Abstract
Background
CXCL13 and CXCR5 are a chemokine and receptor pair whose interaction is critical for naïve B cell trafficking and activation within germinal centers. We sought to determine whether CXCL13 levels are elevated prior to HIV-associated non-Hodgkin B-cell lymphoma (AIDS-NHL), and whether polymorphisms in CXCL13 or CXCR5 are associated with AIDS-NHL risk and CXCL13 levels in a large cohort of HIV-infected men.
Methods
CXCL13 levels were measured in sera from 179 AIDS-NHL cases and 179 controls at three time-points. TagSNPs in CXCL13 (n=16) and CXCR5 (n=11) were genotyped in 183 AIDS-NHL cases and 533 controls. Odds ratios (OR) and 95% confidence intervals (CIs) for the associations between one unit increase in log CXCL13 levels and AIDS-NHL, as well as tagSNP genotypes and AIDS-NHL, were computed using logistic regression. Mixed linear regression was used to estimate mean ratios (MR) for the association between tagSNPs and CXCL13 levels.
Results
CXCL13 levels were elevated >3 years (OR=3.24, 95% CI=1.90–5.54), 1–3 years (OR=3.39, 95% CI=1.94–5.94) and 0–1 year (OR=3.94, 95% CI=1.98–7.81) prior to an AIDS-NHL diagnosis. The minor allele of CXCL13 rs355689 was associated with reduced AIDS-NHL risk (ORTCvsTT=0.65; 95% CI=0.45–0.96) and reduced CXCL13 levels (MRCCvsTT=0.82, 95% CI=0.68–0.99). The minor allele of CXCR5 rs630923 was associated with increased CXCL13 levels (MRAAvsTT=2.40, 95% CI=1.43–4.50).
Conclusions
CXCL13 levels were elevated preceding an AIDS-NHL diagnosis, genetic variation in CXCL13 may contribute to AIDS-NHL risk, and CXCL13 levels may be associated with genetic variation in CXCL13 and CXCR5.
Impact
CXCL13 may serve as a biomarker for early AIDS-NHL detection.
Keywords: Non-Hodgkin Lymphoma, HIV, CXCL13, CXCR5, chemokine
INTRODUCTION
Among HIV-infected people, B cell non-Hodgkin lymphoma (AIDS-NHL) is currently the most commonly diagnosed cancer (1). The depletion of CD4+ T cells during chronic HIV infection contributes to the development of some AIDS-NHL, such as primary central nervous system lymphoma (PCNSL), through the loss of immunoregulatory control over Epstein-Barr virus (EBV) infected B cells (2). Some of the more common non-PCNSL (or systemic) AIDS-NHL subtypes, including diffuse large B cell lymphoma (DLBCL), however, tend to occur in individuals with relatively high CD4+ T cell numbers and a smaller proportion of these tumors are EBV-positive (2, 3). These cancers are thought to arise from downstream effects of chronic B cell activation, which is also a well-documented consequence of chronic HIV infection (4–8).
Chronic B cell activation results in an over-expression of the DNA-editing enzyme activation induced cytidine deaminase (AID), the actions of which are normally reserved for the highly regulated immunoglobulin gene (Ig) diversification reactions of class switch recombination and somatic hypermutation in germinal centers (9). High levels of AID expression can result in aberrant mutations to non-Ig genes and may cause the seminal translocations and mutations seen in AIDS-NHL (10, 11). Studies have shown significant associations between elevated levels of B cell activation-associated biomarkers, including many cytokines and soluble receptors, and AIDS-NHL risk (12–18). Although the exact mechanism driving chronic B cell activation is not known, one possibility that remains to be explored is the potential role of chemokines that regulate B cell trafficking.
CXCL13 (BCA-1, BLC) is a chemokine produced by follicular dendritic and T helper (particularly Th17) cells, in secondary lymphoid organs (19, 20). When bound to its receptor, CXCR5 (BLR1), CXCL13 plays a central role in homeostatic trafficking of antigen-naïve B cells into and within follicles of secondary lymphoid organs. This homing of naïve B cells is necessary for antigen exposure and activation within the germinal center reaction, which are essential elements in the development and structure of secondary lymphoid organs and in the differentiation of B cells into antibody-producing plasma cells (20–22). Aberrant CXCL13 and CXCR5 expression occurs in the setting of HIV infection preventing normal B cell migration, development, and differentiation (22–26). There is also growing evidence that CXCL13 and CXCR5 participate in the pathogenesis of multiple subtypes of lymphomas (27–37). Here we test three hypotheses in studies nested within a large prospective cohort of HIV-infected men: 1) elevated CXCL13 serum levels are associated with increased AIDS-NHL risk, 2) variation in the genes coding for CXCL13 and CXCR5 is associated with increased or decreased AIDS-NHL risk, and 3) variation in the genes coding for CXCL13 and CXCR 5 is associated with CXCL13 serum levels.
MATERIAL AND METHODS
Study Design and Population
The Multicenter AIDS Cohort Study (MACS), established in 1983, includes 6,972 men who have sex with men from four metropolitan areas recruited between 1984 and 2003 (38–41). Participants are re-contacted semiannually for an in-person interview, physical exam, and specimen collection. Antiretroviral use at each visit is summarized according to the Department of Health and Human Services/Kaiser Panel to define HAART usage (42). HIV seropositivity and plasma load, and CD4+ T cell counts, are measured at nearly all study visits, and sera and cell pellets are collected and stored in central repositories (43). Protocols and questionnaires have been approved by the Institutional Review Board from each center and all participants provided written informed consent.
Case and Control Definitions
AIDS-NHL is ascertained in the MACS through self-report with confirmation by pathology records and state cancer registries, or is identified at autopsy. A detailed description of the design and participant selection for the serum marker association study is provided elsewhere (12). Briefly, cases included all MACS participants with AIDS-NHL diagnosed prior to April, 2003 with at least one available pre-diagnosis serum specimen (n=179). One HIV-infected control was selected per case and matched on duration of HIV infection based on known date of infection (21 cases), or date of entry (± 1 year) into MACS for men seroprevalent at enrollment (158 cases), and sample availability at equivalent time-points. Using prospectively collected and stored serum, up to three serum samples were obtained for each participant: that which were collected >3 years pre-NHL (n=147), 1–3 years pre-NHL (n=148), and 0–1 year pre-NHL (n=98) in cases and follow-up matched time-points in controls (n=145, n=145, and n=103, respectively).
Cases in the genetic association study included all HIV-infected MACS participants with AIDS-NHL diagnosed prior to July, 2010 with available cells for DNA extraction (n=183). Among the 183 selected cases, 172 (94%) overlapped with cases included in the serum study, while 11 and 7 cases were unique to the genetic and serum studies, respectively (Figure 1). These differences in case numbers are due to lack of available specimen (DNA or serum) and identification of new cases between 2003 and 2010. Controls in the genetic study were selected from all HIV-infected MACS participants with available specimens and matched individually to each case in up to a 3:1 ratio on recruitment period, race, HIV positive follow-up time (± 1 year), and CD4+ T cell count (in categories of 0–49, 50–99, 100–199, 200–349, 350–499, and 500+) at last measurement pre-AIDS-NHL diagnosis for cases and follow-up matched time-point in controls. Among the 533 selected controls, 50 (9%) were included as controls in the serum study.
Figure 1. Number of cases and controls and overlap between studies.
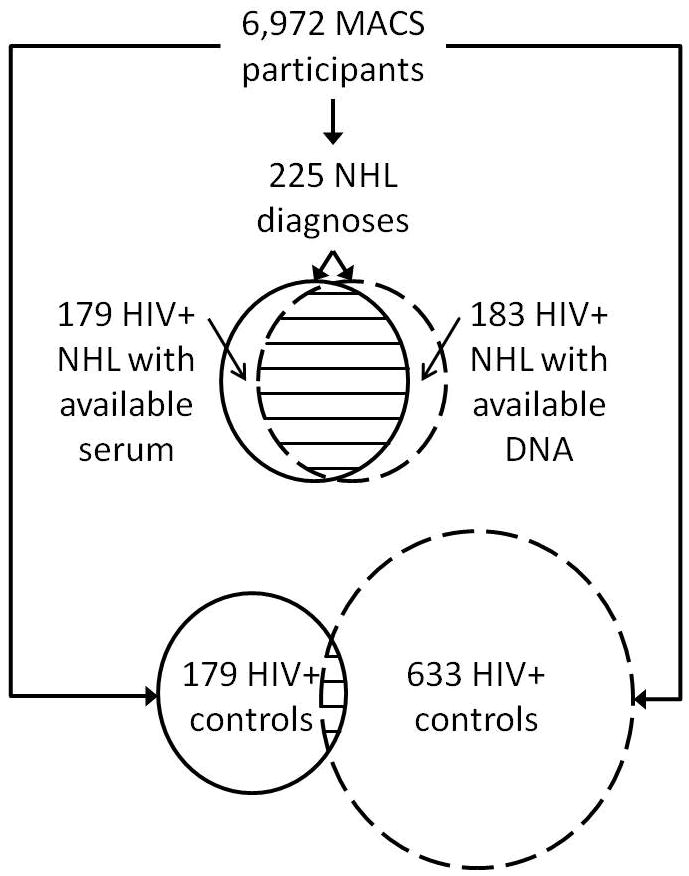
Among the 6,972 participants in the Multicenter AIDS Cohort Study (MACS), 225 have been diagnosed with B cell non-Hodgkin lymphoma (NHL), 179 of whom had available serum and were included in the serum marker association study (top circle with solid line) and 183 of whom had available DNA and were included in the genetic association study (top circle with hashed line); 172 cases were included in both studies. For controls, 179 HIV+ participants were selected for the serum association study (bottom circle with solid line) and 633 HIV+ controls were selected for the genetic association study (bottom circle with hashed line); 50 controls were included in both studies.
