Abstract
Expression of E-cadherin, a hallmark of epithelial-mesenchymal transition (EMT), is often lost due to promoter DNA methylation in basal-like breast cancer (BLBC), which contributes to the metastatic advantage of this disease; however, the underlying mechanism remains unclear. Here we identified that Snail interacted with Suv39H1, a major methyltransferase responsible for H3K9me3 that intimately links to DNA methylation. We demonstrated that the SNAG domain of Snail and the SET domain of Suv39H1 were required for their mutual interactions. We found that H3K9me3 and DNA methylation on the E-cadherin promoter were higher in BLBC cell lines. We showed that Snail interacted with Suv39H1 and recruited it to the E-cadherin promoter for transcriptional repression. Knockdown of Suv39H1 restored E-cadherin expression by blocking H3K9me3 and DNA methylation and resulted in the inhibition of cell migration, invasion and metastasis of BLBC. Our study not only reveals a critical mechanism underlying the epigenetic regulation of EMT, but also paves a way for the development of new treatment strategies against this disease.
Keywords: Metastasis, EMT, Chromatin modifications, Transcription, Snail
INTRODUCTION
Breast cancer is the second leading cause of cancer-related death in women with a yearly toll of more than 40,000 deaths in the United States alone (1). Despite exciting progress toward understanding breast cancer development and progression, and in the development of novel therapeutic strategies, breast cancer–related deaths are primarily due to the "incurable" nature of metastasis. Therefore, novel therapeutic strategies are required to prevent and treat metastatic breast cancer. A better understanding of the molecular events that contribute to tumor invasion and metastasis is crucial to the development of such strategies.
The increased motility and invasiveness of metastatic tumor cells are reminiscent of the events that occur at EMT, which is a characteristic of embryonic development, tissue remodeling, wound healing and metastasis (2–4). In these EMT processes, epithelial cells acquire fibroblast-like properties and exhibit reduced intercellular adhesion and increased motility (5–7). EMT also bestows tumor cells with cancer stem cell-like characters, provides them with resistance to immunosuppression and confers tumor recurrence (8–11). Two distinct phenomena are commonly associated with EMT. First, EMT is provoked by extrinsic signals that cells receive from their microenvironment (4, 12), such as signals from TGF-β, TNF-α and growth factors (13–15). A second phenomenon associated with EMT is that it is a reversible process (4). When breast cancer cells disseminate to distant sites of the body, they no longer encounter the signals that they experienced in the primary tumor, and they can revert to an epithelial state via a mesenchymal-epithelial transition (MET). The phenotypic and cellular plasticity of EMT is determined by a unique gene-expression pattern through transcription factor-mediated chromatin modifications, which mainly consist of histone modifications and DNA methylation (16, 17). Post-translational modifications at the N-termini of histones, including phosphorylation, acetylation, methylation and ubiquitination, alter chromatin structure to control DNA accessibility. These modifications can function individually or in combination to elicit specific effects based on changes in chromatin structure or the recruitment of effector proteins that bind to specific patterns of histone marks. Histone lysine methylation, in particular, has emerged as an important modification due to its central role in transcriptional regulation. For example, methylated lysines 4, 36 and 79 on H3 (denoted H3K4, H3K36 and H3K79) are found in active chromatin, whereas methylated H3K9 and H3K27 are generally associated with gene repression.
Loss of H3K4 methylation and gain of H3K9 methylation appear most closely associated with DNA methylation (18–21), which provides reinforcement as well as establishment of gene silencing. DNA methylation is executed by a family of highly related DNA methyltransferase enzymes (DNMT1, DNMT3a, and DNMT3b) that transfer a methyl group to the cytosine in a CpG dinucleotide which commonly occurs in the promoter region of genes (22, 23). Interestingly, promoter DNA methylation is the common cause of loss of E-cadherin expression in basal-like breast cancer (BLBC, commonly referred to as triple-negative breast cancer), a subtype of breast cancer possesses many EMT characteristics and characterized by an aggressive, metastasis-prone phenotype that associates with poor clinical outcome (24–28). A hallmark of EMT is the loss of E-cadherin expression (6, 29). Snail is key transcriptional repressor for downregulating E-cadherin in EMT (5, 6, 29). Although Snail is highly expressed in BLBC for repressing E-cadherin expression, the sequential event and the molecular mechanism leading to DNA methylation at the E-cadherin promoter in BLBC remain not well characterized. In our previous study, we found that Snail interacted with LSD1 and recruited LSD1 to the E-cadherin promoter to specifically remove H3K4 methylation (30). However, H3K4 demethylation is known to be an initial step in gene repression (31), suggesting that an intermediate step is required to bridge H3K4 demethylation to the DNA methylation on the E-cadherin promoter. Here, we showed that Snail interacted with Suv39H1, a key component of methyltransferase responsible for H3K9me3. We reported that this functional interaction is critical for H3K9me3 and DNA methylation at the E-cadherin promoter in BLBC.
RESULTS
The SNAG domain of Snail interacts with the catalytic domain of Suv39H1
Several transcription factors, such as Snail, Slug, GFI, insulinoma-associated protein IA-1 (Insm1) and Ovo-like 1 (OVOL1), contain a SNAG (Snail/GFI) domain at the N-terminus, which is important for their repressive activity in mammalian cells (32). We previously demonstrated that the SNAG domain adopted a conformation similar to that of the histone H3 tail for interaction with LSD1 (30), suggesting this domain is critical in recruiting chromatin repressive complexes for gene silencing. To further identify the protein complex associated with the SNAG domain of Snail, we generated a SNAG domain peptide (amino acid 1–17) with Biotin conjugation at the C-terminus and incubated this peptide with HeLa nuclear extract proteins. After extensive washing, the final SNAG-peptide protein complexes were separated on SDS-PAGE and subjected to top-down mass spectrometry analysis. Several known proteins associated with the LSD1 complex, such as LSD1, CoREST, HDAC1/2, BHC80, and GAS, were found in the complexes, which validated the specificity of this system. Interestingly, Suv39H1, an H3K9 methyltransferasease, was also identified as a protein that associated with the SNAG peptide.
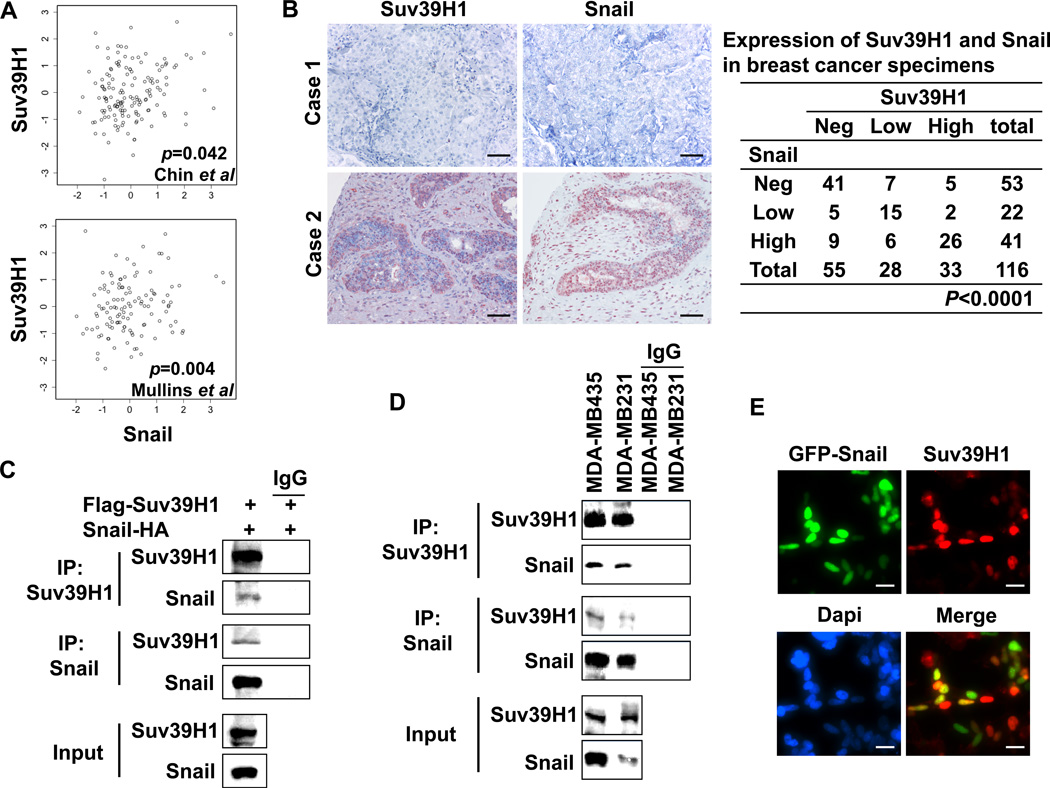
If a gene associated with Snail is biologically meaningful in breast cancer, we expect that their expression pattern may be similar. To this end, we analyzed Suv39H1 expression in two gene expression datasets, which contain 118 and 149 breast tumor samples, respectively (33, 34). We noticed that Suv39H1 expression was significantly correlated with the level of Snail in both gene expression datasets with Pearson’s correlation coefficients of 0.19 (95% confidence interval 0.01 to 0.36) and 0.23 (95% confidence interval 0.07 to 0.38), respectively (Fig. 1A). These bio-informatic analyses suggest that the co-expression of these two molecules is critical for their function in vivo. To further validate this finding, we analyzed the expression of Suv39H1 and Snail via the immunohistochemical staining of 116 breast tumor samples (Fig. 1B). We found that the expression of Suv39H1 significantly correlated with the expression of Snail in breast cancer.
Figure 1.
