SUMMARY
Epigenetic mechanisms regulating lineage differentiation of mammary stem cells (MaSCs) remain poorly understood. Pygo2 is a histone methylation reader and a context-dependent Wnt/β-catenin co-activator. Here we provide evidence for Pygo2’s function in suppressing luminal/alveolar differentiation of MaSC-enriched basal cells. We show that Pygo2-deficient MaSC/basal cells exhibit partial molecular resemblance to luminal cells such as elevated Notch signaling and reduced mammary repopulating capability upon transplantation. Inhibition of Notch signaling suppresses basal-level and Pygo2 deficiency-induced luminal/alveolar differentiation of MaSC/basal cells, whereas activation of Wnt/β-catenin signaling suppresses luminal/alveolar differentiation and Notch3 expression in a Pygo2-dependent manner. We show that Notch3 is a direct target of Pygo2, and that Pygo2 is required for β-catenin binding and maintenance of a poised/repressed chromatin state at the Notch3 locus in MaSC/basal cells. Together, our data support a model where Pygo2-mediated chromatin regulation connects Wnt signaling and Notch signaling to restrict the luminal/alveolar differentiation competence of MaSC/basal cells.
Keywords: mammary stem cell, basal/luminal commitment/differentiation, Pygo2, histone methylation reader, Wnt signaling, Notch signaling, histone modification
INTRODUCTION
Adult stem cells play important roles in tissue homeostasis and regeneration, and serve as cells of origin of some human cancers (Visvader, 2011). Thus, understanding how their lineage differentiation is governed by the interplay between intracellular epigenetic machinery and extracellular signaling will not only provide mechanistic guidance for tissue engineering but also help predict the behavior of heterogeneous tumors. The mouse mammary gland is an excellent model to study adult stem cells, as it is a dynamic ductal epithelial organ that develops/expands during puberty/pregnancy and regresses to virgin-like state after lactation (Gjorevski and Nelson, 2011). Mounting evidence argues for a hierarchical organization within the mammary epithelia, leading to a prevailing model that multipotent mammary stem cells (MaSCs) residing within the basal compartment give rise to all lineage-restricted progenitor cells and their mature progenies (Visvader, 2009). Supporting this model, specific subpopulations of mammary epithelial cells [e.g., MaSC-enriched basal cells (MaSC/basal) marked by Lin−CD29highCD24+ surface marker expression] are able to generate an entire mammary tree that comprises all lineages - the inner layer of ductal and alveolar luminal cells, and the outer layer of basal/myoepithelial cells - upon transplantation into epithelia-cleared fat pads of host mice (Shackleton et al., 2006; Stingl et al., 2006). However, there also exists theoretic and experimental evidence for bi-directional conversion between stem and differentiated mammary epithelial cells (Chaffer et al., 2011; Guo et al., 2012; Gupta et al., 2011). Moreover, recent findings from lineage tracing experiments demonstrate that lineage-restricted basal and luminal progenitor cells drive mammary morphogenesis during postnatal development and pregnancy (van Amerongen et al., 2012; Van Keymeulen et al., 2011). While epigenetic mechanisms likely dictate the restricted lineage progression of MaSC/basal cells under physiological conditions and their impressive plasticity upon transplantation, the actual players that regulate mammary lineage potential remain uncharacterized. Insights into this issue promise to uncover epigenetic principles of lineage differentiation of epithelial stem cells and shed light on the basis of breast cancer heterogeneity.
Central to the “histone code” hypothesis, the complex patterns of histone modifications, such as methylation and acetylation, are recognized and interpreted (“read”) by effector proteins that in turn bring about changes to the chromatin structure that activate or repress transcription (Jenuwein and Allis, 2001). The Pygopus family of highly conserved plant homeo domain (PHD)-containing proteins was initially discovered as transcriptional co-activators of the Wnt/β-catenin signaling pathway (Belenkaya et al., 2002; Jessen et al., 2008; Kramps et al., 2002; Parker et al., 2002; Thompson et al., 2002). Subsequent studies reveal that these proteins possess the ability to directly bind histone H3 that is trimethylated at lysine 4 (H3K4me3, a histone mark associated with active transcription), and thus act as histone methylation readers (Fiedler et al., 2008; Gu et al., 2009; Kessler et al., 2009). Moreover, mammalian Pygo2 participates in “writing” of the histone code by interacting with and recruiting histone-modifying enzymes to target chromatin to facilitate the production of additional active histone marks such as H3K4me3 (Andrews et al., 2009; Chen et al., 2010; Gu et al., 2009; Gu et al., 2012; Nair et al., 2008). Germline deletion of Pygo2 results in defective embryonic mammary morphogenesis, whereas skin/mammary-specific knockout (SSKO) of Pygo2 leads to a transient delay in mammary ductal morphogenesis during puberty (Gu et al., 2009). Whether and how this chromatin effector regulates the lineage potential of adult MaSC/basal cells remain to be addressed.
Wnt/β-catenin and Notch signaling are two fundamental pathways that regulate stem cells in myriad tissues (Reya and Clevers, 2005; Takebe et al., 2011). In mammary epithelia, Wnt/β-catenin signaling is active in and promotes the self-renewal of MaSCs, and its ectopic activation within the MaSC/basal compartment leads to accumulation of basal cells (Roarty and Rosen, 2010; Teuliere et al., 2005; van Amerongen et al., 2012). In contrast, Notch signaling restricts MaSC/basal self-renewal and promotes their commitment/differentiation to a luminal fate (Bouras et al., 2008; Buono et al., 2006; Raouf et al., 2008). Deregulation of both pathways leads to mammary tumorigenesis, with Wnt signaling targeting the MaSC and Notch signaling the luminal cell types (Visvader, 2011). The molecular relationship between these pathways in controlling the decision between self-renewal and differentiation as well as the balance between basal and luminal lineages remains unknown.
In this study, we investigate the involvement of Pygo2 in mammary lineage differentiation. We find Pygo2 to maintain a MaSC/basal fate by suppressing their luminal/alveolar differentiation, and show that this occurs at least in part via suppression of Notch signaling. Our data highlight Pygo2 as an epigenetic regulator that directly links the selfrenewal-promoting Wnt pathway to the luminal-promoting Notch pathway. Finally, we show that Pygo2 acts in MaSC/basal cells to facilitate β -catenin binding to the Notch3 locus and to maintain Notch3 in a “bivalent” chromatin structure (Bernstein et al., 2006; Wei et al., 2009).
RESULTS
Reduced presence of MaSC/basal and luminal progenitor cells in Pygo2-deficient mammary epithelia
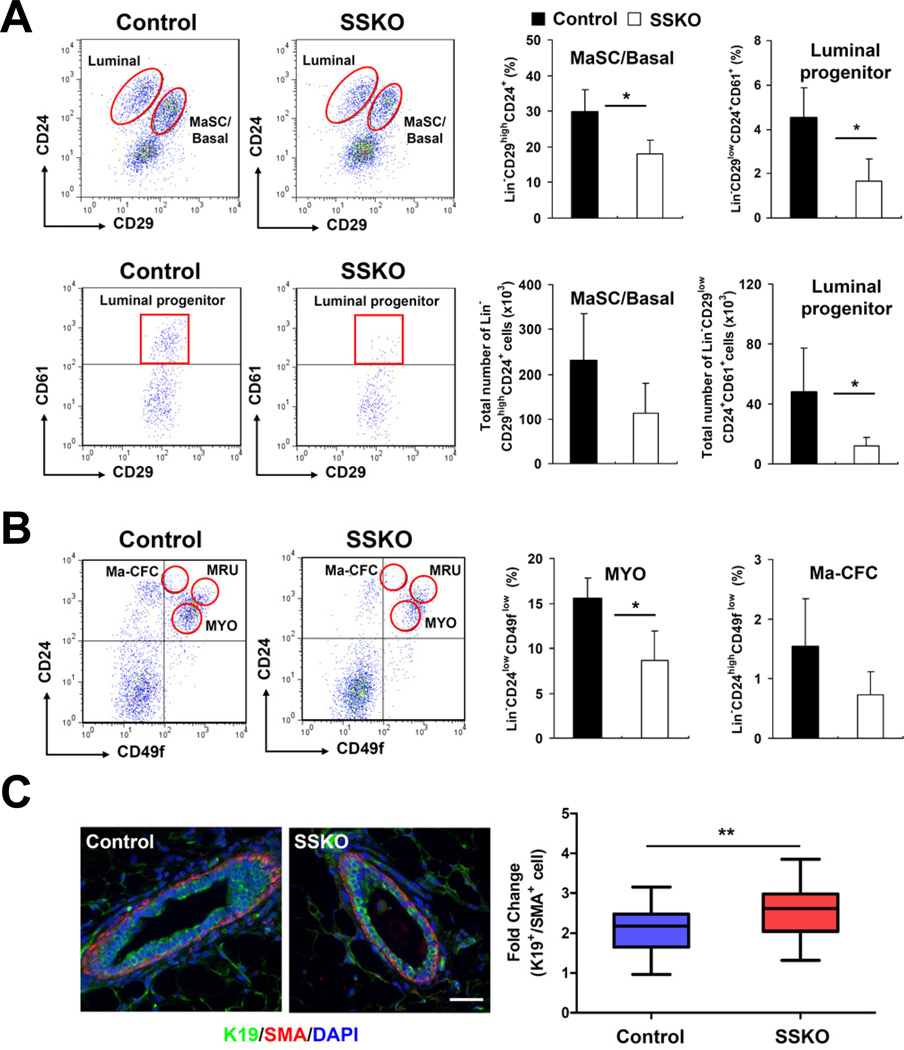
Consistent with our previous finding (Gu et al., 2009), fluorescence activated cell sorting (FACS) analysis revealed a reduced number of MaSC-enriched Lin−CD29highCD24+ basal/myoepithelial (MaSC/basal) cells relative to the total Lin− population in adult Pygo2 SSKO mammary epithelia (Figure 1A). The overall size of the bulk luminal population (Lin− CD29lowCD24+) varied from mouse to mouse, but the relative presence of the CD61+ luminal/alveolar progenitor cell pool (Asselin-Labat et al., 2007) within this population was consistently and significantly lower in SSKO glands than the controls (Figure 1A). Furthermore, we observed a significant reduction in the relative size of the Lin−CD24lowCD49flow population previously shown to encompass mature basal/myoepithelial cells (MYO), whereas the difference in the size of the Lin−CD24highCD49flow population, known to contain mammary colony-forming cells (Ma-CFCs) (Stingl et al., 2006), between WT and SSKO was insignificant (Figure 1B). Together, our results demonstrate that Pygo2 loss associates with smaller pools of bulk basal/myoepithelial cells, as well as of luminal/alveolar progenitor cells relative to the mature luminal population.