To investigate the association between genetic variation in CXCL13 and CXCR5 and CXCL13 serum levels, we included all the cases that were included in both the genetic study and serum study (n=172). Since the overlap of controls was low (n=50), we genotyped additional DNA specimens from serum study controls. In total, there were 172 cases and 174 controls with both CXCL13 serum levels and CXCL13/CXCR5 genotyping results included in this analysis.
Tissue Samples
Archival tissue blocks from biopsy, surgery, or autopsy were obtained for 99 AIDS-NHL cases. EBV detection was performed using EBER in situ hybridization or LMP1 immunohistochemistry in 87 cases, as described previously (44). A sample was considered positive if positive for EBER or LMP1.
Serum CXCL13 Determination
Serum levels of CXCL13 were measured in sera by ELISA (R&D Systems, Minneapolis, MN) using an automated plate washer and VersaMax microplate reader and software (Molecular Devices, Sunnyvale, CA), according to manufacturer’s protocol. No samples were below the lower detection limit of 7.8 pg/ml. The interassay coefficient of variation was 8%. Data for additional cytokines and immune markers (sCD30, sCD27, IL6, TNFα, IL10, neopterin, and IP10) were included in multivariate regression models. Methods for measurement of these markers and their association with AIDS-NHL risk have been described previously (12, 45).
CXCL13 and CXCR5 TagSNP Selection and Genotype Determination
All SNPs within coding and non-coding regions of CXCL13 and CXCR5 plus 10 kilobases of flanking sequence on each end of each gene were identified using the European descent genotype data from the International HAPMAP Project (46). We considered only SNPs with a minor allele frequency (MAF) ≥5% which included 40 out of 50 SNPs in CXCL13 and 18 out of 20 SNPs in CXCR5. A pairwise r2≥0.80 was used to delineate groups of highly correlated SNPs within each gene, and one SNP (i.e. tagSNP) per group was selected for genotyping (47). Priority was given to potential tagSNPs with high Illumina design scores and those located in exons or other putative functional regions. Genomic DNA was extracted from cell pellets using the QIAmp DNA Blood Mini Kit according to manufacturer’s protocol (Qiagen Inc., Valencia, CA). Genotyping was performed using a customized GoldenGate® assay according to manufacturer’s protocol (Illumina Inc., San Diego, CA). Two to three positive controls (samples known to be heterozygous or homozygous for each allele based on sequencing) and negative controls (wells containing no DNA) were included in each reaction plate. Quality control (QC) replicate DNA aliquots for 5% of study subjects were distributed throughout the reaction plates. Analysis of these 44 replicate pairs revealed a high concordance proportion of 99.97%. Laboratory personnel were blinded to all research information about the samples, including the identities of the QC replicate aliquots.
One CXCR5 tagSNP (rs676925) failed to be genotyped. All other tagSNP genotypes were tested for consistency with Hardy-Weinberg equilibrium (HWE) in the control sample. Two CXCL13 tagSNPs (rs17002760 and rs171388) had HWE p-values <0.001 and were excluded from further analysis. All other tagSNPs had an HWE p-value >0.01, MAF ≥5% in our study population, and call rates greater than 93% (average call rate=98.6%). All pairwise r2 values between tagSNPs were <0.60.
Statistical Analysis
Means and frequencies were calculated for select covariates separately for cases and controls. CD4+ T cell slope was defined as the change in number of CD4+ T cells per year and was calculated using pre-HAART CD4+ T cell counts for men with at least two measurements. Only CD4+ T cell counts starting with the third HIV positive study visit were included in the calculation. For participants who seroconverted during follow-up, HIV RNA at set point refers to the average HIV RNA measurement 12–24.5 months after seroconversion such that the interim slope approximated zero. For seroprevalent men recruited in 1984, HIV at set point was estimated as the measurement obtained at study visit 3 or 4. Unmatched comparisons of mean natural log transformed (loge) CXCL13 levels at each time-point between cases and controls was performed using t-tests, reported as back-transformed geometric means. Spearman’s correlation coefficients (ρ) were calculated for all pairwise associations between biomarkers at each sampling time-point for cases and controls separately.
Odds ratios (ORs) and 95% confidence intervals (95% CIs) for the association between loge CXCL13 (continuous variable) at each time-point and AIDS-NHL risk were computed using random effects multivariate logistic regression models, and included 179 cases and 179 controls. The ORs represent risk of AIDS-NHL associated with one unit increase in loge CXCL13. The matching by design between each case/control pair was incorporated into the models by adding a random effect term allowing for correlation within the sets and assuming independence between sets. Covariates were included in the models if they were strong predictors of AIDS-NHL risk and plausibly related to HIV disease progression or CXCL13 levels. These included age at AIDS-NHL diagnosis in cases or reference date in controls, absolute CD4+ T cell counts from each visit where CXCL13 was measured, loge HIV RNA levels at set point, having an AIDS diagnosis prior to AIDS-NHL diagnosis or reference date, and having been treated with HAART prior to AIDS-NHL diagnosis or reference date. Reference date in controls was determined by duration of HIV-positive follow-up time of the case in the matched set. Multiple imputation was used to estimate missing covariate data (48). Additionally, multivariate models were estimated including loge CXCL13 levels, covariates, and seven other serum biomarkers related to B cell activation, which were previously found to be strongly associated with AIDS-NHL (sCD30, sCD27, IL6, TNFα, IL10, neopterin, and IP10) (12, 45).
To estimate the association between genotypes of CXCL13 and CXCR5 tagSNPs and AIDS-NHL risk, we computed multivariate ORs and 95% CIs using random effects multivariate logistic regression models, which included 183 cases and 533 controls, and were adjusted for age at AIDS-NHL diagnosis or reference date, absolute CD4+ T cell counts from the visit closest and prior to AIDS-NHL diagnosis date or reference date, loge HIV viral RNA levels at set point, having an AIDS diagnosis prior to AIDS-NHL diagnosis or reference date, having exposure to HAART prior to AIDS-NHL diagnosis or reference date, and race/ethnicity.
The association between genotypes of tagSNPs and mean loge CXCL13 levels was modeled using repeated measures (mixed) linear regression to estimate mean ratios (MR) and 95% CI, and included 172 cases and 174 controls and the following covariates as fixed effects: age at AIDS-NHL diagnosis or reference date, absolute CD4+ T cell counts from each visit where CXCL13 was measured, loge HIV viral RNA levels at set point, having an AIDS diagnosis prior to AIDS-NHL diagnosis or reference date, having exposure to HAART prior to AIDS-NHL diagnosis or reference date, and race/ethnicity. Two random effect variables were included, one to account for the non-independence of observations on the same subject across the three visits and a second to account for the non-independence between each case and its matched control. The variance of loge CXCL13 explained by each tagSNP was estimated by subtracting the r2 value for a model that included the parameters for each tagSNP from the r2 for a model that excluded the tagSNP.
RESULTS
Cases and controls were of similar age, and the majority were White, non-Hispanic (Table 1). Due to differences in control matching between the serum marker study and the genetic association study, there were larger differences between cases and controls from the serum study compared to the genetic study for several variables related to HIV disease progression, including median HIV RNA at setpoint, having a prior AIDS illness, CD4+ T cell slope, and CD4+ T cell count. Less than 10% of the study population was treated with HAART. The majority of tumors were systemic (68%), most of which were DLBCL. Fewer than 50% of cases had adequate tumor tissue available for EBV testing. A majority of tested specimens were EBV positive (68%), and a larger proportion of PCNSL tumors (89%) were EBV-positive compared to systemic tumors (58%).
Table 1.
