Abstract
Human brain cells rely on a specific subset of microRNAs (miRNAs or miRs) to shape their gene expression patterns, and this is mediated through microRNA effects on messenger RNA (mRNA) speciation and complexity. In recent studies (a) in short post-mortem interval Alzheimer’ disease (AD) brain tissues versus age-matched controls, and (b) in pro-inflammatory cytokine- and Aβ42 peptide-stressed human neuronal-glial (HNG) cells in primary culture, we have identified several brain-abundant miRNA species found to be significantly up-regulated, including miR-125b and miR-146a. Both of these nuclear factor kappa B (NF-κB)-activated, 22 nucleotide small non-coding RNAs (sncRNAs) target the mRNA of the key, innate-immune- and inflammation-related regulatory protein, complement factor-H (CFH; chr 1q32), resulting in significant decreases in CFH expression (p< 0.01, ANOVA). Our results further indicate that HNG cells respond to IL-1β+Aβ42-peptide-induced stress by significant NF-κB-modulated up-regulation of miRNA-125b- and miRNA-146a. The complex interactive signaling of NF-κB, miR-125b, miR-146a, and perhaps other miRNAs, further illustrate interplay between inducible transcription factors and multiple pro-inflammatory sncRNAs that regulate CFH expression. The novel concept of miRNA actions involving mRNA target convergence and divergence are proposed and discussed. The combinatorial use of NF-κB inhibitors with anti-miRNAs (AMs; antagomirs) may have potential against CFH-driven pathogenic signaling in neurodegenerative disease, and may redirect our therapeutic perspectives to novel treatment strategies that have not yet been considered.
Keywords: 15-lipoxygenase (15-LOX), Alzheimer’s disease, Cell-cycle-dependent kinase N2A (CDKN2A), Complement factor H (CFH), Interleukin-1beta associated kinase-1 (IRAK-1), IRAK-2, microRNA (miRNA), miR-125b, miR-146a, miR-155, Neurodegeneration, NF-κB signaling, Synapsin-2 (SYN-2), Synaptogenesis
Introduction
Human neurogenetic regulatory mechanisms are highly interactive and exceedingly complex and vary considerably in different brain cell types and across anatomical regions [1, 2]. A major post-transcriptional regulator of brain gene expression patterns has been recently uncovered in the discovery of microRNA (miRNA or miR), and the mechanism of miR’s critical action on shaping the human brain transcriptome [3-5]. Indeed these small non-coding RNAs (sncRNAs) have taken prominent positions in our understanding of the control of both homeostatic and pathological gene expression in human brain health and disease [6-8]. As several human neurodegenerative conditions such as Alzheimer’s disease (AD) are intimately associated with alterations in innate immunity and wide-spectrum increases in pro-inflammatory signaling, we chose to focus our current research on a key, innate-immune- and inflammation-related regulatory protein, complement factor-H (CFH; chr 1q32) in areas of the human brain targeted by AD pathology, namely, the temporal lobe association neocortex, Brodmann area A22. Interestingly, altered CFH signaling is not only implicated in the AD process but also appears to be involved in age-related macular degeneration (AMD), a common and progressive degeneration of the aging human retina [9]. The information in this brief review will summarize our current understanding of the regulation of CFH by multiple miRNAs in AD and related human neurodegenerative disorders.