Figure 1. Loss of Pygo2 results in reduced numbers of MaSC/basal and luminal progenitor cells.
(A-B) FACS analysis of mammary glands of 8-12-week-old virgin control and Pygo2 SSKO mice using the Lin/CD29/CD24/CD61 (A) or Lin/CD24/CD49f (B) surface marker set. Left, representative FACS profiles. Right, quantitative results. MRU, mammary repopulating unit. (C) Immunofluorescence analysis of K19+ luminal or SMA+ basal cells in mammary ducts from 4-8-week old control and SSKO mice. n>4 per genotype used for all quantifications. Values are means ± standard deviations (SD); * p<0.05; ** p<0.01. Bar = 25 µm.
To determine whether there is any lineage imbalance in Pygo2-deficient mammary epithelia, we stained control and SSKO mammary ducts using K19, a luminal-enriched keratin, and smooth muscle actin (SMA), a basal/myoepithelial marker (Bartek et al., 1985; Gudjonsson et al., 2002; Gugliotta et al., 1988; Sun et al., 2010). Quantification of K19+ and SMA+ cells revealed a statistically significant increase in the ratio between luminal and basal/myoepithelial cells in ductal sections from SSKO mice (p < 0.05; Figure 1C).
Pygo2 suppresses luminal/alveolar differentiation of sorted MaSC/basal cells
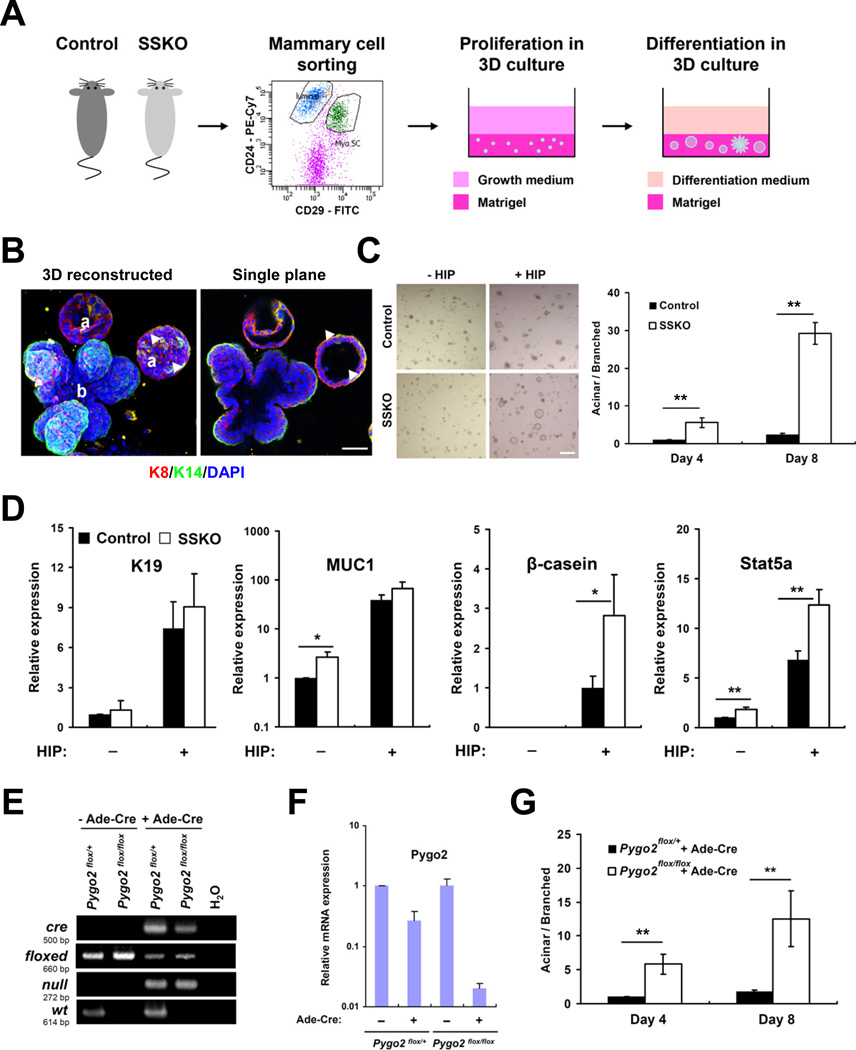
We next asked whether MaSC/basal cells were reduced in the absence of Pygo2 as a consequence of direct, precocious differentiation towards a luminal state. MaSC/basal cells isolated from control and Pygo2 SSKO glands were compared for their differentiation potential using a 3D-Matrigel assay (Shackleton et al., 2006) (Figure 2A). Two major, morphologically distinct types of colonies were observed in the control culture: branched and acinar-like (Figure 2A,B), which were likely derived from MaSC/basal progenitor and luminal/alveolar-restricted progenitor cells, respectively (Dontu et al., 2003; Petersen et al., 1992; Shackleton et al., 2006). As expected (Shackleton et al., 2006), both types of colonies expressed basal marker K14 and luminal marker K8 (Figure 2B). Interestingly, the ratio between acinar and branched colonies was significantly higher in SSKO than the control culture (Figure 2C). Consistently, the mRNA levels of MUC1, β-casein, and Stat5a, but not of K19, were also significantly up-regulated in the SSKO culture (Figure 2D). Therefore, it appears that Pygo2-deficient MaSC/basal cells are prone to adopt a luminal/alveolar fate and/or undergo luminal/alveolar differentiation ex vivo.
Figure 2. Pygo2 loss facilitates luminal/alveolar differentiation of MaSC/basal cells in 3D-Matrigel culture.
(A) Experimental procedure. (B) Examples of branched (b) and acinar-like (a) colonies produced after differentiation induction with HIP (see Experimental Procedures for details). Patterns of K8 (red) and K14 (green) expression as analyzed by confocal microscopy are shown in 3D reconstructed (left) and single plane (right) images. Arrowheads indicate a few K14-positive basal cells bordering an acinar colony. (C) Morphology (left) and quantification (right) of colonies produced by control and SSKO (n=3 each) MaSC/basal cells 6 (left), 4 and 8 (right) days after differentiation induction. (D) RT-qPCR analysis of expression of the indicated genes in control and SSKO (n=3 each) colonies produced 6 days after HIP induction. (E) Genotyping analysis of uninfected or Ade-Cre-infected Pygo2flox/+ and Pygo2flox/flox MaSC/basal cells. (F) RT-qPCR analysis of Pygo2 expression in cells 3 days after infection. (G) Effect of acute Pygo2 deletion on colony formation. Shown are results of quantitative analysis of Ade-Cre-infected MaSC/basal cells from 4 pairs of Pygo2flox/+ and Pygo2flox/flox mice. Values are means ± SD; * p<0.05; ** p<0.01. Bar = 50 µm in (B) and 300 µm in (C). See also Figure S1.
To address the immediate effect of Pygo2 loss on MaSC/basal differentiation, we performed 3D-Matrigel assay on sorted MaSC/basal cells from Pygo2flox/flox and Pygo2flox/+ mice following infection with adenoviruses expressing Cre-IRES-GFP (Ade-Cre) (Figure S1A). Efficient deletion of Pygo2 was verified by recombination at the floxed Pygo2 locus (Figure 2E), the significant reduction of Pygo2 mRNA in Pygo2flox/flox comparing to Pygo2flox/+ cells (Figure 2F), and the loss of Pygo2 protein in GFP-positive Pygo2flox/flox cells (Figure S1B). Acute depletion of Pygo2 led to a significant increase in the ratio between acinar and branched colonies (Figure 2G), indicating that Pygo2 plays a direct role in promoting the luminal/alveolar differentiation of MaSC/basal cells.