Select characteristics of HIV positive cases and controls from the Multicenter AIDS Cohort Study
| Serum marker association study | Genetic association study | |||
|---|---|---|---|---|
|
| ||||
| AIDS-NHL cases | HIV+ controls | AIDS-NHL cases | HIV+ controls | |
|
| ||||
| N | 179 | 179 | 183 | 533 |
| Baseline visit year, N (%) | ||||
| 1984–1985 | 158 (88) | 160 (89) | 163 (89) | 479 (90) |
| 1987–1991 | 21 (12) | 19 (11) | 20 (11) | 54 (10) |
| Reference year, N (%)a | ||||
| 1984–1995 | 155 (86) | 155 (86) | 155 (85) | 450 (84) |
| 1996–2001 | 23 (13) | 24 (13) | 22 (12) | 67 (13) |
| 2002–2006 | 1 (1) | 0 | 6 (3) | 6 (3) |
| MACS site, N (%) | ||||
| Baltimore | 44 (25) | 37 (21) | 45 (25) | 119 (22) |
| Chicago | 42 (23) | 33 (18) | 44 (24) | 130 (25) |
| Pittsburgh | 22 (12) | 39 (22) | 22 (12) | 86 (16) |
| Los Angeles | 71 (40) | 70 (39) | 72 (39) | 198 (37) |
| Age, median years (range)a | 41 (24–60) | 39 (24–60) | 41 (24–60) | 40 (24–70) |
| Race/Ethnicity, N (%) | ||||
| White, non-Hispanic | 149 (83) | 156 (87) | 153 (84) | 483 (90) |
| White, Hispanic | 19 (11) | 8 (4) | 18 (10) | 25 (5) |
| Black, non-Hispanic | 11 (6) | 10 (6) | 12 (6) | 21 (4) |
| Other | 0 | 5 (3) | 0 | 4 (1) |
| HIV RNA at set point, median (range) | 31,133 (400– 960,960) | 14,467 (300– 237,951) | 31,564 (400– 960,960) | 20,698 (300– 672,810) |
| Prior AIDS illness, N (%)b | 94 (53) | 20 (11) | 95 (52) | 220 (41) |
| CD4+ T cell slope, median cells per year (range)c | −74 (−283 to 178) | −44 (−188 to 84) | −67 (−283 to 178) | −60 (−370 to 812) |
| CD4+ T cell count, median cells/mm3 (range)d | 74 (0–707) | 468 (4–1255) | 79 (2–923) | 87 (1–1361) |
| Prior HAART exposure, N (%)b | 8 (4) | 8 (4) | 11 (6) | 46 (9) |
| Tumor Subtype, N (%) | ||||
| Systemic | 121 (68) | – | 125 (68) | – |
| Diffuse large B cell | 60 (50) | – | 65 (52) | – |
| Burkitt lymphoma/BL-like | 21 (17) | – | 21 (17) | – |
| Other subtypes | 6 ( 5) | – | 7 (6) | – |
| Not specified | 34 (28) | – | 32 (26) | – |
| Central nervous system | 58 (32) | – | 58 (32) | – |
| Tumor EBV status, N (%) | ||||
| Not tested | 92 (51) | – | 97 (53) | – |
| Tested | 87 (49) | – | 86 (47) | – |
| Negative | 28 (32) | – | 28 (32) | – |
| Positivee | 59 (68) | – | 58 (67) | – |
At time of AIDS-NHL diagnosis in cases and reference date in controls (determined by matching HIV positive follow-up time to that of the case in the matched set)
At least 30 days prior to AIDS-NHL diagnosis date in cases and reference date in controls
Calculated from all available pre-HAART CD4+ T cell count data
For the serum marker association study cell counts from at 0–1 years prior to AIDS-NHL diagnosis in cases and reference date in controls. For the genetic association study, cell counts from the visit most closely preceding AIDS-NHL diagnosis in cases and reference date in controls
34/59 (58%) of systemic AIDS-NHL tested were EBV positive compared to 25/28 (89%) of PCNSL
Mean CXCL13 levels were consistent among the three time-points in the controls (74, 80, and 83 pg/mL, Figure 2). Mean CXCL13 levels were significantly higher in cases compared to controls at each time-point (p<0.001) and increased as time approached AIDS-NHL diagnosis date (123, 153, 178 pg/mL). In the adjusted models, a one unit increase in loge CXCL13 levels was significantly associated with AIDS-NHL risk at all three time-points (>3 years: OR=3.24, 95% CI=1.90–5.54; 1–3 years: OR=3.39, 95% CI=1.94–5.94; 0–1 year: OR=3.94, 95% CI=1.98–7.81, Table 2). In stratified analyses, the association between CXCL13 levels and AIDS-NHL appeared to be stronger for systemic lymphoma compared to PCNSL at the >3 years and 1–3 years time-points, although the differences in ORs were not statistically significant. At the time-point furthest from AIDS-NHL diagnosis, the association between CXCL13 and AIDS-NHL was stronger for EBV-negative compared to EBV-positive AIDS-NHLs. As time approached NHL diagnosis date, higher ORs were observed for EBV-positive tumors, although the differences in ORs by EBV status were not statistically significant.
Figure 2. CXCL13 levels in HIV positive non-Hodgkin lymphoma cases and controls at three time-points.
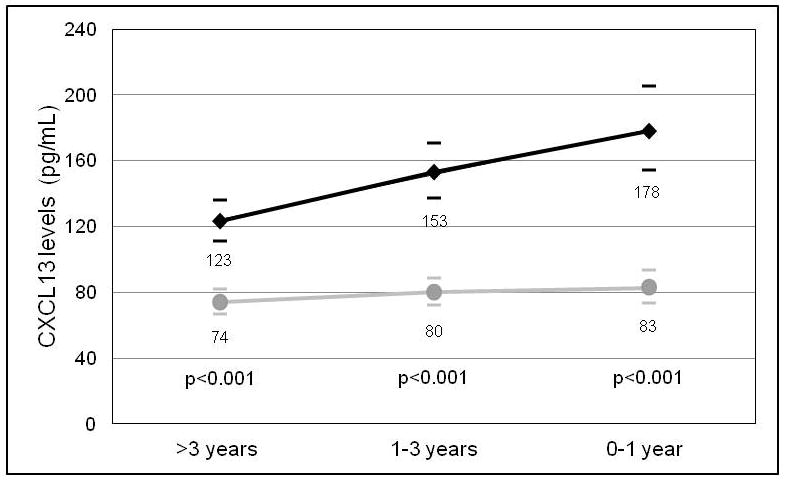
Geometric mean values of serum CXCL13 (pg/mL) in HIV positive non-Hodgkin lymphoma cases (black diamonds) and HIV positive controls (grey circles), with 95 % confidence intervals (bars) at three time-points (>3 years, 1–3 years, or 0–1 year) preceding cancer diagnosis date in the cases, or reference date in the controls. P-values were calculated for the difference in the means between cases and controls at each time-point.
Table 2.
Association between CXCL13 serum levels and HIV-associated non-Hodgkin lymphoma risk at three time-points
| Model | >3 years
|
1–3 years
|
0–1 year
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| N | ORa | 95% CI | N | OR | 95% CI | N | OR | 95% CI | |
|
|
|
|
|||||||
| Crude | 292 | 3.77 | (2.45–5.84) | 293 | 4.88 | (3.12–7.67) | 201 | 5.88 | (3.36–10.3) |
| Adjustedb | |||||||||
| All cases and controls | 292 | 3.24 | (1.90–5.54) | 293 | 3.39 | (1.94–5.94) | 201 | 3.94 | (1.98–7.81) |
| Systemic | 198 | 4.06 | (2.12–7.78) | 190 | 3.84 | (2.01–7.37) | 147 | 3.59 | (1.68–7.72) |
| Central nervous system | 94 | 1.92 | (0.68–5.43) | 103 | 2.28 | (0.62–8.41) | 54 | NEd | |
| p-valuec | 0.24 | 0.49 | |||||||
| EBV negative | 42 | 6.56 | (1.15–37.4) | 49 | 3.80 | (1.04–14.0) | 41 | 4.23 | (0.78–23.0) |
| EBV positive | 93 | 3.22 | (1.24–8.38) | 105 | 4.41 | (1.33–14.7) | 68 | 8.98 | (1.66–48.8) |
| p-valuec | 0.49 | 0.87 | 0.54 | ||||||
OR indicates odds ratio; CI, confidence interval
The ORs represent risk of AIDS-NHL associated with one unit increase in loge CXCL13
Adjusted for age at AIDS-NHL diagnosis in cases or reference date in controls, absolute CD4+ T cell counts from each visit where CXCL13 was measured, loge HIV viral RNA levels at set point, having an AIDS diagnosis prior to AIDS-NHL diagnosis or reference date, and having exposure to HAART prior to AIDS-NHL diagnosis or reference date
P-value for the difference in ORs between each strata
Model did not converge
In multivariate models that included CXCL13 levels and seven other serum biomarkers, the highest statistically significant OR was observed for CXCL13 (OR=2.68, 95% CI=1.38–5.26) at the >3 years time-point (Table 3). At the 1–3 year time-point, none of the biomarkers were independently significantly associated with AIDS-NHL, however in strength of association CXCL13 was second highest following neopterin. At the 0–1 year time-point, neopterin was the most strongly associated with AIDS-NHL risk (OR=5.18, 95% CI=1.36–19.8). Many of these biomarkers were positively correlated, however no two exhibited correlation greater than ρ=0.72, with most between ρ=0.20 and ρ=0.50 (Supplementary Table 1).
Table 3.