Global Gene Expression Patterns in AD
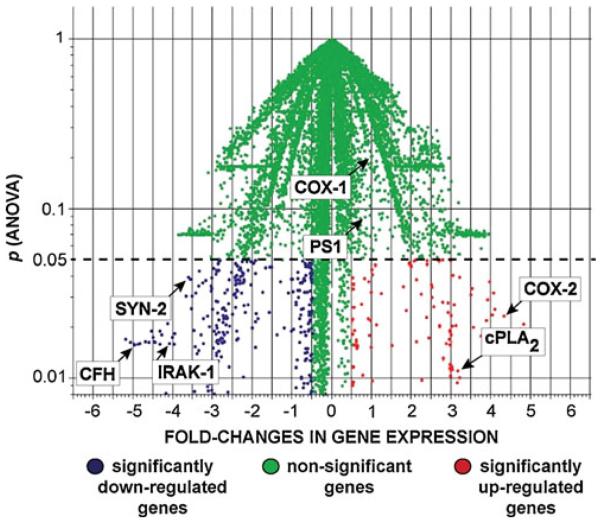
When one distills the extensive, archived work in several major studies on global genome-wide gene expression patterns in AD, at least three independent investigations show that in AD affected brain regions, compared with healthy age-matched controls, about one third of all expressed genes are significantly up-regulated and two thirds are down-regulated [1, 10-12]. Compared with the genes whose expression are not significantly altered in AD, such as presenilin-1 (PS1) and cyclooxygenase-1 (COX-1), many of the up-regulated genes are involved in pro-apoptotic and pro-inflammatory signaling (Fig. 1) [4-8, 10-12]. Consistently up-regulated genes include the stress-induced and NF-kB-regulated cyclooxygenase-2 (COX-2) and cytoplasmic phospholipase A2 (cPLA2), the latter representing the first committed step in the arachidonic acid cycle [13-15] (Fig. 1). At the other end of this altered gene expression spectrum are the significantly down-regulated genes in AD that include synapsin-2 (SYN-2; an essential synaptic glycoprotein), 15-lipoxygenase (15-LOX; an enzyme that converts the essential omega-3 fatty acid docosahexaenoic acid into neuroprotectin D1, or NPD1), the interleukin 1-beta receptor associated kinase (IRAK-1; an essential cytokine signaling adaptor-receptor) and complement factor H (CFH). These down-regulated genes are involved, respectively, in synaptogenesis, neurotrophic support, cytokine-mediated inflammatory signaling, and the innate and immune response, the latter in part mediated by the Toll-like receptor (TLR) signaling system which has been recently shown to be selectively targeted by the AD process [7, 16-24].
Fig. 1.
Significantly altered gene expression in Alzheimer’s disease (AD). Palm tree plot utilizing Genespring algorithms (Silicon Genetics Corporation, Redwood City, CA), indicates fold-changes in gene expression versus statistical significance (p, ANOVA) of significantly up- and down-regulated genes in AD versus age-matched controls [4-8]. Non-significant genes (green) indicate genes whose expression in AD versus age-matched controls does not change, such as cycloxygenase-1 (COX-1) and presenilin 1 (PS1). “Outlier” genes showing the most significant down-regulation and up-regulation are sequestered, respectively, to the lowest left- and right-most quadrants of this plot [22, 23]. Significantly up-regulated genes include those encoding the inducible cyclooxygenase-2 (COX-2) and cytosolic phospholipase A2 (cPLA2;red) [11-13]; the most significantly down-regulated genes include complement factor H (CFH) [6, 18, 20, 55], synapsin-2 (SYN-2) [16, 17], and interleukin-1β associated kinase-1 (IRAK-1; blue) [7, 59]. Other significantly down-regulated genes include the cell-cycle kinase regulator (CDKN2A) [54] and 15-lipoxygenase (15-LOX) [14, 23], omitted here for clarity. N=72 age-matched control and AD superior temporal neocortical samples (Brodmann area A22); the control and AD groups each consisted of 14 male and 22 female samples; further details on the pathology of these samples have been recently published [6, 7]
MicroRNAs
MicroRNAs represent an evolutionarily conserved group of 20–23 nucleotide, single-stranded, small non-coding RNA (sncRNA) molecules, and represent a recently discovered family of labile, heterogeneous, regulatory sncRNAs [2, 3, 25, 26]. Their primary mode of action is the recognition, via base pair complementarity, with the 3′ un-translated regions (3′-UTRs) of specific messenger RNA (mRNA) targets. Depending on 3′-UTR sequence complementarity within an RNA-induced silencing complex, productive miR-mRNA interaction results in either reduction or inhibition in the translational efficiency of the target mRNA [3, 25, 26]. Ribosome profiling has indicated that mammalian miRs predominantly act to decrease the abundance of their target mRNA levels, and in doing so quench specific mRNA transcription to down-regulate expression of that gene [3-8, 26]. While the potential contribution of sncRNA to brain genetic function has been known for at least 20 years [27], more recently there been an explosion of effort into molecular and genetic research involving the neurobiological and pathological functions of these miRs and sncRNAs in central nervous system (CNS) development, injury, aging, health and disease [28-38].