Pygo2-deficient MaSC/basal cells exhibit a partial transcriptional drift towards a mature luminal state and reduced repopulating ability upon transplantation
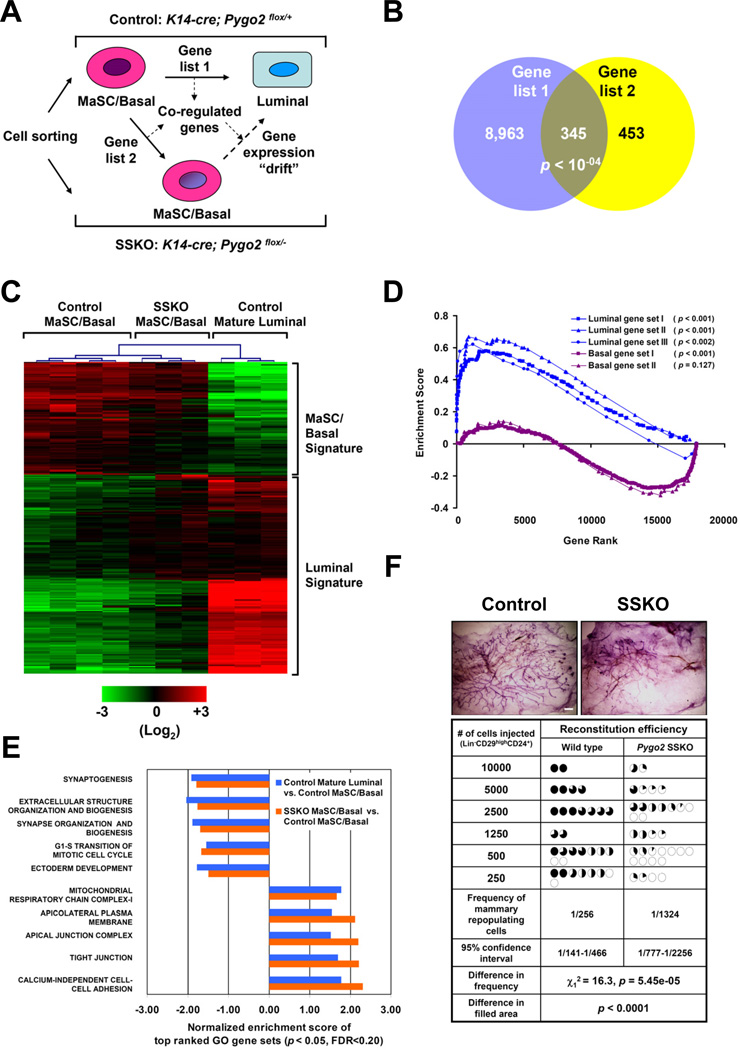
Next we tested the hypothesis that Pygo2 suppresses luminal/alveolar differentiation of MaSC/basal cells by earmarking their differentiation competence. First, we found Pygo2 mRNA to be present in decreasing abundance from MaSC/basal (Lin−CD29highCD24+), luminal progenitor (Lin−CD29lowCD24+CD61+), to mature luminal cells (Lin−CD29lowCD24+CD61−) (Figure S2A,B). Next, we asked whether Pygo2-deficient MaSC/basal cells differ molecularly from their control counterparts despite the fact that they are identified by the same surface markers. Interestingly, MaSC/basal-specific genes SMA and p63 showed a trend of reduced expression in Pygo2-deficient MaSC/basal cells compared to the control, whereas K19, the expression of which normally increased from control MaSC/basal to luminal cells, was slightly elevated in Pygo2-deficient MaSC/basal and luminal progenitor cells (Figure S2B). To evaluate comprehensively whether Pygo2-deleted MaSC/basal cells exhibit luminal-like molecular features, we performed DNA microarray analysis to compare gene expression in three sorted populations: control MaSC/basal, Pygo2 SSKO MaSC/basal, and control mature luminal cells (Figure 3A). List 1 (9,308 genes) included genes whose expression was significantly changed from control MaSC/basal to control luminal cells (Figure 3B). List 2 (798 genes) included genes whose expression is consistently changed in MaSC/basal cells due to Pygo2 deficiency (Figure 3B). A comparison of the two lists revealed that ~43% of the Pygo2-sensitive genes were part of the luminal gene signature (termed “co-regulated”; Figure S2C). Interestingly, co-regulation became more prominent when higher stringency cut-offs were applied. Co-regulated genes included known luminal markers/regulators, such as K19 (KRT19), amphiregulin (AREG), and forkhead box protein A1 (FOXA1), the up-regulation of which in SSKO MaSC/basal cells was confirmed by RT-qPCR (Figure 3C, Figure S2D–Figure S2F). Overall, our molecular analysis portraits a scenario where the expression of a subset of genes in Pygo2-deficient MaSC/basal cells is of intermediate levels between normal MaSC/basal and luminal states.
Figure 3. Transcriptional profiling of control and Pygo2SSKO mammary populations.
(A) Schematic diagram outlining the strategy to identify co-regulated genes. (B) Venn diagram illustrating the overlap between lists 1 (p < 0.05) and 2 (p < 0.05). (C) Heat map of the co-regulated genes. (D) GSEA indicates enhanced luminal and diminished basal gene signatures in Pygo2 SSKO MaSC/basal cells. (E) GO analysis of top ranked gene sets. (F) Summary of limiting dilution transplantation results. Top panel shows the representative whole mount analysis of mammary glands regenerated from control and Pygo2 SSKO MaSC/basal cells. Bar = 1 mm. Bottom table summarizes the transplantation results from 3 independent experiments. Each circle represents one transplanted fat pad, and the percentage of darkness in the circle indicates the proportion of the fat pad that is filled by the transplant. See also Figure S2 and Table S1.
Gene Set Enrichment Analysis (GSEA) (Mootha et al., 2003; Subramanian et al., 2005) confirmed that, compared to control MaSC/basal cells, Pygo2-deleted MaSC/basal cells exhibited a significant up-regulation of several published mammary luminal gene signatures (Gupta et al., 2011; Huper and Marks, 2007; Lim et al., 2010) (Figure 3D, Table S1). In contrast, GSEA indicated a specific down-regulation of basal gene signatures in SSKO MaSC/basal cells. Furthermore, comparing to control MaSC/basal cells, SSKO MaSC/basal and control mature luminal cells displayed similar alterations in the expression of several gene ontology (GO) groups, including decreased expression of G1-S cell cycle genes (Gu et al., 2009) and increased expression of cell adhesion genes (Figure 3E). Taken together, our results are consistent with a partial drift of gene expression in Pygo2-deficient MaSC/basal cells towards a mature luminal state.
To determine the significance of this molecular drift, we performed limiting dilution transplantation assays using Lin−CD29highCD24+ MaSC/basal cells isolated from control and Pygo2 SSKO mammary glands. Compared to the control, SSKO MaSC/basal cells displayed a significantly reduced rate of successful transplantation and less extensive mammary outgrowth (Figure 3F). We estimated that Pygo2 loss led to a ~5-fold reduction in repopulating frequency. Therefore, Pygo2-deficient MaSC/basal cells are compromised in their ability to repopulate the epithelia-cleared mammary fat pad.
Pygo2 suppresses Notch signaling in MaSC/basal cells
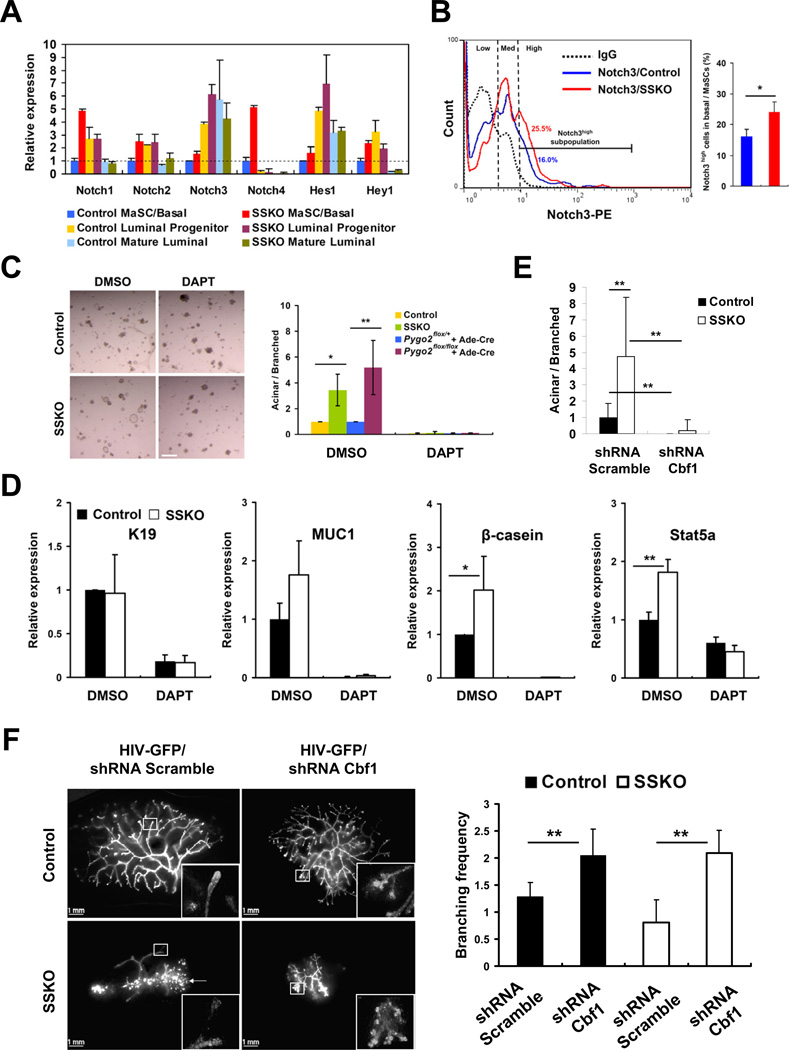
Among the genes up-regulated in Pygo2-deficient MaSC/basal cells were several Notch signaling components, including Notch3, Dll4, and Hes1 (Figure S3A). RT-qPCR analysis using independently sorted cell populations confirmed higher expression of Notch3 and Hes1 in SSKO MaSC/basal and luminal progenitor cells compared to the control counterparts (Figure 4A). The expression of Notch1, Notch2, Notch4, and Hey1 was also up-regulated in SSKO MaSC/basal cells, but not in SSKO luminal progenitor cells (Figure 4A). Moreover, Notch3 and Hes1, but not Notch1, Notch2, Notch4, and Hey1, showed higher expression in control mature luminal cells than control MaSC/basal cells, opposite of that of Pygo2 (Figure 4A, Figure S2B, Figure S3A). These results demonstrate elevated Notch pathway activity in Pygo2-deficient MaSC/basal cells, and identify a tight inverse correlation between Notch3 and Pygo2 expression. Supporting a cell-autonomous, repressive effect of Pygo2 on Notch3, overexpression and depletion (using two different shRNAs) of Pygo2 in MCF10A mammary epithelial cells resulted in reduced and elevated, respectively, levels of Notch3 mRNA, whereas no effect was seen for Notch1 and Notch2 (Figure S3B).