Multivariate analysis of CXCL13 and other biomarkers related to B cell activation and HIV-associated non-Hodgkin lymphoma risk
| >3 years
|
1–3 years
|
0–1 year
|
||||
|---|---|---|---|---|---|---|
| ORa | 95% CI | ORa | 95% CI | ORa | 95% CI | |
|
|
|
|
||||
| CXCL13 | 2.68 | (1.38–5.26) | 1.73 | (0.84–3.58) | 1.63 | (0.71–3.82) |
| sCD30 | 1.00 | (0.40–2.54) | 1.58 | (0.67–3.78) | 2.50 | (0.76–8.24) |
| sCD27 | 0.39 | (0.10–1.51) | 0.70 | (0.22–2.35) | 0.29 | (0.07–1.34) |
| IL6 | 1.88 | (1.17–3.03) | 0.94 | (0.52–1.74) | 1.64 | (0.85–3.22) |
| TNFα | 1.62 | (0.98–2.69) | 1.54 | (0.84–2.83) | 1.29 | (0.80–2.11) |
| IL10 | 1.09 | (0.86–1.38) | 1.27 | (0.95–1.71) | 1.38 | (0.94–2.05) |
| Neopterin | 1.47 | (0.50–4.40) | 2.41 | (0.83–7.03) | 5.18 | (1.36–19.8) |
| IP10 | 1.14 | (0.55–2.39) | 1.28 | (0.61–2.70) | 1.73 | (0.72–4.18) |
OR indicates odds ratio; CI, confidence interval
The ORs represent risk of AIDS-NHL associated with one unit increase in the log transformed biomarker. ORs are adjusted for age at AIDS-NHL diagnosis in cases or reference date in controls, absolute CD4+ T cell counts from each visit where the biomarkers were measured, loge HIV viral RNA levels at set point, and having an AIDS diagnosis prior to AIDS-NHL diagnosis or reference date, and having exposure to HAART prior to AIDS-NHL diagnosis or reference date
AIDS-NHL risk was inversely associated with carriership of the minor allele of CXCL13 rs355689; OR=0.66, 95%, CI=0.46–0.97 for heterozygotes and OR=0.87, 95% CI=0.5–1.55 for homozygotes, compared to major allele homozygotes (Table 4). CXCL13 levels were inversely associated with carriership of the minor allele of rs355689; MR=0.82, 95% CI=0.68–0.99 for minor allele homozygotes compared to major allele homozygotes (Figure 3, Supplementary Table 2). Two additional SNPs were associated with CXCL13 levels, CXCL13 rs17002733 and CXCR5 rs630923 (Figure 3), although these risk estimates were based on only a few minor allele homozygote study subjects. The percentage of variation in loge CXCL13 levels explained by rs355689, rs17002733, and rs630923 was 2.3%, 4.2%, and 3.2%, respectively. CXCL13 levels were strongly associated with AIDS-NHL risk at all three time-points within each genotype strata for rs355689, rs1700273, and rs630923 (Supplementary Table 3).
Table 4.
Association between CXCL13 and CXCR5 tagSNPs and HIV-associated non-Hodgkin lymphoma risk
| HUGO gene name | dbSNP reference sequence number | SNP context | Genotype | Cases
|
Controls
|
ORa | 95% CI | ||
|---|---|---|---|---|---|---|---|---|---|
| N | % | N | % | ||||||
| CXCL13 | rs1442691 | 5′ flanking | TT | 157 | 85.8 | 471 | 89.5 | 1.00 | |
| TC | 24 | 13.1 | 52 | 9.9 | 1.23 | (0.71–2.14) | |||
| CC | 2 | 1.1 | 3 | 0.6 | 1.61 | (0.23–11.3) | |||
| CXCL13 | rs2053526 | 5′ flanking | AA | 72 | 40.0 | 195 | 37.7 | 1.00 | |
| AG | 73 | 40.6 | 219 | 42.4 | 0.86 | (0.59–1.28) | |||
| GG | 35 | 19.4 | 103 | 19.9 | 0.76 | (0.46–1.27) | |||
| CXCL13 | rs6852819 | 5′ flanking | GG | 118 | 64.5 | 348 | 66.5 | 1.00 | |
| TG | 58 | 31.7 | 156 | 29.8 | 1.07 | (0.73–1.57) | |||
| TT | 7 | 3.8 | 19 | 3.6 | 1.04 | (0.42–2.63) | |||
| CXCL13 | rs17406477 | Intron | GG | 149 | 81.9 | 430 | 81.4 | 1.00 | |
| AG | 32 | 17.6 | 94 | 17.8 | 0.93 | (0.59–1.48) | |||
| AA | 1 | 0.5 | 4 | 0.8 | 0.90 | (0.10–8.50) | |||
| CXCL13 | rs355679 | Intron | GG | 122 | 67.0 | 348 | 66.7 | 1.00 | |
| TG | 57 | 31.3 | 156 | 29.9 | 1.00 | (0.69–1.47) | |||
| TT | 3 | 1.6 | 18 | 3.4 | 0.41 | (0.12–1.46) | |||
| CXCL13 | rs2119976 | Intron | CC | 155 | 84.7 | 460 | 88.0 | 1.00 | |
| AC | 28 | 15.3 | 59 | 11.3 | 1.00 | (0.69–1.47) | |||
| AA | 0 | 0.0 | 4 | 0.8 | NE† | ||||
| CXCL13 | rs189587 | Intron | GG | 64 | 35.2 | 185 | 35.3 | 1.00 | |
| AG | 88 | 48.4 | 254 | 48.5 | 0.89 | (0.61–1.31) | |||
| AA | 30 | 16.5 | 85 | 16.2 | 0.93 | (0.55–1.59) | |||
| CXCL13 | rs355686 | Intron | CC | 111 | 60.7 | 333 | 63.4 | 1.00 | |
| TC | 67 | 36.6 | 165 | 31.4 | 1.20 | (0.83–1.73) | |||
| TT | 5 | 2.7 | 27 | 5.1 | 0.57 | (0.21–1.54) | |||
| CXCL13 | rs355689 | Intron | TT | 102 | 56.4 | 256 | 49.2 | 1.00 | |
| TC | 58 | 32.0 | 206 | 39.6 | 0.65 | (0.45–0.96) | |||
| CC | 21 | 11.6 | 58 | 11.2 | 0.84 | (0.47–1.49) | |||
| CXCL13 | rs17002733 | Intron | CC | 150 | 82.0 | 406 | 78.1 | 1.00 | |
| TC | 29 | 15.8 | 108 | 20.8 | 0.67 | (0.42–1.07) | |||
| TT | 4 | 2.2 | 6 | 1.2 | 1.48 | (0.39–5.66) | |||
| CXCL13 | rs171388 | Intron | CC | 126 | 69.2 | 348 | 65.7 | 1.00 | |
| CT | 49 | 26.9 | 166 | 31.3 | 0.74 | (0.50–1.10) | |||
| TT | 7 | 3.9 | 16 | 3.0 | 0.91 | (0.34–2.46) | |||
| CXCL13 | rs355661 | Intron | AA | 162 | 89.5 | 464 | 88.7 | 1.00 | |
| AG | 18 | 9.9 | 59 | 11.3 | 0.79 | (0.44–1.41) | |||
| GG | 1 | 0.6 | 0 | 0.0 | NEb | ||||
| CXCL13 | rs1052563 | 3′ UTR | TT | 150 | 82.4 | 414 | 78.6 | 1.00 | |
| TC | 28 | 15.4 | 108 | 20.5 | 0.66 | (0.41–1.06) | |||
| CC | 4 | 2.2 | 5 | 0.9 | 3.05 | (0.78–12.0) | |||
| CXCL13 | rs10022693 | 3′ flanking | TT | 91 | 49.7 | 233 | 46.3 | 1.00 | |
| TC | 70 | 38.3 | 206 | 41.0 | 0.79 | (0.54–1.15) | |||
| CC | 22 | 12.0 | 64 | 12.7 | 0.72 | (0.41–1.32) | |||
| CXCL13 | rs17002760 | 3′ flanking | AA | 97 | 53.6 | 282 | 54.5 | 1.00 | |
| AT | 68 | 37.6 | 198 | 38.3 | 0.92 | (0.64–1.34) | |||
| TT | 16 | 8.8 | 37 | 7.2 | 1.23 | (0.64–2.40) | |||
| CXCL13 | rs1566485 | 3′ flanking | GG | 69 | 38.3 | 165 | 32.3 | 1.00 | |
| TG | 75 | 41.7 | 242 | 47.4 | 0.69 | (0.47–1.02) | |||
| TT | 36 | 20.0 | 104 | 20.4 | 0.77 | (0.47–1.26) | |||
| CXCR5 | rs11217077 | 5′ flanking | TT | 109 | 59.6 | 309 | 59.2 | 1.00 | |
| TC | 66 | 36.1 | 190 | 36.4 | 1.03 | (0.72–1.50) | |||
| CC | 8 | 4.4 | 23 | 4.4 | 1.11 | (0.48–2.64) | |||
| CXCR5 | rs4938576 | 5′ flanking | GG | 63 | 34.8 | 163 | 31.7 | 1.00 | |
| TG | 86 | 47.5 | 267 | 51.8 | 0.75 | (0.52–1.12) | |||
| TT | 32 | 17.7 | 85 | 16.5 | 0.94 | (0.56–1.60) | |||
| CXCR5 | rs6589706 | 5′ flanking | GG | 54 | 29.8 | 150 | 28.9 | 1.00 | |
| AG | 95 | 52.5 | 263 | 50.7 | 0.98 | (0.65–1.48) | |||
| AA | 32 | 17.7 | 106 | 20.4 | 0.87 | (0.52–1.48) | |||
| CXCR5 | rs11217078 | 5′ flanking | TT | 71 | 38.8 | 220 | 42.9 | 1.00 | |
| TC | 97 | 53.0 | 240 | 46.8 | 1.25 | (0.87–1.81) | |||
| CC | 15 | 8.2 | 53 | 10.3 | 1.00 | (0.52–1.92) | |||
| CXCR5 | rs10892306 | 5′ flanking | TT | 153 | 84.5 | 454 | 87.8 | 1.00 | |
| TA | 26 | 14.4 | 60 | 11.6 | 1.09 | (0.65–1.84) | |||
| AA | 2 | 1.1 | 3 | 0.6 | 1.51 | (0.24–9.81) | |||
| CXCR5 | rs630923 | 5′ flanking | CC | 133 | 73.9 | 366 | 71.8 | 1.00 | |
| AC | 46 | 25.6 | 134 | 26.3 | 0.93 | (0.63–1.41) | |||
| AA | 1 | 0.6 | 10 | 2.0 | 0.26 | (0.04–2.12) | |||
| CXCR5 | rs10892307 | 5′ UTR | CC | 126 | 68.9 | 378 | 72.8 | 1.00 | |
| GC | 50 | 27.3 | 130 | 25.0 | 1.19 | (0.81–1.78) | |||
| GG | 7 | 3.8 | 11 | 2.1 | 2.12 | (0.77–5.84) | |||
| CXCR5 | rs523604 | Intron | AA | 43 | 24.4 | 148 | 29.7 | 1.00 | |
| AG | 92 | 52.3 | 255 | 51.2 | 1.16 | (0.76–1.79) | |||
| GG | 41 | 23.3 | 95 | 19.1 | 1.48 | (0.88–2.50) | |||
| CXCR5 | rs1623316 | Intron | CC | 74 | 41.6 | 214 | 41.8 | 1.00 | |
| GC | 73 | 41.0 | 222 | 43.4 | 0.89 | (0.61–1.31) | |||
| GG | 31 | 17.4 | 76 | 14.8 | 1.08 | (0.65–1.81) | |||
| CXCR5 | rs2230321 | SYN | CC | 172 | 94.0 | 497 | 74.3 | 1.00 | |
| TC | 11 | 6.0 | 172 | 25.7 | 1.43 | (0.63–3.29) | |||
| CXCR5 | rs3922 | 3′ UTR | TT | 57 | 31.8 | 154 | 30.0 | 1.00 | |
| TC | 91 | 50.8 | 268 | 52.2 | 0.89 | (0.61–1.33) | |||
| CC | 31 | 17.3 | 91 | 17.7 | 0.89 | (0.53–1.52) | |||
OR indicates odds ratio; CI, confidence interval
Adjusted for age at AIDS-NHL diagnosis in cases or reference date in controls, absolute CD4+ T cell counts from the visit closest to (and prior to) AIDS-NHL diagnosis in cases and reference date in controls, loge HIV viral RNA levels at set point, having an AIDS diagnosis prior to AIDS-NHL diagnosis or reference date, having exposure to HAART prior to AIDS-NHL diagnosis or reference date, and race/ethnicity
Model did not converge
Figure 3. CXCL13 levels by CXCL13 and CXCR5 tagSNPs.
