Abstract
A series of fluorescent phosphatidylserine and phosphatidylcholine derivatives were prepared and evaluated by cell microscopy for ability to translocate across mammalian plasma membranes via the putative aminophospholipid flippase. Phosphatidylserine derivatives, with either a neutral 7-nitrobenz-2-oxa-1,3-diazol-4-yl (NBD) or a coumarin fluorophore appended to the 2-acyl chain, entered the cytosol of all three cell lines tested and control experiments showed that the translocation was due to flippase activity. In contrast, a phosphatidylserine conjugate containing a charged and polar carboxyfluorescein was not translocated and remained in the cell plasma membrane. The phosphatidylserine-coumarin derivative exhibits bright fluorescence and higher photostability than the NBD analogues, and thus is a promising new fluorescent probe for extended-imaging studies of flippase action in living cells using laser confocal microscopes.
Keywords: Fluorescent probes, cell microscopy, phosphatidylserine, aminophospholipid flippase
Introduction
The distribution of phospholipids across the plasma membrane of mammalian cells is asymmetric. The zwitterionic phospholipids, phosphatidylcholine (PC) and sphingomyelin (SM), are enriched in the outer leaflet of the bilayer membrane, while the aminophospholipids phosphatidylserine (PS) and phosphatidylethanolamine (PE) are located on the cytoplasmic leaflet [1,2]. This asymmetric distribution is critical for various biological functions including clearance of apoptotic cells [3], signal transduction [4] and blood clotting [5,6]. Maintenance of phospholipid distribution requires the concerted action of several membrane transport proteins called phospholipid translocases. These translocases can be divided into three categories: bidirectional non-energy dependent “scramblases”, and energy dependent “translocases” that move phospholipids toward or away from the inner leaflet of the membrane [7,8,9,10]. One of the most studied translocases is the aminophospholipid flippase, which selectively transports PS, and to a much lesser extent PE, to the inner leaflet in the plasma membrane of human erythrocytes and most nucleated cells. Until recently, it has proven difficult to reconstitute this putative transport system in artificial membranes and most studies in the literature have employed intact cells [11,12,13,14].
Substrate selectivity studies of the aminophospholipid flippase indicate a strong preference for the phosphatidylserine head group structure, with insensitivity to the composition of the acyl chains [15,16]. Synthetic PS derivatives with acyl chains of varying length are accepted, as are derivatives containing spin or fluorescent probes in this lipophilic region [15,16,17,18,19,20]. The most studied fluorescent substrate for the aminophospholipid flippase is compound 1, a PS derivative with a 2-acyl chain containing a small and uncharged 7-nitrobenz-2-oxa-1,3-diazol-4-yl (NBD) fluorophore [21,22]. While PS-NBD conjugates are valuable for spectroscopy experiments and “single-acquisition” fluorescence microscopy studies, they have several photophysical limitations. The emission band of the NBD fluorophore is quite broad, which discourages multicolor imaging. The quantum yield changes substantially with solvent polarity and the fluorophore self-quenches upon aggregation [23]. In addition, the fluorophore readily photobleaches, which means that PS-NBD conjugates are not suitable for intravital microscopy or long-term image acquisition studies using high intensity confocal microscopes [24]. It is clear that future studies of phospholipid translocase activity in living cells and tissue would be greatly facilitated by the development of new, high performance, fluorescently labeled PS substrates for the aminophospholipid flippase, that are compatible with confocal one-photon and two-photon microscopy methods.
The goal of this present study was to determine if the putative aminophospholipid flippase(s) in nucleated mammalian cells could translocate phosphatidylserine derivatives with attached fluorophores that were slightly larger and more polar than NBD. This information would help to define the promiscuity of the aminophospholipid flippase and guide the development of more useful, fluorescent PS substrates. Recently, we reported new synthetic chemistry methods that efficiently attach a fluorophore to the end of the 2-acyl chain of a glycerophospholipid [25], and here we have employed this chemistry to prepare PS and PC derivatives 2–7 with three different fluorophores; NBD, coumarin, and carboxyfluorescein (Scheme 1). Using cell microscopy, we find that the endogenous aminophospholipid flippase activity in three different mammalian cell lines readily translocates the PS derivatives 2 and 4 with neutral NBD and coumarin fluorophores, respectively, but not the PS derivative 6 with a charged and highly polar carboxyfluorescein fluorophore. The coumarin fluorophore is quite bright and can be observed selectively in the blue channel of fluorescent microscopes and flow cytometers. Thus, fluorescent PS-coumarin conjugate 4 is a new translocation substrate for the aminophospholipid flippase and should be useful in modern confocal microscopy studies.
Scheme 1.
Structures of fluorescent PS and PC conjugates and synthetic intermediates.
Materials and methods
Materials
Culture media and bovine serum albumin (BSA) were purchased from Sigma Aldrich (St. Louis, MO). ER Tracker Red and Mito Tracker Red were purchased from Invitrogen (Carlsbad, CA). PS-NBD probe 1 was purchased from Avanti Polar Lipids (Alabaster, AL). Compounds 2–5 were prepared by the methods described previously [25]. V79 (ATCC: CCL-93), CHO (ATCC: CCL-61), and A549 (ATCC: CCL-185) cells were certified and obtained from the ATCC.