Suv39H1 interacts with Snail and correlates with its expression in breast cancer. (A) A statistically significant correlation in gene expression between Suv39H1 and Snail was found in two datasets from Chin et al and Mullins et al (33, 34) with Pearson’s correlation coefficients of 0.19 (95% confidence interval 0.01 to 0.36) and 0.23 (95% confidence interval 0.07 to 0.38), respectively. (B) The 116 surgical specimens of breast cancer were immuno-stained using antibodies against Snail, Suv39H1, and the control serum (data not shown). Statistical analysis is shown in the left panel and representative stainings from the same tumor samples are shown in the right panel (Scale bar = 100 µm). (C) HEK293 cells were transiently co-expressed with Flag-tagged Suv39H1 and HA-tagged Snail. Cell extracts were immunoprecipitated separately with IgG or specific antibodies against Flag or HA, and the associated Suv39H1 and Snail were examined by Western blotting, respectively. (D) Endogenous Snail and Suv39H1 were immunoprecipitated from MDA-MB435 and MDA-MB231 cells and bound endogenous Snail and Suv39H1 were examined by Western blotting, respectively. (E) GFP-tagged Snail was expressed in HEK293 cells. After fixation, the cellular localization of Snail (green) and Suv39H1 (red) was examined by immunofluorescent staining. Scale bar = 10 µm.
To validate the interaction of Snail with Suv39H1, we co-expressed Snail-HA and Flag-Suv39H1 in HEK293 cells and performed a co-immunoprecipitation experiment. After immunoprecipitating Suv39H1, we detected the associated Snail, and vice versa (Fig. 1C). We also immunoprecipitated endogenous Snail and Suv39H1 from BLBC MDA-MB 231 and MDA-MB435 cells and detected the presence of endogenous Suv39H1 and Snail, respectively (Fig. 1D). Consistent with the interaction and the correlated expression of these two molecules in breast cancer, when GFP-Snail was expressed in HEK293 cells, we found that Snail was co-localized with endogenous Suv39H1 in the nucleus (Fig. 1E). Taken together, our results indicate that Snail interacts with Suv39H1 in vivo and their expression is correlated in breast cancer.
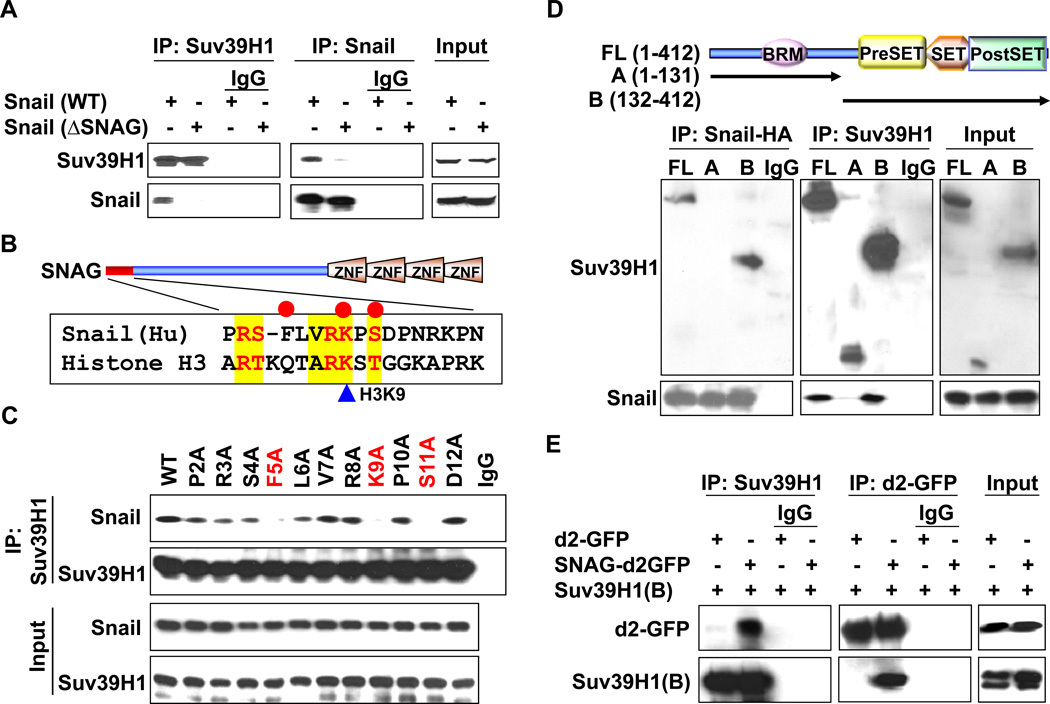
We previously showed that the SNAG domain adopted a conformation similar to that of the Histone H3 tail for recruiting the chromatin modifying enzyme LSD1 (30). Because Suv39H1 was identified as a protein associated with the SNAG peptide of Snail, we performed a Co-IP experiment to assess whether the SNAG domain of Snail is required for the interaction with Suv39H1. We found that deletion of the SNAG domain significantly reduced the interaction of Snail with Suv39H1, indicating that the SNAG domain is required for the interaction of Snail with Suv39H1 (Fig. 2A). We noticed that the sequence of the SNAG domain is highly similar to that of the N-terminus of histone H3 that surrounds the lysine 9 residue, a substrate of Suv39H1 (Fig. 2B). To identify the critical residues within the SNAG domain required for interaction with Suv39H1, we performed alanine-scan mutagenesis on the SNAG domain of Snail (Fig. 2C). Among the 12 Snail mutants screened, we found that mutations at Phe5, Lys9, and Ser11 completely lost their ability to interact with Suv39H1. The loss of interaction of these mutants with Suv39H1 was not due to the variation of protein stability as about equal amounts of Snail was immunoprecipitated (input lysates on Fig. 2C). Together, these results indicate that the SNAG domain, particularly several key amino acid residues surrounding lysine 9, was required for the interaction of Snail with Suv39H1.
Figure 2.
The SNAG domain of Snail and the SET domain of Suv39H1 are required for their mutual interactions. (A) Flag-tagged Suv39H1 and HA-tagged wild type or SNAG-deleted Snail were co-expressed in HEK293 cells. After immunoprecipitation, bound Snail or Suv39H1 was examined by Western blotting. (B) Schematic diagram showing the functional domains of Snail and sequence alignment of the SNAG domain of Snail with histone H3. The consensus sequence is highlighted in yellow and the critical interacting residues are marked with red cycle on top. (C) WT or mutant Snail was co-expressed with Suv39H1 in HEK293 cells. After immunoprecipitating Suv39H1, bound Snail was examined by Western blotting. Input lysates are shown in the bottom panel. (D) Schematic diagram showing the structure of Suv39H1 and the two different deletion constructs used in this study (top panel). HEK293 cells were transiently co-expressed with plasmids encoding Flag-tagged full-length or deletion mutants of Suv39H1 and HA-tagged Snail. Extracts were immunoprecipitated with IgG or specific antibodies against Flag or HA, and bound Suv39H1 or Snail was examined by Western blotting. (E) d2-GFP or SNAG-d2-GFP was co-expressed with the catalytic domain of Suv39H1 (B) in HEK293 cells. After immunoprecipitation of d2-GFP or Suv39H1, bound Suv39H1 and GFP were examined by Western blotting, respectively.
Suv39H1 contains several functional domains, including a N-terminal bromo domain, a catalytic region containing a Pre-SET, SET and Post-SET domain that is responsible for its enzymatic methyltransferase activity (Top panel, Fig. 2D). To identify the region responsible for the interaction with Snail, we generated two deletion mutants of Suv39H1; the N-terminal region of Suv39H1 [Suv39H1(A); amino acids 1–131] that contains the bromo domain of Suv39H1 and the C-terminal region of Suv39H1 [Suv39H1(B); amino acids 132–412] that includes the conservative Pre-SET, SET and Post-SET domains (Top panel, Fig. 2D). When these two deletion mutants of Suv39H1 were co-expressed with Snail in HEK293 cells, we found that Suv39H1(B) was able to interact with Snail, indicating that the catalytic region of Suv39H1 was responsible for its interaction with Snail (Fig. 2D). We also co-expressed d2-GFP or SNAG-d2-GFP with Suv39H1(B) in HEK293 cells. Immunoprecipitation of SNAG-d2-GFP, but not d2-GFP, maintained the association of Suv39H1(B), indicating that the SNAG domain is sufficient for Snail to interact with the catalytic domain of Suv39H1 (Fig. 2E).