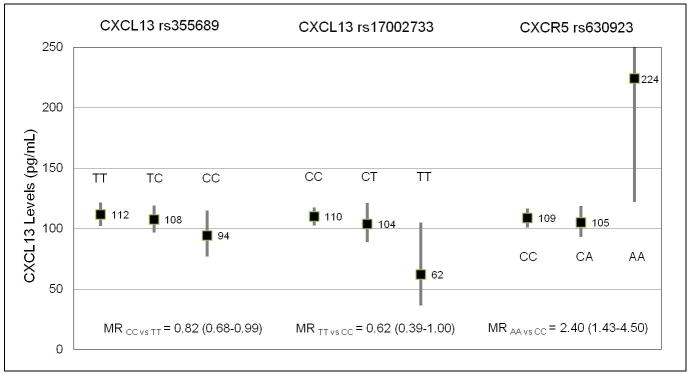
Geometric mean values of serum CXCL13 (pg/mL) in cases and controls by genotypes (black boxes) with 95% confidence intervals (bars) for three tagSNPs: CXCL13 rs355689, CXCL13 rs17002733, and CXCR5 rs630923. Adjusted mean ratios (MRs) and 95% confidence intervals are indicated for each tagSNP for homozygote minor allele carriers versus major allele homozygote carriers.
DISCUSSION
There is accumulating evidence that CXCL13 and CXCR5 are involved in the pathogenesis of lymphoma in general, and in particular, of extranodal lymphomas which comprise the majority of AIDS-NHL (3). Here, we show that CXCL13 serum levels predict B cell non-Hodgkin lymphoma risk in a cohort of HIV-infected men. The consistent, significant elevation of CXCL13 at three separate time-points across three or more years prior to the diagnosis of AIDS-NHL, and the significant association between CXCL13 and AIDS-NHL after adjustment for AIDS-NHL risk factors and other biomarkers, suggests that CXCL13 may be one of the drivers of the chronic B cell activation phenotype observed in AIDS-NHL.
In a healthy system, CXCL13 is constitutively expressed by T helper cells in lymphoid follicles where it regulates B cell homing (20). CXCL13 has been detected in follicular lymphomas and extranodal lymphomas occurring in a variety of organs including the brain, eyes, stomach, and skin (29, 32)(30)(33)(34). The source of CXCL13 in these lymphomas appears to vary by tumor site and includes malignant B cells, neighboring cells in the tumor environment, and cells produced elsewhere in the body. Malignant B cells of extranodal lymphomas express high levels of CXCR5, (27, 28, 30, 32, 35), the CXCL13 receptor which is normally only expressed in lymphoid follicles (49).
There are at least three possible explanations for our observation that CXCL13 is elevated prior to AIDS-NHL. One possibility is that CXCL13 and CXCR5 drive the formation of ectopic germinal centers, permitting chronic B cell activation and the accumulation of mutations and translocations which contribute to the pathogenesis of lymphoma (50). Alternatively, CXCL13 may be responsible for attracting CXCR5-expressing malignant B cells to these extranodal sites leading to the formation of tumors (27, 37). Lastly, it is possible that malignant B cells from the developing tumors, and/or tumor-infiltrating macrophages and/or T cells, are secreting CXCL13, and that CXCL13 is not contributing to the etiology of these tumors, but may be promoting their growth. However, the strong and consistent association between CXCL13 and AIDS-NHL risk for over three years suggests an etiological role of CXCL13 in AIDS-NHL.
Despite the accumulating in vitro evidence for CXCL13 and CXCR5 in the pathogenesis of lymphoma, there is only one prior epidemiologic study on CXCL13 and lymphoma, in HIV-infected or uninfected populations. Widney et. al. reported that CXCL13 serum levels are elevated up to 2.5 years (mean of 8.2 months) prior to cancer diagnosis in a small sample (n=46) of AIDS-NHL cases when compared to similarly sized groups of either AIDS controls without cancer, or HIV-infected controls (27). Our study confirms and extends this observation utilizing a larger sample of AIDS-NHL cases and HIV-infected controls sampled at three time-points prior to cancer diagnosis.
In stratified models, CXCL13 was significantly associated with systemic lymphoma but not PCNSL, and the point estimates were also higher for systemic lymphoma. These findings are consistent with what has been observed for other B cell activation associated biomarkers and AIDS-NHL (12). Among those cases with known EBV tumor status (n=87, 49%), CXCL13 was more strongly associated with EBV-negative tumors compared to EBV positive tumors at the time-point furthest from NHL diagnosis. In our study, 58% of systemic tumors and 89% of PCNSL tumors were EBV-positive. These findings are consistent with the hypothesis that systemic AIDS-NHLs are more likely to arise from the downstream effects of chronic B cell activation occurring over several years while PCNSL tumors arise principally due to the oncogenic properties of uncontrolled EBV reactivation in severely immunocompromised individuals (3, 15).
Mean CXCL13 levels in cases, but not controls, increased with time to diagnosis, while the ORs remained consistent. Previous studies have shown that CXCL13 serum levels are elevated in HIV-infected patients compared to healthy controls (24), and increase with HIV disease progression (25). Thus, our observation regarding increasing CXCL13 levels with time to diagnosis may reflect a higher rate of HIV disease progression in cases, which is accounted for in the CD4+ T cell count- and HIV RNA level–adjusted OR estimates. A previous study showed that B cell migration towards CXCL13 was higher in HIV-infected patients with low CD4+ T cell counts (<350 cells/μL) compared to patients with higher cell counts (>350 cells/μL) or healthy controls, suggesting an increased functional effect of CXCL13 in advanced HIV disease (24). Alternatively, the developing tumor may be responsible for the increasing CXCL13 levels with time to diagnosis in cases. However, the strong and consistent association between CXCL13 and AIDS-NHL risk for over three years argues against this possibility.
The seven other biomarkers (sCD30, sCD27, IL6, TNFα, IL10, neopterin, and IP10) included in the multivariate biomarker model were selected from over 30 biomarkers with strong (adjusted ORs~2.0 or greater), statistically significant and consistent associations with AIDS-NHL risk at multiple time-points (12, 45). Though mostly positively correlated, these biomarkers were not strongly correlated, which likely reflects their diverse biological functions. All of these biomarkers are thought to induce or result from B cell immune activation, yet they are secreted from, expressed on, and bind to receptors on various cell subsets including Th2 cells, Th17 cells, follicular helper T cells, and monocytes/macrophages (Mo/MΦ).
Th17 cells are a recently-identified subset of pro-inflammatory CD4+ T cells, distinct from Th1 or Th2 cells, which have an established pathogenic role in autoimmune disease and an emerging role in cancer initiation, particularly in the setting of infection (51). Th17 cells express the retinoic acid-related orphan receptor, (ROR)γt, which promotes the gene expression pattern that characterizes the Th17 phenotype (52). There is increasing evidence in support of the hypothesis that Th17 cells drive chronic B cell immune activation and lymphomagenesis and our findings add support to this hypothesis (45, 53). CXCL13, IL6, and TNFα are key examples of Th17-associated chemokines/cytokines (54), and these biomarkers exhibited the strongest association with AIDS-NHL in our study at the >3 year pre-NHL time-point in the multivariate biomarker model.