Synthesis
PS-carboxyfluorescein (6)
To a solution of 8 [25] (0.04 g, 0.04 mmol) in CHCl3 (10 mL) was added DBU (100 μL) and the reaction was stirred for 30 min at ambient temperature. Fluorescein isothiocyanate (FITC) (0.02 g, 0.05 mmol) was then added and the resulting mixture was allowed to stir overnight. The solvent was then evaporated and the crude residue purified using flash chromatography (4:1 CHCl3:MeOH eluent system) to afford the protected PS-carboxyfluorescein (31 mg, 67%) as an orange solid: 1H NMR (2:1 CDCl3:CD3OD) δ 0.68 (t, J = 6.9 Hz, 3H), 1.07 (m, 38H), 1.22 (s, 9H), 1.26 (s, 9H), 1.41 (m, 6H), 2.11 (m, 4H), 3.43 (m, 2H), 3.75 (m, 2H), 3.91 (m, 4H), 4.06 (m, 1H), 4.17 (m, 2H), 5.01 (m, 1H), 6.41 (m, 2H), 6.50 (s, 2H), 6.86 (d, J = 8.4 Hz, 2H), 6.94 (d, J = 8.4 Hz, 1H), 7.85 (m, 2H). 13C NMR (2:1 CDCl3:CD3OD) δ 14.0, 22.7, 24.7, 24.9, 26.8, 27.8, 28.2, 28.9, 29.2, 29.3, 29.4, 29.5, 29.7, 29.7, 31.9, 34.1, 44.7, 55.0, 62.5, 63.8, 65.7, 70.3, 77.6, 80.2, 82.6, 103.1, 114.3, 131.0, 140.5, 169.7, 173.6, 173.9, 180.7. LRMS (FAB−) calcd for C64H93N3O17PS− (M−): 1238.6 found 1238.7. A solution of protected PS-carboxyfluorescein (0.02 g, 0.02 mmol) CH2Cl2 (4 mL) and TFA (2 mL) was allowed to stir at room temperature for 12 h. The reaction was then concentrated to a crude residue which was taken up in 2:1 CHCl3:MeOH and washed with saturated NaHCO3 and water. The organic layer was then concentrated and purified using flash chromatography (65:25:4 CHCl3:MeOH:H2O) to afford 6 (15 mg, 85%): Rf = 0.05, Dittmer-Lester and ninhydrin stain positive. The NMR spectrum extremely broadened due to extensive aggregation in deuterated solvents; however, the PS headgroup was clearly deprotected because the t-Bu singlets at 1.20–1.30 ppm were absent. HRMS (FAB−) calcd for C55H77N3O15PS− (M−) 1082.4818, found 1082.4785.
PC-carboxyfluorescein (7)
To a solution of 9 [25] (0.04 g, 0.04 mmol) in CHCl3 (10 mL) was added DBU (100 μL) and the reaction was stirred for 30 min at ambient temperature. FITC (0.02 g, 0.06 mmol) was then added and the resulting mixture was allowed to stir overnight. The solvent was removed using reduced pressure and the crude residue purified using flash chromatography (65:30:5 CHCl3:MeOH:H2O eluent system) to afford PC-carboxyfluorescein (31 mg, 65%) as an orange solid: 1H NMR (2:1 CDCl3:CD3OD) δ 0.63 (t, J = 6.9 Hz, 3H), 1.05 (m, 34H), 1.48 (m, 6H), 2.08 (m, 4H), 2.98 (s, 9H), 3.09 (m, 2H), 3.39 (m, 2H), 3.77 (t, J = 6.3 Hz, 2H), 3.92 (dd, J = 12.3 Hz, 6.9 Hz, 1H), 4.03 (m, 2H), 4.17 (dd, J = 12.3 Hz, 3.3 Hz, 1H), 4.99 (m, 1H), 6.35 (m, 2H), 6.45 (m, 2H), 6.56 (m, 2H), 6.87 (m, 1H), 7.70 (m, 1H), 8.02 (s, 1H). 13C NMR (2:1 CDCl3:CD3OD) δ 13.7, 19.0, 22.4, 23.5, 24.6, 26.2, 26.7, 28.6, 28.7, 28.8, 28.9, 29.1, 29.2, 29.2, 29.3, 29.4, 31.6, 32.3, 33.8, 33.9, 37.8, 44.3, 53.8, 54.2, 58.9, 62.3, 63.4, 66.1, 70.0, 70.1, 102.6, 111.3, 114.6, 125.5, 129.5, 141.0, 153.9, 165.9, 170.2, 173.3, 173.7, 180.5. HRMS (FAB+) calcd for C57H84N3O13PS (M + H+) 1082.5541, found 1082.5544.