Figure 4. Pygo2 deficiency-induced luminal/alveolar differentiation of MaSC/basal cells requires Notch signaling.
(A) RT-qPCR analysis of Notch pathway gene expression in sorted populations from control and Pygo2 SSKO mice (n=2 each). (B) Representative FACS profiles of surface Notch3 protein expression in control and SSKO MaSC/basal cells. The Notch3high subpopulation was quantified on the right (n=3 per genotype). (C) Morphology (left) and quantification (right) of colonies produced by DMSO- or DAPT-treated MaSC/basal cells from control and SSKO mice (n=3 each), or Ade-Cre-infected MaSC/basal cells from Pygo2flox/+ and Pygo2flox/flox mice (n=4 each). Bar = 300 µm. (D) RT-qPCR analysis of colonies formed as in (C). n=3 per genotype. Note that the difference in K19 expression caused by Pygo2 deficiency is no longer as prominent after culturing MaSC/basal cells in Matrigel as that in freshly sorted cells. (E) Quantification of colonies produced by MaSC/basal cells from control and SSKO mice (n=3 each) with infection of control scramble shRNA or Cbf-1 shRNA lentiviruses. (F) Effect of Cbf-1 knockdown on mammary outgrowths from control and Pygo2 SSKO MaSC/basal cells transplanted in vivo. Transplants were visualized by GFP fluorescence (left), and branching frequency was calculated as the number of total branching points per total ductal length (mm) in randomly selected high resolution fields from multiple transplants (n=2–6 each) (right). Arrow indicates colonized GFP-positive cells with no outgrowth. Values are means ± SD. * p<0.05; ** p<0.01. See also Figure S3.
To ask whether Notch3 up-regulation occurred in the entire SSKO MaSC/basal population or only a subset of the cells, we performed FACS analysis on mammary cells harvested from control and SSKO mice using Notch3 antibody together with the Lin/CD29/CD24 marker set. Whereas the control MaSC/basal population contained two subpopulations of cells that can be described as Notch3low and Notch3medium, the Pygo2-deficient MaSC/basal population contained a more abundant Notch3medium subpopulation and a distinct Notch3high subpopulation (Figure 4B, left). Quantification of Notch3high cells revealed a significant increase in SSKO samples (Figure 4B, right). These data provide in vivo evidence that in the absence of Pygo2, both Notch3low and Notch3medium cells within the MaSC/basal population gain higher Notch3 protein expression.
To determine the functional relevance of increased Notch signaling in Pygo2-deficient MaSC/basal cells, we first treated these cells in 3D-Matrigel culture with DAPT, a γ-secretase inhibitor that blocks the activating, proteolytic cleavage of Notch receptors (Dovey et al., 2001; Geling et al., 2002). As expected (Bouras et al., 2008), DAPT reduced the expression of targets of Notch signaling (Figure S3C) and suppressed the formation of acinar colonies in control MaSC/basal culture (Figure 4C). Importantly, DAPT also completely suppressed the greatly enhanced acinar colony formation in Pygo2-deficient MaSC/basal culture, regardless of whether deficiency was engineered chronically or acutely (Figure 4C). This cellular phenotype correlated well with a significant suppression of luminal/milk genes K19, MUC1, β-casein, and Stat5a (Figure 4D).
We next depleted Cbf-1, a transcription factor essential for Notch signaling, in control and Pygo2 SSKO MaSC/basal cells by infecting them with lentiviruses expressing Cbf-1-specific shRNA and GFP (Bouras et al., 2008) (Figure S3D). Compared to treatment with control viruses expressing a scramble shRNA and GFP, and similar to DAPT treatment, Cbf-1 knockdown led to a significant decrease of both basal-level and Pygo2 deficiency-induced formation of acinar colonies in 3D-Matrigel culture (Figure 4E, Figure S3E). Upon transplantation, Cbf-1-depleted control MaSC/basal cells produced mammary outgrowths with increased but disorganized branching and aberrant terminal buds (Figure 4F), defects attributed to Notch signaling’s restrictive role of a MaSC/basal state (Bouras et al., 2008). Importantly, albeit smaller, the transplants generated by Pygo2 SSKO MaSC/basal cells also showed elevated branching frequency and aberrant terminal buds upon Cbf-1 knockdown (Figure 4F). Taken together, these results show that inhibition of Notch signaling is able to reverse the elevated luminal/alveolar differentiation in Pygo2 SSKO MaSC/basal cells.
Activation of Wnt/β-catenin signaling suppresses luminal/alveolar differentiation and Notch signaling in a Pygo2-dependent manner
Given the context-dependent role of Pygo2 as a Wnt co-activator, we next tested whether 1) forced activation of Wnt/β-catenin signaling in MaSC/basal cells also suppresses their luminal/alveolar differentiation or Notch signaling; and 2) if so, whether these inhibitory effects depend on Pygo2.
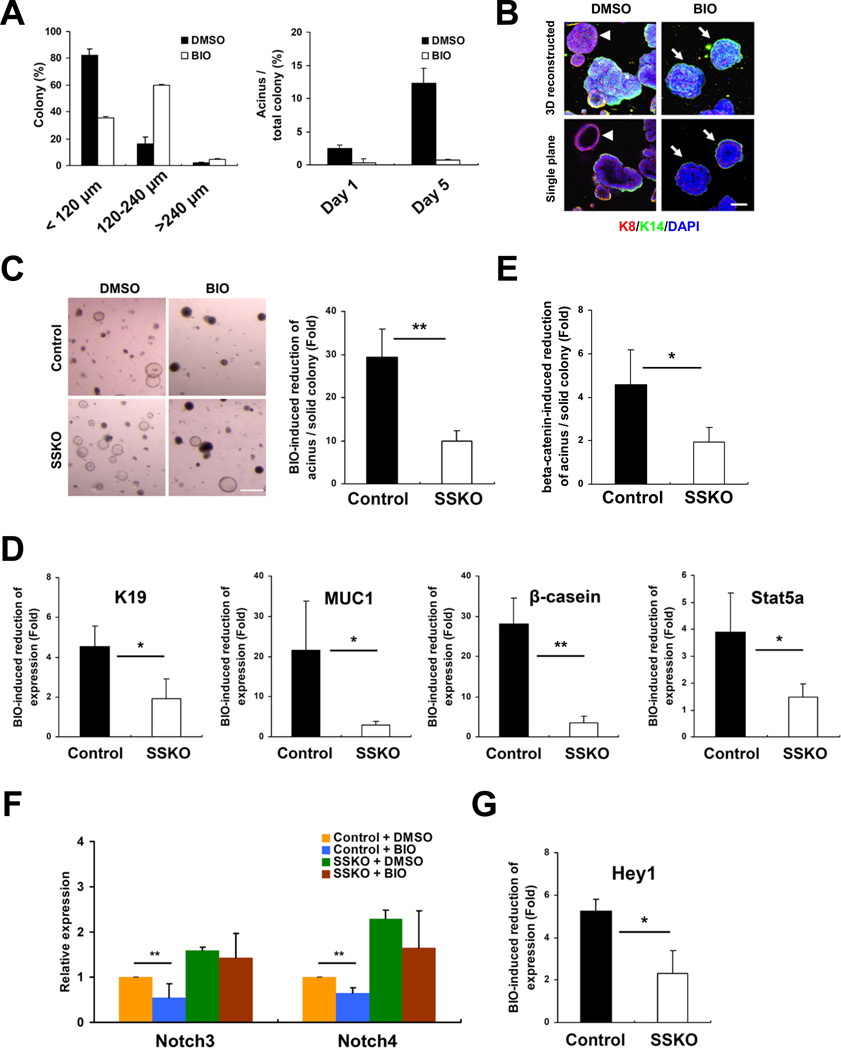
Application of BIO, a GSK3 inhibitor that activates Wnt/β-catenin signaling (Sato et al., 2004), to control MaSC/basal cells resulted in elevated expression of Wnt target Axin2 and more rapid colony growth (Figure 5A, Figure S4A,B). However, there was a dramatic reduction of acinar colonies in BIO-treated cultures; the colonies that were produced were mostly solid, lacked branches, and composed of a K14-positive outer layer and multiple layers of K8-positive inner cells (Figure 5A,B, Figure S4B). The extent of BIO-induced acinar colony reduction was significantly compromised when Pygo2 was absent (Figure 5C). BIO also reduced the expression of K19, MUC1, β-casein, and Stat5a in control MaSC/basal cells, and the fold reduction was considerably smaller in SSKO MaSC/basal cells (Figure 5D). Finally, lentiviral expression of ∆N-β-catenin, a stabilized form of β-catenin mimicking activated Wnt signaling (Gat et al., 1998), caused a less significant reduction in the formation of acinar colonies in SSKO than in control MaSC/basal 3D-Matrigel cultures (Figure 5E, Figure S4C,D). These results suggest that Pygo2 is required for the maximal inhibitory effect of Wnt/β-catenin signaling on luminal/alveolar differentiation of MaSC/basal cells.
Figure 5. Pygo2-dependent Wnt/β-catenin suppression of induced luminal/alveolar differentiation andNotch3 expression in MaSC/basal cells.
(A) Quantification of the size (left) and type (right) of colonies derived from BIO (0.5 µM)-treated MaSC/basal cells. Percent of acinar-like colonies per total colonies was calculated for cultures 1 and 5 days after HIP induction (mean ± SD; n=2 mice). (B) Immunofluorescent detection of K8- (red) and K14 (green)-positive cells in colonies produced by DMSO- or BIO-treated MaSC/basal cells. Arrow, solid colony; arrowhead, acinus. (C) Morphology (left) and quantification (right) of colonies produced by DMSO- or BIO-treated control and SSKO (n=3 each) MaSC/basal cells. (D, F–G) RT-qPCR analysis of the indicated genes in colonies formed as in (C). (E) Quantification of colonies produced by MaSC/basal cells from control and SSKO mice (n=3 each) with infection of control GFP or GFP/∆N-β-catenin lentiviruses. Values are means ± SD. * p<0.05; ** p<0.01. Bar = 50 µm in (B), and 300 µm in (C). See also Figure S4.
