Abstract
Plant growth is coordinately regulated by environmental and hormonal signals. Brassinosteroid (BR) plays essential roles in growth regulation by light and temperature, but the interactions between BR and these environmental signals remain poorly understood at the molecular level. Here, we show that direct interaction between the dark- and heat-activated transcription factor phytochrome-interacting factor4 (PIF4) and the BR-activated transcription factor BZR1 integrates the hormonal and environmental signals. BZR1 and PIF4 interact with each other in vitro and in vivo, bind to nearly two thousand common target genes, and synergistically regulate many of these target genes, including the PRE family HLH factors required for promoting cell elongation. Genetic analysis indicates that BZR1 and PIFs are interdependent in promoting cell elongation in response to BR, darkness, or heat. These results show that the BZR1-PIF4 interaction controls a core transcription network, allowing plant growth co-regulation by the steroid and environmental signals.
In plants, cell elongation and seedling morphogenesis are controlled by multiple environmental factors and endogenous hormones, including light, temperature, BR, GA, and auxin. How these signals regulate largely overlapping cellular and physiological responses through distinct signalling pathways remains an outstanding question in plant biology1, 2. In particular, BR is required for hypocotyl elongation responses to dark, shade, and high temperature in Arabidopsis3–7. How BR mediates or modulates the environmental responses is poorly understood at the molecular level.
Light switches the developmental program of seedlings from skotomorphogenesis to photomorphogenesis, causing inhibition of hypocotyl elongation, cotyledon opening and expansion, chloroplast development, and switch from heterotrophic to photoautotrophic growth. Light acts through a suite of photoreceptors, among which the red light-activated photoreceptor phytochromes directly interact with the bHLH transcription factors named phytochrome interacting factors (PIFs)8, 9. PIFs accumulate in the dark to promote skotomorphogenesis, but are phosphorylated and degraded upon light activation of phytochromes9–12. The activities of PIFs are also regulated by the growth hormone GA (Ref. 13, 14), the circadian clock, and temperature7, 15–17. Thus, PIFs are considered key transcription factors that integrate multiple hormonal and environmental signals.
BR is known to play a key role in light and temperature regulation of plant development, as the BR mutants, such as de-etiolated 2 (det2), show light-grown morphology and express light-induced genes in the dark3, 4, 18, 19, and are defective in hypocotyl elongation responses to high temperature and shade5–7, 20. BR signalling is mediated by the BRI1 receptor kinase and its downstream signal transduction cascade that leads to dephosphorylation and activation of the BZR1 family transcription factors21, 22. Constitutive active forms of BZR1, due to either increased dephosphorylation by PP2A (bzr1-1D) or reduced binding by the 14-3-3 proteins (BZR1-S173A), suppresses the photomorphogenesis-in-the-dark phenotypes and most of gene expression changes of the BR-deficient or insensitive mutants, indicating that BR promotes skotomorphogenesis through BZR1 (Ref. 19, 22–24).
The crosstalk between light- and BR-pathways is believed to occur at or downstream of BZR1, because light does not significantly affect the levels of BR and BZR1(Ref. 25). Indeed BZR1 modulates the expression levels of many light-signaling components19, 25, 26. In addition, genome-wide protein-DNA interaction analysis revealed BZR1 binding to the promoters of a significant portion of light-regulated genes19, suggesting that BR and light signals converge at the promoters of common target genes through direct interaction between BZR1 and some light-signalling transcription factors. Here, we demonstrate that PIF4 directly interacts with BZR1, and together they program a central transcriptional network that controls cell elongation and seedling photomorphogenesis. Our genetic evidence indicates that this interaction is a key mechanism for BR-dependent growth responses to light and temperature.
RESULTS
BZR1 interacts with PIF4
To test if BZR1 interacts with any light-signalling transcription factors, we performed transient bimolecular fluorescence complementation (BiFC) assays in tobacco (Nicotiana benthamiana), by co-expressing a BZR1 fused to the C-terminal half of yellow florescence protein (BZR1-cYFP) and light-signalling transcription factors fused to the N-terminal half of YFP (nYFP). As shown in Fig. 1a, strong YFP fluorescence signal was observed in the nucleus when BZR1-cYFP was co-transformed with PIF4-nYFP and PIF1-nYFP, indicating specific BZR1 interactions with PIF4 and PIF1. Co-immunoprecipitation assays using transgenic plants expressing BZR1-CFP and PIF4-myc from their native promoters confirmed their interaction in vivo (Fig. 1b). In vitro pull down assays showed that MBP-BZR1 interacted with GST-PIF4 and GST-PIF1 but not GST alone (Fig. 1c), and GST-PIF4 interacted with full length MBP-BZR1, MBP-BZR2, and the N-terminal DNA-binding domain but not the C-terminal fragment of BZR1 (Fig. 1d,e,g), whereas MBP-BZR1N interacted with both the N-terminal fragment and bHLH domain but not the C-terminal fragment of PIF4 (Fig. 1f,h). The results indicate that BZR1 and PIF4 interact through their DNA-binding domains and the N-terminal domain of PIF4.
Figure 1. BZR1 interacts with PIF4.
