA120475-S1 to S.C.K. and U01CA141464 to G.T. B.J.A. is funded by W81XWH-10-0325 (Department of Defense) and U54 RR-25216.
Footnotes
The mTOR-allosteric inhibitor, RAD001, in combination with a PI3K/mTOR ATP-site competitive inhibitor, BEZ235, causes gene reprogramming, autophagy and tumor regression, in a mouse model approximating human HCC with poor prognosis, leading to an investigator Phase 1B-2 clinical trial.
Author contributions: H.E.T. conceived, designed, performed, and analyzed most experiments. C.A.M. conceived and designed autophagy experiments. L.S.C. provided conceptual and technical input. J.P. contributed to mitophagy data. J.B.A. and E.A.C. contributed to designing and running the microarray samples. B.J.A. and L.M. performed microarray analyses. K.T., T.M., and A.I. quantified IHC sections. S.M.M., S.S.T., and P.S.M. provided conceptual input. G.T. and S.C.K. conceived and supervised all experiments. The manuscript was written by H.E.T., C.A.M., G.T., and S.C.K.
Competing interests: S.M.M. and L.M. are Novartis employees. P.S.M. has served on an advisory board for Celgene, and G.T. has received consultancy fees from Novartis. All other authors declare that they have no competing interests.
Data and materials availability: The microarray data have been deposited in the Gene Expression Omnibus database (accession number GSE37129).
REFERENCES AND NOTES
- 1.El-Serag HB, Rudolph KL. Hepatocellular carcinoma: Epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 2.Hung H. Treatment modalities for hepatocellular carcinoma. Curr. Cancer Drug Targets. 2005;5:131–138. doi: 10.2174/1568009053202063. [DOI] [PubMed] [Google Scholar]
- 3.Blonski W, Kotlyar DS, Forde KA. Non-viral causes of hepatocellular carcinoma. World J. Gastroenterol. 2010;16:3603–3615. doi: 10.3748/wjg.v16.i29.3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thomas MB, Jaffe D, Choti MM, Belghiti J, Curley S, Fong Y, Gores G, Kerlan R, Merle P, O’Neil B, Poon R, Schwartz L, Tepper J, Yao F, Haller D, Mooney M, Venook A. Hepatocellular carcinoma: Consensus recommendations of the National Cancer Institute Clinical Trials Planning Meeting. J. Clin. Oncol. 2010;28:3994–4005. doi: 10.1200/JCO.2010.28.7805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.El-Serag HB. Epidemiology of hepatocellular carcinoma in USA. Hepatol. Res. 2007;37(Suppl. 2):S88–S94. doi: 10.1111/j.1872-034X.2007.00168.x. [DOI] [PubMed] [Google Scholar]
- 6.Cheng AL, Kang YK, Chen Z, Tsao CJ, Qin S, Kim JS, Luo R, Feng J, Ye S, Yang TS, Xu J, Sun Y, Liang H, Liu J, Wang J, Tak WY, Pan H, Burock K, Zou J, Voliotis D, Guan Z. Efficacy and safety of sorafenib in patients in the Asia-Pacific region with advanced hepatocellular carcinoma: A phase III randomised, double-blind, placebo-controlled trial. Lancet Oncol. 2009;10:25–34. doi: 10.1016/S1470-2045(08)70285-7. [DOI] [PubMed] [Google Scholar]
- 7.Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, de Oliveira AC, Santoro A, Raoul JL, Forner A, Schwartz M, Porta C, Zeuzem S, Bolondi L, Greten TF, Galle PR, Seitz JF, Borbath I, Häussinger D, Giannaris T, Shan M, Moscovici M, Voliotis D, Bruix J. SHARP Investigators Study Group, Sorafenib in advanced hepatocellular carcinoma. N. Engl. J. Med. 2008;359:378–390. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- 8.Park EJ, Lee JH, Yu GY, He G, Ali SR, Holzer RG, Osterreicher CH, Takahashi H, Karin M. Dietary and genetic obesity promote liver inflammation and tumorigenesis by enhancing IL-6 and TNF expression. Cell. 2010;140:197–208. doi: 10.1016/j.cell.2009.