BIO exerted different effects on Notch genes in control MaSC/basal 3D culture: it repressed the expression of Notch3 and, to a less extent, Notch4, but not Notch1 and Notch2 (Figure 5F and data not shown). Importantly, the repression of Notch3 did not occur in Pygo2 SSKO MaSC/basal 3D culture (Figure 5F). Similarly, BIO inhibited the expression of Hey1, and this effect was partially reversed by loss of Pygo2 (Figure 5G). Thus, Pygo2 is required for BIO inhibition of Notch3 expression and signaling. In interesting contrast, Axin2 expression in MaSC/basal 3D cultures was reduced by Pygo2 loss regardless of BIO presence (Figure S4A).
Pygo2 recruits β-catenin to, and maintains a bivalent domain at, the Notch3 locus in MaSC/basal cells
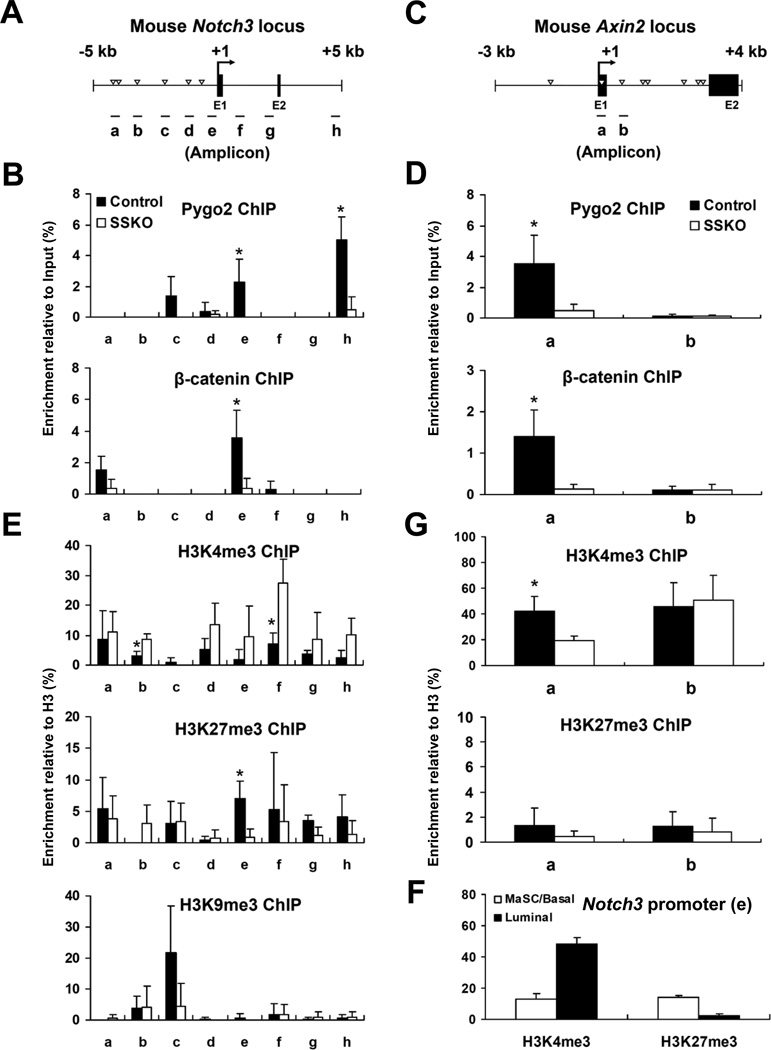
To date, a few genes have been identified as being directly repressed by Wnt/β-catenin signaling and none by Pygo2 (Hoverter and Waterman, 2008; Jessen et al., 2008). We therefore determined whether Notch3 is a direct target of Pygo2 or β-catenin. We performed micro chromatin immunoprecipitation (microChIP) (Dahl and Collas, 2008) that allowed the determination of protein binding to chromatin using purified MaSC/basal cells. With Pygo2 antibody and multiple primer sets spanning a −5 kb to +5 kb region relative to the putative Notch3 transcriptional start site (TSS), we detected appreciable signals at three sites: c and e upstream of TSS, and h within a putative enhancer/repressor at ~+5 kb (http://genome.ucsc.edu/ENCODE/) (Figure 6A,B). ChIP signals were significantly reduced in Pygo2 SSKO MaSC/basal cells, demonstrating specificity. Interestingly, the strongest binding site, h, does not contain known LEF/TCF consensus sequence, whereas several putative sites (e.g., a, b, d) did not exhibit Pygo2 occupancy. Thus, a LEF/TCF consensus sequence is not essential for Pygo2 occupancy.
Figure 6. Pygo2 regulates β-catenin binding and chromatin configuration at theNotch3 locus.
(A,C) Schematic diagrams showing the mouse Notch3 (A) and Axin2 (C) loci. Triangles, putative LEF/TCF-binding sites; short lines, amplicons (a-h for Notch3 and a-b for Axin2) in qPCR analysis. (B,D-G) Results of microChIP-qPCR analysis of Pygo2 and β-catenin binding (B, D), and histone modifications (E-G) at the Notch3 (B,E-F) and Axin2 (D,G) genes. Shown in (B,D,E,G) are average values from 3 pairs of control land SSKO mice ± SD. * p<0.05 (between control and SSKO). Shown in (F) are average values from 2 WT mice ± SD at Notch3 amplicon e. All signals were acquired with removal of IgG background first and then normalization to inputs. Histone modification signals were further normalized to histone H3.
β-catenin also bound to site e, but not to c and h (Figure 6B). Importantly, binding to e was markedly reduced in Pygo2 SSKO MaSC/basal cells, suggesting that Pygo2 is required for recruiting β-catenin and/or stabilizing its binding to the Notch3 promoter. This dependence was surprising, given the proposed model that Pygopus proteins are recruited to chromatin by a molecular chain composed of LEF/TCF, β-catenin, and adaptor Lgs/Bcl9 (Stadeli and Basler, 2005). To ask whether this finding is applicable to other Wnt targets, we examined Pygo2 and β-catenin binding to the Axin2 locus. In MaSC/basal cells, both Pygo2 and β-catenin bound to site a, which is near the TSS and contains a LEF/TCF consensus motif, but not site b, which lies downstream (Jho et al., 2002) (Figure 6C,D). Importantly, binding of both proteins was reduced to a background level when Pygo2 was absent.
Given the known involvement of Pygo2 in histone modification, we examined H3K4me3 as well as two repressive histone marks, H3K27me3 (tri-methylated lysine 27 of histone H3) and H3K9me3 (tri-methylated lysine 9 of histone H3), at the Notch3 locus in control and Pygo2-deficient cells. In control MaSC/basal cells, both H3K4me3 and H3K27me3 were found across the Notch3 locus, with extensive low-level peaks particularly from around the TSS to +5 kb (e-h; Figure 6E). This chromatin configuration is reminiscent of the “bivalent domain” found at promoters of many developmental genes that need to be silenced in embryonic stem (ES) cells but poised for prompt activation upon lineage differentiation (Bernstein et al., 2006; Mikkelsen et al., 2007). A strong peak of H3K9me3 was observed at site c (Figure 6E). In control bulk luminal cells, site e displayed a high level of H3K4me3 and a low level of H3K27me3 (Figure 6F). Thus, the Notch3 locus appears to be in a poised but silenced state in MaSC/basal cells, and is resolved to an active chromatin configuration in luminal cells. In contrast, the Axin2 locus is in a predominantly active state in MaSC/basal cells, as evident by the strong H3K4me3 but background-level H3K27me3 signals at sites a and b (Figure 6G).
In Pygo2 SSKO MaSC/basal cells, there was a general increase in H3K4me3 across the Notch3 locus, with the normally broad but weak H3K4me3 signals between TSS and the downstream enhancer now being replaced by prominent peaks centered around f (Figure 6E). In contrast, both H3K27me3 and H3K9me3 levels were decreased, especially at the proximal gene regulatory region (Figure 6E). Thus, it appears that with Pygo2 loss, the bivalent domain at the Notch3 locus in MaSC/basal cells was resolved to an active chromatin configuration, reminiscent of that in normal luminal cells. This is in contrast to the Axin2 locus, where Pygo2 loss resulted in a significant reduction in H3K4me3 at site a, to which both Pygo2 and β-catenin binds, but not site b (Figure 6G). Collectively, our data suggest that Pygo2 functions in MaSC/basal cells to maintain the Notch3 locus in a bivalent chromatin state to prevent it from being prematurely resolved to an active state compatible with luminal differentiation.
DISCUSSION
Our study identifies Pygo2 as an epigenetic gatekeeper of the MaSC/basal fate, and adds Pygo2 to a small list of known chromatin regulators of mammary epithelial stem/progenitor cells (Liu et al., 2006; Pal et al., 2013; Pietersen et al., 2008). That Pygo2 is expressed and suppresses the luminal-like gene signature in MaSC/basal cells provides at least one epigenetic mechanism by which these cells maintain their lineage identity during development (van Amerongen et al., 2012; Van Keymeulen et al., 2011). To our knowledge, this is the first to show that a histone methylation reader (i.e., Pygo2) regulates the lineage potential of adult epithelial stem cells. With the recent success in identifying small molecule inhibitors that block the activities of histone readers (Chung, 2012), understanding the biological function of this class of chromatin effectors opens new doors to direct the lineage differentiation potential for tissue engineering and treat heterogeneous human cancers with a stem/progenitor cell origin. In fact, we observed that Pygo2 loss leads to a restriction of the lineage potential of tumor initiating cells in the MMTV-Wnt1 mammary tumor model (Watanabe et al., 2013).