(a) BiFC assay shows PIFs interaction with BZR1 in tobacco leaf cells. Images show overlay of fluorescence and light view. (b) Co-immunoprecipitation assay of BZR1 with PIF4. Plants expressing BZR1-CFP from native BZR1 promoter and PIF4-myc from native PIF4 promoter or plants expressing only PIF4-myc were incubated at 28°C for 8 hr to accumulate PIF4 protein and treated with 100 nM BL for 1.5 hr. BZR1-CFP was immunoprecipitated using anti-GFP antibody and immunoblotted using anti-myc or anti-GFP antibody. (c) PIFs directly interact with BZR1 in vitro. GST-PIF1 and GST-PIF4 were pulled down by MBP-BZR1 immobilized on maltose agarose beads and eluted and analyzed by immunoblotting using anti-GST antibody. (d) PIF4 interacts with both BZR1 and BZR2 in vitro. MBP-BZR1 and MBP-BZR2 were pulled down by GST-PIF4 immobilized on glutathione-agarose beads and then eluted and analyzed by immunoblotting using anti-MBP antibody. (e) N-terminal DNA binding domain of BZR1 interacts with PIF4 in vitro. Various fragments of BZR1 fused to MBP were pulled down by GST-PIF4 and then eluted, analyzed by immunoblotting using anti-MBP antibody. (f) Both N-terminal and bHLH domain of PIF4 interact with BZR1 in vitro. Various fragments of PIF4 fused to GST were pulled down by MBP-BZR1N and eluted, analyzed by immunoblotting using anti-GST antibody. (g,h) Box diagrams of the various fragments of BZR1 (g) and PIF4 (h) used in the pull-down assays in panels e and f. The black boxes indicate the DNA binding domains. The numbers indicate the amino acids.
BZR1 and PIFs act interdependently in promoting hypocotyl elongation
We examined whether BR response requires PIFs. Members of the PIF family play overlapping roles in promoting skotomorphogenesis and cell elongation. The quadruple mutant (pifq) lacking four PIFs (PIF1/PIL5, PIF3, PIF4 and PIF5/PIL6) exhibits a constitutive de-etiolation phenotype in the dark, characterized by short hypocotyl and open cotyledons, similar to the BR-deficient or insensitive mutants11, 12. The pifq mutant was less sensitive to exogenous brassinolide (BL, the most active form of BR) and more sensitive to BR biosynthesis inhibitor, brassinazole (BRZ) (Fig. 2a,b), suggesting that the loss of PIFs compromises BR response. However, pif4 single mutant responded to BRZ similar to wild type (Fig. 2b), indicating redundant functions of PIFs with regard to BR response. The bzr1-1D gain-of-function mutation causes constitutive dephosphorylation of BZR1 by PP2A(Ref. 22) and a BRZ-resistant phenotype23. However, the pifq;bzr1-1D quintuple mutant had similar short hypocotyl as pifq grown on the medium with or without BRZ (Fig. 2c,d), suggesting that PIFs are required for the BZR1 promotion of hypocotyl elongation in the dark.
Figure 2. BZR1 and PIFs act interdependently in promoting hypocotyl elongation.
(a) The pifq mutant is hyposensitive to BL compared to wild type (Col-0). Seedlings were grown on various concentrations of BL under white light for 7 days before hypocotyl lengths were measured. Error bars indicate s.d. (n=10 plants) and ** : P < 0.01. (b) The pifq mutant, but not pif4, is hypersensitive to BRZ. Seedlings were grown on various concentrations of BRZ in the dark for 5 days. Error bars indicate s.d. (n=12 plants) and ** : P < 0.01. (c) bzr1-1D cannot promote etiolation in the dark in the pifq mutant. Seedlings were grown on medium either with (+BRZ) or without (−BRZ) 2 µM BRZ in the dark for 5 days. Scale bar, 5 mm. (d) Quantification data of panel (c). Numbers indicate ratio of average hypocotyl length of bzr1-1D to that of control. Error bars indicate s.d. (n=10 plants) and ** : P < 0.01. (e) PIF4 is required for BZR1 promotion of hypocotyl elongation under light. Seedlings were grown on the mock (M) or 2 µM BRZ (BRZ) medium under red light for 5 days. Numbers indicate ratios of hypocotyl length of bzr1-1D to that of control. Error bars indicate s.d. (n=10 plants) and ** : P < 0.01. (f) Both PIF-4OX and bzr1-1D are required to promote hypocotyl elongation in the bri1-116 (bri1) mutant background under light. Seedlings were grown in the dark or under red light for 5 days. Scale bar, 5 mm. (g) Quantification data of panel (f) (light). Numbers indicate ratio of average hypocotyl length of multiple mutant including bri1-116 to that of bri1-116. Error bars indicate s.d. (n=10 plants) and ** : P < 0.01.
In contrast to the etiolation-promoting effect observed in the dark-grown bzr1-1D seedlings, the light-grown bzr1-1D mutant plants are weak dwarfs, with shorter hypocotyls and petioles than wild type23, 27 (Fig. 2e, f). It seems that a light-inactivated factor, possibly PIFs, is required for BZR1 promotion of cell elongation. Indeed bzr1-1D increased hypocotyl elongation in the PIF4-overexpression (PIF4-OX) background in under light (Fig. 2e), suggesting that BZR1’s function of promoting cell elongation requires PIF4.
To test whether PIF4 promotion of cell elongation also requires BR signalling and active BZR1, we introduced PIF4-OX into the bri1-116 single mutant, in which BZR1 is phosphorylated and inactive, and the bri1-116;bzr1-1D double mutant, in which BZR1 is active. While bzr1-1D suppressed the bri1-116 de-etiolation phenotype in the dark (Fig. 2f), the light-grown bri1-116;bzr1-1D showed similar short hypocotyls as bri1-116 (Fig. 2f,g)23, consistent with bzr1-1D being unable to promote cell elongation under light. Overexpression of PIF4 increased the hypocotyl length of bzr1-1D;bri1-116 but not of bri1-116 under light (Fig. 2f,g), demonstrating that both PIF4 and BZR1 are required for hypocotyl elongation. The pifq mutant showed normal BZR1 accumulation and phosphorylation status both before and after BR treatment (Supplementary Fig. S1), indicating that PIF4 does not affect BR signalling upstream of BZR1. Together these results demonstrate that the BZR1-PIF complex is required for cell elongation and skotomorphogenesis; both degradation of PIFs by light signalling and inactivation of BZR1 by reduced BR signalling decrease the BZR1-PIF dimer formation and promote photomorphogenesis.