Coumarin triazole dye (10)
The known precursor 3-azidocoumarin dye [26] (21 mg, 80 μmol) and 1-octyne (27 mg, 240 μmol) were dissolved in dichloromethane (4 mL) and mixed with an aqueous solution (4 mL) of CuSO4·5H2O (11 mg) and sodium L-ascorbate (13 mg). The reaction was allowed to stir overnight at room temperature and monitored by TLC. After completion the reaction was extracted with chloroform and washed with EDTA to remove the copper salts. The organic phase was evaporated and subjected to flash chromatography (0–3% MeOH in CHCl3) to yield 10 as a dark yellow oil (24.3 mg, 82%). 1H-NMR (CDCl3): δ 0.89 (t, J = 7.2 Hz, 3 H), 1.24 (t, J = 7.1 Hz, 6 H), 1.36 (m, 6 H), 1.73 (m, 2 H), 2.78 (t, J = 7.8 Hz, 2 H), 3.45 (q, J = 7.2 Hz, 4 H), 6.55 (d, J = 2.4 Hz, 1 H), 6.67 (dd, J = 8.9, 2.4 Hz, 1 H), 7.40 (d, J = 8.9 Hz, 1 H), 8.28 (s, 1 H), 8.37 (s, 1 H). 13C NMR (CDCl3) δ 12.6, 14.2, 22.7, 25.9, 29.1, 29.5, 31.8, 45.2, 97.2, 107.4, 110.2, 117.5, 121.80, 130.1, 134.5, 148.5, 151.6, 155.1,157.3. HRMS (ESI) calcd for C21H29N4O2 (M + H+) 369.2285, found 369.2290. UV-Vis: λmax (abs, CHCl3) = 415 nm, (em, CHCl3) = 461 nm, log ε (415 nm, CHCl3) = 4.56, Φ (ex 341nm, CHCl3) = 0.46 ± 5%, in reference to Coumarin 6 (CHCl3, Φ = 0.93). λmax (abs, ethanol) = 410 nm, (em, EtOH) = 480 nm, Φ (ex 357nm, EtOH) = 0.44 ± 5%, in reference to Coumarin 6 (EtOH, Φ = 0.87), λmax (abs, 1:1 water:EtOH) = 420 nm, (em, 1:1 water: EtOH) = 490 nm, Φ (ex 357nm, 1:1 water: EtOH) = 0.38 ± 5%, in reference to Coumarin 6 (EtOH, Φ = 0.87).
Fluorescent phospholipid preparation
The fluorescent phospholipids were dissolved in 2:1 CHCl3:MeOH mixture and dried by vacuum. The film was resuspended in TES Buffer (5 mM TES, 145 mM NaCl, pH 7.4) to give a stock probe concentration of 1 mM.
Cell culture
Monolayer cultures of V79 cells were grown in DMEM media supplemented with 10% fetal bovine serum (FBS), penicillin, streptomycin, and 2.25 mM L-glutamine. Monolayer cultures of CHO and A549 cells were grown in HF-12K media supplemented with 10% fetal bovine serum (FBS), penicillin, streptomycin, and 2.25 mM L-glutamine. Cells were grown in plastic culture flasks at 37 °C in 5% CO2 atmosphere, passed in 1:5 dilutions and seeded at 1:4 dilutions on chambered coverslips for microscopy studies.
Cell microscopy
Brightfield and fluorescence microscopy was performed on a Nikon TE-2000U epifluorescence microscope equipped with filters that allowed detection of each probe. Filter sets were obtained from Nikon and included UV-2E/C filter (ex: 340/80, em: 435/85), EN GFP HQ filter (ex: 450/90, em: 500/50), Cy3 filter (ex: 535/50 nm, em: 610/75), and Cy5 filter (ex: 620/60, em: 700/75). Fluorescence images were captured using Metamorph software (Universal) and analyzed using ImageJ 1.40.
For probe translocation studies, cells were seeded on chambered coverslips at a 1:4 dilution following passage. Cells were grown to 80% confluency. The old media was removed and replaced with serum free media. The fluorescent phospholipids were added to the cells at a final concentration of 10 μM. The mixture was incubated at 37 °C for 1 hour followed by three washes with serum free media and then analysis by fluorescence microscopy.
Metabolic stability of fluorescent probes
Probes 6 and 7 were added to separate samples of cells to give a final concentration of 10 μM. In each case, cells were incubated at 37 °C for 15 min then trypsinized. The cells were transferred to an eppendorf tube and centrifuged for 1 min at 5000 g. The supernatant was removed and the fluorescent pellet was resuspended in 50 μL of H2O, then transferred to 500 μL of 3 % BSA and allowed to incubate on ice for 5 min. Following incubation and BSA-mediated extraction of all the fluorescent probe from the cells, the mixture was centrifuged for 30 sec at 12000 g and the pellet was discarded. The supernatant containing the BSA and all of the extracted probe was added to MeOH:CHCl3 (2:1, v/v), vortexed, and allowed to incubate in the dark at room temperature for 30 min. After incubation, the solution was centrifuged for 10 min at 2000 g, which allowed separation of the solvent layers. The cellular phospholipid components in the organic layer were analyzed by thin-layer chromatography using CHCl3:MeOH:H2O (65:25:4 by volume) as the eluent and fluorescence emission to visualize the plate. Comparison of the chromatogram with a control plate containing pure material showed that cell incubation produced negligible (< 10%) loss in probe purity.
Aminophospholipid flippase inhibition studies
ATP depletion was achieved by incubating cells in 0.1% sodium azide and 50 mM 2′-deoxyglucose in glucose free media for 30 minutes at 37 °C. Cells were then treated with 10 μM of fluorescent phospholipid probe and incubated for 30 minutes at 37 °C. After incubation, the cells were washed three times with serum free media and imaged by fluorescence microscopy. Chemical inhibition of aminophospholipid flippase activity was achieved by treating cells with 10 mM of iodoacetamide in serum free media followed by incubation at 37 °C for 30 min. The media was removed and 10 μM of phospholipid probe was introduced and the cells incubated at 37 °C for 1 hour. After incubation, cells were washed three times with serum free media and imaged by fluorescence microscopy.