Our discovery of the Pygo2 deficiency-induced partial drift of MaSC/basal gene expression towards a mature luminal state is particularly interesting, as a dramatic differentiation phenotype of these Pygo2-deficient MaSC/basal cells is only manifest upon switching to lactogenic conditions that induce luminal/alveolar differentiation. This presents a previously unrecognized mode of molecular regulation, namely “prospective” regulation, of mammary stem/progenitor cells, where a chromatin effector earmarks their differentiation competence. It is important to note that the Lin−CD29hiCD24+ MaSC/basal population is inherently heterogeneous in its cellular constituents. Our detection of two distinct subpopulations of Notch3-expressing cells within this population is consistent with the reported finding of low and high expression of Hey1 in Axin2+ and Axin2−, respectively, fractions of the Lin−CD29highCD24+ pool (Zeng and Nusse, 2010). Interestingly, Pygo2 loss results in a general upward shift in Notch3 expression of the entire MaSC/basal population while maintaining two distinct subpopulations. As the Wnt-responsive Axin2+ cells are further enriched for MaSC cells, our findings are consistent with Pygo2 repressing Notch3 expression in not only MaSCs but also bulk basal cells.
Our study provides evidence for a functional and mechanistic connection between two fundamentally important signaling pathways, Wnt and Notch, in negotiating the choice between MaSC/basal and luminal lineages within the mammary epithelia. Specifically, our findings highlight Pygo2 as an epigenetic proponent of the self-renewal/basal-promoting Wnt/β-catenin signaling, and a direct epigenetic suppressor of the luminal-promoting Notch signaling. As these two pathways regulate the fates of stem cells in myriad tissues as well as in tumors, our study adds to the general knowledge of how developmental signaling pathways cross-talk with each other (Shahi et al., 2011). Of note, the negative regulation of Notch3 by Wnt signaling and Pygo2 (this study) contrasts the activation of Notch2 by Wnt signaling in colorectal cancer cells, underscoring the cell/tissue-type specificity of Wnt-Notch cross-talks (Duncan et al., 2005; Ungerback et al., 2011).
Our work identifies Notch3 as a direct target of transcriptional repression by Wnt/β-catenin and Pygo2, and as such also assigns a novel repressive role for Pygo2 in gene expression. Intriguingly, Pygo2 is required for β-catenin binding to both Axin2 and Notch3 gene regulatory regions. While this may be in apparent conflict with the LEF/TCF-β-catenin-Lgs/Bcl9-Pygo chain-of-adaptor model, it supports and refines an alternative model that Pygo2 facilitates β-catenin nuclear retention (Stadeli and Basler, 2005; Townsley et al., 2004). Reconciling our data with both models, Pygo2 and β-catenin may stabilize each other’s occupancy at target loci via multivalent interactions between Pygo2 and chromatin via H3K4me3, β-catenin and LEF/TCF sites via LEF/TCF, and Pygo2 and β-catenin via Lgs/Bcl9 (Gu et al., 2009). Overall, the functional interplay between Wnt/β-catenin signaling and Pygo2 in MaSC/basal cells appears complex: Pygo2 facilitates Wnt-induced alteration in some genes (e.g., Notch3), but activates or represses other genes (e.g., Axin2, Notch1/2) regardless of the Wnt activation status. This new mechanistic insight is entirely consistent with the finding of both Wntdependent and Wnt-independent biological functions of Pygopus proteins (Jessen et al., 2008).
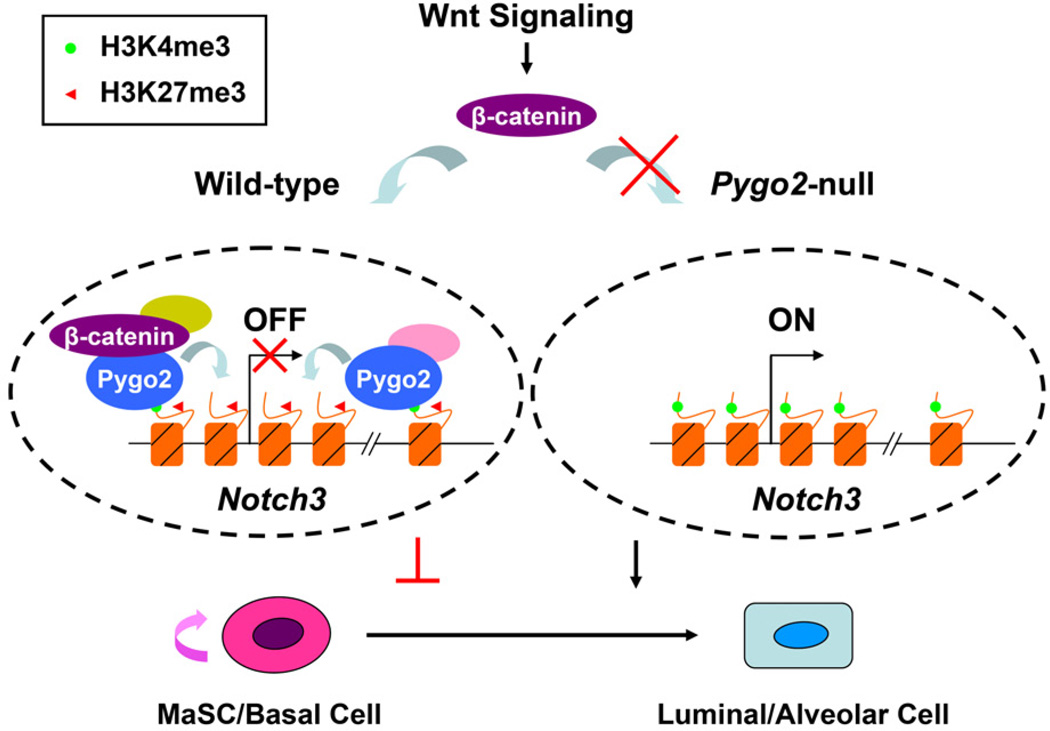
Our work offers a new example of how a histone code reader can generate different chromatin and transcriptional outcomes at different target loci. Specifically, Pygo2 is required for optimal H3K4me3 at the Axin2 promoter, but maintains a bivalent chromatin configuration at a key lineage-regulatory locus, Notch3, in MaSC/basal cells. Bivalent domains have been found in not only ES but also adult cells including the mammary epithelial lineage (Cui et al., 2009; Lien et al., 2011; Maruyama et al., 2011; Pal et al., 2013); however, their functional significance remains to be characterized. Our discovery that loss of a bivalent status at the Notch3 locus correlates with altered lineage potential now provides important but indirect evidence for the significance of a bivalent chromatin configuration. We postulate that in normal MaSC/basal cells, Pygo2 helps to maintain the Notch3 locus in a poised yet repressed state to maintain the low level of Notch3 expression (Raafat et al., 2011) and to prevent the cells from prematurely adopting a luminal/alveolar fate (Figure 7, left). When Pygo2 is absent, the Notch3 chromatin undergoes premature resolution towards an active and luminal-like state to support gene expression, which in turn primes MaSC/basal cells for luminal/alveolar differentiation upon extrinsic differentiation cues (Figure 7, right). Future studies will investigate the molecular mechanism by which Pygo2 regulates histone modifications at the Notch3 locus.
Figure 7. Working model on Pygo2 regulation of mammary epithelial lineage differentiation.
Pygo2 binding facilitates the association of β-catenin and a bivalent chromatin configuration at the Notch3 locus in MaSC/basal cells. In Pygo2-null MaSC/basal cells, Notch3 chromatin shifts precociously from having dominant repressive H3K27me3 marks to prominent permissive H3K4me3 marks, allowing derepression/activation of Notch3. See text for details.
EXPERIMENTAL PROCEDURES
Mouse strains
Pygo2 SSKO female mice were generated in congenic C57BL/6 background by crossing K14-cre; Pygo2+/− males with Pygo2flox/flox females as previously described (Gu et al., 2009). Pygo2flox/flox and Pygo2flox/+ female littermates were generated by crossing Pygo2flox/+ males with Pygo2flox/flox females.
Flow cytometry, sorting, and 3D-Matrigel differentiation assay
Mammary cells from 8-12-week old virgin females were immuno-labeled with specific antibodies (CD31-APC, CD45-APC, TER119-APC, CD24-PE-Cy7, CD29-FTIC, CD49f-FITC, CD61-PE or Notch3-PE), and analyzed by LSRII (BD Biosciences) or sorted by FACSAriaII (BD Biosciences). Data analysis was performed by Flowjo 7.6.1.
3D-Matrigel assay was performed as previously reported (Shackleton et al., 2006). Briefly, sorted MaSC/basal cells were resuspended in growth factor reduced Matrigel (BD Biosciences) and plated onto 8-well chamber slide (Thermo Fisher Scientific). Cells were cultured in a mammary epithelial growth medium for a week, and induced to undergo luminal/alveolar differentiation by medium switch to DMEM/F12 containing HIP (hydrocortisone, insulin, prolactin) and 1% FBS.
To acutely delete Pygo2, sorted Pygo2flox/flox and control Pygo2flox/+ MaSC/basal cells were infected in suspension with Ade-Cre (Vector Biolabs) before embedding in Matrigel. To modulate signaling pathways, growing cells/colonies were pretreated with DMSO, DAPT (Calbiochem) or BIO (Sigma-Aldrich) for 24 hours in growth medium before switching to differentiation medium (referred to as Day 0) containing the same agent.
Cleared fat pad transplantation
FACS-sorted MaSC/basal cells were injected into the cleared fat pad of 3-week old female C57BL/6 mice. Cbf-1 knockdown was performed by transduction with shCbf-1-expressing lentiviruses before transplantation. Outgrowths were analyzed eight weeks after transplantation. GFP fluorescence was visualized with a Leica MZFLIII fluorescent dissecting scope or Nikon E600 fluorescent microscope. Statistical analysis of the take rate was performed using the ELDA Web-based tool (http://bioinf.wehi.edu.au/software/elda/), and the statistical difference in filled fat pad area was estimated by one-way ANOVA.