It has also recently been observed that microbial translocation causes systemic immune activation in HIV and is associated with HIV progression (55–57). Neopterin, which is produced exclusively by macrophages, was the biomarker most strongly associated with AIDS-NHL risk at the visit closest to AIDS-NHL diagnosis date, suggesting that Mo/MΦ activation, possibly due to microbial translocation, coincides with the presentation of AIDS-NHL, but may not necessarily be driving the process.
Our study population was composed nearly entirely of men who were unexposed to HAART prior to AIDS-NHL diagnosis or reference date. HAART, which results in HIV RNA suppression and partial restoration of immune competence, may alter the association between CXCL13 and AIDS-NHL. In recent work, serum CXCL13 levels were significantly reduced in HIV-infected men following HAART initiation compared to their pre-HAART levels, yet still remained higher in comparison to HIV-uninfected controls (58). Thus, this incomplete normalization of CXCL13 by HAART suggests that there is an ongoing environment of chronic B cell activation that could predispose to AIDS-NHL for HAART users and non-users alike.
We did not find strong evidence for an association between genetic variation in either CXCL13 or CXCR5 and AIDS-NHL risk. This does not rule out the possibility that rare variants, structural variation, or other SNPs in these genes not captured by our tagSNPs, could be associated with AIDS-NHL. Interestingly, the minor allele of rs355689 was associated with both a reduced risk of AIDS-NHL and also a lower level of CXCL13. Rs355689 is an intronic SNP and is not in LD with any other known common SNPs in CXCL13 or in neighboring genes according to HAPMAP (59), nor is it predicted to be located in a splicing domain (60). This SNP may be marking some unknown functional variation in CXCL13, or these findings may be due to chance. Furthermore, rs355689 does not appear to modify the association between CXCL13 levels and AIDS-NHL risk (Supplementary Table 3).
Polymorphisms in chemokines and their receptors are important in HIV risk and progression, as well as in the development of AIDS-NHL. The delta 32 deletion polymorphism of chemokine receptor CCR5, protective against HIV-1, is associated with decreased risk of AIDS-NHL, while the stromal cell-derived factor 1 chemokine polymorphism (SDF1-3′A), associated with variable progression rates of HIV, is associated with increased risk of AIDS-NHL (61, 62). A recent study examined the association between four CXCR5 SNPs and NHL in an HIV-uninfected Chinese population (63). Three of the SNPs (rs78440425, rs148351692, and rs80202369) have minor allele frequencies less than 1% in Caucasians and rs6421571 was greater than 10kb upstream of the CXCR5 transcription start site, thus none of these SNPs were genotyped or represented by tagSNPs in our study. Additionally, rs6421571 was not in LD with any of the tagSNPs in our study. The authors reported a significant increased risk of NHL associated with the minor allele of rs80202369 and rs6421571 which raises the possibility that there may be functional variation in or near CXCR5 with consequences for NHL development.
Two other SNPs were significantly associated with CXCL13 levels. Minor allele homozygote carriers for CXCL13 rs17002733 had significantly reduced levels of CXCL13 compared to major allele homozygote carriers. Similar to rs355689, rs17002733 is an intronic SNP, which is neither in LD with any other known SNPs nor has any predicted functional consequence (59, 60). Minor allele homozygote carriers for CXCR5 rs630923 had significantly increased levels (greater than 2-fold) of CXCL13 compared to major allele homozygote carriers. Interestingly, rs630923 is located in the 5′ flanking region and is predicted to be involved in transcriptional regulation by affecting transcription factor binding sites (60). Homozygotes for the rs630923 minor allele may have decreased cell surface expression of CXCR5 leading to an excess of CXCL13 serum levels. Disruption of B cell homing into follicles could cause a reduction in B cell activation. The near 4-fold reduction of AIDS-NHL risk observed for rs630923 minor allele homozygote carriers (non-statistically significant) supports this theory and our hypothesis that AIDS-NHL develops from chronic B cell hyperactivation. However, given that the minor allele homozygote carriers represent a small number of cases and controls in our study, and the confidence intervals surrounding the MR and OR for this SNP are very wide, we cannot rule out the possibility of chance. The percent of variation in CXCL13 explained by our SNPs is small, particularly when compared to the large percent variation explained by variables related to HIV disease progression: 4.2% for rs17002733 compared to 31.7% for CD4+ T cell count, for example. This suggests that individual variation in other risk factors may be driving the differential expression in CXCL13 between cases and controls.
The main strength of this study is the inclusion of a large sample of known AIDS-NHL cases with detailed longitudinal covariate data and specimen availability. In addition to CXCL13, this study population has been characterized with respect to a number of important biomarkers related to AIDS-NHL, permitting us to evaluate our data in this context. Although we controlled for HIV progression with HIV RNA levels and CD4+ T cell numbers, our serum marker ORs could be affected by residual confounding by HIV progression. However, in preliminary analyses, we also included CD4+ T cell slope in our regression models and did not observe any appreciable difference in the ORs (64). For our genetic association study, the inclusion of tagSNPs allowed us to capture all known common variation in our candidate genes. However, due to the sample size, these analyses were underpowered to detect weak or moderate genetic effects. Complementing our tagSNP-AIDS-NHL association testing was the evaluation of potential functional consequence of genetic variation on CXCL13 levels.
As HIV-infected individuals are living longer and to older ages, chronic B cell activation will continue to increase risk of AIDS-NHL. We have identified CXCL13 as an important and novel early pre-diagnosis biomarker for AIDS-NHL that potentially may be used in risk assessment, early detection, or prevention of AIDS-NHL. Our study also raises the possibility that genetic variation in CXCL13 and CXCR5 could influence CXCL13 levels and AIDS-NHL risk, although these findings require validation due to the small sample size.
Supplementary Material
Acknowledgments
Dr. Jeffrey Gornbein (UCLA) provided consultation on the statistical analyses. Mr. Joe DeYoung at the UCLA Neuroscience Genomics Core genotyped the CXCL13/CXCR5 SNPs. The authors would like to thank the MACS participants, without whom this and many other studies would not be possible.
GRANT SUPPORT
This work was supported by grants from the National Institutes of Health (K07-CA-140360, R25-CA-87949, an NCI supplement to U01-AI-035040, a UCLA CFAR grant P30-AI-028697, R01-CA-057152, R01-CA-121195, and the UCLA Older Americans Independence Center grant P30-AG-02874). The Multicenter AIDS Cohort Study (MACS) is funded by the National Institute of Allergy and Infectious Diseases, with additional supplemental funding from the National Cancer Institute (UO1-AI-35042, UL1-RR025005 [GCRC], UO1-AI-35043, UO1-AI-35039, UO1-AI-35040, UO1-AI-35041). Samples and data in this manuscript were collected by the MACS, with centers (Principal Investigators) at The Johns Hopkins Bloomberg School of Public Health (Joseph B. Margolick, Lisa P. Jacobson), Howard Brown Health Center, Feinberg School of Medicine, Northwestern University, and Cook County Bureau of Health Services (John P. Phair, Steven M. Wolinsky), University of California, Los Angeles (Roger Detels, Otoniel Martínez-Maza), and University of Pittsburgh (Charles R. Rinaldo). Some of this work was carried out in the facilities of the UCLA AIDS Institute, which were supported, in part, by funds from the James B. Pendleton Charitable Trust and the McCarthy Family Foundation.
Footnotes
Conflicts of Interest: None of the authors have a conflict of interest to report.
AUTHORSHIP
Contribution: S.K.H. and O.M.M. were responsible for the conception and design, acquisition of data, analysis and interpretation of data, and writing and revision of the manuscript. W.Z., S.C.C., L.M., D.W., and E.V. were responsible for generation of laboratory data, and reviewing/revising of the manuscript. E.C.B. was responsible for the conception and design and reviewing/revising of the manuscript. D.C. and M.S. participated in data analysis and reviewing/revising of the manuscript. L.P.J., J.H.B., S.W., C.R.R., R.F.A., R.D., and Z.F.Z. were responsible for the conception and design, acquisition of data, and reviewing/revising the manuscript.