BZR1 and PIF4 bind to overlapping genomic targets
To understand the functions of BZR1-PIF4 interaction in regulating genome expression, chromatin-immunoprecipitation-sequencing (ChIP-Seq) was performed using transgenic plants expressing PIF4-myc from PIF4 promoter (pPIF4::PIF4-myc) in the pifq mutant background, with the non-transgenic wild-type plants as a negative control. PIF4 binding to known targets (HFR1 and PIL1)14 was detected in the PIF4-myc ChIP-DNA sample but not in the control (Supplementary Fig. S2a). Analyses of the ChIP-seq data with two statistical software CisGenome28 and PRI-CAT(Ref. 29) identified 3510 and 4573 PIF4 binding peaks, respectively (Fig. 3a). Among them, 3186 peaks were identified by both statistical methods and thus considered as the high-confidence PIF4 binding peaks and used for further analysis. About 55% PIF4-binding peaks were associated with at least one PIF-regulated gene whose expression is affected in the pif4;pif5 or pifq mutants according to previously published microarray11, 12, 30–32 or our RNA-Seq data (Fig. 3a). The 3186 high-confidence PIF4 binding peaks were linked to 4363 neighbor genes which were considered as high-confidence PIF4 target genes (Supplementary Table S1), of which 1537 were PIF-regulated genes (58% genes down-regulated and 35% genes up-regulated by PIFs) and thus considered as PIF-regulated PIF4-target genes (PRPT) (Fig. 3b, Supplementary Fig. S2b, and Supplementary Table S2,3).
Figure 3. BZR1 and PIF4 share a large number of genomic targets.
(a) Total PIF4 binding peaks identified by ChIP-Seq analyses using two statistical methods. Numbers indicate percentage of binding peaks associated with at least one PIF-regulated gene. (b) Venn diagram shows significant overlap between PIF4-binding target genes and PIF-regulated genes identified by microarray analysis of pif4;pif5 or pifq and RNA-Seq analysis of pifq. (c) Distribution of PIF4 binding peaks (Frequency) relative to gene structure (−5 kb to +1 kb downstream of 3’ end). (d) Frequency of shown cis-elements around the PIF4 binding sites. Sequence logo shows the most enriched motifs in the PIF4 binding regions. (e) Venn diagrams of PIF4 target genes, BZR1 target genes identified by previous ChIP-chip assay and all light-regulated genes from the light-treatment microarray assays25. (f) Spatial distribution of PIF4 and BZR1 binding peaks along the promoter (−5000 bp to start), coding (start-end, gray), and 3’ (end to +1000 bp) regions of common target genes. Frequency of peak pairs is represented with a color scale. Counts are numbers of peak pairs in certain area. (g) Representative PIF4 and BZR1 binding peaks in the promoters of common target genes (IAA19 and SAUR15). (h) ChIP-reChIP analysis shows that BZR1 and PIF4 co-occupy common targets. The chromatin of 35s-BZR1-myc;35s-PIF4-YFP double transgenic plants was first immunoprecipitated using anti-myc antibody, and then using anti-GFP antibody, and the precipitated DNA was quantified by qPCR. Enrichment of DNA was calculated as ratio between BZR1-myc;PIF4-YFP and wild type control, normalized to that of PP2A coding region as an internal control. Error bars indicate s.d. of three independent experiments (n=3).
Most of the PIF4-binding peaks were in the promoter regions (within −5 kb from transcription start site) (Fig. 3c), consistent with PIF4 function as a transcription factor. The distributions of PIF4 binding sites were similar between PIF-activated or PIF-repressed genes, suggesting that the up or down-regulation is independent of the binding location (Fig. 3c). Consistent with previous studies showing PIFs binding to G-box element33, 34, the G-box motif (CACGTG) is the most enriched cis-element in the PIF4 binding sites, and the E-box motif (CACATG) was also enriched but to a less degree, whereas the frequency of CTATAG motif as the negative control was not increased near the PIF4 binding peaks (Fig. 3d and Supplementary Fig. S2c). Similar cis-element enrichment was found in PIF-activated or repressed genes (Supplementary Fig. S2c).
The target genes of PIF4 include over half (51.7%) of the BZR1 target genes identified by chromatin-immunoprecipitation microarray19 (Fig. 3e). These target genes of both BZR1 and PIF4 contain a higher portion of light-regulated genes than the target genes of either BZR1 or PIF4 alone (Fig. 3e). When plotted along the gene structure, most of the binding peaks of BZR1 and PIF4 were clustered along the diagonal line (Fig. 3f), suggesting that they bind to nearby or same cis-elements (Fig. 3g). Motif analysis showed that the G-box (CACGTG), which contains two inverted repeats of the core of BZR1 binding site (CGTG)27, was the most enriched motif in the regions bound by both BZR1 and PIF4 (Supplementary Fig. S2d,e). To test if BZR1 and PIF4 bind to same promoter elements at the same time or exclusive of each other, chromatin from transgenic Arabidopsis expressing both BZR1-myc and PIF4-YFP was immunoprecipitated sequentially using anti-myc and anti-YFP antibodies, and then analyzed by qPCR. The results show a high enrichment of the BZR1 and PIF4 common target promoters, indicating that BZR1 and PIF4 co-occupy these promoters in vivo (Fig. 3h). These results, together with in vitro data showing direct interaction between their DNA-binding domains, suggest that BZR1 and PIF4 form a heterodimer to bind to promoters of the common target genes (Fig. 1).