Micro chromatin immunoprecipitation
MicroChIP was performed according to the protocol described previously (Dahl and Collas, 2008) using 5×104 sorted cells. The primers for qPCR were summarized in Table S2.
Additional details for the above procedures, as well as procedures for lentiviral expression/shRNA, Western blot, RNA isolation, RT-qPCR, indirect immunofluorescence, whole mount staining, and microarray analysis are described in Extended Experimental Procedures.
Supplementary Material
HIGHLIGHTS.
A histone methylation reader regulates lineage potential of adult mammary stem cells
A reader generates different chromatin/transcriptional outcomes at different loci
Pygo2 is required for maximal beta-catenin occupancy at target chromatin
Pygo2 mechanistically connects Wnt and Notch signaling in mammary lineage choice
ACKNOWLEDGEMENTS
We thank Bogi Andersen for discussions, and the UCI Genomics High Throughput Facility (GHTF) and Sue and Bill Gross Stem Cell Research Center Core Facility for expert service. This work was supported by NIH grant R01-GM083089 and Susan G. Komen grant KG110897 (to X.D.). B. G. and K. W. were supported by CBCRP Postdoctoral Fellowship (14FB-0129) and U.S. DOD BCRP Postdoctoral Fellowship (W81XWH-10-1-0383), respectively.
ABBREVIATIONS
- Pygo2
Pygopus 2
- MaSC
mammary stem cell
- LRP5/6
low-density lipoprotein-related protein 5/6
- LEF/TCF
lymphoid enhancer factor/T-cell factor
- PHD
plant homeo domain
- H3K4me2/3
di-/tri-methylated lysine 4 of histone H3
- H3K27me3
tri-methylated lysine 27 of histone H3
- 3D
3-dimensional
- K14
Keratin 14
- SSKO
skin/mammary-specific knockout
- MYO
myoepithelial
- FACS
fluorescence activated cell sorting
- Ma-CFC
mammary colony-forming cell
- SMA
smooth muscle actin
- FDR
false discovery rate
- RT-qPCR
reverse transcription-quantitative PCR
- FoxA1
forkhead box protein A1
- Areg
amphiregulin
- GSEA
gene set enrichment analysis
- GO
gene ontology
- BIO
6-bromoindirubin-3'-oxime
- ChIP
chromatin immunoprecipitation
- TSS
transcriptional start site
- HSC
hematopoietic stem cell
- PcG
Polycomb Group;
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Andrews PG, He Z, Popadiuk C, Kao KR. The transcriptional activity of Pygopus is enhanced by its interaction with cAMP-response-element-binding protein (CREB)-binding protein. Biochem J. 2009;422:493–501. doi: 10.1042/BJ20090134. [DOI] [PubMed] [Google Scholar]
- Asselin-Labat ML, Sutherland KD, Barker H, Thomas R, Shackleton M, Forrest NC, Hartley L, Robb L, Grosveld FG, van der Wees J, et al. Gata-3 is an essential regulator of mammary-gland morphogenesis and luminal-cell differentiation. Nat Cell Biol. 2007;9:201–209. doi: 10.1038/ncb1530. [DOI] [PubMed] [Google Scholar]
- Bartek J, Taylor-Papadimitriou J, Miller N, Millis R. Patterns of expression of keratin 19 as detected with monoclonal antibodies in human breast tissues and tumours. Int J Cancer. 1985;36:299–306. [PubMed] [Google Scholar]
- Belenkaya TY, Han C, Standley HJ, Lin X, Houston DW, Heasman J. pygopus Encodes a nuclear protein essential for wingless/Wnt signaling. Development. 2002;129:4089–4101. doi: 10.1242/dev.129.17.4089. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Bouras T, Pal B, Vaillant F, Harburg G, Asselin-Labat ML, Oakes SR, Lindeman GJ, Visvader JE. Notch signaling regulates mammary stem cell function and luminal cell-fate commitment. Cell Stem Cell. 2008;3:429–441. doi: 10.1016/j.stem.2008.08.001. [DOI] [PubMed] [Google Scholar]
- Buono KD, Robinson GW, Martin C, Shi S, Stanley P, Tanigaki K, Honjo T, Hennighausen L. The canonical Notch/RBP-J signaling pathway controls the balance of cell lineages in mammary epithelium during pregnancy. Dev Biol. 2006;293:565–580. doi: 10.1016/j.ydbio.2006.02.043. [DOI] [PubMed] [Google Scholar]
- Chaffer CL, Brueckmann I, Scheel C, Kaestli AJ, Wiggins PA, Rodrigues LO, Brooks M, Reinhardt F, Su Y, Polyak K, et al. Normal and neoplastic nonstem cells can spontaneously convert to a stem-like state. Proc Natl Acad Sci U S A. 2011;108:7950–7955. doi: 10.1073/pnas.1102454108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Luo Q, Yuan Y, Huang X, Cai W, Li C, Wei T, Zhang L, Yang M, Liu Q, et al. Pygo2 associates with MLL2 histone methyltransferase and GCN5 histone acetyltransferase complexes to augment Wnt target gene expression and breast cancer stem-like cell expansion. Mol Cell Biol. 2010;30:5621–5635. doi: 10.1128/MCB.00465-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung CW. Small molecule bromodomain inhibitors: extending the druggable genome. Prog Med Chem. 2012;51:1–55. doi: 10.1016/B978-0-12-396493-9.00001-7. [DOI] [PubMed] [Google Scholar]
- Cui K, Zang C, Roh TY, Schones DE, Childs RW, Peng W, Zhao K. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell. 2009;4:80–93. doi: 10.1016/j.stem.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl JA, Collas P. A rapid micro chromatin immunoprecipitation assay (microChIP) Nat Protoc. 2008;3:1032–1045. doi: 10.1038/nprot.2008.68. [DOI] [PubMed] [Google Scholar]
- Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ, Wicha MS. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dovey HF, John V, Anderson JP, Chen LZ, de Saint Andrieu P, Fang LY, Freedman SB, Folmer B, Goldbach E, Holsztynska EJ, et al. Functional gamma-secretase inhibitors reduce beta-amyloid peptide levels in brain. J Neurochem. 2001;76:173–181. doi: 10.1046/j.1471-4159.2001.00012.x. [DOI] [PubMed] [Google Scholar]
- Duncan AW, Rattis FM, DiMascio LN, Congdon KL, Pazianos G, Zhao C, Yoon K, Cook JM, Willert K, Gaiano N, et al. Integration of Notch and Wnt signaling in hematopoietic stem cell maintenance. Nat Immunol. 2005;6:314–322. doi: 10.1038/ni1164. [DOI] [PubMed] [Google Scholar]
- Fiedler M, Sanchez-Barrena MJ, Nekrasov M, Mieszczanek J, Rybin V, Muller J, Evans P, Bienz M. Decoding of methylated histone H3 tail by the Pygo-BCL9 Wnt signaling complex. Mol Cell. 2008;30:507–518. doi: 10.1016/j.molcel.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gat U, DasGupta R, Degenstein L, Fuchs E. De Novo hair follicle morphogenesis and hair tumors in mice expressing a truncated beta-catenin in skin. Cell. 1998;95:605–614. doi: 10.1016/s0092-8674(00)81631-1. [DOI] [PubMed] [Google Scholar]
- Geling A, Steiner H, Willem M, Bally-Cuif L, Haass C. A gamma-secretase inhibitor blocks Notch signaling in vivo and causes a severe neurogenic phenotype in zebrafish. EMBO Rep. 2002;3:688–694. doi: 10.1093/embo-reports/kvf124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gjorevski N, Nelson CM. Integrated morphodynamic signalling of the mammary gland. Nat Rev Mol Cell Biol. 2011;12:581–593. doi: 10.1038/nrm3168. [DOI] [PubMed] [Google Scholar]
- Gu B, Sun P, Yuan Y, Moraes RC, Li A, Teng A, Agrawal A, Rheaume C, Bilanchone V, Veltmaat JM, et al. Pygo2 expands mammary progenitor cells by facilitating histone H3 K4 methylation. J Cell Biol. 2009;185:811–826. doi: 10.1083/jcb.200810133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu B, Watanabe K, Dai X. Pygo2 regulates histone gene expression and H3 K56 acetylation in human mammary epithelial cells. Cell Cycle. 2012;11:79–87. doi: 10.4161/cc.11.1.18402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudjonsson T, Villadsen R, Nielsen HL, Ronnov-Jessen L, Bissell MJ, Petersen OW. Isolation, immortalization, and characterization of a human breast epithelial cell line with stem cell properties. Genes Dev. 2002;16:693–706. doi: 10.1101/gad.952602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gugliotta P, Sapino A, Macri L, Skalli O, Gabbiani G, Bussolati G. Specific demonstration of myoepithelial cells by anti-alpha smooth muscle actin antibody. J Histochem Cytochem. 1988;36:659–663. doi: 10.1177/36.6.3367051. [DOI] [PubMed] [Google Scholar]
- Guo W, Keckesova Z, Donaher JL, Shibue T, Tischler V, Reinhardt F, Itzkovitz S, Noske A, Zurrer-Hardi U, Bell G, et al. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell. 2012;148:1015–1028. doi: 10.1016/j.cell.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta PB, Fillmore CM, Jiang G, Shapira SD, Tao K, Kuperwasser C, Lander ES. Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell. 2011;146:633–644. doi: 10.1016/j.cell.2011.07.026. [DOI] [PubMed] [Google Scholar]
- Hoverter NP, Waterman ML. A Wnt-fall for gene regulation: repression. Sci Signal. 2008;1:e43. doi: 10.1126/scisignal.139pe43. [DOI] [PubMed] [Google Scholar]