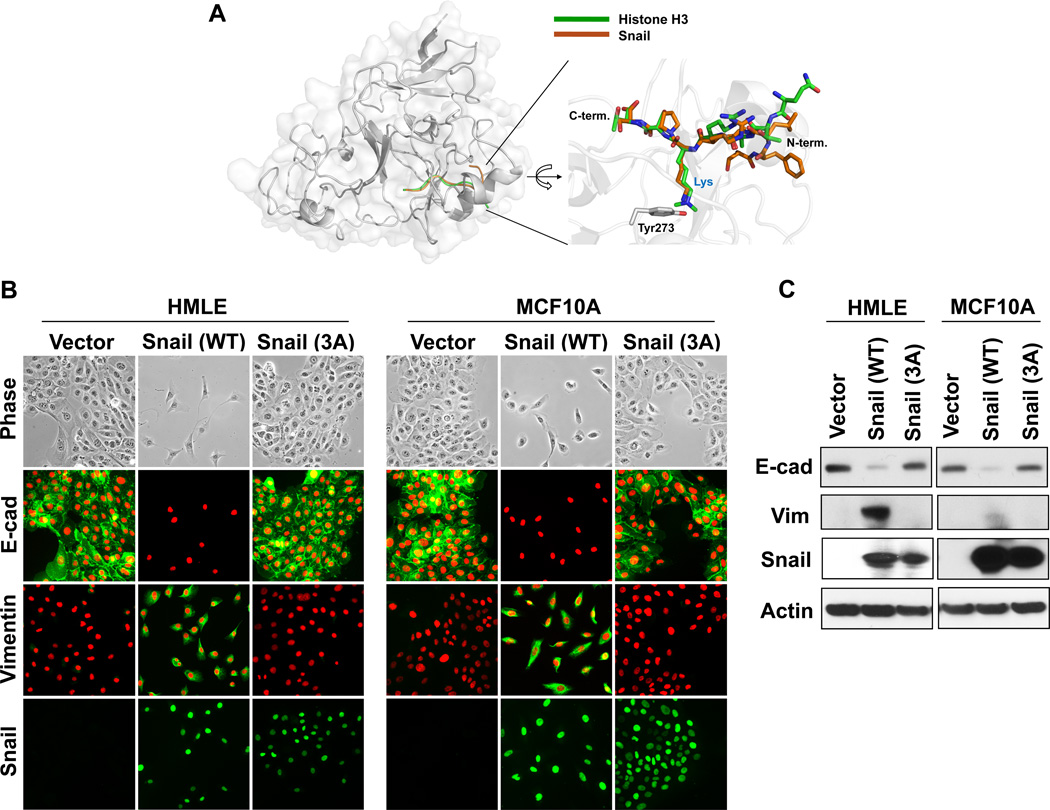
Methylation of H3K9 in humans relies mostly on members of the Suv39 family, namely Suv39H1, Suv39H2, GLP, G9a, SETDB1 and SETDB2 (35, 36); all of these enzymes have a highly conserved catalytic region containing Pro-SET, SET and Post-SET domains. Although the crystal structure of Suv39H1 is not available, the 3D structures of Suv39H2, GLP and G9a have been determined recently (37); these Suv39 family H3K9 methyltransferases have a similar ternary structure within their catalytic region. In particular, the 3D structure of GLP associated with histone H3 peptide revealed a detailed mechanism of interaction (37). A highly conserved tyrosine residue in the SET domain of GLP forms a hydrogen-bond with the ε-amine nitrogen of the H3K9 peptide (37). Based on the template structure of the GLP-histone H3 peptide complex, we built a 3D model of Suv39H1 with the SNAG domain of Snail by comparative protein structure modeling. We found that the SNAG domain of Snail adopted a conformation superimposed by the histone H3 tail at the catalytic cavity of Suv39H1 (Fig. 3A). Strikingly, Lys9 of the SNAG domain of Snail participated in similar critical contacts with the conserved tyrosine residue of Suv39H1 as compared to those of the histone H3 tail. Consistent with these analyses, a Snail mutant [Snail(3A)-F5/K9/S11] with mutation of the critical residues important to the interaction with Suv39H1 completely lost the ability to induce EMT in two model cell lines, HMLE and MCF10A cells (Fig. 3B & 3C) (38–40). Together, our results indicate that (1) expression of Suv39H1 correlates with Snail in breast cancer; (2) the SNAG domain of Snail and the catalytic domain of Suv39H1 are required for their interaction; and (3) mutation of the critical residues in Snail required to interact with Suv39H1 abolishes EMT induction.
Figure 3.
Interaction of Snail with Suv39H1 is required for EMT induction. (A) A model structure of Suv39H1-Snail complex. The homolog (53% sequence identity) structure of GLP-Histone H3 peptide complex (PDB access code 2RFI) was used for the construction of this model (see Materials and Methods for details). The Snail and histone 3 peptides are shown in brown and green, respectively. A magnified view of the binding pocket is shown in the inset where the Suv39H1-Snail complex (brown) is superposed onto the GLP-histone H3 peptide complex structure (green). This ball-and-stick model highlights the position of the substrate lysine residue of Snail/H3 and the catalytic residue (Tyr273) of Suv39H1. In the GLP-histone H3 peptide complex structure, the lysine residue is modified as a form of N-dimethyl lysine to mimic the reaction product. (B) Wild-type and mutant Snail-3A (in which F5/K9/S11 were mutated to alanine) expressed in HMLE and MCF10A cells, the induction of EMT were analyzed by morphological changes and immunofluorescent staining of the expression of E-cadherin and vimentin. (C) Protein lysate from (B) were also examined for the expression of E-cadherin, vimentin and Snail by Western blotting.
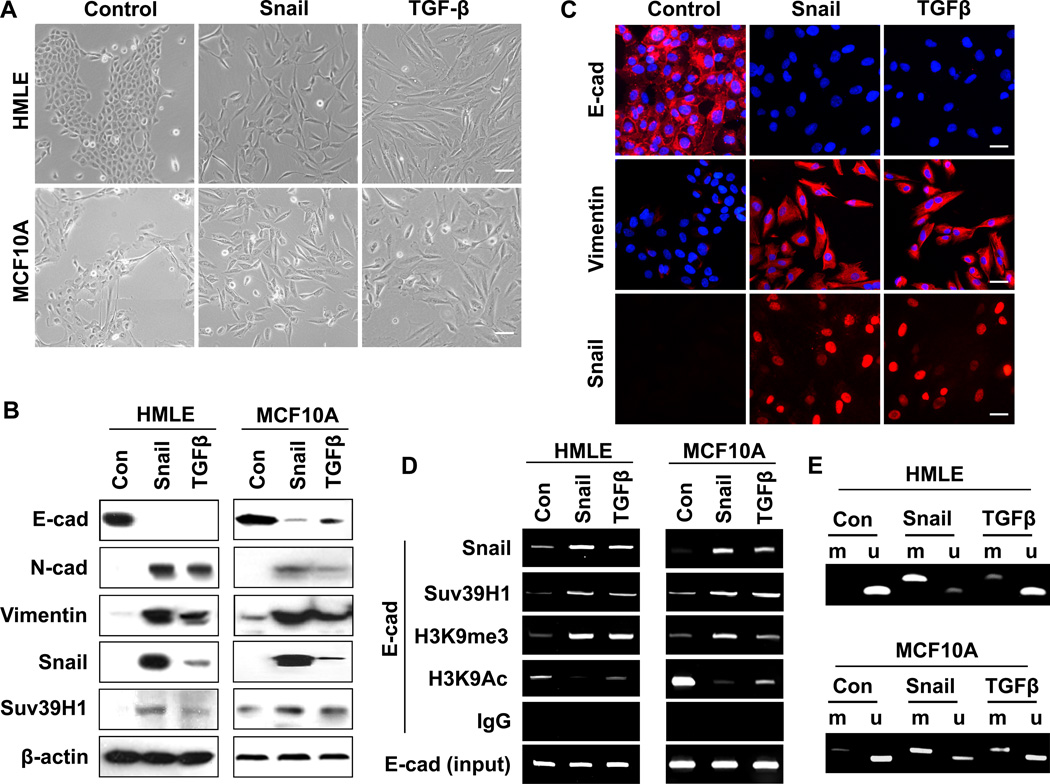
Increased association of Suv39H1 and the corresponding H3K9me3 on the E-cadherin promoter occur in EMT and are commonly found in BLBC cell lines
The majority of the BLBC cells are “locked” in the mesenchymal state by DNA methylation at the E-cadherin promoter, thus making it difficult to study the initial and dynamic event of chromatin modification leading to E-cadherin promoter DNA methylation in this disease (41). To test whether Suv39H1 is involved in the dynamic process of EMT induction, we began by investigating the association of Suv39H1 and the corresponding H3K9me3 at the E-cadherin promoter during EMT induced by Snail expression or TGF-β in HMLE and MCF10A cells. As expected, expression of Snail or TGF-β treatment (5 ng/ml), which resulted in the induction of Snail, induced EMT in these two model cell lines as characterized by the acquisition of fibroblastic mesenchymal morphology (Fig. 4A), downregulation of the epithelial markers E-cadherin, and upregulation of mesenchymal markers vimentin and N-cadherin (Fig. 4B and 4C). This EMT event also resulted in a slight upregulation of Suv39H1 (Fig. 4B). We noticed that H3K9me3 was significantly increased at the E-cadherin promoter in two cell lines tested (Fig. 4D), and this increased H3K9me3 was accompanied with decreased H3K9 acetylation of the E-cadherin promoter in these cells (Fig. 4D). The elevated H3K9me3 on the E-cadherin promoter is likely due to Suv39H1 because the association of Suv39H1 on the E-cadherin promoter was significantly increased in both cell lines (Fig. 4D). Consistent with the critical role of H3K9me3 in mediating DNA promoter methylation, we noticed that the E-cadherin promoter was DNA methylated in EMT induced by Snail expression and TGF-β in both cell lines (Fig. 4E). Together, these results suggest that the cooperative association of Suv39H1 and the corresponding upregulation of H3K9me3 at the E-cadherin promoter play a critical role in silencing the expression of E-cadherin.
Figure 4.
H3K9me3 is involved in Snail and TGF-β induced EMT. (A) HMLE and MCF10A cells were infected with retrovirus Snail or treated with TGF-β1 (5 ng/ml), respectively. Cell morphological changes associated with EMT are shown in the phase contrast images, Scale bar = 20 µm. (B) Expression of E-cadherin, N-cadherin, vimentin, Snail and Suv39H1 in the cells above was analyzed by Western blotting. Actin served as a loading control. (C) HMLE cells were treated as indicated in (A), expression of E-cadherin, Vimentin and Snail in these cells was analyzed by immunofluorescent staining. Nuclei were visualized with DAPI staining (blue). Scale bar = 10 µm. (D) HMLE and MCF10A cells were treated as in (A), association of Snail and Suv39H1 and the corresponding H3K9me3 and H3K9Ac at the E-cadherin promoter in these cell lines were analyzed by the CHIP assay as described in the Materials and Methods section. (E) HMLE and MCF10A were treated as above, EMT-mediated E-cadherin promoter methylation was examined by methylation-specific PCR (MSP).