BZR1 and PIF4 co-regulate light- and hormone-responsive genes
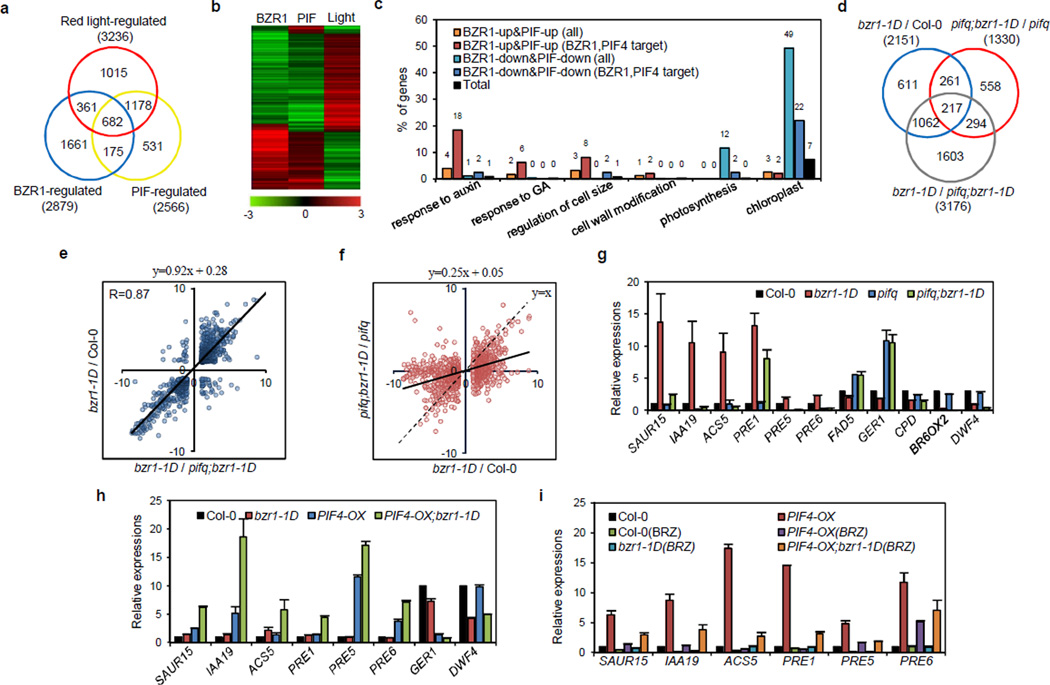
Of the 2879 BZR1-regulated genes, 857 (335 expected randomly) were regulated by PIFs (Fig. 4a). While 80% (682) of the BZR1 and PIF co-regulated genes were also regulated by light, only 18% of BZR1-only-regulated and 69% of PIF-only-regulated genes were regulated by light, indicating that the co-regulated genes are more likely to be light responsive than genes regulated by only BZR1 or PIF4 (Fig. 4a). BZR1 and PIF4 regulated most genes in a similar direction and their effects were opposite to that of light (Fig. 4b), consistent with their roles as negative regulators of photomorphogenesis.
Figure 4. BZR1 and PIF4 regulate different genes interdependently and independently.
(a) Venn diagram shows significant overlap between BZR1-regulated and PIF-regulated and red light-regulated genes. (b) Heat map of 682 BZR1, PIF and light co-regulated genes. Scale bar shows fold changes (log2 value). (c) Gene ontology (GO) analyses of BZR1 and PIF4 targets, and BZR1-regulated, PIF4-regulated genes. Numbers indicate percentage of genes belonging to each GO category. Total: Arabidopsis total genes. (d) Comparison of BZR1-regulated genes (bzr1-1D vs Col-0 and pifq;bzr1-1D vs pifq) and PIF-regulated genes (bzr1-1D vs pifq;bzr1-1D) identified by RNA-Seq analysis. Differentially expressed genes were defined by 2-fold difference between samples with p-value < 0.01. (e) Scatter plot of log2 fold change values in bzr1-1D/Col-0 or bzr1-1D/pifq;bzr1-1D RNA-Seq data for 1279 BZR1 and PIF co-regulated genes. (f) Scatter plot of log2 fold change values of 2151 BZR1-regulated genes in the wild type vs pifq mutant background. (g, h) qRT-PCR analysis of the expression levels of BZR1 and PIF co-regulated genes and BR biosynthesis genes. Seedlings were grown on 2 µM BRZ medium in the dark (g) or under red light (h) for 5 days. (i) PIF4 activation of target gene expression requires BR signalling and BZR1. qRT-PCR analysis of seedlings grown on media without or with 2 µM BRZ (BRZ) under red light for 5 days. All gene expression levels were normalized to that of PP2A and are shown relative to the expression levels in wild type (Col-0). All error bars indicate s.d. of three independent experiments (n=3).
Consistent with previously reports of PIF4 functions in the environmental and hormonal responses such as light, high temperature, circadian rhythm, shade avoidance, auxin and GA responses9, 13–15, 17, 32, gene ontology (GO) analyses showed that all these PIF4-mediated responses are highly represented by the PIF4 target genes (Supplementary Fig. S3), indicating that PIF4 directly regulates these responses. In particular, genes involved in cell size regulation and responses to auxin and GA showed highest enrichment in genes that were activated directly by BZR1 and PIF4 (Fig. 4c). In contrast, genes encoding photosynthetic and chloroplast proteins were most enriched among the genes that are repressed by BZR1 and PIFs but are not direct targets (Fig. 4c). These results indicate that the BZR1-PIF4 heterodimer tends to directly regulate the genes involved in auxin-, and GA-responses and cell elongation, but indirectly regulate the genes encoding photosynthetic and chloroplast proteins through downstream transcription factors.