Results and Discussion
Synthetic chemistry and fluorophore photostability
The syntheses of NBD and coumarin conjugates 2–5 have been recently reported [25]. The new carboxyfluorescein probes 6 and 7 were prepared from the protected PS and PC precursors 8 and 9 by first removing the Fmoc protecting group and reacting the liberated amine with fluorescein isothiocyanate, followed by removal of the remaining protecting groups in the case of 6 [25]. Coumarins are well-known as bright and relatively stable blue-emitting fluorescencent dyes [27], and the 3-triazole-coumarin fluorophore in probes 4 and 5 has been utilized several times in recent years for various chemical biology studies [26,28] and also for two-photon microscopy [29,30]. To quantify these favorable photophysical properties we prepared the organic soluble, non self-aggregating control dye 10 and found that it is very weakly solvatochromatic, with a large Stokes shift, and that its quantum yield hardly changes with solvent polarity. For example, the relevant photophysical properties in chloroform are λabs 415 nm, log ε 4.56, λem 461 nm, Φ 0.46, and in 1:1 water:ethanol are λabs 420 nm, log ε 4.55, λem 490 nm, Φ 0.38. A photostability study in ethanol solvent found that the absorption spectrum of coumarin dye 10 was unaffected by 30 minutes of intense irradiation from a 150 W Xenon lamp, whereas the same conditions lead to significant photobleaching of the NBD dye 3. The superior photostability of the coumarin fluorophore was also apparent from the greatly decreased rates of photobleaching observed during the following cell microscopy studies.
Fluorescent microscopy of phospholipid translocation
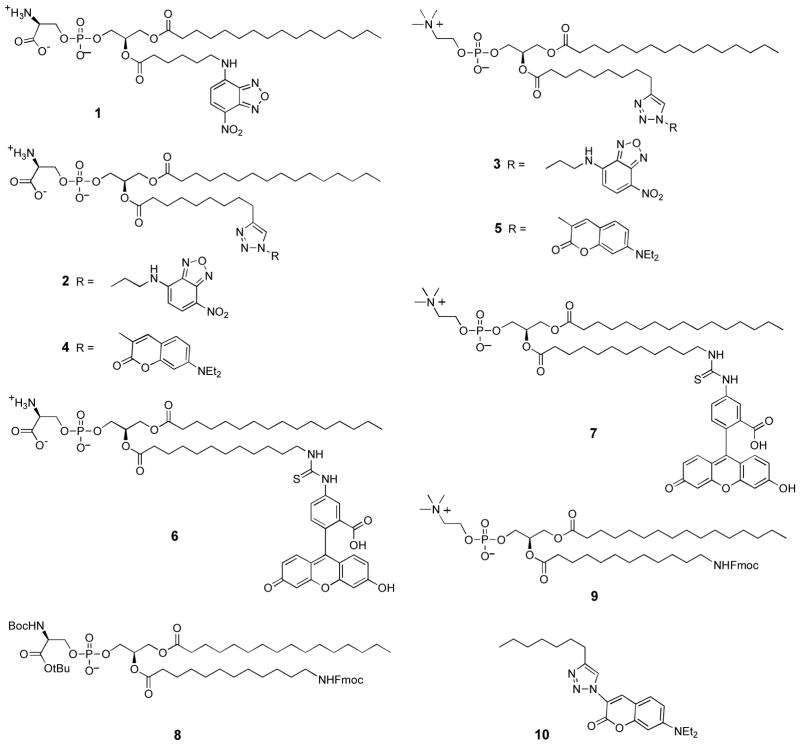
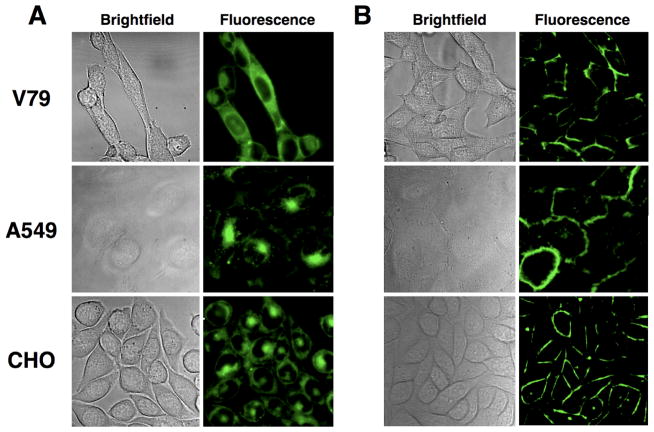
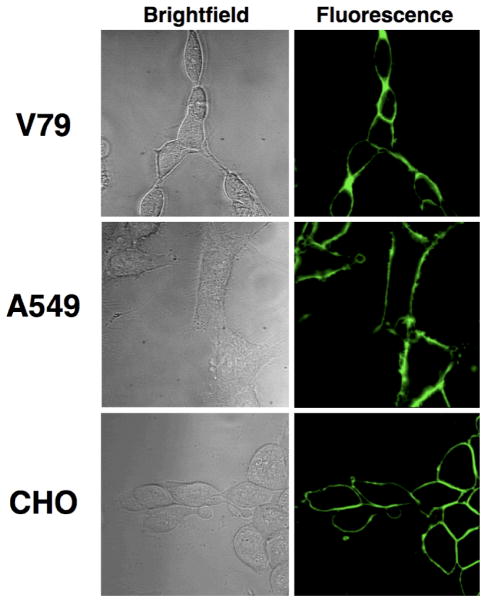
Fluorescence microscopy was used to evaluate the ability of each phosphoplipid probe to penetrate three eukaryotic cell lines: Chinese hamster lung fibroblasts (V79), Chinese hamster ovary cells (CHO), and human lung carcinoma cells (A549). Control studies using the commercially available PS-NBD probe 1 showed that each cell line exhibited aminophospholipid flippase activity and was able to transport 1 into the cytosol (see for example Figure 1A). Not surprisingly, the structurally similar PS-NBD probe 2 with a triazole linker was also internalized (Figure 1B), whereas the PC-NBD probe 3 remained in the cell plasma membrane (Figure 1C), which agrees with the known selectivity of the aminophospholipid flippase to not translocate PC derivatives [31]. The same phospholipid head group selectivity was obtained with coumarin probes 4 and 5. As shown in Figure 2, PS-coumarin 4 was strongly internalized after incubation for 1 hour at 37 °C; whereas, PC-coumarin 5 remained concentrated at the plasma membrane. With each cell line, the rate and extent of cell internalization of PS-coumarin 