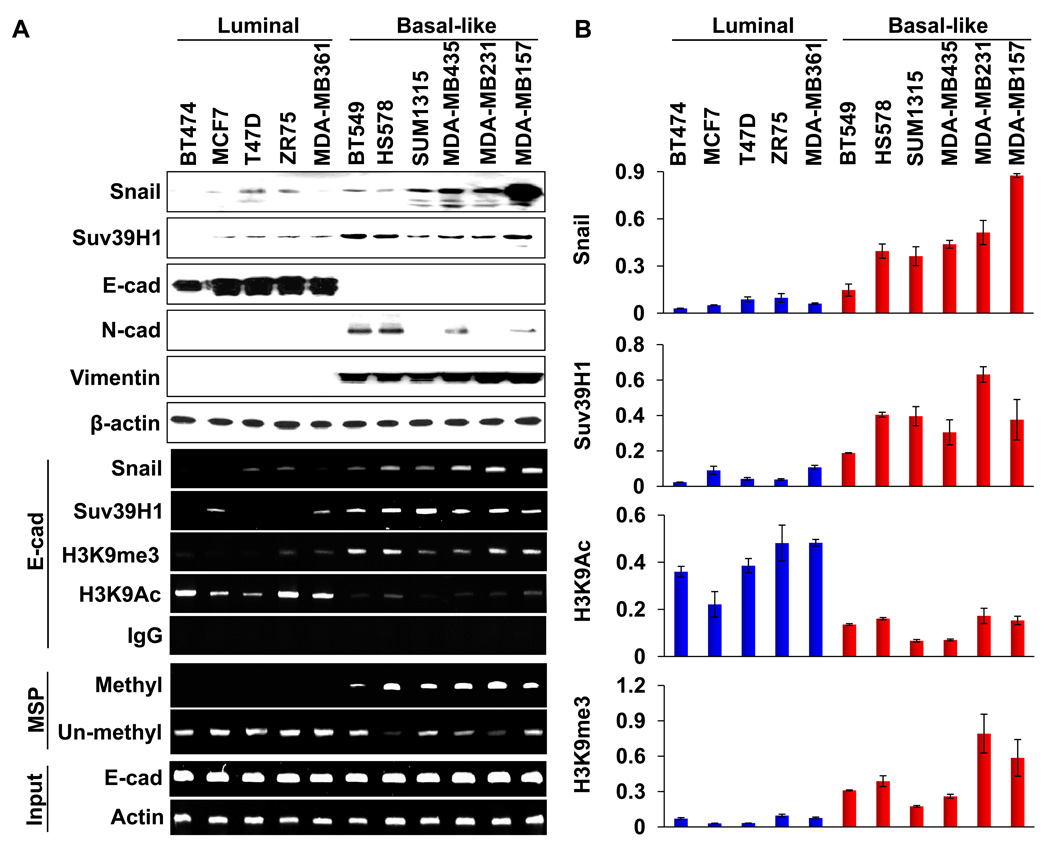
BLBC associates with a poor clinical outcome as it contains many EMT characteristics and has a significantly reduced level of E-cadherin expression (26, 27, 42). Having observed that the Snail-Suv39H1 complex is involved in E-cadherin silencing in two model cell lines, we hypothesized that this complex may also be responsible for the loss of E-cadherin expression in BLBC. To test this idea, we compared the expression of Snail, Suv39H1, and EMT markers in five luminal and six BLBC cell lines (43, 44). Consistent with previous reports, six of these BLBC cell lines lost E-cadherin expression and gained the expression of Snail, N-cadherin and vimentin (Fig. 5A). We noticed that the expression of Suv39H1 was elevated in BLBC cell lines when comparing that from luminal cell lines (Fig. 5A). In addition, when we performed ChIP analysis on the E-cadherin promoter, we detected a dramatic elevation of H3K9me3 at the E-cadherin promoter in most of the BLBC cell lines (Fig. 5A and 5B). Furthermore, H3K9 acetylation at the E-cadherin promoter was lower in BLBC cell lines than that in luminal breast cancer cell lines (Fig. 5A and 5B). The elevation of H3K9me3 at the E-cadherin promoter is likely due to the association of the Snail-Suv39H1 complex because the occupancy of Snail and Suv39H1 at the E-cadherin promoter was also significantly higher in BLBC cell lines in comparison to that of luminal breast cancer cell lines (Fig. 5A and 5B). We also assessed E-cadherin promoter DNA methylation in both subtypes using methylation-specific PCR (MSP). Again, all BLBC cell lines showed DNA methylation at the E-cadherin promoter, whereas no methylation at the E-cadherin promoter was observed in all luminal breast cancer cell lines (bottom two panels, Fig. 5A). Together, these observations clearly indicate that Suv39H1 is involved in H3K9me3 and DNA methylation on the E-cadherin promoter in EMT and breast cancer.
Figure 5.
Suv39H1-related repressive marks are enriched at the E-cadherin promoter in BLBC cell lines. (A) Cell extracts were prepared from five luminal and six basal-like subtypes of human breast cancer cell lines, and expressions of Snail, Suv39H1 and other EMT markers were analyzed by Western blotting (top panels). The association of Snail, Suv39H1 and the levels of H3K9me3 and H3K9 acetylation at the E-cadherin promoter in various cell lines was analyzed with the ChIP assay (middle panels). Methylation of the E-cadherin promoter in various breast cell lines was examined by MSP (bottom two panels). (B) Quantitative real-time PCR was performed to analyze CHIP samples from (A). Results from three independent experiments are presented (mean ± SD from three separate experiments).
Knockdown of Suv39H1 expression inhibits H3K9me3 and DNA methylation on the E-cadherin promoter and suppresses cell migration and invasion
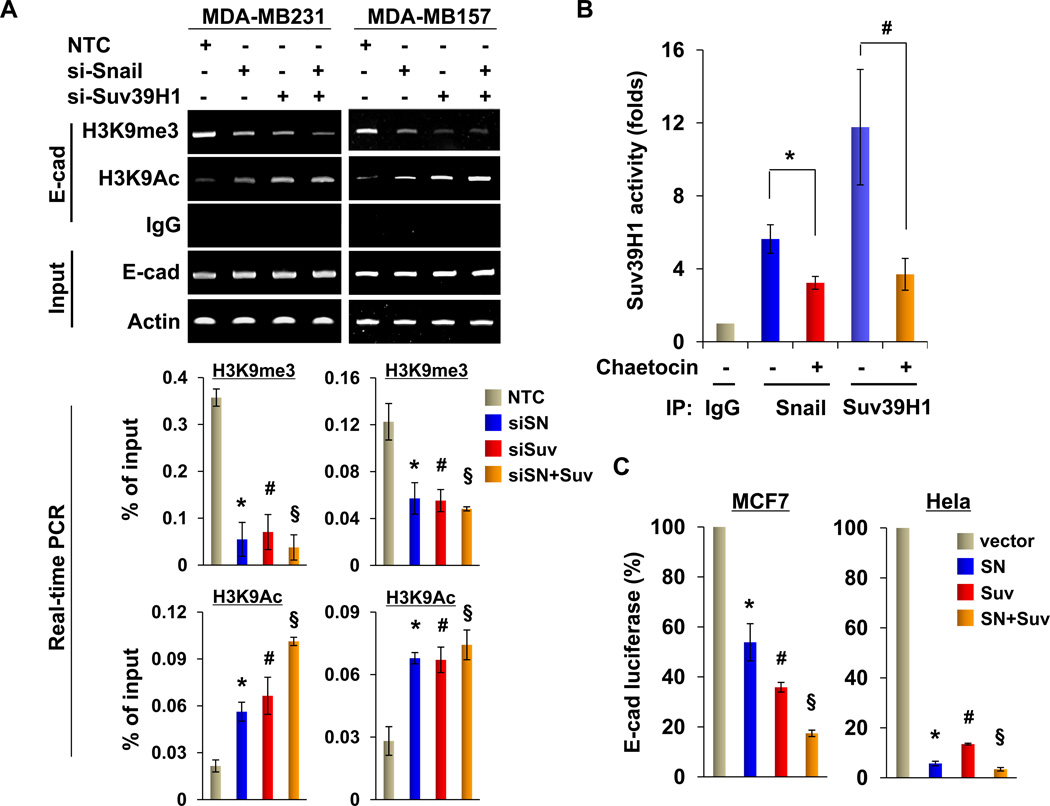
To further validate that the Snail-Suv39H1 complex is associated with the E-cadherin promoter and mediates the transcriptional repression of the E-cadherin gene in BLBC, we knocked down the expression of Snail and Suv39H1 in MDA-MB231 and MDA-MB157 cells. We found that knockdown of Snail or Suv39H1 expression reduced the level of H3K9me3 and increased H3K9 acetylation at the E-cadherin promoter (Fig. 6A; quantitative real-time PCR results are presented on the bottom panel). We also immunoprecipitated the Snail complex and performed Suv39H1 enzymatic assays. The Snail complex contained H3K9me3 methyltransferase activity in vitro and this enzymatic activity could be suppressed by a specific Suv39H1 inhibitor Chaetocin (45) (Fig. 6B). These results further support the findings that Snail interacts with Suv39H1 and is required to recruit Suv39H1 to the E-cadherin promoter for H3K9me3. Conversely, when Snail and Suv39H1 were co-expressed with the E-cadherin promoter luciferase reporter in MCF7 and HeLa cells, we found that expression of either one of them suppressed E-cadherin promoter luciferase activity; expression of both of these molecules further enhanced this suppression (Fig. 6C). These data indicate a cooperative role for Suv39H1 and Snail in the suppression of E-cadherin expression and induction of EMT.
Figure 6.
Suv39H1 is recruited to the E-cadherin promoter for epigenetic silencing of E-cadherin expression. (A) Snail, Suv39H1 or non-target control (NTC) siRNA was expressed in MDA-MB157 and MDA-MB231 cells, and H3K9me3 and H3K9 acetylation at the E-cadherin promoter was analyzed by the ChIP assay (top panel). Quantitative real-time PCR was also performed to analyze ChIP samples above and results are presented in the bottom panels (mean ± SD from three separate experiments). *, # and § P < 0.01 for MDA-MB231 or MDA-MB157 cells transfected with NTC compared with cells with knockdown of Snail, Suv39H1 or both expression. (B) An in vitro Suv39H1 methylation assay was carried out as described in the Materials and Methods section. Statistical analysis of the methyltransferase activity, from three independent experiments, is shown in the bar graph. *P < 0.05 for treatment with or without Chaetocin in immunoprecipitation sample using Snail antibody. #P < 0.01 for treatment with or without Chaetocin in immunoprecipitation sample using Suv39H1 antibody. (C) Snail, Suv39H1 or both were co-expressed with the E-cadherin promoter luciferase construct in MCF7 and HeLa cells. After 48 h, luciferase activities were normalized and determined (mean ± SD in three separate experiments). *, # and § P < 0.01 for MCF7 or Hela cells transfected with vector compared with corresponding cells with Snail, Suv39H1 or both expression.