BZR1 and PIF4 regulate different genes interdependently and independently
We performed RNA-Seq analysis to determine whether genes are controlled independently or interdependently by PIFs and BZR1. Wild type, bzr1-1D, pifq, and pifq;bzr1-1D plants were grown on the medium containing 2 µM BRZ (to inactivate wild type BZR1) in the dark for 5 days. RNA-Seq analyses identified 2151 genes affected >2-fold by bzr1-1D mutation compared to wild type (BZR1-regulated genes) (Fig. 4d and Supplementary Table S4), and 3176 genes affected by the pifq mutation in the bzr1-1D background (pifq;bzr1-1D vs bzr1-1D, PIF-regulated genes) (Fig. 4d and Supplementary Table S5). About 59% of (1279) of the BZR1-regulated genes are also regulated by PIFs, mostly in the same direction (correlation coefficient = 0.87) (Fig. 4e). The overall effects of bzr1-1D on the gene expression are significantly diminished in the pifq background compared to the wild type background (Fig. 4f), with differential expression of about 78% of the 2151 bzr1-1D-affected genes abolished or reversed in the pifq background (Fig. 4d,f). Among the 3176 genes affected by pifq in the bzr1-1D background, 2039 genes (64%) were unaffected or affected in opposite ways by pifq in the wild type background, which lacks BZR1 activity when grown on the BRZ medium (Supplementary Fig. S5). These results demonstrate that BZR1 and PIFs regulate a large portion of downstream genes interdependently of each other but also regulate some genes independently.
We performed qRT-PCR analysis of several BZR1-PIF4 common target genes in the bzr1-1D, pifq and bzr1-1D;pifq mutants to confirm their independent or interdependent regulation by BZR1 and PIF4. As shown in Figure 4g, the expression levels of the BZR1-activated SAUR15, IAA19, PREs and ACS5 and BZR1-repressed genes GER1 and FAD5 were less affected by bzr1-1D in the pifq background than in the wild type background, suggesting that the optimal BZR1 transcriptional activity on these target genes requires PIFs. In contrast to pifq, PIF4 overexpression had similar effects on these genes as bzr1-1D, and together PIF4-OX and bzr1-1D showed synergistic effects (Fig. 4h). On the other hand, inhibiting BR synthesis using BRZ diminished the effects of PIF4-OX on these genes, whereas bzr1-1D partly overcame the inhibitory effects of BRZ (Fig. 4i), indicating that BZR1 is required for the PIF4 regulation of these genes. These results confirm that BZR1 and PIF4 cooperatively regulate these common target genes. The qRT-PCR analysis also confirmed the PIF-independent actions of BZR1 on the BR biosynthesis genes CPD, BR6OX2 and DWF4 (Fig. 4g–h).
PREs promote cell elongation downstream of BZR1 and PIF4
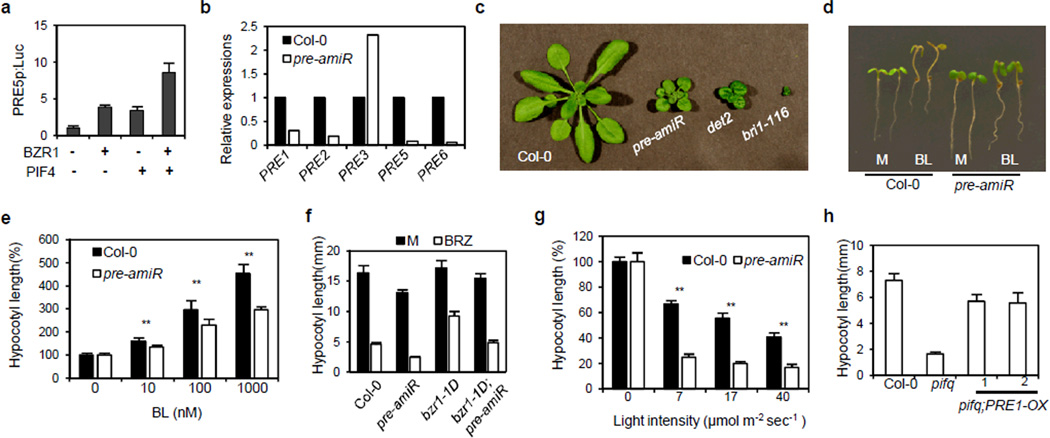
Among the BZR1-PIF4-coregulated targets (Fig. 4g), the PRE family of small HLH proteins has been characterized as positive regulators of cell elongation35, 36. Among the six PRE members, PRE1, PRE5 and PRE6 were identified as PIF4 targets in the ChIP-Seq analysis (Supplementary Fig. S6a), and confirmed by ChIP-qPCR to be direct targets of both BZR1 and PIF4 (Supplementary Fig. S6b,c). The expression levels of PREs were dramatically reduced in pifq but only slightly reduced in the pif4 single mutant, suggesting that PIF4 and additional PIFs redundantly activate these PRE genes (Supplementary Fig. S6d). Expression of these PREs were activated by bzr1-1D in a PIF-dependent manner (Fig. 4g), synergistically activated by bzr1-1D and PIF4-OX (Fig. 4h), and activated by PIF4-OX in a BR/BZR1-dependent manner (Fig. 4i). Transient assays of a pPRE5::Luc reporter gene further confirmed that BZR1 and PIF4 synergistically activate PRE5 (Fig. 5a).
Figure 5. PREs promote hypocotyl elongation downstream of BZR1 and PIF4.
(a) Transient gene expression assays of co-regulation of PRE5 expression by BZR1 and PIF4. The PRE5 promoter (2 kb) fused to firefly luciferase reporter gene was co-transfected with 35S::BZR1, 35S::PIF4, or both, into Arabidopsis mesophyll protoplast. The firefly luciferase activities were normalized by renilla luciferase as an internal control. Error bars indicate s.d. of three independent experiments (n=3). (b) Expression levels of PRE1 to PRE6 in the pre-amiR plants. PRE4 was not detected in this condition. Similar results were obtained in two independent experiments. (c) The pre-amiR plants show dwarfism. Four-week-old plants were photographed. (d, e) The pre-amiR plants had reduced sensitivity to BL. Seedlings were grown on various concentration of BL medium under light for 5days. Representative seedlings grown on either mock (M) or 100 nM BL (BL) media are shown in panel (d). Error bars indicate s.d. (n=10 plants) and ** : P < 0.01. (f) The pre-amiR plants are hypersensitive to BRZ. Seedlings were grown either on mock (M) or 2 µM BRZ in the dark for 5 days. Error bars indicate s.d. (n=10 plants). (g) The pre-amiR plants are hypersensitive to light. Seedlings were grown in various intensities of red light for 5 days. Error bars indicate s.d. (n=10 plants) and ** : P < 0.01. (h) PRE1-OX suppresses the short-hypocotyl phenotype of the pifq mutant. Seedlings were grown under red light for 5 days. Error bars indicate s.d. (n=15 plants).