References
- 1.Grulich AE, Li YM, McDonald AM, Correll PK, Law MG, Kaldor JM. Decreasing rates of Kaposi’s sarcoma and non-Hodgkin’s lymphoma in the era of potent combination antiretroviral therapy. Aids. 2001;15:629–33. doi: 10.1097/00002030-200103300-00013. [DOI] [PubMed] [Google Scholar]
- 2.Epeldegui M, Widney DP, Martinez-Maza O. Pathogenesis of AIDS lymphoma: role of oncogenic viruses and B cell activation-associated molecular lesions. Curr Opin Oncol. 2006;18:444–8. doi: 10.1097/01.cco.0000239882.23839.e5. [DOI] [PubMed] [Google Scholar]
- 3.Martinez-Maza O, Breen EC. B-cell activation and lymphoma in patients with HIV. Curr Opin Oncol. 2002;14:528–32. doi: 10.1097/00001622-200209000-00009. [DOI] [PubMed] [Google Scholar]
- 4.Lane HC, Masur H, Edgar LC, Whalen G, Rook AH, Fauci AS. Abnormalities of B-Cell Activation and Immunoregulation in Patients with the Acquired Immunodeficiency Syndrome. New England Journal of Medicine. 1983;309:453–8. doi: 10.1056/NEJM198308253090803. [DOI] [PubMed] [Google Scholar]
- 5.Amadori A, Zamarchi R, Ciminale V, Delmistro A, Siervo S, Alberti A, et al. HIV-1-specific Bcell activation - a major constituent of spontaneous b-cell activation during HIV-1 infection. Journal of Immunology. 1989;143:2146–52. [PubMed] [Google Scholar]
- 6.Amadori A, Chiecobianchi L. B-cell activation and HIV-1 infection - deeds and misdeeds. Immunology Today. 1990;11:374–9. doi: 10.1016/0167-5699(90)90144-x. [DOI] [PubMed] [Google Scholar]
- 7.Macchia D, Almerigogna F, Parronchi P, Ravina A, Maggi E, Romagnani S. Membrane tumor-necrosis-factor-alpha is involved in the polyclonal B-cell activation-induced by HIV-infected human T-cells. Nature. 1993;363:464–6. doi: 10.1038/363464a0. [DOI] [PubMed] [Google Scholar]
- 8.Martinezmaza O, Crabb E, Mitsuyasu RT, Fahey JL, Giorgi JV. Infection with the human-immunodeficiency-virus (HIV) is associated with an invivo increase in lymphocyte-B activation and immaturity. Journal of Immunology. 1987;138:3720–4. [PubMed] [Google Scholar]
- 9.Xu ZM, Pone EJ, Al-Qahtani A, Park SR, Zan H, Casali P. Regulation of aicda expression and AID activity: Relevance to somatic Hypermutation and class switch DNA recombination. Crit Rev Immunol. 2007;27:367–97. doi: 10.1615/critrevimmunol.v27.i4.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okazaki I, Hiai H, Kakazu N, Yamada S, Muramatsu M, Kinoshita K, et al. Constitutive expression of AID leads to tumorigenesis. Journal of Experimental Medicine. 2003;197:1173–81. doi: 10.1084/jem.20030275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Komeno Y, Kitaura J, Watanabe-Okochi N, Kato N, Oki T, Nakahara F, et al. AID-induced T-lymphoma or B-leukemia/lymphoma in a mouse BMT model. Leukemia. 2010;24:1018–24. doi: 10.1038/leu.2010.40. [DOI] [PubMed] [Google Scholar]
- 12.Breen EC, Hussain SK, Magpantay L, Jacobson LP, Detels R, Rabkin CS, et al. B-Cell Stimulatory Cytokines and Markers of Immune Activation Are Elevated Several Years Prior to the Diagnosis of Systemic AIDS-Associated Non-Hodgkin B-Cell Lymphoma. Cancer Epidemiology Biomarkers & Prevention. 2011;20:1303–14. doi: 10.1158/1055-9965.EPI-11-0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Przybylski GK, Goldman J, Ng VL, McGrath MS, Herndier BG, Schenkein DP, et al. Evidence for early B-Cell activation preceding the development of Epstein-Barr virus-negative acquired immunodeficiency syndrome-related lymphoma. Blood. 1996;88:4620–9. [PubMed] [Google Scholar]
- 14.Fisher SG, Fisher RI. The emerging concept of antigen-driven lymphomas: epidemiology and treatment implications. Curr Opin Oncol. 2006;18:417–24. doi: 10.1097/01.cco.0000239878.31463.0b. [DOI] [PubMed] [Google Scholar]
- 15.Grulich AE, Wan XN, Law MG, Milliken ST, Lewis CR, Garsia RJ, et al. B-cell stimulation and prolonged immune deficiency are risk factors for non-Hodgkin’s lymphoma in people with AIDS. AIDS. 2000;14:133–40. doi: 10.1097/00002030-200001280-00008. [DOI] [PubMed] [Google Scholar]
- 16.Landgren O, Goedert JJ, Rabkin CS, Wilson WH, Dunleavy K, Kyle RA, et al. Circulating Serum Free Light Chains As Predictive Markers of AIDS-Related Lymphoma. J Clin Oncol. 2010;28:773–9. doi: 10.1200/JCO.2009.25.1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rabkin CS, Engels EA, Landgren O, Schuurman R, Camargo MC, Pfeiffer R, et al. Circulating cytokine levels, Epstein-Barr viremia, and risk of acquired immunodeficiency syndrome-related non-Hodgkin lymphoma. American Journal of Hematology. 2011;86:875–8. doi: 10.1002/ajh.22119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Purdue MP, Lan Q, Martinez-Maza O, Oken MM, Hocking W, Huang WY, et al. A prospective study of serum soluble CD30 concentration and risk of non-Hodgkin lymphoma. Blood. 2009;114:2730–2. doi: 10.1182/blood-2009-04-217521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takagi R, Higashi T, Hashimoto K, Nakano K, Mizuno Y, Okazaki Y, et al. B cell chemoattractant CXCL13 is preferentially expressed by human Th17 cell clones. Journal of Immunology. 2008;181:186–9. doi: 10.4049/jimmunol.181.1.186. [DOI] [PubMed] [Google Scholar]
- 20.Ansel KM, Ngo VN, Hyman PL, Luther SA, Forster R, Sedgwick JD, et al. A chemokine-driven positive feedback loop organizes lymphoid follicles. Nature. 2000;406:309–14. doi: 10.1038/35018581. [DOI] [PubMed] [Google Scholar]
- 21.Legler DF, Loetscher M, Roos RS, Clark-Lewis I, Baggiolini M, Moser B. B cell-attracting chemokine 1, a human CXC chemokine expressed in lymphoid tissues, selectively attracts B lymphocytes via BLR1/CXCR5. Journal of Experimental Medicine. 1998;187:655–60. doi: 10.1084/jem.187.4.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reif K, Ekland EH, Ohl L, Nakano H, Lipp M, Forster R, et al. Balanced responsiveness to chemoattractants from adjacent zones determines B-cell position. Nature. 2002;416:94–9. doi: 10.1038/416094a. [DOI] [PubMed] [Google Scholar]
- 23.Forster R, Schweigard G, Johann S, Emrich T, Kremmer E, Nerl C, et al. Abnormal expression of the B-cell homing chemokine receptor BLR1 during the progression of acquired immunodeficiency syndrome. Blood. 1997;90:520–5. [PubMed] [Google Scholar]
- 24.Cagigi A, Mowafi F, Dang LVP, Tenner-Racz K, Atlas A, Grutzmeier S, et al. Altered expression of the receptor-ligand pair CXCR5/CXCL13 in B cells during chronic HIV-1 infection. Blood. 2008;112:4401–10. doi: 10.1182/blood-2008-02-140426. [DOI] [PubMed] [Google Scholar]
- 25.Widney DP, Breen EC, Boscardin WJ, Kitchen SG, Alcantar JM, Smith JB, et al. Serum levels of the homeostatic B cell chemokine, CXCL13, are elevated during HIV infection. Journal of Interferon and Cytokine Research. 2005;25:702–6. doi: 10.1089/jir.2005.25.702. [DOI] [PubMed] [Google Scholar]
- 26.Forster R, Mattis AE, Kremmer E, Wolf E, Brem G, Lipp M. A putative chemokine receptor, BLR1, directs B cell migration to defined lymphoid organs and specific anatomic compartments of the spleen. Cell. 1996;87:1037–47. doi: 10.1016/s0092-8674(00)81798-5. [DOI] [PubMed] [Google Scholar]
- 27.Widney DP, Gui D, Popoviciu LM, Said JW, Breen EC, Huang X, et al. Expression and Function of the Chemokine, CXCL13, and Its Receptor, CXCR5, in Aids-Associated Non-Hodgkin's Lymphoma. AIDS Research and Treatment. 2010;2010 doi: 10.1155/2010/164586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deutsch AJA, Agelsreiter A, Steinbauer E, Fruhwirth M, Kerl H, Behann-Schmid C, et al. Distinct signatures of B-cell homeostatic and activation-dependent chemokine receptors in the development and progression of extragastric MALT lymphomas. J Pathol. 2008;215:431–44. doi: 10.1002/path.2372. [DOI] [PubMed] [Google Scholar]
- 29.Smith JR, Braziel RM, Paoletti S, Lipp M, Uguccioni M, Rosenbaum JT. Expression of B-cell-attracting chemokine 1 (CXCL13) by malignant lymphocytes and vascular endothelium in primary central nervous system lymphoma. Blood. 2003;101:815–21. doi: 10.1182/blood-2002-05-1576. [DOI] [PubMed] [Google Scholar]
- 30.Chan CC, Shen DF, Hackett JJ, Buggage RR, Tuaillon N. Expression of chemokine receptors, CXCR4 and CXCR5, and chemokines, BLC and SDF-1, in the eyes of patients with primary Intraocular lymphoma. Ophthalmology. 2003;110:421–6. doi: 10.1016/S0161-6420(02)01737-2. [DOI] [PubMed] [Google Scholar]
- 31.Husson H, Freedman AS, Cardoso AA, Schultze J, Munoz O, Strola G, et al. CXCL13 (BCA-1) is produced by follicular lymphoma cells: role in the accumulation of malignant B cells. British Journal of Haematology. 2002;119:492–5. doi: 10.1046/j.1365-2141.2002.03832.x. [DOI] [PubMed] [Google Scholar]