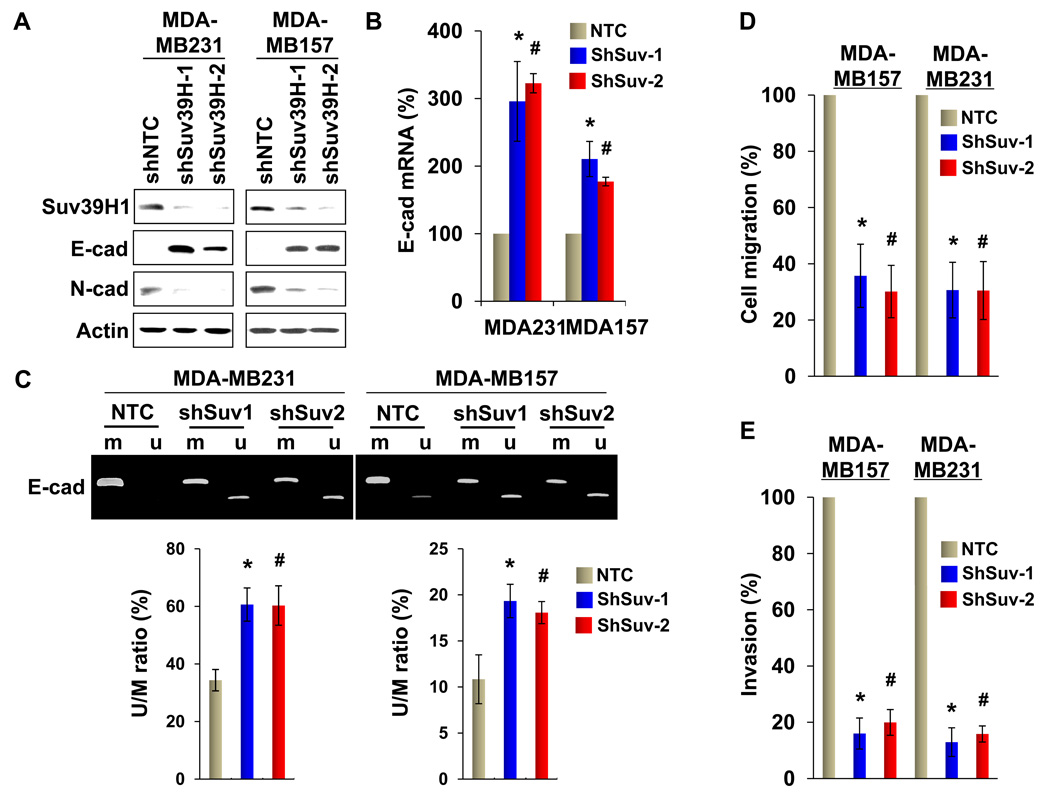
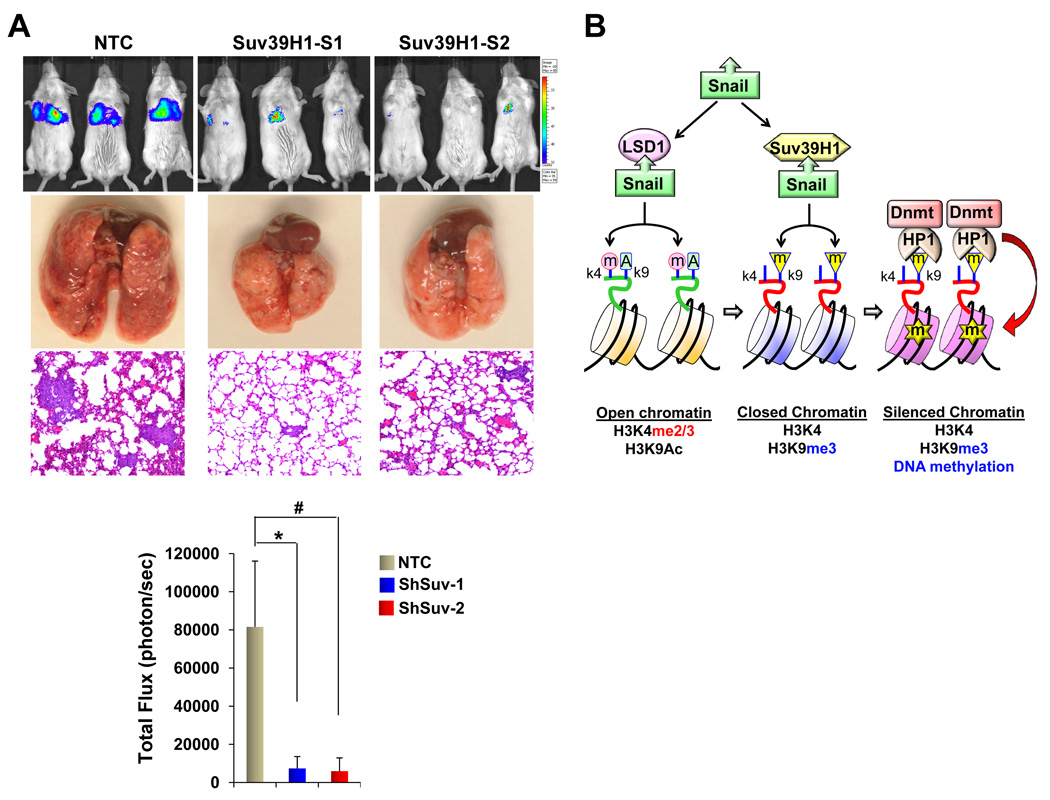
We also established stable transfectants with knockdown of Suv39H1 expression in MDA-MB231 and MDA-MB157 cells. We achieved about 80–90% knockdown efficiency of endogenous Suv39H1 using two independent shRNAs (Fig. 7A). Knockdown of Suv39H1 expression in both clones caused a significant restoration of E-cadherin expression and a dramatic downregulation of N-cadherin expression in MDA-MB231 and MDA-MB157 cells (Fig. 7A). In line with this result, knockdown of Suv39H1 expression upregulated E-cadherin mRNA as measured by quantitative real-time PCR in both cell lines (Fig. 7B). Consistent with the upregulation of E-cadherin mRNA and protein, de-methylated E-cadherin promoter was significantly increased, whereas DNA methylation on the E-cadherin promoter was decreased in cells with knockdown of Suv39H1 expression (Fig. 7C). Although we did not observe notable changes in cell growth or proliferation in vitro as measured by cell count and the MTT assay, respectively (data not shown), knockdown of Suv39H1 expression greatly inhibited the migratory ability and invasiveness (Fig. 7D & 7E) of MDA-MB231 and MDA-MB157 cells. We also extended our findings in a xenograft metastasis model in which MDA-MB231 cells were used to generate pulmonary metastases. We found that knockdown of Suv39H1 expression significantly suppressed lung metastases (Fig. 8A). Together, these results support our finding that Suv39H1 is a critical factor controlling the silencing of E-cadherin at EMT and BLBC.
Figure 7.
Knockdown of Suv39H1 derepresses E-cadherin expression and inhibits cell migration and invasion. (A) MDA-MB231 and MDA-MB157 cells stably expressing control vector or Suv39H1 shRNA were examined for the expression of Suv39H1, E-cadherin and N-cadherin by Western blotting. Actin served as a loading control. (B) Endogenous mRNA of E-cadherin from the cells above was examined by real-time PCR. (C) E-cadherin promoter methylation from cells with knockdown of Suv39H1 expression was examined by MSP. Samples from MSP analyses were also analyzed by quantitative real-time PCR and the ratio of un-methylated vs methylated DNA was plotted (mean ± SD from three separate experiments) (bottom panel). (D) The migratory ability of MDA-MB231 and MDA-MB157 cells and their corresponding stable transfectants with knockdown of Suv39H1 expression was analyzed by wound healing assay. After 48 h, a scratch (“wound”) was induced in a cell layer and cell culture was continued. Wound closures were photographed at 0 and 24 hr. Statistical analysis for the cell migration is shown on the bar graph (mean ± SD from three independent experiments). (E) MDA-MB231 and MDA-MB157 cells and their corresponding stable transfectants with knockdown of Suv39H1 expression was analyzed with a modified Boyden Chamber invasion assay as described in the Materials and Methods. The percentage of invasive cells is shown in the bar graph (mean ± SD in three separate experiments). For panels (C) to (E), * and # P < 0.01 for MDA-MB231 or MDA-MB157 cells transfected with control vector compared with corresponding two independent clones with knockdown of Suv39H1 expression.
Figure 8.
Knockdown of Suv39H1 inhibits metastasis of MDA-MB231 cells. (A) MDA-MB231 cells with stably expressing control vector or Suv39H1 shRNA were injected into the tail vein of ICR-SCID mice. After 9 wks, the development of lung metastases was recorded using luciferase bioluminescence imaging and quantified by measuring photon flux. Values are the mean of six animals ± SEM. Three representative mice from each group are shown. Mice were also sacrificed; lung metastatic nodules were examined macroscopically or detected in paraffin-embedded sections stained with hematoxylin and eosin. For lung metastases, * and # P < 0.01 for MDA-MB231 cells transfected with control vector compared with corresponding two independent stable clones with knockdown of Suv39H1 expression. (B) A proposed model to illustrate the interaction of Snail with LSD1, Suv39H1 and Dnmts leading to E-cadherin promoter methylation and EMT induction. The SNAG domain of Snail assembles a histone H3-like structure to interact with and recruit LSD1 and Suv39H1 to the E-cadherin promoter for sequential H3K4 demethylation and H3K9 methylation, respectively. Suv39H1-mediated H3K9me3 provides a docking site for the HP1-Dnmt1 complex, which results in DNA methylation at the E-cadherin promoter.