We generated a transgenic line in which four PREs (PRE1, 2, 5, 6) were knocked-down using artificial micro RNA (pre-amiR) (Fig. 5b). The pre-amiR plants showed dwarf phenotypes similar to BR-deficient or -insensitive mutants (Fig. 5c), and a reduced response to BR treatment (Fig. 5d,e). Suppression of PREs also increased the sensitivity to BRZ in both wild type and bzr1-1D background (Fig. 5f). Knockdown of PREs also enhanced the plant sensitivity to light (Fig. 5g), whereas overexpression of PRE1 suppressed the dwarf phenotype of pifq (Fig. 5h). These results demonstrate that PREs are key components promoting cell elongation downstream of BZR1 and PIF4.
High temperature promotion of hypocotyl elongation requires both BZR1 and PIF4
BR is required for Arabidopsis hypocotyl elongation in response to high temperature7, which is mediated by an increase of PIF4 expression7. We thus tested whether BZR1 acts together with PIF4 to promote hypocotyl elongation under high temperature. The high temperature-induce hypocotyl elongation was abolished by the BR biosynthesis inhibitor propiconazole (PPZ) in wild type but not in the bzr1-1D mutant (Fig. 6a,b), indicating that BR activation of BZR1 is required for the high temperature-induced hypocotyl elongation. The pifq;bzr1-1D mutant showed no response to high temperature, consistent with PIF4’s essential role in hypocotyl response to high temperature (Fig. 6a,b). As reported previously7, high temperature increased the PIF4 protein accumulation (Fig. 6c). In contrast, BZR1 was not obviously affected by temperature (Fig. 6d), indicating that high temperature mainly increase the level of PIF4, which promotes hypocotyl elongation in a BZR1-dependent manner. Consistent with a BZR1-dependent regulation, the expressions of several BZR1 and PIF4 co-target genes including PREs were increased by high temperature in wild type and bzr1-1D;bri1-116 but not in bri1-116 (Fig. 6e). The hypocotyl lengths of pre-amiR plants showed greatly reduced response to high temperature (Fig. 6f), supporting a key role of PREs in hypocotyl elongation downstream of BZR1 and PIF4.
Figure 6. High temperature promotion of hypocotyl elongation requires both BZR1 and PIF4.
(a, b) Both BZR1 and PIF4 are required for high temperature promotion of hypocotyl elongation. Seedlings were grown either on mock (−PPZ) or 2 µM PPZ (+PPZ) at 20°C or 28°C for 7 days. Numbers indicate ratios of hypocotyl lengths of seedlings at 28°C to 20°C. Error bars indicate s.d. (n=10 plants). (c) PIF4 protein accumulates at high temperature. Seedlings expressing PIF4-myc from native PIF4 promoter were grown at 20°C for 5 days and transferred to 28°C for 1, 4 and 24 hr. (d) BZR1 accumulation and phosphorylation status are not significantly affected at high temperature. Seedlings expressing BZR1-CFP from native BZR1 promoter were grown at 20°C for 5 days and transferred to 28°C for 1, 4 and 24 hr. (e) Both BZR1 and PIF4 are required for high temperature-induced gene expressions. Seedlings were grown at 20°C for 4 days and transferred to 28°C for 24 hr (28) or kept at 20°C (20). Relative gene expression levels were normalized to that of PP2A. Similar results were obtained in two independent experiments. (f) The pre-amiR plants are defective in the high temperature promotion of hypocotyl elongation. Seedlings were grown at 20°C or 28°C for 7 days. Error bars indicate s.d. (n=10 plants).
DISCUSSION
Signalling crosstalk is considered important for cellular decision making, but few examples of direct crosstalk between signalling pathways have been elucidated in plants, although plants are expected to have more complex signalling system than animals2. Genetic and physiological studies have suggested a close relationship between BR- and light-signalling pathways, yet evidence for direct crosstalk has been elusive. Here we show that light-regulated PIF4 interacts directly with BR-regulated BZR1. Together they bind to thousands of common target genes in the genome, and interdependently control gene expressions and seedling morphogenesis. The functional interdependence between PIF4 and BZR1 explains why BR is required for cell elongation responses to multiple environmental signals that activate PIFs, including darkness, shade and high temperature3, 5–8, 17. This relation is also consistent with the light-dependent phenotype of bzr1-1D27. The interaction seems conserved among additional members of the BZR1 and PIF families. Although bes1-D was reported to show a light-independent long-hypocotyl phenotype37, the difference could be due to different genetic background or a possibly light-independent activity of BES1. Our strong genetic and molecular evidence indicates that BZR1-PIF4 interaction is a key mechanism for coordination of growth regulation by BR, light, and temperature.
Most of the BZR1-PIF4 co-regulated target genes were positively regulated by both BZR1 and PIFs, suggesting that BZR1-PIF4 heterodimer functions as a transcription activator. However, the interactions between BZR1 and PIF4 appear to be promoter specific, and BZR1 and PIF4 each also controls large numbers of unique targets. This potentially allows differential regulation of various processes by BR and environmental signals. The genome-wide analysis confirmed that the major functions of BZR1-PIF4 are activating genes involved in cell elongation while repressing the transcription pathways for chloroplast development. In addition to cell wall-related genes, BZR1-PIF4 promotion of cell elongation seems to also require downstream PRE family of HLH factors, which are homologous to the human Inhibitor of DNA-binding (Id) protein35, 36. Previous studies have shown major function of the PREs in promoting growth by antagonizing other bHLH factors including IBH1, AIF1, and PAR1 (Ref. 35, 36, 38). We show here that PREs are co-activated by BZR1 and PIF4, and they are required for the BZR1/PIF4-mediated hypocotyl elongation responses to BR, dark, and high temperature. A recent study showed that PAR1 interacts with PIF4 (Ref.39); therefore, the transcription activation of PREs may also activate a feedback loop further activating PIF4. BZR1 and PIFs appear to act at the center of a complex transcriptional network that controls cellular growth and development of photosynthetic apparatus.