- 32.Brunn A, Montesinos-Rongen M, Strack A, Reifenberger G, Mawrin C, Schaller C, et al. Expression pattern and cellular sources of chemokines in primary central nervous system lymphoma. Acta Neuropathol. 2007;114:271–6. doi: 10.1007/s00401-007-0258-x. [DOI] [PubMed] [Google Scholar]
- 33.Mazzucchelli L, Blaser A, Kappeler A, Scharli P, Laissue JA, Baggiolini M, et al. BCA-1 is highly expressed in Helicobacter pylori-induced mucosa-associated lymphoid tissue and gastric lymphoma. Journal of Clinical Investigation. 1999;104:R49–R54. doi: 10.1172/JCI7830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ortonne N, Dupuis J, Plonquet A, Martin N, Copie-Bergman C, Bagot M, et al. Characterization of CXCL13(+) neoplastic T cells in cutaneous lesions of angioimmunoblastic T-cell lymphoma (AITL) American Journal of Surgical Pathology. 2007;31:1068–76. doi: 10.1097/PAS.0b013e31802df4ef. [DOI] [PubMed] [Google Scholar]
- 35.Kurtova AV, Tamayo AT, Ford RJ, Burger JA. Mantle cell lymphoma cells express high levels of CXCR4, CXCR5, and VLA-4 (CD49d): importance for interactions with the stromal microenvironment and specific targeting. Blood. 2009;113:4604–13. doi: 10.1182/blood-2008-10-185827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tun HW, Personett D, Baskerville KA, Menke DM, Jaeckle KA, Kreinest P, et al. Pathway analysis of primary central nervous system lymphoma. Blood. 2008;111:3200–10. doi: 10.1182/blood-2007-10-119099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trentin L, Cabrelle A, Facco M, Carollo D, Miorin M, Tosoni A, et al. Homeostatic chemokines drive migration of malignant B cells in patients with non-Hodgkin lymphomas. Blood. 2004;104:502–8. doi: 10.1182/blood-2003-09-3103. [DOI] [PubMed] [Google Scholar]
- 38.Dudley J, Jin S, Hoover D, Metz S, Thackeray R, Chmiel J. The multicenter AIDS cohort study - retention after 9-1/2 years. American Journal of Epidemiology. 1995;142:323–30. doi: 10.1093/oxfordjournals.aje.a117638. [DOI] [PubMed] [Google Scholar]
- 39.Kaslow RA, Ostrow DG, Detels R, Phair JP, Polk BF, Rinaldo CR. The multicenter AIDS cohort study - rationale, organization, and selected characteristics of the participants. American Journal of Epidemiology. 1987;126:310–8. doi: 10.1093/aje/126.2.310. [DOI] [PubMed] [Google Scholar]
- 40.Silvestre AJ, Hylton JB, Johnson LM, Houston C, Witt M, Jacobson L, et al. Recruiting minority men who have sex with men for HIV research: Results from a 4-city campaign. American Journal of Public Health. 2006;96:1020–7. doi: 10.2105/AJPH.2005.072801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Detels R, Jacobson L, Margolick J, Martinez-Maza O, Muñoz A, Phair J, et al. The multicenter AIDS Cohort Study, 1983 to …. Public Health. 2012;126:196–8. doi: 10.1016/j.puhe.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Panel on Clinical Practices for the Treatment of HIV infection. DHHS/Henry J Kaiser Family Foundation; Nov 3, 2008. Guidelines for the use of antiretroviral agents in HIV-infected adults and adolescents. revision. [Google Scholar]
- 43.Giorgi JV, Cheng H-L, Margolick JB, Bauer KD, Ferbas J, Waxdal M, et al. Quality control in the flow cytometric measurement of T-lymphocyte subsets: The Multicenter AIDS Cohort Study experience. Clinical Immunology and Immunopathology. 1990;55:173–86. doi: 10.1016/0090-1229(90)90096-9. [DOI] [PubMed] [Google Scholar]
- 44.Murray PG, Constandinou CM, Crocker J, Young LS, Ambinder RF. Analysis of Major Histocompatibility Complex Class I, TAP Expression, and LMP2 Epitope Sequence in Epstein-Barr Virus–Positive Hodgkin’s Disease. Blood. 1998;92:2477–83. [PubMed] [Google Scholar]
- 45.Vendrame E, Hussain SK, Breen EC, Magpantay L, Widney DP, Jacobson LP, Detels R, Martinez-Maza O. Cytokines and biomarkers associated with inflammation and immune activation are elevated preceding the diagnosis of AIDS-associated non-Hodgkin B cell lymphoma. Manuscript in preperation. [Google Scholar]
- 46.Altshuler D, Brooks LD, Chakravarti A, Collins FS, Daly MJ, Donnelly P. A haplotype map of the human genome. Nature. 2005;437:1299–320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet. 2004;74:106–20. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Desai M, Kubo J, Esserman D, Terry MB. The Handling of Missing Data in Molecular Epidemiology Studies. Cancer Epidemiology Biomarkers & Prevention. 2011;20:1571–9. doi: 10.1158/1055-9965.EPI-10-1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hargreaves DC, Hyman PL, Lu TT, Ngo VN, Bidgol A, Suzuki G, et al. A coordinated change in chemokine responsiveness guides plasma cell movements. Journal of Experimental Medicine. 2001;194:45–56. doi: 10.1084/jem.194.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Amft N, Curnow SJ, Scheel-Toellner D, Devadas A, Oates J, Crocker J, et al. Ectopic expression of the B cell-attracting chemokine BCA-1 (CXCL13) on endothelial cells and within lymphoid follicles contributes to the establishment of germinal center-like structures in Sjogren’s syndrome. Arthritis Rheum. 2001;44:2633–41. doi: 10.1002/1529-0131(200111)44:11<2633::aid-art443>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 51.Wilke CM, Bishop K, Fox D, Zou WP. Deciphering the role of Th17 cells in human disease. Trends in Immunology. 2011;32:603–11. doi: 10.1016/j.it.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ivanov, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, et al. The orphan nuclear receptor ROR gamma t directs the differentiation program of proinflammatory IL-17(+) T helper cells. Cell. 2006;126:1121–33. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 53.Galand C, Donnou S, Crozet L, Brunet S, Touitou V, Ouakrim H, et al. Th17 Cells Are Involved in the Local Control of Tumor Progression in Primary Intraocular Lymphoma. Plos One. 2011:6. doi: 10.1371/journal.pone.0024622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Louten J, Boniface K, Malefyt RD. Development and function of T(H)17 cells in health and disease. Journal of Allergy and Clinical Immunology. 2009;123:1004–11. doi: 10.1016/j.jaci.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 55.Brenchley JM, Price DA, Schacker TW, Asher TE, Silvestri G, Rao S, et al. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nature Medicine. 2006;12:1365–71. doi: 10.1038/nm1511. [DOI] [PubMed] [Google Scholar]
- 56.Marchetti G, Cozzi-Lepri A, Merlini E, Bellistri GM, Castagna A, Galli M, et al. Microbial translocation predicts disease progression of HIV-infected antiretroviral-naive patients with high CD4(+) cell count. AIDS. 2011;25:1385–94. doi: 10.1097/QAD.0b013e3283471d10. [DOI] [PubMed] [Google Scholar]
- 57.Nowroozalizadeh S, Mansson F, da Silva Z, Repits J, Dabo B, Pereira C, et al. Microbial Translocation Correlates with the Severity of Both HIV-1 and HIV-2 Infections. J Infect Dis. 2010;201:1150–4. doi: 10.1086/651430. [DOI] [PubMed] [Google Scholar]
- 58.Regidor DL, Detels R, Breen EC, Widney DP, Jacobson LP, Palella F, et al. Effect of highly active antiretroviral therapy on biomarkers of B-lymphocyte activation and inflammation. AIDS. 2011;25:303–14. doi: 10.1097/QAD.0b013e32834273ad. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–U3. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu ZL, Taylor JA. SNPinfo: integrating GWAS and candidate gene information into functional SNP selection for genetic association studies. Nucleic Acids Res. 2009;37:W600–W5. doi: 10.1093/nar/gkp290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rabkin CS, Yang Q, Goedert JJ, Nguyen G, Mitsuya H, Sei S. Chemokine and chemokine receptor gene variants and risk of non-Hodgkin’s lymphoma in human immunodeficiency virus-1-infected individuals. Blood. 1999;93:1838–42. [PubMed] [Google Scholar]
- 62.O’Brien SJ, Moore JP. The effect of genetic variation in chemokines and their receptors on HIV transmission and progression to AIDS. Immunological Reviews. 2000;177:99–111. doi: 10.1034/j.1600-065x.2000.17710.x. [DOI] [PubMed] [Google Scholar]
- 63.Song H, Tong D, Cha Z, Bai J. C-X-C chemokine receptor type 5 gene polymorphisms are associated with non-Hodgkin lymphoma. Molecular Biology Reports. 2012;39:8629–35. doi: 10.1007/s11033-012-1717-6. [DOI] [PubMed] [Google Scholar]
- 64.Mellors JW, Margolick JB, Phair JP, Rinaldo CR, Detels R, Jacobson LP, et al. Prognostic value of HIV-1 RNA, CD4 cell count, and CD4 cell count slope for progression to AIDS and death in untreated HIV-1 infection. Jama-Journal of the American Medical Association. 2007;297:2349–50. doi: 10.1001/jama.297.21.2349. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