DISCUSSION
In this study, we show that Snail interacts with Suv39H1 and their interaction is critical for H3K9me3 and DNA methylation on the E-cadherin promoter in BLBC. Our study has provided several new insights into the chromatin modification in EMT and breast cancer metastasis. First, our study has identified an important mechanism underlying the sequential epigenetic modification on the E-cadherin promoter and reinforces the notion that EMT is an epigenetically regulated event. Chromatin is broadly divided in two defined states: euchromatin and heterochromatin. Euchromatin generally contains H3K4me2/3 while heterochromatin is enriched with H3K9me2/3 marks. Interestingly, H3K9me2/3-enriched histones are devoid of H3K4me2/3 and histones depleted of H3K4me2/3 have elevated H3K9me2/3, which is because H3K9 methyltransferases (including G9A/GLP and Suv39H1) fail to bind and methylate H3K4me2/3 substrates (18, 36). In our previous study, we found that Snail interacted with LSD1 and recruited LSD1 to the E-cadherin promoter to specifically remove methylation from H3K4me2, which is known to be an initial and critical step in gene repression in drosophila (31). Thus, demethylation of H3K4 by LSD1 provides a condition that favors Snail-mediated association of Suv39H1 on the E-cadherin promoter and facilitates the corresponding H3K9me3. Our study suggested that the sequential interaction of Snail with Suv39H1 is the second step leading to the reinforcement of gene repression and the consequent DNA methylation on the E-cadherin promoter (Fig. 8B). The individual interactions of Snail with LSD1 and Suv39H1 are both through its SNAG domain, which is required for recruiting these two molecules to the E-cadherin promoter. Forneris et al showed that the histone H3 tail adopted different configurations to interact with various individual chromatin modifying enzymes (46), highlighting a remarkable structural plasticity of the H3 tail in chromatin modification. We showed that the sequence of the SNAG domain is similar to that of the histone H3 tail and it can interact with LSD1 and Suv39H1, respectively. The mechanism that governs the differential binding of the SNAG domain with LSD1 and Suv39H1, separately, to achieve a synergistic modification event on the E-cadherin promoter remains an interesting area to explore in future.
Second, our study indicates that the interaction of Snail with Suv39H1 plays a critical role in E-cadherin promoter DNA methylation. DNA is globally hypomethylated in repeat-rich intergenic regions in cancer cells, but a subset of tumor suppressor genes, such as p16, RB1, GSTP1 and E-cadherin, gain aberrantly hypermethylation at the CpG islands of their promoters. For example, the E-cadherin promoter is commonly methylated in BLBC cells (Fig. 5). The existence of epigenetic interactions between histone modifications and DNA methylation leading to transcriptional silencing is well established; however, the precise sequence of events is not clear. H3K9 methylation plays a critical role leading to promoter DNA methylation in gene repression and heterochromatin formation. The lysine ε-amine group of H3K9 can be mono-, di-, or trimethylated, and each degree of modification contributes an additional hierarchy in methyllysine-mediated signaling (47). In mammals, heterochromatic regions are highly trimethylated on H3K9, whereas silent euchromatin regions are enriched for mono- and dimethylated H3K9 (47). SETDB1, which is essential for early embryonic development (48), performs mono- or dimethylation of H3K9, whereas G9a and GLP are mainly responsible for dimethylation of H3K9 in euchromatin (49, 50). Suv39H1 is responsible for the trimethylation of H3K9, which contributes mainly to the establishment of silenced heterochromatin (51, 52). At the enzymatic level, Suv39H1 prefers mono- or dimethylated H3K9 as a primary substrate (51, 52), which suggests a cooperation with an H3K9 mono- or dimethylase. Consistent with this, Suv39H1, G9a, GLP, and SETDB1 form a big complex to regulate H3K9 methylation both in euchromatin and heterochromatin, and the protein stability of one individual H3K9 methyltransferase affects the stability of the other two methyltransferases in cells, suggesting that the integrity of the H3K9 methyltransferases is interdependent (53). All of these H3K9 methyltransferases contain a conserved SET domain and sequence alignment of the SET domain of these HMTs indicates that the enzymatic domains of Suv39H1 and G9a/GLP are highly homologous (54).
Although H3K9me2 (mediated by G9a or GLP) and H3K9me3 (mediated by Suv39H1) commonly link to de novo gene silencing by promoting promoter DNA methylation, their mechanisms of action are different. G9a interacts with Dnmt directly and recruits it to target gene promoters for DNA methylation, whereas Suv39H1 creates a H3K9me3 binding and docking site for the adaptor molecule HP1, which in turn recruits DNMT and HDAC to catalyze DNA methylation and histone de-acetylation, respectively (55). In our study, we found that increased H3K9me3 is accompanied with the de-acetylation of H3K9 and increased DNA methylation on the E-cadherin promoter. Knockdown of Suv39H1 reduces H3K9me3 and increases H3K9 acetylation and thus results in the de-methylation and activation of E-cadherin expression. This result suggests that the recruitment of Suv39H1 by Snail is critical for E-cadherin promoter DNA methylation (Fig. 8B).
Third, our study suggests that Suv39H1 is a potential therapeutic target for treating and preventing breast cancer metastasis. The availability of multiple genetic approaches has offered an unprecedented opportunity for systematic, genome-wide discovery of genetic mutations involved in tumorigenesis. In addition to the identification of the well-studied oncogenes and tumor suppressor genes, such as Ras, PI3K/Akt, PTEN, and mTOR, mutations in chromatin modifying enzymes/regulators that lead to altered gene expression and tumorigenesis have recently been identified. For example, mutations have been found in MEN1, DAXX and ATRX in pancreatic neuroendocrine tumors (56). In addition, nearly half of all clear cell ovarian tumors contain mutations in the ARID1A gene (57, 58), which encodes the BAF250 subunit of the human SWI-SNFα chromatin-remodeling complex. Furthermore, overexpression of EZH2, an enzyme responsible for H3K27me3 has been found in prostate, breast and various tumor types (59). In this study, we found that the level of Suv39H1 is elevated in BLBC. Snail interacts with Suv39H1 and recruits it to the E-cadherin promoter for gene silencing. Thus, blocking the interaction and function of the Snail-Suv39H1 complex may represent a new approach for treating and preventing metastatic breast cancer.
In summary, we demonstrated that the SNAG domain of Snail and the SET domain of Suv39H1 were responsible for their mutual interactions. We also elucidated that Suv39H1 was recruited by Snail to the E-cadherin promoter for transcriptional repression. Knockdown of Suv39H1 expression restored E-cadherin expression by blocking Suv39H1-mediated chromatin modification and inhibited cell migration, invasion and metastasis of BLBC. Our study not only reveals a critical mechanism underlying the epigenetic regulation of EMT, but also offers a novel treatment strategy against the metastatic breast cancer.
MATERIALS AND METHODS
Plasmids, siRNA, and Antibodies
Suv39H1 shRNA expression plasmids were purchased from MISSION shRNA at Sigma-Aldrich (St Louis, MO). Smartpool siRNA against human Suv39H1 (siRNA-1) and Snail (siRNA-1) were from Dharmacon (Chicago, IL). Human Suv39H1was amplified from a HeLa cDNA library and subcloned into pcDNA3-Flag. Deletion mutants of Snail and Suv39H1 were constructed as described previously (30, 60). The expression plasmid for human Snail was described previously (15, 30). All sequences were verified by automated DNA sequencing.
Antibodies HA, Flag, and actin were purchased from Sigma-Aldrich (St. Louis, MO). Antibodies for E-cadherin and β-actin were from BD Transduction (San Jose, CA). N-cadherin and Suv39H1 antibodies were from Upstate (Charlottesville, VA) and Abcam Laboratories (Carlsbad, CA), respectively.
Cell Culture
MCF10A, HMLE and SUM1315 cells were grown in special medium following manufacturer’s instruction. All other cancer cell lines were grown in Dulbecco's modified Eagle's medium (DMEM)/F12 supplemented with 10% fetal bovine serum (FBS) except breast cancer cell lines T47D and ZR75 were grown in RPMI1640 plus 10% FBS. For establishing stable transfectants with knockdown of Suv39H1 expression, the breast cancer MDA-MB157 and MDA-MB231 cell lines were transfected with Suv39H1 shRNA; stable clones were selected with puromycin (1 µg/ml) for 4 wks.
Immunostaining, Immunoprecipitation, and Immunoblotting
Experiments were performed as described previously (61). For immunofluorescent staining, cells were grown on chamber slides, fixed with 4% paraformaldehyde, and incubated with primary antibodies. Secondary antibodies used were Texas red-conjugated goat anti-mouse, FITC-conjugated goat anti-rabbit, or Alexa Fluor 350 goat anti-rabbit (Molecular Probe, Carlsbad, CA). Immunoprecipitation and immunoblotting were performed as described previously (15).
Immunohistochemistry (IHC)
IHC staining was performed as we described previously (62). The immunoreactivity of these antibodies was scored according to the staining intensity (0 to 3+), and fraction of positive staining (0, <25%, 25–50% and >50%). The mean fraction of positive tumor cells was determined in at least nine areas at ×10 or ×20 magnifications. For staining intensity, a score of 0 was defined as undetectable staining in <10% of tumor cells, a score of 1+ was defined as faint staining in >10% of tumor cells, a score of 2+ was defined as weak to moderate staining in >10% of tumor cells, and a score of 3+ was defined as intense staining in >10% of tumor cells. For the percentage of positively stained tumor cells: 0, no staining; 1+, <25% cells stained; 2+, 20–49% cells stained; and 3+, >50% cells stained. For analysis, Snail and Suv39H1 expressions were further classified as low (intensity score 1+ and percentage score 1+) or high (both scores at 2+ and 3+). The slides were read independently by two investigators without prior knowledge. If differences between observers occurred, both investigators used a multiheaded microscope to reexamine the slides.