Interestingly, BZR1 and PIF4 tend to regulate photosynthetic/chloroplast genes indirectly through their target transcription factors such as GLK1 and GLK2, which are key transcription factors regulating expression of photosynthetic apparatus40–42. GLK1 is a target gene of both BZR1 and PIF4, whereas GLK2 is a PIF4 target; both are repressed by BZR1, BZR2/BES1, and PIFs12, 19, 43. Of the 120 GLK1/2-activated genes identified by microarray analysis, 55 genes (46%) were repressed by BZR1 and/or PIFs (Supplementary Table S6), consistent with the repression of GLK1/2 by BZR1 and PIFs.
Previous studies have shown that BZR1 inhibits photomorphogenesis by transcriptional repression of light-signalling components19, including phytochrome B as well as GATA2 and BZS1 transcription factors19, 25, 26. As such, BZR1 not only directly cooperates with the negative regulators (PIFs) but also transcriptionally represses the positive regulators of photomorphogenesis, providing two levels of control. These interactions at multiple levels support the importance of steroid regulation of light sensitivity. In addition to light and temperature, circadian rhythm and gibberellin also control the activities of PIF factors13–15, and thus the responses to these signals may also depend on BR/BZR1. The BZR1-PIF4 module therefore seems to be the core of a complex network that integrates multiple endogenous and environmental signals.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the Stanford Center for Genomics and Personalized Medicine (SCGPM) service center lead by Drs. Michael Snyder and Arend Sidow for the sequencing service, Ziming Wen for performing the sequencing, Yang Bai for help with genomic data analysis, and Yaqi Hao for experimental assistance. Research was primarily supported by a grant from NIH (R01GM066258).
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
AUTHOR CONTRIBUTIONS
E.O. and J.Z. performed experiments. E.O. analyzed data, and wrote the manuscript. Z.W. designed experiments, analyzed data and wrote the manuscript.
The authors declare no competing financial interests.
REFERENCES
- 1.Depuydt S, Hardtke CS. Hormone signalling crosstalk in plant growth regulation. Curr Biol. 2011;21:R365–R373. doi: 10.1016/j.cub.2011.03.013. [DOI] [PubMed] [Google Scholar]
- 2.Vert G, Chory J. Crosstalk in cellular signaling: background noise or the real thing? Dev Cell. 2011;21:985–991. doi: 10.1016/j.devcel.2011.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li J, Nagpal P, Vitart V, McMorris TC, Chory J. A role for brassinosteroids in light-dependent development of Arabidopsis. Science. 1996;272:398–401. doi: 10.1126/science.272.5260.398. [DOI] [PubMed] [Google Scholar]
- 4.Szekeres M, et al. Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell. 1996;85:171–182. doi: 10.1016/s0092-8674(00)81094-6. [DOI] [PubMed] [Google Scholar]
- 5.Nemhauser JL, Mockler TC, Chory J. Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biol. 2004;2:E258. doi: 10.1371/journal.pbio.0020258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kozuka T, et al. Involvement of auxin and brassinosteroid in the regulation of petiole elongation under the shade. Plant Physiol. 2010;153:1608–1618. doi: 10.1104/pp.110.156802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stavang JA, et al. Hormonal regulation of temperature-induced growth in Arabidopsis. Plant J. 2009;60:589–601. doi: 10.1111/j.1365-313X.2009.03983.x. [DOI] [PubMed] [Google Scholar]
- 8.Leivar P, Quail PH. PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci. 2011;16:19–28. doi: 10.1016/j.tplants.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen M, Chory J. Phytochrome signaling mechanisms and the control of plant development. Trends Cell Biol. 2011;21:664–671. doi: 10.1016/j.tcb.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Al-Sady B, Ni W, Kircher S, Schafer E, Quail PH. Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol Cell. 2006;23:439–446. doi: 10.1016/j.molcel.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 11.Shin J, et al. Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proc Natl Acad Sci U S A. 2009;106:7660–7665. doi: 10.1073/pnas.0812219106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leivar P, et al. Definition of early transcriptional circuitry involved in light-induced reversal of PIF-imposed repression of photomorphogenesis in young Arabidopsis seedlings. Plant Cell. 2009;21:3535–3553. doi: 10.1105/tpc.109.070672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feng S, et al. Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature. 2008;451:475–479. doi: 10.1038/nature06448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Lucas M, et al. A molecular framework for light and gibberellin control of cell elongation. Nature. 2008;451:480–484. doi: 10.1038/nature06520. [DOI] [PubMed] [Google Scholar]
- 15.Nozue K, et al. Rhythmic growth explained by coincidence between internal and external cues. Nature. 2007;448:358–361. doi: 10.1038/nature05946. [DOI] [PubMed] [Google Scholar]
- 16.Foreman J, et al. Light receptor action is critical for maintaining plant biomass at warm ambient temperatures. Plant J. 2011;65:441–452. doi: 10.1111/j.1365-313X.2010.04434.x. [DOI] [PubMed] [Google Scholar]
- 17.Koini MA, et al. High temperature-mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Curr Biol. 2009;19:408–413. doi: 10.1016/j.cub.2009.01.046. [DOI] [PubMed] [Google Scholar]
- 18.Song L, et al. Genome-wide analysis revealed the complex regulatory network of brassinosteroid effects in photomorphogenesis. Mol Plant. 2009;2:755–772. doi: 10.1093/mp/ssp039. [DOI] [PubMed] [Google Scholar]