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nobukuni T, Joaquin M, Roccio M, Dann SG, Kim SY, Gulati P, Byfield MP, Backer JM, Natt F, Bos JL, Zwartkruis FJ, Thomas G. Amino acids mediate mTOR/raptor signaling through activation of class 3 phosphatidylinositol 3OH-kinase. Proc. Natl. Acad. Sci. U.S.A. 2005;102:14238–14243. doi: 10.1073/pnas.0506925102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Newgard CB, An J, Bain JR, Muehlbauer MJ, Stevens RD, Lien LF, Haqq AM, Shah SH, Arlotto M, Slentz CA, Rochon J, Gallup D, Ilkayeva O, Wenner BR, Yancy WE, Eisenson H, Musante G, Surwit R, Millington DS, Butler MD, Svetkey LP. A branched-chain amino acid related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. 2009;9:311–326. doi: 10.1016/j.cmet.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Villanueva A, Llovet JM. Targeted therapies for hepatocellular carcinoma. Gastroenterology. 2011;140:1410–1426. doi: 10.1053/j.gastro.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vander Haar E, Lee SI, Bandhakavi S, Griffin TJ, Kim DH. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat. Cell Biol. 2007;9:316–323. doi: 10.1038/ncb1547. [DOI] [PubMed] [Google Scholar]
- 13.Sancak Y, Thoreen CC, Peterson TR, Lindquist RA, Kang SA, Spooner E, Carr SA, Sabatini DM. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol. Cell. 2007;25:903–915. doi: 10.1016/j.molcel.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 14.Zoncu R, Efeyan A, Sabatini DM. mTOR: From growth signal integration to cancer, diabetes and ageing. Nat. Rev. Mol. Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jung CH, Ro SH, Cao J, Otto NM, Kim DH. mTOR regulation of autophagy. FEBS Lett. 2010;584:1287–1295. doi: 10.1016/j.febslet.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beuvink I, Boulay A, Fumagalli S, Zilbermann F, Ruetz S, O’Reilly T, Natt F, Hall J, Lane HA, Thomas G. The mTOR inhibitor RAD001 sensitizes tumor cells to DNA-damaged induced apoptosis through inhibition of p21 translation. Cell. 2005;120:747–759. doi: 10.1016/j.cell.2004.12.040. [DOI] [PubMed] [Google Scholar]
- 17.Radimerski T, Montagne J, Hemmings-Mieszczak M, Thomas G. Lethality of Drosophila lacking TSC tumor suppressor function rescued by reducing dS6K signaling. Genes Dev. 2002;16:2627–2632. doi: 10.1101/gad.239102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Um SH, D’Alessio D, Thomas G. Nutrient overload, insulin resistance, and ribosomal protein S6 kinase 1, S6K1. Cell Metab. 2006;3:393–402. doi: 10.1016/j.cmet.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 19.Dowling RJ, Topisirovic I, Alain T, Bidinosti M, Fonseca BD, Petroulakis E, Wang X, Larsson O, Selvaraj A, Liu Y, Kozma SC, Thomas G, Sonenberg N. mTORC1-mediated cell proliferation, but not cell growth, controlled by the 4E-BPs. Science. 2010;328:1172–1176. doi: 10.1126/science.1187532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee JS, Chu IS, Heo J, Calvisi DF, Sun Z, Roskams T, Durnez A, Demetris AJ, Thorgeirsson SS. Classification and prediction of survival in hepatocellular carcinoma by gene expression profiling. Hepatology. 2004;40:667–676. doi: 10.1002/hep.20375. [DOI] [PubMed] [Google Scholar]
- 21.Lee JS, Chu IS, Mikaelyan A, Calvisi DF, Heo J, Reddy JK, Thorgeirsson SS. Application of comparative functional genomics to identify best-fit mouse models to study human cancer. Nat. Genet. 2004;36:1306–1311. doi: 10.1038/ng1481. [DOI] [PubMed] [Google Scholar]
- 22.Chou TC. Drug combination studies and their synergy quantification using the Chou- Talalay method. Cancer Res. 2010;70:440–446. doi: 10.1158/0008-5472.CAN-09-1947. [DOI] [PubMed] [Google Scholar]
- 23.Ladu S, Calvisi DF, Conner EA, Farina M, Factor VM, Thorgeirsson SS. E2F1 inhibits c-Myc-driven apoptosis via PIK3CA/Akt/mTOR and COX-2 in a mouse model of human liver cancer. Gastroenterology. 2008;135:1322–1332. doi: 10.1053/j.gastro.2008.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kalender A, Selvaraj A, Kim SY, Gulati P, Brûlé S, Viollet B, Kemp BE, Bardeesy N, Dennis P, Schlager JJ, Marette A, Kozma SC, Thomas G. Metformin, independent of AMPK, inhibits mTORC1 in a rag GTPase-dependent manner. Cell Metab. 2010;11:390–401. doi: 10.1016/j.cmet.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell. 2007;12:9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 26.Fausto N. Mouse liver tumorigenesis: Models, mechanisms, and relevance to human disease. Semin. Liver Dis. 1999;19:243–252. doi: 10.1055/s-2007-1007114. [DOI] [PubMed] [Google Scholar]