DNA Methylation Analysis
Genomic DNA (~0.75 µg) was treated with sodium bisulfite using the EpiTect system (Qiagen) by following the manufacturer’s protocol. Bisulfite-converted DNAs (~50 ng) were used as templates for PCR amplification of the CpG islands in the CDH1 promoter. Methylation-specific PCR (MSP) was performed on bisulfate-modified DNA as described by Herman et al (63).
Quantitative Real-Time PCR
Total RNA was isolated using the RNeasy Mini kit (Qiagen) according to the manufacturer’s instructions. Specific quantitative real-time PCR experiments were performed using SYBR Green Power Master Mix following manufacturer’s protocol (Applied Biosystems).
Invasion Assay
Invasion assays were performed in Boyden chambers with coated Matrigel as instructed by the manufacturer (BD Biosciences, San Jose, CA). The invasive cancer cells were stained with DAPI and visualized with a fluorescence microscope. All experiments were performed at least twice in triplicate. Statistical analysis was performed using the Student's t-test; a p value of <0.05 was considered significant.
Luciferase Reporter Assay
Cells were grown to 50% confluence in 6-well dishes and then co-transfected with reporter gene constructs and, to control for transfection efficiency, pTK-RL (Promega) using Fugene 6 (Roche, Indianapolis, IN). Cell extracts were prepared 40 h after transfection, and luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI). All experiments were performed three times in triplicate.
Chromatin Immunoprecipitation (ChIP)
ChIP assays were performed as described previously (30). Briefly, cells were cross-linked with disuccimidyl glutarate (Pierce, Rockford, IL) and formaldehyde at room temperature. Cells were subjected to lysis with L1 buffer (50 mM Tris, 2 mM EDTA, 0.1% IGEPAL, 10% glycerol, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride (PMSF) and protease inhibitor mixture, pH 8.0) on ice. After centrifugation, the nuclear pellet was resuspended in ChIP lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris and protease inhibitor mixture, pH 8.0). The cell lysate was subjected to sonication and then incubated with 4 µg of antibody overnight, followed by incubation with a 50% slurry of protein A-agarose/salmon sperm DNA (Upstate Biotechnology, Lake placid, NY) for 3 h at 4°C. Bound DNA-protein complexes were eluted and cross-links were reversed after a series of washes. Purified DNA was resuspended in TE buffer (10 mM Tris-HCl and 1 mM EDTA, pH 8.0) for PCR. The primers for the E-cadherin promoter were: 5′-ACTCCAGGCTAGAGGGTCACC-3′ and 5′-CCGCAAGCTCACAGGTGCTTTGCAGTTCC-3′.
Suv39H1 Methyltransferase Assay
HEK293T cells were co-transfected with constructs encoding Suv39H1 and Snail. After immunoprecipitation of Snail or Suv39H1, the complexes were analyzed for H3K9 methyltransferase activity using the H3K9 Methyltransferase Assay Kit (Epigentek, Brooklyn, NY, USA) following the manufacturer’s protocols. The enzymatic activities of Suv39H1 were detected with a microplate reader at 450 nm and fluorescent plate reader at 450 nm, respectively.
Bio-informatic analysis
Publicly available microarray with patients’ clinical data from two studies, Chin et al (http://www.ebi.ac.uk/arrayexpress/experiments/E-TABM-158) and Mullins et al (GSE6130) (33, 34), were utilized for the analysis. For each data set, expression values of Suv39H1 and Snail were standardized to zero mean and unit variance. Pearson’s correlation coefficient was calculated to quantify the correlation between expression patterns of the two genes. Mann-Whitney U test was used to study whether Suv39H1 expression is associated with triple negative status. The same test was also used for the data set of Mullins et al to study the association between Suv39H1 expression and breast tumor intrinsic subtype, which was assigned by the PAM50 classifier.
Protein model structure
A three-dimensional (3D) model of the Suv39H1-Snail complex was built by comparative protein structure modeling using the program MODELLER 9v3 (64). The input consisted of the template structure and the alignment of the target sequence with this structure. The output was a 3D model of the target including all non-hydrogen atoms. This model was derived by minimizing violations of distance and dihedral angle restraints extracted from the template structure while maintaining favorable interactions. The template structure was the homolog (53% sequence identity) GLP-Histone H3 peptide complex structure (Protein Data Bank, 2RFI). The figure was prepared with PyMol (65).
Experimental Lung Metastasis Model
Female ICR-SCID mice (6–8 wks old) were purchased from Taconic (Germantown, NY) and maintained and treated under specific pathogen-free conditions. Mice were injected with MDA-MB-231-luc-D3H1 (1×106 cells/mouse) cells and their corresponding stable clones with knockdown of Suv39H1 expression via tail vein (6 mice/group). Mice were imaged from dorsal and ventral views once per week. All mice (6/6) injected with MDA-MB231 cells developed the expected lung metastatic lesions with luciferase signals beginning to appear at week 5. Visible lung metastatic nodules were examined macroscopically or detected in paraffin-embedded sections stained with hematoxylin and eosin. Data were analyzed using the Student's t-test; a p value <0.05 was considered significant.
Statistical Analysis
The experiments were repeated at least two times. Results are expressed as mean ± SD or SEM as indicated. An independent Student’s t-test was performed to analyze the luciferase assay; a two-tailed Student’s t-test was used to compare the intergroup. p < 0.05 was considered statistically significant.
ACKNOWLEDGMENTS
We thank Dr. Nathan L. Vanderford for critical reading and editing of this manuscript. This work was supported by grants from NIH (RO1CA125454), Susan G Komen Foundation (KG081310), and Mary Kay Ash Foundation (to B.P. Zhou).
Abbreviations
- BLBC
Basal-like Breast Cancer
- ChIP
Chromatin Immunoprecipitation
- EMT
Epithelial-mesenchymal Transition
- LSD1
Lysine-specific Demethylase 1
- MET
Mesenchymal-epithelial Transition
- Suv39H1
Suppressor of Variegation 3–9 Homolog 1
Footnotes
CONFLICT OF INTEREST
The authors declare that there is no conflict any interest.
REFERENCES
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, et al. Cancer statistics, 2008. CA Cancer J Clin. 2008 Mar-Apr;58(2):71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Nieto MA. The snail superfamily of zinc-finger transcription factors. Nat Rev Mol Cell Biol. 2002 Mar;3(3):155–166. doi: 10.1038/nrm757. [DOI] [PubMed] [Google Scholar]
- 3.Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer. 2007 Jun;7(6):415–428. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- 4.Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol. 2006 Feb;7(2):131–142. doi: 10.1038/nrm1835. [DOI] [PubMed] [Google Scholar]
- 5.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009 Jun;119(6):1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009 Nov 25;139(5):871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 7.Wu Y, Zhou BP. New insights of epithelial-mesenchymal transition in cancer metastasis. Acta Biochim Biophys Sin (Shanghai) 2008 Jul;40(7):643–650. doi: 10.1111/j.1745-7270.2008.00443.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li X, Lewis MT, Huang J, Gutierrez C, Osborne CK, Wu MF, et al. Intrinsic resistance of tumorigenic breast cancer cells to chemotherapy. J Natl Cancer Inst. 2008 May 7;100(9):672–679. doi: 10.1093/jnci/djn123. [DOI] [PubMed] [Google Scholar]
- 9.Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008 May 16;133(4):704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moody SE, Perez D, Pan TC, Sarkisian CJ, Portocarrero CP, Sterner CJ, et al. The transcriptional repressor Snail promotes mammary tumor recurrence. Cancer Cell. 2005 Sep;8(3):197–209. doi: 10.1016/j.ccr.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 11.Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009 Apr;9(4):265–273. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- 12.Yang J, Weinberg RA. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell. 2008 Jun;14(6):818–829. doi: 10.1016/j.devcel.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 13.Julien S, Puig I, Caretti E, Bonaventure J, Nelles L, van Roy F, et al. Activation of NF-kappaB by Akt upregulates Snail expression and induces epithelium mesenchyme transition. Oncogene. 2007 Nov 22;26(53):7445–7456. doi: 10.1038/sj.onc.1210546. [DOI] [PubMed] [Google Scholar]
- 14.Lopez-Novoa JM, Nieto MA. Inflammation and EMT: an alliance towards organ fibrosis and cancer progression. EMBO Mol Med. 2009 Sep;1(6–7):303–314. doi: 10.1002/emmm.200900043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu Y, Deng J, Rychahou PG, Qiu S, Evers BM, Zhou BP. Stabilization of snail by NF-kappaB is required for inflammation-induced cell migration and invasion. Cancer Cell. 2009 May 5;15(5):416–428. doi: 10.1016/j.ccr.2009.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007 May 24;447(7143):433–440. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- 17.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007 Feb 23;128(4):683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng X, Blumenthal RM. Coordinated chromatin control: structural and functional linkage of DNA and histone methylation. Biochemistry. 2010 Apr 13;49(14):2999–3008. doi: 10.1021/bi100213t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eissenberg JC, Shilatifard A. Histone H3 lysine 4 (H3K4) methylation in development and differentiation. Dev Biol. 2010 Mar 15;339(2):240–249. doi: 10.1016/j.ydbio.2009.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laurent L, Wong E, Li G, Huynh T, Tsirigos A, Ong CT, et al. Dynamic changes in the human methylome during differentiation. Genome Res. 2010 Mar;20(3):320–331. doi: 10.1101/gr.101907.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell. 2007 Jan 12;25(1):15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 22.Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet. 2009 May;10(5):295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 23.McCabe MT, Brandes JC, Vertino PM. Cancer DNA methylation: molecular mechanisms and clinical implications. Clin Cancer Res. 2009 Jun 15;15(12):3927–3937. doi: 10.1158/1078-0432.CCR-08-2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bergamaschi A, Hjortland GO, Triulzi T, Sorlie T, Johnsen H, Ree AH, et al. Molecular profiling and characterization of luminal-like and basal-like in vivo breast cancer xenograft models. Mol Oncol. 2009 Dec;3(5–6):469–482. doi: 10.1016/j.molonc.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blick T, Widodo E, Hugo H, Waltham M, Lenburg ME, Neve RM, et al. Epithelial mesenchymal transition traits in human breast cancer cell lines. Clin Exp Metastasis. 2008;25(6):629–642. doi: 10.1007/s10585-008-9170-6. [DOI] [PubMed] [Google Scholar]