4 was similar to that observed with PS-NBD probes 1 and 2. As expected for amphiphilic compounds, it was possible to remove the probes 1–5 from inside the cells and also the plasma membrane by a back extraction process that washed the stained cells with excess bovine serum albumin (BSA) solution. The intracellular distribution of probe 4 appeared to be cell line dependent. Separate colocalization experiments with ER Tracker Red and Mito Tracker Red indicated that 4 preferred the endoplasmic reticulum in A549 cells, the mitochondria in CHO cells, and a mixture of both organelle locations in V79 cells (Supplemental Information). A possible explanation for the difference in staining patterns is different rates of intracellular metabolism of the PS-coumarin 4; specifically, decarboxylation of the PS head group to form a PE head group [18,32]. In the case of the A549 cells, treatment with PS-coumarin 4 also produced punctate staining in the cytosol, suggesting partial localization in endosomes (Supplemental Information). In addition to the normal endogenous mechanisms for probe uptake by endocytosis, it is possible that flippase-mediated transport of the probe from the outer to the inner leaflet of the plasma membrane can generate invaginations in the membrane resulting in endocytic vesicles [33].
Figure 1.
Internalization of phospholipid NBD derivatives in eukaryotic cells. Cells were incubated with 10 μM of either 1 (A), 2 (B), or 3 (C) at 37 °C for 1 h, then washed and imaged at 60X.
Figure 2.
Internalization of PS-coumarin probe 4 (A) and membrane localization of PC-coumarin probe 5 (B) in three mammalian cell lines. Probe 4 or 5 (10 μM) were incubated with mammalian cells at 37 °C for 1 h, then washed and imaged at 60X.
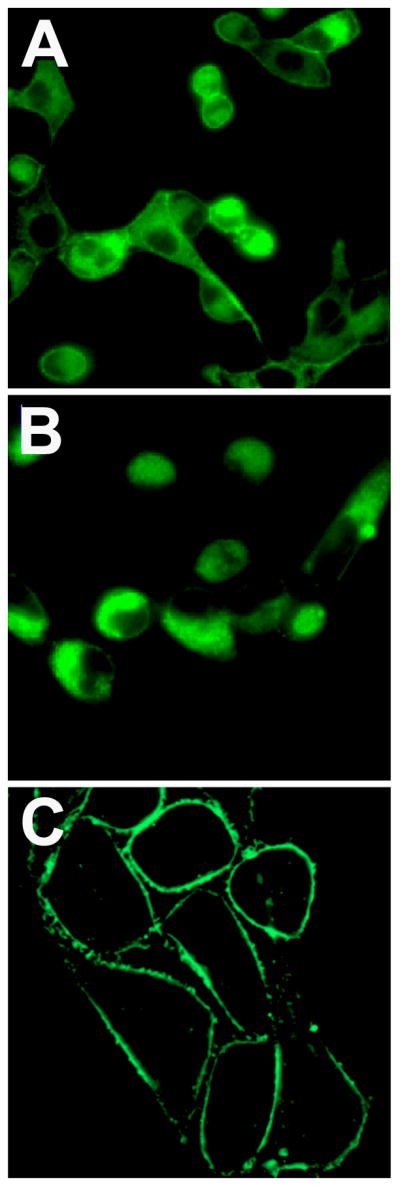
Identical fluorescence microscopy analysis of cells treated separately with carboxyfluorescein conjugates 6 and 7 indicated that they are not substrates for the aminophosphalipid flippase. When incubated at 37 °C for 1 hour, both 6 and 7 localized to the plasma membranes of all three cell types (Figure 3). Back extraction of the probes by washing with a solution of BSA allowed thin layer chromatography analysis of probe biochemical stability and showed that the recovered probes had not undergone any substantial metabolic decomposition. The lack of flippase promoted translocation with PS-carboxyfluorescein probe 6 is attributed to the charge and hydrophilicity of the attached carboxyfluorescein dye. The size of the dye is not likely a major factor since PS conjugates with attached lipophilic pyrene fluorophores have been reported to translocate across mammalian plasma membranes [32,34].
Figure 3.
Plasma membrane localization of carboxyfluorescein probes 6 (A) and 7 (B). Separate samples of the probes (10 μM) were incubated with mammalian cells at 37 °C for 1 h, then washed and imaged at 60X.