- 27.Vesselinovitch SD, Koka M, Mihailovich N, Rao KV. Carcinogenicity of diethylnitrosamine in newborn, infant, and adult mice. J. Cancer Res. Clin. Oncol. 1984;108:60–65. doi: 10.1007/BF00390974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takamura A, Komatsu M, Hara T, Sakamoto A, Kishi C, Waguri S, Eishi Y, Hino O, Tanaka K, Mizushima N. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 2011;25:795–800. doi: 10.1101/gad.2016211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Inami Y, Waguri S, Sakamoto A, Kouno T, Nakada K, Hino O, Watanabe S, Ando J, Iwadate M, Yamamoto M, Lee MS, Tanaka K, Komatsu M. Persistent activation of Nrf2 through p62 in hepatocellular carcinoma cells. J. Cell Biol. 2011;193:275–284. doi: 10.1083/jcb.201102031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fan QW, Cheng C, Hackett C, Feldman M, Houseman BT, Nicolaides T, Haas-Kogan D, James CD, Oakes SA, Debnath J, Shokat KM, Weiss WA. Akt and autophagy cooperate to promote survival of drug-resistant glioma. Sci. Signal. 2010;3:ra81. doi: 10.1126/scisignal.2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramírez-Valle F, Braunstein S, Zavadil J, Formenti SC, Schneider RJ. eIF4GI links nutrient sensing by mTOR to cell proliferation and inhibition of autophagy. J. Cell Biol. 2008;181:293–307. doi: 10.1083/jcb.200710215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Balakumaran BS, Porrello A, Hsu DS, Glover W, Foye A, Leung JY, Sullivan BA, Hahn WC, Loda M, Febbo PG. MYC activity mitigates response to rapamycin in prostate cancer through eukaryotic initiation factor 4E–binding protein 1–mediated inhibition of autophagy. Cancer Res. 2009;69:7803–7810. doi: 10.1158/0008-5472.CAN-09-0910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mercer CA, Kaliappan A, Dennis PB. A novel, human Atg13 binding protein, Atg101, interacts with ULK1 and is essential for macroautophagy. Autophagy. 2009;5:649–662. doi: 10.4161/auto.5.5.8249. [DOI] [PubMed] [Google Scholar]
- 35.Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, Yoshimori T, Yamamoto A. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat. Cell Biol. 2009;11:1433–1437. doi: 10.1038/ncb1991. [DOI] [PubMed] [Google Scholar]
- 37.Kim I, Lemasters JJ. Mitochondrial degradation by autophagy (mitophagy) in GFP-LC3 transgenic hepatocytes during nutrient deprivation. Am. J. Physiol. Cell Physiol. 2011;300:C308–C317. doi: 10.1152/ajpcell.00056.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McMahon LP, Choi KM, Lin TA, Abraham RT, Lawrence JC., Jr The rapamycin binding domain governs substrate selectivity by the mammalian target of rapamycin. Mol. Cell. Biol. 2002;22:7428–7438. doi: 10.1128/MCB.22.21.7428-7438.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, Reichling LJ, Sim T, Sabatini DM, Gray NS. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J. Biol. Chem. 2009;284:8023–8032. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feldman ME, Apsel B, Uotila A, Loewith R, Knight ZA, Ruggero D, Shokat KM. Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS Biol. 2009;7:e38. doi: 10.1371/journal.pbio.1000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nyfeler B, Bergman P, Triantafellow E, Wilson CJ, Zhu Y, Radetich B, Finan PM, Klionsky DJ, Murphy LO. Relieving autophagy and 4EBP1 from rapamycin resistance. Mol. Cell. Biol. 2011;31:2867–2876. doi: 10.1128/MCB.05430-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yip CK, Murata K, Walz T, Sabatini DM, Kang SA. Structure of the human mTOR complex I and its implications for rapamycin inhibition. Mol. Cell. 2010;38:768–774. doi: 10.1016/j.molcel.