- Huper G, Marks JR. Isogenic normal basal and luminal mammary epithelial isolated by a novel method show a differential response to ionizing radiation. Cancer Res. 2007;67:2990–3001. doi: 10.1158/0008-5472.CAN-06-4065. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Jessen S, Gu B, Dai X. Pygopus and the Wnt signaling pathway: a diverse set of connections. Bioessays. 2008;30:448–456. doi: 10.1002/bies.20757. [DOI] [PubMed] [Google Scholar]
- Jho EH, Zhang T, Domon C, Joo CK, Freund JN, Costantini F. Wnt/beta-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol Cell Biol. 2002;22:1172–1183. doi: 10.1128/MCB.22.4.1172-1183.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler R, Hausmann G, Basler K. The PHD domain is required to link Drosophila Pygopus to Legless/beta-catenin and not to histone H3. Mech Dev. 2009;126:752–759. doi: 10.1016/j.mod.2009.04.003. [DOI] [PubMed] [Google Scholar]
- Kramps T, Peter O, Brunner E, Nellen D, Froesch B, Chatterjee S, Murone M, Zullig S, Basler K. Wnt/wingless signaling requires BCL9/legless-mediated recruitment of pygopus to the nuclear beta-catenin-TCF complex. Cell. 2002;109:47–60. doi: 10.1016/s0092-8674(02)00679-7. [DOI] [PubMed] [Google Scholar]
- Lien WH, Guo X, Polak L, Lawton LN, Young RA, Zheng D, Fuchs E. Genome-wide maps of histone modifications unwind in vivo chromatin states of the hair follicle lineage. Cell Stem Cell. 2011;9:219–232. doi: 10.1016/j.stem.2011.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim E, Wu D, Pal B, Bouras T, Asselin-Labat ML, Vaillant F, Yagita H, Lindeman GJ, Smyth GK, Visvader JE. Transcriptome analyses of mouse and human mammary cell subpopulations reveal multiple conserved genes and pathways. Breast Cancer Res. 2010;12:R21. doi: 10.1186/bcr2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Dontu G, Mantle ID, Patel S, Ahn NS, Jackson KW, Suri P, Wicha MS. Hedgehog signaling and Bmi-1 regulate self-renewal of normal and malignant human mammary stem cells. Cancer Res. 2006;66:6063–6071. doi: 10.1158/0008-5472.CAN-06-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama R, Choudhury S, Kowalczyk A, Bessarabova M, Beresford-Smith B, Conway T, Kaspi A, Wu Z, Nikolskaya T, Merino VF, et al. Epigenetic regulation of cell type-specific expression patterns in the human mammary epithelium. PLoS Genet. 2011;7:e1001369. doi: 10.1371/journal.pgen.1001369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- Nair M, Nagamori I, Sun P, Mishra DP, Rheaume C, Li B, Sassone-Corsi P, Dai X. Nuclear regulator Pygo2 controls spermiogenesis and histone H3 acetylation. Dev Biol. 2008;320:446–455. doi: 10.1016/j.ydbio.2008.05.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal B, Bouras T, Shi W, Vaillant F, Sheridan JM, Fu N, Breslin K, Jiang K, Ritchie ME, Young M, et al. Global Changes in the Mammary Epigenome Are Induced by Hormonal Cues and Coordinated by Ezh2. Cell Rep. 2013 doi: 10.1016/j.celrep.2012.12.020. [DOI] [PubMed] [Google Scholar]
- Parker DS, Jemison J, Cadigan KM. Pygopus, a nuclear PHD-finger protein required for Wingless signaling in Drosophila. Development. 2002;129:2565–2576. doi: 10.1242/dev.129.11.2565. [DOI] [PubMed] [Google Scholar]
- Petersen OW, Ronnov-Jessen L, Howlett AR, Bissell MJ. Interaction with basement membrane serves to rapidly distinguish growth and differentiation pattern of normal and malignant human breast epithelial cells. Proc Natl Acad Sci U S A. 1992;89:9064–9068. doi: 10.1073/pnas.89.19.9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietersen AM, Evers B, Prasad AA, Tanger E, Cornelissen-Steijger P, Jonkers J, van Lohuizen M. Bmi1 regulates stem cells and proliferation and differentiation of committed cells in mammary epithelium. Curr Biol. 2008;18:1094–1099. doi: 10.1016/j.cub.2008.06.070. [DOI] [PubMed] [Google Scholar]
- Raafat A, Goldhar AS, Klauzinska M, Xu K, Amirjazil I, McCurdy D, Lashin K, Salomon D, Vonderhaar BK, Egan S, et al. Expression of Notch receptors, ligands, and target genes during development of the mouse mammary gland. J Cell Physiol. 2011;226:1940–1952. doi: 10.1002/jcp.22526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raouf A, Zhao Y, To K, Stingl J, Delaney A, Barbara M, Iscove N, Jones S, McKinney S, Emerman J, et al. Transcriptome analysis of the normal human mammary cell commitment and differentiation process. Cell Stem Cell. 2008;3:109–118. doi: 10.1016/j.stem.2008.05.018. [DOI] [PubMed] [Google Scholar]
- Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434:843–850. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- Roarty K, Rosen JM. Wnt and mammary stem cells: hormones cannot fly wingless. Curr Opin Pharmacol. 2010;10:643–649. doi: 10.1016/j.coph.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato N, Meijer L, Skaltsounis L, Greengard P, Brivanlou AH. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat Med. 2004;10:55–63. doi: 10.1038/nm979. [DOI] [PubMed] [Google Scholar]
- Shackleton M, Vaillant F, Simpson KJ, Stingl J, Smyth GK, Asselin-Labat ML, Wu L, Lindeman GJ, Visvader JE. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439:84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- Shahi P, Seethammagari MR, Valdez JM, Xin L, Spencer DM. Wnt and Notch pathways have interrelated opposing roles on prostate progenitor cell proliferation and differentiation. Stem Cells. 2011;29:678–688. doi: 10.1002/stem.606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadeli R, Basler K. Dissecting nuclear Wingless signalling: recruitment of the transcriptional co-activator Pygopus by a chain of adaptor proteins. Mech Dev. 2005;122:1171–1182. doi: 10.1016/j.mod.2005.07.004. [DOI] [PubMed] [Google Scholar]
- Stingl J, Eirew P, Ricketson I, Shackleton M, Vaillant F, Choi D, Li HI, Eaves CJ. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439:993–997. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun P, Yuan Y, Li A, Li B, Dai X. Cytokeratin expression during mouse embryonic and early postnatal mammary gland development. Histochem Cell Biol. 2010;133:213–221. doi: 10.1007/s00418-009-0662-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takebe N, Harris PJ, Warren RQ, Ivy SP. Targeting cancer stem cells by inhibiting Wnt, Notch, and Hedgehog pathways. Nat Rev Clin Oncol. 2011;8:97–106. doi: 10.1038/nrclinonc.2010.196. [DOI] [PubMed] [Google Scholar]
- Teuliere J, Faraldo MM, Deugnier MA, Shtutman M, Ben-Ze'ev A, Thiery JP, Glukhova MA. Targeted activation of beta-catenin signaling in basal mammary epithelial cells affects mammary development and leads to hyperplasia. Development. 2005;132:267–277. doi: 10.1242/dev.01583. [DOI] [PubMed] [Google Scholar]
- Thompson B, Townsley F, Rosin-Arbesfeld R, Musisi H, Bienz M. A new nuclear component of the Wnt signalling pathway. Nat Cell Biol. 2002;4:367–373. doi: 10.1038/ncb786. [DOI] [PubMed] [Google Scholar]
- Townsley FM, Cliffe A, Bienz M. Pygopus and Legless target Armadillo/beta-catenin to the nucleus to enable its transcriptional co-activator function. Nat Cell Biol. 2004;6:626–633. doi: 10.1038/ncb1141. [DOI] [PubMed] [Google Scholar]
- Ungerback J, Elander N, Grunberg J, Sigvardsson M, Soderkvist P. The Notch-2 gene is regulated by Wnt signaling in cultured colorectal cancer cells. PLoS One. 2011;6:e17957. doi: 10.1371/journal.pone.0017957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Amerongen R, Bowman AN, Nusse R. Developmental stage and time dictate the fate of Wnt/beta-catenin-responsive stem cells in the mammary gland. Cell Stem Cell. 2012;11:387–400. doi: 10.1016/j.stem.2012.05.023. [DOI] [PubMed] [Google Scholar]
- Van Keymeulen A, Rocha AS, Ousset M, Beck B, Bouvencourt G, Rock J, Sharma N, Dekoninck S, Blanpain C. Distinct stem cells contribute to mammary gland development and maintenance. Nature. 2011;479:189–193. doi: 10.1038/nature10573. [DOI] [PubMed] [Google Scholar]
- Visvader JE. Keeping abreast of the mammary epithelial hierarchy and breast tumorigenesis. Genes Dev. 2009;23:2563–2577. doi: 10.1101/gad.1849509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvader JE. Cells of origin in cancer. Nature. 2011;469:314–322. doi: 10.1038/nature09781. [DOI] [PubMed] [Google Scholar]
- Watanabe K, Fallahi M, Dai X. Chromatin effector Pygo2 regulates mammary tumor initiation and heterogeneity in MMTV-Wnt1 mice. Oncogene. 2013 doi: 10.1038/onc.2012.620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei G, Wei L, Zhu J, Zang C, Hu-Li J, Yao Z, Cui K, Kanno Y, Roh TY, Watford WT, et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 2009;30:155–167. doi: 10.1016/j.immuni.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng YA, Nusse R. Wnt proteins are self-renewal factors for mammary stem cells and promote their long-term expansion in culture. Cell Stem Cell. 2010;6:568–577. doi: 10.1016/j.stem.2010.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.