Pathogenically up-regulated miRs can be considered a novel epigenetic mechanism that down-regulates their target mRNAs and hence their expression. Up-regulated miRs in neurodegenerative disorders such as AD may help explain the large number (roughly two thirds) of all brain gene messenger RNAs (mRNAs) observed to be progressively down-regulated in anatomical regions sensitive to the AD process [1, 8-11]. Interestingly, oligonucleotide and bioin-formatics sequence analysis indicates that a 22 nucleotide single stranded sncRNA, which is the average size of a typical brain miR and composed of four different ribonucleotides, could have over 1013 possible sequence combinations, so the fact that there typically only about 103 different miRs in any cell suggests a very high developmental and evolutionary selection pressure to utilize only very specific miR oligonucleotide sequences that will yield biologically useful miR-mRNA interactions. Further, miRs are highly developmental stage, tissue, and cell specific, even in adjacent cell types, and in human brain cells high abundance miRs number considerably less than 102 individual miR species [5, 31-35, unpublished observations]. The small size of miRs and recent identification of miR-binding proteins that prolong naked miR half-life may provide a novel means of epigenetic “genetic signal storage” or “genetic memory” [4-8, 33, 34]. Further, the recent discovery of miR-containing micro-vesicles further suggests that miRs may provide a novel means for paracrine and related forms of inter-cellular, intertissue and perhaps even systemic communication [25-29, 39]. As for other nuclear transcribed genes, the expression of a cell’s miR repertoire is regulated by multiple transcription factors, are transcribed as pre-miRs, and are not only under the transcriptional control of DNA binding proteins, transcription factors and RNA polymerase II (RNAPII) and RNAPIII enzymes but further by miR-modifying enzymes in the nucleus and cytoplasm that include DGCR8, Exportin 5, Drosha, Dicer, Argonaute, and others [2-8, 28-38].
In essence, brain miRs regulate the post-transcriptional stability or translational efficiency of target mRNAs to function as natural negative regulators of gene expression [2-8, 28-38]. Of the ~1,250 or so human miRs identified to date, only a specific subset of less than 100, including about 25–30 prominent species, are highly expressed in the human brain, and these appear to be extremely critical to the regulation of normal brain cell functions. While miRs are known to be dynamically regulated during neural development, differentiation, and aging [11-18], the roles of miRs in inflammatory neurodegenerative diseases such as AD are not very well understood. Recent studies show that of the total miR population expressed in the healthy aging brain, only a selective subset appear to be involved in the AD process, and that altered miR-mediated processing of mRNA populations contribute to atypical mRNA abundances, altered gene expression, pro-inflammatory signaling, altered synaptogenesis and amyloidogenesis, including secretase-mediated neurodegenerative aspects of AD pathology [31-35] (Fig. 2).
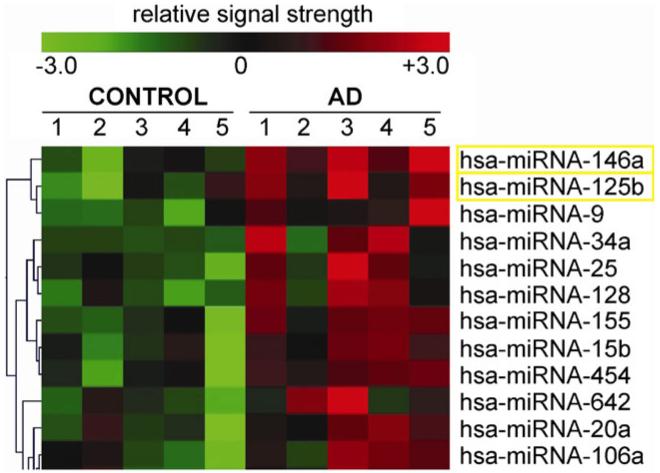
Fig. 2.
Significantly up-regulated miRs in AD versus age-matched controls. In this selective sampling, all control (N=5) and AD (N=5) neocortical samples were obtained from the superior temporal neocortex (Brodmann area A22) and had post-mortem intervals (PMI; death to brain freezing interval) of 2 h or less [4-8, 11, 12]. Controls were age-matched to moderate cases of AD; further details on the pathology of these samples have been recently published [6, 7]. There were no significant differences in age, PMI, or RNA yield or quality between the two brain groups [6-8]. Of the 12 different Homo sapien micro-RNAs (hsa-miR) shown, miR-146a and miR-125b exhibited the greatest up-regulation compared with age-matched controls (p<0.01, ANOVA); the seventh most up-regulated miR was found to be miR-155 (p<0.05, ANOVA) which can also target the CFH mRNA 3′-UTR (see Fig. 4) [22]. We cannot exclude the participation of other human brain-enriched miRs or sncRNAs which may additionally contribute to the neuropathological mechanisms which define the AD process
Because few miRs have been functionally linked to specific neurochemical and AD-relevant pathways, these ongoing studies have been undertaken to further understand the involvement of specific brain-enriched miRs in the molecular-genetic mechanisms that drive inflammatory signaling and AD-type change. As aformentioned, how certain mRNA populations, encoded by brain-essential genes are significantly reduced in light of global elevations of the pro-inflammatory transcription factor NF-κB, may be explained in part through the negative regulatory actions of specific miR species. This manuscript further reviews the evidence that up-regulation of at least two NF-κB-mediated brain-enriched miRs, miR-125b and miR-146a, down-regulates a specific mRNA target encoding CFH, a complement-immune repressor protein known to contribute to the regulation of the brain’s innate immune and inflammatory response (Table 1).