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.García-Martínez JM, Moran J, Clarke RG, Gray A, Cosulich SC, Chresta CM, Alessi DR. Ku-0063794 is a specific inhibitor of the mammalian target of rapamycin (mTOR) Biochem. J. 2009;421:29–42. doi: 10.1042/BJ20090489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sarbassov DD, Ali SM, Sengupta S, Sheen JH, Hsu PP, Bagley AF, Markhard AL, Sabatini DM. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol. Cell. 2006;22:159–168. doi: 10.1016/j.molcel.2006.03.029. [DOI] [PubMed] [Google Scholar]
- 45.Guertin DA, Stevens DM, Saitoh M, Kinkel S, Crosby K, Sheen JH, Mullholland DJ, Magnuson MA, Wu H, Sabatini DM. mTOR complex 2 is required for the development of prostate cancer induced by Pten loss in mice. Cancer Cell. 2009;15:148–159. doi: 10.1016/j.ccr.2008.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mazzoletti M, Bortolin F, Brunelli L, Pastorelli R, Di Giandomenico S, Erba E, Ubezio P, Broggini M. Combination of PI3K/mTOR inhibitors: Antitumor activity and molecular correlates. Cancer Res. 2011;71:4573–4584. doi: 10.1158/0008-5472.CAN-10-4322. [DOI] [PubMed] [Google Scholar]
- 47.Xu CX, Li Y, Yue P, Owonikoko TK, Ramalingam SS, Khuri FR, Sun SY. The combination of RAD001 and NVP-BEZ235 exerts synergistic anticancer activity against nonsmall cell lung cancer in vitro and in vivo. PLoS One. 2011;6:e20899. doi: 10.1371/journal.pone.0020899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Blommaart EF, Luiken JJ, Blommaart PJ, van Woerkom GM, Meijer AJ. Phosphorylation of ribosomal protein S6 is inhibitory for autophagy in isolated rat hepatocytes. J. Biol. Chem. 1995;270:2320–2326. doi: 10.1074/jbc.270.5.2320. [DOI] [PubMed] [Google Scholar]
- 49.Mathew R, Karantza-Wadsworth V, White E. Role of autophagy in cancer. Nat. Rev. Cancer. 2007;7:961–967. doi: 10.1038/nrc2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.He G, Yu GY, Temkin V, Ogata H, Kuntzen C, Sakurai T, Sieghart W, Peck-Radosavljevic M, Leffert HL, Karin M. Hepatocyte IKKb/NF-kB inhibits tumor promotion and progression by preventing oxidative stress-driven STAT3 activation. Cancer Cell. 2010;17:286–297. doi: 10.1016/j.ccr.2009.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carnevalli LS, Masuda K, Frigerio F, Le Bacquer O, Um SH, Gandin V, Topisirovic I, Sonenberg N, Thomas G, Kozma SC. S6K1 plays a critical role in early adipocyte differentiation. Dev. Cell. 2010;18:763–774. doi: 10.1016/j.devcel.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cloughesy TF, Yoshimoto K, Nghiemphu P, Brown K, Dang J, Zhu S, Hsueh T, Chen Y, Wang W, Youngkin D, Liau L, Martin N, Becker D, Bergsneider M, Lai A, Green R, Oglesby T, Koleto M, Trent J, Horvath S, Mischel PS, Mellinghoff IK, Sawyers CL. Antitumor activity of rapamycin in a phase I trial for patients with recurrent PTEN-deficient glioblastoma. PLoS Med. 2008;5:e8. doi: 10.1371/journal.pmed.0050008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 54.R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing; 2011. [Google Scholar]
- 55.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B. 1995;57:89–300. [Google Scholar]
- 56.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: A knowledge based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Andersen JB, Factor VM, Marquardt JU, Raggi C, Lee YH, Seo D, Conner EA, Thorgeirsson SS. An integrated genomic and epigenomic approach predicts therapeutic response to zebularine in human liver cancer. Sci. Transl. Med. 2010;2:54ra77. doi: 10.1126/scitranslmed.3001338. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.