Fluorescent microscopy of inhibited phospholipid translocation
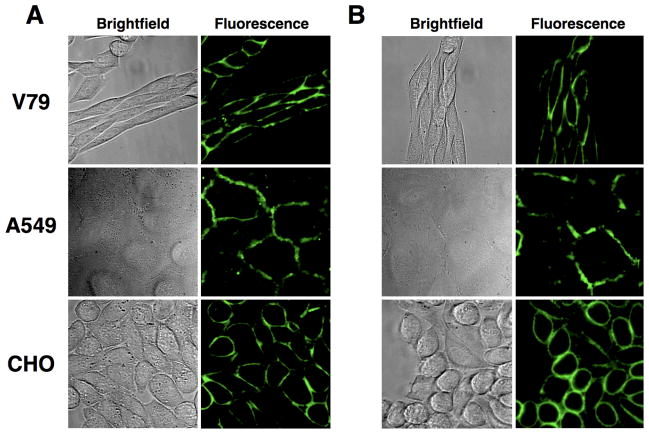
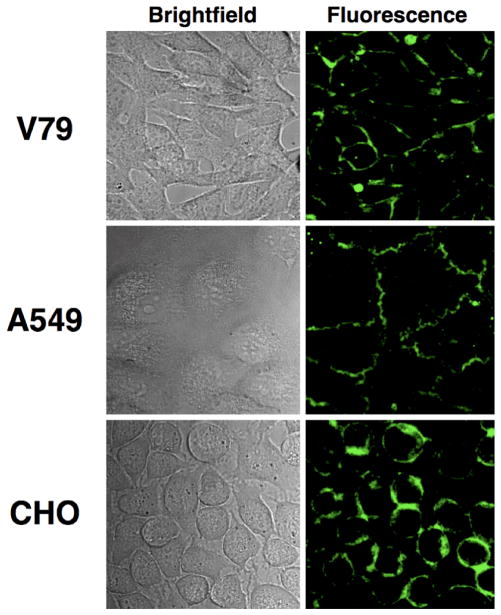
Cell entry of PS-NBD probes 1, 2 and PS-coumarin probe 4 was greatly inhibited when the incubation was conducted at 4 °C, evidence that the membrane translocation is due to an active transport mechanism. Two sets of additional microscopy experiments were conducted to demonstrate that the selective cell entry of PS-coumarin probe 4 was due to putative aminophospholipid flippase activity, and not a chemical process such as probe hydrolysis. There is growing evidence that the aminophospholipid flippase is a P4 ATPase, and cell culture conditions that deplete ATP levels are known to inhibit translocation [11,14,35,36]. As shown in Figure 4, incubating separate samples of the three cell lines with a mixture of 2′-deoxyglucose and sodium azide for 30 minutes at 37 °C (standard conditions to achieve intracellular ATP depletion) prior to the addition of PS-coumarin 4 prevented translocation of 4 such that most of the fluorescent signal remained concentrated in the plasma membrane. A second set of experiments inhibited probe translocation activity by direct chemical treatment. It is known that the aminophospholipid flippase is inhibited by sulfhydryl reactive reagents [37], and as shown in Figure 5, pre-treatment of the three cell lines with iodoacetamide (10 μM) prevented cell entry of PS-coumarin probe 4 after a subsequent incubation at 37 °C for 1 hour.
Figure 4.
Inhibition of PS-coumarin probe 4 translocation by ATP depletion. Cells were incubated with 50 mM 2′-deoxyglucose and 0.1% sodium azide in glucose free media for 30 min at 37 °C then treated with 10 μM of 4. After incubation at 37 °C for 30 min, cells were washed and imaged at 60X.
Figure 5.
Inhibition of PS-coumarin probe 4 translocation by covalent sulfhydryl modification. Cells were incubated with 10 mM of iodoacetamide in media at 37 °C for 30 min, then treated with 10 μM of 4. After incubation at 37 °C for 30 min, cells were washed and imaged at 60X.
Conclusion
The new fluorescent PS-coumarin probe 4 readily enters the cytosol of all three mammalian cell lines that were tested, and there is strong evidence that the process is due to selective translocation by the endogenous aminophospholipid flippase in the cell plasma membrane. PS-coumarin 4 exhibits bright fluorescence and much better photostability than the commercially available PS-NBD 1. Thus, it should be useful as a blue-emitting fluorescent translocation substrate for extended-imaging studies of flippase action in living cells using laser confocal microscopy. Furthermore, PS-coumarin 4 may have value as an energy donor in FRET experiments with NBD acceptor probes [38]. The PS-carboxyfluorescein probe 6 is not a translocation substrate, indicating that the aminophospholipid flippase does not translocate PS derivatives with charged and highly polar groups attached to the 2-acyl chain. Together, these results suggest that the flippase catalyzed translocation process does not involve phospholipid passage through a non-selective hydrophilic channel. This mechanistic picture is in agreement with the latest literature suggesting that the flippase is a P4 ATPase, perhaps associated with other proteins [11,12,39].
Supplementary Material
Acknowledgments
This work was supported by NIH grants GM059078 (B.D.S.), T32GM075762 (B.A.S.), and the Walther Cancer Research Center.
Footnotes
Electronic supplementary material. The online version of this article contains supplementary material, which is available to authorized users.