- 19.Sun Y, et al. Integration of Brassinosteroid Signal Transduction with the Transcription Network for Plant Growth Regulation in Arabidopsis. Dev Cell. 2010;19:765–777. doi: 10.1016/j.devcel.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gray WM, Ostin A, Sandberg G, Romano CP, Estelle M. High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proc Natl Acad Sci U S A. 1998;95:7197–7202. doi: 10.1073/pnas.95.12.7197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim TW, Wang ZY. Brassinosteroid Signal Transduction from Receptor Kinases to Transcription Factors. Annu Rev Plant Biol. 2010;61:681–704. doi: 10.1146/annurev.arplant.043008.092057. [DOI] [PubMed] [Google Scholar]
- 22.Tang W, et al. PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat Cell Bio. 2011;13:124–131. doi: 10.1038/ncb2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang ZY, et al. Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev Cell. 2002;2:505–513. doi: 10.1016/s1534-5807(02)00153-3. [DOI] [PubMed] [Google Scholar]
- 24.Gampala SS, et al. An essential role for 14-3-3 proteins in brassinosteroid signal transduction in Arabidopsis. Dev Cell. 2007;13:177–189. doi: 10.1016/j.devcel.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo X-M, et al. Integration of light and brassinosteroid signaling pathways by a GATA transcription factor in Arabidopsis. Dev Cell. 2010;19:872–883. doi: 10.1016/j.devcel.2010.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fan XY, et al. BZS1, a B-box Protein, Promotes Photomorphogenesis Downstream of Both Brassinosteroid and Light Signaling Pathways. Mol Plant. 2012;5:65–74. doi: 10.1093/mp/sss041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He J-X, et al. BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science. 2005;307:1634–1638. doi: 10.1126/science.1107580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ji H, Jiang H, Ma W, Wong WH. Using CisGenome to analyze ChIP-chip and ChIP-seq data. Curr Protoc Bioinformatics. 2011;Chapter 2(Unit2):13. doi: 10.1002/0471250953.bi0213s33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muino JM, Hoogstraat M, van Ham RC, van Dijk AD. PRI-CAT: a web-tool for the analysis, storage and visualization of plant ChIP-seq experiments. Nucleic Acids Res. 2011;39:W524–W527. doi: 10.1093/nar/gkr373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richter R, Behringer C, Muller IK, Schwechheimer C. The GATA-type transcription factors GNC and GNL/CGA1 repress gibberellin signaling downstream from DELLA proteins and PHYTOCHROME-INTERACTING FACTORS. Genes Dev. 2010;24:2093–2104. doi: 10.1101/gad.594910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lorrain S, Trevisan M, Pradervand S, Fankhauser C. Phytochrome interacting factors 4 and 5 redundantly limit seedling de-etiolation in continuous far-red light. Plant J. 2009;60:449–461. doi: 10.1111/j.1365-313X.2009.03971.x. [DOI] [PubMed] [Google Scholar]
- 32.Nozue K, Harmer SL, Maloof JN. Genomic analysis of circadian clock-, light-, and growth-correlated genes reveals PHYTOCHROME-INTERACTING FACTOR5 as a modulator of auxin signaling in Arabidopsis. Plant Physiol. 2011;156:357–372. doi: 10.1104/pp.111.172684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martinez-Garcia JF, Huq E, Quail PH. Direct targeting of light signals to a promoter element-bound transcription factor. Science. 2000;288:859–863. doi: 10.1126/science.288.5467.859. [DOI] [PubMed] [Google Scholar]
- 34.Oh E, et al. Genome-wide analysis of genes targeted by PHYTOCHROME INTERACTING FACTOR 3-LIKE5 during seed germination in Arabidopsis. Plant Cell. 2009;21:403–419. doi: 10.1105/tpc.108.064691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee S, et al. Overexpression of PRE1 and its homologous genes activates Gibberellin-dependent responses in Arabidopsis thaliana. Plant Cell Physiol. 2006;47:591–600. doi: 10.1093/pcp/pcj026. [DOI] [PubMed] [Google Scholar]
- 36.Zhang LY, et al. Antagonistic HLH/bHLH Transcription Factors Mediate Brassinosteroid Regulation of Cell Elongation and Plant Development in Rice and Arabidopsis. Plant Cell. 2009;21:3767–3780. doi: 10.1105/tpc.109.070441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yin Y, et al. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell. 2002;109:181–191. doi: 10.1016/s0092-8674(02)00721-3. [DOI] [PubMed] [Google Scholar]
- 38.Wang H, Zhu Y, Fujioka S, Asami T, Li J. Regulation of Arabidopsis Brassinosteroid Signaling by Atypical Basic Helix-Loop-Helix Proteins. Plant Cell. 2009;21:3781–3791. doi: 10.1105/tpc.109.072504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hao Y, Oh E, Choi G, Liang Z, Wang ZY. Interactions between HLH and bHLH Factors Modulate Light-Regulated Plant Development. Mol Plant. 2012;5:162–171. doi: 10.1093/mp/sss011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Waters MT, Moylan EC, Langdale JA. GLK transcription factors regulate chloroplast development in a cell-autonomous manner. Plant J. 2008;56:432–444. doi: 10.1111/j.1365-313X.2008.03616.x. [DOI] [PubMed] [Google Scholar]
- 41.Waters MT, et al. GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis. Plant Cell. 2009;21:1109–1128. doi: 10.1105/tpc.108.065250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fitter DW, Martin DJ, Copley MJ, Scotland RW, Langdale JA. GLK gene pairs regulate chloroplast development in diverse plant species. Plant J. 2002;31:713–727. doi: 10.1046/j.1365-313x.2002.01390.x. [DOI] [PubMed] [Google Scholar]
- 43.Yu X, et al. A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. Plant J. 2011;65:634–646. doi: 10.1111/j.1365-313X.2010.04449.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.