- 26.Hennessy BT, Gonzalez-Angulo AM, Stemke-Hale K, Gilcrease MZ, Krishnamurthy S, Lee JS, et al. Characterization of a naturally occurring breast cancer subset enriched in epithelial-to-mesenchymal transition and stem cell characteristics. Cancer Res. 2009 May 15;69(10):4116–4124. doi: 10.1158/0008-5472.CAN-08-3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sarrio D, Rodriguez-Pinilla SM, Hardisson D, Cano A, Moreno-Bueno G, Palacios J. Epithelial-mesenchymal transition in breast cancer relates to the basal-like phenotype. Cancer Res. 2008 Feb 15;68(4):989–997. doi: 10.1158/0008-5472.CAN-07-2017. [DOI] [PubMed] [Google Scholar]
- 28.Storci G, Sansone P, Trere D, Tavolari S, Taffurelli M, Ceccarelli C, et al. The basal-like breast carcinoma phenotype is regulated by SLUG gene expression. J Pathol. 2008 Jan;214(1):25–37. doi: 10.1002/path.2254. [DOI] [PubMed] [Google Scholar]
- 29.Wang Y, Zhou BP. Epithelial-mesenchymal transition in breast cancer progression and metastasis. Chin J Cancer. 2011 Sep;30(9):603–611. doi: 10.5732/cjc.011.10226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin Y, Wu Y, Li J, Dong C, Ye X, Chi YI, et al. The SNAG domain of Snail1 functions as a molecular hook for recruiting lysine-specific demethylase 1. EMBO J. 2010 Jun 2;29(11):1803–1816. doi: 10.1038/emboj.2010.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rudolph T, Yonezawa M, Lein S, Heidrich K, Kubicek S, Schafer C, et al. Heterochromatin formation in Drosophila is initiated through active removal of H3K4 methylation by the LSD1 homolog SU(VAR)3-3. Mol Cell. 2007 Apr 13;26(1):103–115. doi: 10.1016/j.molcel.2007.02.025. [DOI] [PubMed] [Google Scholar]
- 32.Barrallo-Gimeno A, Nieto MA. Evolutionary history of the Snail/Scratch superfamily. Trends Genet. 2009 Jun;25(6):248–252. doi: 10.1016/j.tig.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 33.Chin K, DeVries S, Fridlyand J, Spellman PT, Roydasgupta R, Kuo WL, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006 Dec;10(6):529–541. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 34.Mullins M, Perreard L, Quackenbush JF, Gauthier N, Bayer S, Ellis M, et al. Agreement in breast cancer classification between microarray and quantitative reverse transcription PCR from fresh-frozen and formalin-fixed, paraffin-embedded tissues. Clin Chem. 2007 Jul;53(7):1273–1279. doi: 10.1373/clinchem.2006.083725. [DOI] [PubMed] [Google Scholar]
- 35.Dillon SC, Zhang X, Trievel RC, Cheng X. The SET-domain protein superfamily: protein lysine methyltransferases. Genome Biol. 2005;6(8):227. doi: 10.1186/gb-2005-6-8-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shinkai Y, Tachibana M. H3K9 methyltransferase G9a and the related molecule GLP. Genes Dev. 2011 Apr 15;25(8):781–788. doi: 10.1101/gad.2027411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu H, Min J, Lunin VV, Antoshenko T, Dombrovski L, Zeng H, et al. Structural biology of human H3K9 methyltransferases. PLoS One. 2010;5(1):e8570. doi: 10.1371/journal.pone.0008570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bindels S, Mestdagt M, Vandewalle C, Jacobs N, Volders L, Noel A, et al. Regulation of vimentin by SIP1 in human epithelial breast tumor cells. Oncogene. 2006 Aug 17;25(36):4975–4985. doi: 10.1038/sj.onc.1209511. [DOI] [PubMed] [Google Scholar]
- 39.Xie L, Law BK, Aakre ME, Edgerton M, Shyr Y, Bhowmick NA, et al. Transforming growth factor beta-regulated gene expression in a mouse mammary gland epithelial cell line. Breast Cancer Res. 2003;5(6):R187–R198. doi: 10.1186/bcr640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004 Jun 25;117(7):927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 41.Lombaerts M, van Wezel T, Philippo K, Dierssen JW, Zimmerman RM, Oosting J, et al. E-cadherin transcriptional downregulation by promoter methylation but not mutation is related to epithelial-to-mesenchymal transition in breast cancer cell lines. Br J Cancer. 2006 Mar 13;94(5):661–671. doi: 10.1038/sj.bjc.6602996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kreike B, van Kouwenhove M, Horlings H, Weigelt B, Peterse H, Bartelink H, et al. Gene expression profiling and histopathological characterization of triple-negative/basal-like breast carcinomas. Breast Cancer Res. 2007;9(5):R65. doi: 10.1186/bcr1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006 Dec;10(6):515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010 Sep 2;12(5):R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Greiner D, Bonaldi T, Eskeland R, Roemer E, Imhof A. Identification of a specific inhibitor of the histone methyltransferase SU(VAR)3-9. Nat Chem Biol. 2005 Aug;1(3):143–145. doi: 10.1038/nchembio721. [DOI] [PubMed] [Google Scholar]
- 46.Forneris F, Binda C, Battaglioli E, Mattevi A. LSD1: oxidative chemistry for multifaceted functions in chromatin regulation. Trends Biochem Sci. 2008 Apr;33(4):181–189. doi: 10.1016/j.tibs.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 47.Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol. 2005 Nov;6(11):838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- 48.Dodge JE, Kang YK, Beppu H, Lei H, Li E. Histone H3-K9 methyltransferase ESET is essential for early development. Mol Cell Biol. 2004 Mar;24(6):2478–2486. doi: 10.1128/MCB.24.6.2478-2486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tachibana M, Sugimoto K, Nozaki M, Ueda J, Ohta T, Ohki M, et al. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 2002 Jul 15;16(14):1779–1791. doi: 10.1101/gad.989402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tachibana M, Ueda J, Fukuda M, Takeda N, Ohta T, Iwanari H, et al. Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes Dev. 2005 Apr 1;19(7):815–826. doi: 10.1101/gad.1284005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peters AH, Kubicek S, Mechtler K, O'Sullivan RJ, Derijck AA, Perez-Burgos L, et al. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell. 2003 Dec;12(6):1577–1589. doi: 10.1016/s1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
- 52.Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000 Aug 10;406(6796):593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 53.Fritsch L, Robin P, Mathieu JR, Souidi M, Hinaux H, Rougeulle C, et al. A subset of the histone H3 lysine 9 methyltransferases Suv39h1, G9a, GLP, and SETDB1 participate in a multimeric complex. Mol Cell. 2010 Jan 15;37(1):46–56. doi: 10.1016/j.molcel.2009.12.017. [DOI] [PubMed] [Google Scholar]
- 54.Kouzarides T. Chromatin modifications and their function. Cell. 2007 Feb 23;128(4):693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 55.Fuks F. DNA methylation and histone modifications: teaming up to silence genes. Curr Opin Genet Dev. 2005 Oct;15(5):490–495. doi: 10.1016/j.gde.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 56.Jiao Y, Shi C, Edil BH, de Wilde RF, Klimstra DS, Maitra A, et al. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science. 2011 Mar 4;331(6021):1199–1203. doi: 10.1126/science.1200609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jones S, Wang TL, Shih Ie M, Mao TL, Nakayama K, Roden R, et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science. 2010 Oct 8;330(6001):228–231. doi: 10.1126/science.1196333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wiegand KC, Shah SP, Al-Agha OM, Zhao Y, Tse K, Zeng T, et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. N Engl J Med. 2010 Oct 14;363(16):1532–1543. doi: 10.1056/NEJMoa1008433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011 Jan 20;469(7330):343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu Y, Evers BM, Zhou BP. Small C-terminal domain phosphatase enhances snail activity through dephosphorylation. J Biol Chem. 2009 Jan 2;284(1):640–648. doi: 10.1074/jbc.M806916200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhou BP, Deng J, Xia W, Xu J, Li YM, Gunduz M, et al. Dual regulation of Snail by GSK-3beta-mediated phosphorylation in control of epithelial-mesenchymal transition. Nat Cell Biol. 2004 Oct;6(10):931–940. doi: 10.1038/ncb1173. [DOI] [PubMed] [Google Scholar]
- 62.Xia W, Chen JS, Zhou X, Sun PR, Lee DF, Liao Y, et al. Phosphorylation/cytoplasmic localization of p21Cip1/WAF1 is associated with HER2/neu overexpression and provides a novel combination predictor for poor prognosis in breast cancer patients. Clin Cancer Res. 2004 Jun 1;10(11):3815–3824. doi: 10.1158/1078-0432.CCR-03-0527. [DOI] [PubMed] [Google Scholar]
- 63.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A. 1996 Sep 3;93(18):9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY, et al. Comparative protein structure modeling using Modeller. Curr Protoc Bioinformatics. 2006 Oct;Chapter 5(Unit 5):6. doi: 10.1002/0471250953.bi0506s15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.DeLano WL. Unraveling hot spots in binding interfaces: progress and challenges. Curr Opin Struct Biol. 2002 Feb;12(1):14–20. doi: 10.1016/s0959-440x(02)00283-x. [DOI] [PubMed] [Google Scholar]