References
- 1.Daleke DL. Phospholipid flippases. J Biol Chem. 2007;282(2):821–825. doi: 10.1074/jbc.R600035200. [DOI] [PubMed] [Google Scholar]
- 2.Leventis PA, Grinstein S. The distribution and function of phosphatidylserine in cellular membranes. Annu Rev Biophys. 2010;39:407–427. doi: 10.1146/annurev.biophys.093008.131234. [DOI] [PubMed] [Google Scholar]
- 3.Fadok VA, de Cathelineau A, Daleke DL, Henson PM, Bratton DL. Loss of phospholipid asymmetry and surface exposure of phosphatidylserine is required for phagocytosis of apoptotic cells by macrophages and fibroblasts. J Biol Chem. 2001;276(2):1071–1077. doi: 10.1074/jbc.M003649200. [DOI] [PubMed] [Google Scholar]
- 4.Daleke DL, Lyles JV. Identification and purification of aminophospholipid flippases. Biochim Biophys Acta. 2000;1486(1):108–127. doi: 10.1016/s1388-1981(00)00052-4. [DOI] [PubMed] [Google Scholar]
- 5.Lentz BR. Exposure of platelet membrane phosphatidylserine regulates blood coagulation. Prog Lipid Res. 2003;42(5):423–438. doi: 10.1016/s0163-7827(03)00025-0. [DOI] [PubMed] [Google Scholar]
- 6.Zwaal RFA, Schroit AJ. Pathophysiologic implications of membrane phospholipid asymmetry in blood cells. Blood. 1997;89(4):1121–1132. [PubMed] [Google Scholar]
- 7.Daleke DL. Regulation of phosphatidylserine asymmetry in the erythrocyte membrane. Curr Opin Hematol. 2008;15(3):191–195. doi: 10.1097/MOH.0b013e3282f97af7. [DOI] [PubMed] [Google Scholar]
- 8.Sahu SK, Gummadi SN, Manoj N, Aradhyam GP. Phospholipid scramblases: an overview. Arch Biochem Biophys. 2007;462(1):103–114. doi: 10.1016/j.abb.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 9.Pomorski T, Holthuis JCM, Herrmann A, van Meer G. Tracking down lipid flippases and their biological functions. J Cell Sci. 2004;117(6):805–1813. doi: 10.1242/jcs.01055. [DOI] [PubMed] [Google Scholar]
- 10.Bevers EM, Comfurius P, Dekkers DW, Zwaal RFA. Lipid translocation across the plasma membrane of mammalian cells. Biochim Biophys Acta. 1999;1439(3):317–330. doi: 10.1016/s1388-1981(99)00110-9. [DOI] [PubMed] [Google Scholar]
- 11.Zhou X, Graham TR. Reconstitution of phospholipid translocase activity with purified Drs2p, a type-IV P-type ATPase from budding yeast. Proc Natl Acad Sci USA. 2009;106(39):16586–16591. doi: 10.1073/pnas.0904293106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coleman JA, Kwok MCM, Molday RS. Localization, purification, and functional reconstitution of the P4-ATPase Atp8a2, a phosphatidylserine flippase in photoreceptor disc membranes. J Biol Chem. 2009;284(47):32670–32679. doi: 10.1074/jbc.M109.047415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poulsen LR, López-Marqués RL, Palmgren MG. Flippases: still more questions than answers. Cell Mol Life Sci. 2008;65(20):3119–3125. doi: 10.1007/s00018-008-8341-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paulusma CC, Elferink RPJO. P4 ATPases – The physiological relevance of lipid flipping transporters. FEBS Lett. 2010;584(13):2708–2716. doi: 10.1016/j.febslet.2010.04.071. [DOI] [PubMed] [Google Scholar]
- 15.Smriti, Nemergut EC, Daleke DL. ATP-dependent transport of phosphatidylserine analogues in human erythrocytes. Biochemistry. 2007;46(8):2249–2259. doi: 10.1021/bi061333x. [DOI] [PubMed] [Google Scholar]
- 16.Daleke DL. Regulation of transbilayer plasma membrane phospholipid asymmetry. J Lipid Res. 2003;44(2):233–242. doi: 10.1194/jlr.R200019-JLR200. [DOI] [PubMed] [Google Scholar]
- 17.Devaux PF, Fellmann P, Hervé P. Investigation on lipid asymmetry using lipid probes: Comparison between spin-labeled lipids and fluorescent lipids. Chem Phys Lipids. 2002;116(1–2):115–134. doi: 10.1016/s0009-3084(02)00023-3. [DOI] [PubMed] [Google Scholar]
- 18.Pomorski T, Müller P, Zimmerman B, Burger K, Devaux PF, Herrman A. Transbilayer movement of fluorescent and spin-labeled phospholipids in the plasma membrane of human fibroblasts: a quantitative approach. J Cell Sci. 1996;109(3):687–698. doi: 10.1242/jcs.109.3.687. [DOI] [PubMed] [Google Scholar]
- 19.Morrot G, Hervé P, Zachowski A, Fellmann P, Devaux PF. Aminophospholipid translocase of human erythrocytes: phospholipid substrate specificity and effect of cholesterol. Biochemistry. 1989;28(8):3456–3462. doi: 10.1021/bi00434a046. [DOI] [PubMed] [Google Scholar]
- 20.Schroit AJ, Madsen J, Ruoho AE. Radioiodinated, photoactivatable phosphatidylcholine and phosphatidylserine: transfer properties and differential photoreactive interaction with human erythrocyte membrane proteins. Biochemistry. 1987;26(7):1812–1819. doi: 10.1021/bi00381a004. [DOI] [PubMed] [Google Scholar]
- 21.Chattopadhyay A. Chemistry and biology of N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)-labeled lipids: fluorescent probes of biological and model membranes. Chem Phys Lipids. 1990;53(1):1–15. doi: 10.1016/0009-3084(90)90128-e. [DOI] [PubMed] [Google Scholar]
- 22.Monti JA, Christian ST, Shaw WA. Synthesis and properties of a highly fluorescent derivative of phosphatidylethanolamine. J Lipid Res. 1978;19(2):222–228. [PubMed] [Google Scholar]
- 23.Brown SR, Brennan JD, Krull UJ. Self-quenching of nitrobenzoxadiazole labeled phospholipids in lipid membranes. J Chem Phys. 1994;100(8):6018–6027. [Google Scholar]
- 24.Kay JG, Grinstein S. Sensing phosphatidylserine in cellular membranes. Sensors. 2011;11(2):1744–1755. doi: 10.3390/s110201744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lampkins AJ, O’Neil EJ, Smith BD. Bio-orthogonal phosphatidylserine conjugates for delivery and imaging applications. J Org Chem. 2008;73(16):6053–6058. doi: 10.1021/jo8011336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sivakumar K, Xie F, Cash BM, Long S, Barnhill HN, Wang Q. A fluorogenic 1,3-dipolar cycloaddition reaction of 3-azidocoumarins and acetylenes. Org Lett. 2004;6(24):4603–4606. doi: 10.1021/ol047955x. [DOI] [PubMed] [Google Scholar]
- 27.Davis LD, Raines RT. Bright ideas for chemical biology. ACS Chem Biol. 2008;3(3):142–155. doi: 10.1021/cb700248m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jølck RI, Sun H, Berg RH, Andreson TL. Catalyst-free conjugation and in situ quantification of nanoparticle ligand surface density using fluorogenic Cu-free chemistry. Chem Eur J. 2011;17(12):3326–3331. doi: 10.1002/chem.201003131. [DOI] [PubMed] [Google Scholar]
- 29.Fischer A, Cremer CC, Stelzer EHK. Fluorescence of coumarins and xanthenes after two-photon absorption with a pulsed titanium–sapphire laser. Appl Optics. 1995;34(12):1989–2003. doi: 10.1364/AO.34.001989. [DOI] [PubMed] [Google Scholar]
- 30.Beatty KE, Xie F, Wang Q, Tirrell DA. Selective dye-labelling of newly synthesized proteins in bacterial cells. J Am Chem Soc. 2005;127(41):14150–14151. doi: 10.1021/ja054643w. [DOI] [PubMed] [Google Scholar]
- 31.Seigneuret M, Devaux PF. ATP-dependent asymmetric distribution of spin-labeled phospholipids in the erythrocyte membrane: relation to shape changes. Proc Natl Acad Sci USA. 1984;81(12):3751–3755. doi: 10.1073/pnas.81.12.3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tanhuanpää K, Somerharju P. γ-Cyclodextrins greatly enhance translocation of hydrophobic fluorescent phospholipids from vesicles to cells in culture: importance of molecular hydrophobicity in phospholipid trafficking studies. J Biol Chem. 1999;274(50):35359–35366. doi: 10.1074/jbc.274.50.35359. [DOI] [PubMed] [Google Scholar]
- 33.Devaux PF, Hermann A, Ohlwein N, Kozlov MM. How lipid flippases can modulate membrane structure. Biochim Biophys Acta. 2008;1778(7–8):1591–1600. doi: 10.1016/j.bbamem.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 34.Heikinheimo L, Somerharju P. Translocation of pyrene-labeled phosphatidylserine from the plasma membrane to mitochondria diminishes systematically with molecular hydrophobicity: implications on the maintenance of high phosphatidylserine content in the inner leaflet of the plasma membrane. Biochim Biophys Acta. 2002;1591(1–3):75–85. doi: 10.1016/s0167-4889(02)00253-7. [DOI] [PubMed] [Google Scholar]
- 35.Folmer DE, Elferink RP, Paulusma CC. P4 ATPases - Lipid flippases and their role in disease. Biochim Biophys Acta. 2009;1791(7):628–635. doi: 10.1016/j.bbalip.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 36.Zachowski A, Favre E, Cribier S, Hervé P, Devaux PF. Outside-inside translocation of aminophospholipids in the human erythrocyte membrane is mediated by a specific enzyme. Biochemistry. 1986;25(9):2585–2590. doi: 10.1021/bi00357a046. [DOI] [PubMed] [Google Scholar]
- 37.Connor J, Schroit AJ. Aminophospholipid translocation in erythrocytes: evidence for the involvement of a specific transporter and an endofacial protein. Biochemistry. 1990;29(1):37–43. doi: 10.1021/bi00453a005. [DOI] [PubMed] [Google Scholar]
- 38.Lovell JF, Billen LP, Bindner S, Shamas-Din A, Fradin C, Leber B, Andrews DW. Membrane binding of tBid initiates an ordered series of events culminating in membrane permeabilization by Bax. Cell. 2008;135(6):1074–1084. doi: 10.1016/j.cell.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 39.Puts CF, Holthuis JCM. Mechanism and significance of P4 ATPase-catalyzed lipid transport: Lessons from a Na+/K+-pump. Biochim Biophys Acta. 2009;1791(7):603–611. doi: 10.1016/j.bbalip.2009.02.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.