Table 1.
Selected brain-relevant RNA (mRNA) targets of miR-125b and miR-146a
| Human brain (miR) |
Human brain mRNA target |
mRNA encoded function |
Free energy of association (kcal/mol) |
Functional consequence | Reference |
|---|---|---|---|---|---|
| miR-125b | CDKN2A | Cell cycle regulator | −29.5 | Astrogliosis and glial cell proliferation |
[54] |
| miR-125b | SYN-2 | Synaptic phosphoprotein | −24.6 | Synaptic deficits and defective synaptogenesis |
[16, 17] |
| miR-125b | 15-LOX | 15-lipoxygenase; conversion of DHA to NPD1 |
−21.7 | Neurotrophic deficits | [14, 23, 57] |
| miR-146a | CFH | Complement activation repressor |
−24.5 | Altered innate immune response and neuroinflammation |
[6, 18-20, 50, 55] |
| miR-146a | IRAK-1 | regulator of IL-1β signaling |
−25.7 | Compensatory up- regulation of IRAK-2 and sustained NF-kB signaling |
[7, 58, 59] |
| miR-146a | TSPAN12 | transmembrane protein; regulator of βAPP cleavage |
−26.4 | Aberrant βAPP processing and amyloidogenesis |
[60, 61] |
Table indicates human brain miR species, high potential mRNA targets in the brain, the known neurobiological function of that mRNA, the free energy of association, the functional consequence of this interaction, and appropriate primary source references. The lower the number of the free energy of association the higher the probability of a productive miR-mRNA interaction; very stable free energies of association (≤−21 kcal/mol) between miRs and their mRNA 3’-UTR targets may be predictive for selective miR-mRNA targeting and down-regulation of gene expression [4, 6, 7, 22]. Other brain-abundant miR-mRNA pairs such as miR-155-CFH may be involved that additionally contribute to the AD process or other neurodegenerative brain diseases [22, 40]
Complement Factor H
CFH (encoded at chr 1q32; also known as AC3bINA, adrenomedullin binding protein-1, AM binding protein-1factor H, β1H globulin, C3b inactivator accelerator, H factor, and H factor-1) is an important member of the regulator of complement activation (RCA) group of proteins encoded within the RCA gene locus on chromosome 21 (chr 1q21–1q32). CFH is a highly soluble, hydrophilic 155 kDa regulatory glycoprotein normally secreted by the liver, and reaches blood plasma concentrations of 500–800 μg/ml in the systemic circulation. Hence, CFH is the second most abundant plasma protein after human serum albumin, and normally performs a systemic sentinel function against unscheduled or spontaneous immune system activation [6, 18, 19]. At the current time it is not clear if CFH is permeable to the blood-brain barrier [6, 18-20]. Such large glycosylated serum proteins as CFH are usually prevented from access to CNS compartments, and the brain may have an independent CFH supply secreted by neurons, astroglia and/or microglial cells [2, 12, 19]. CFH normally acts as a critical complement and innate immune system repressor, as a specific inhibitor of the C3 to C3b transition in the complement pathway [6-8, 18, 20]. Systemic CFH deficits are conducive to excessive and pathogenic complement pathway activation associated with increased complement activity on otherwise healthy host cells, autoimmunity, host tissue damage, and a sustained or chronic inflammatory response [7, 9, 10, 18-20]. CFH has been shown to be significantly down-regulated in AD brain, and predominantly by miR-125b and miR-146a with important ancillary contributions by miR-155, depending on brain cell type [2, 5-8, 22, 30, 32, 37, 38, 40]. Recently, for the first time it was demonstrated that at least two differently up-regulated miRs target the same region of the CFH mRNA 3′-UTR to coordinately down-regulate the expression of this important immune- and inflammatory regulatory factor [22, 40]. This genetic repressor function of two independent miRs targeting the same CFH mRNA 3′UTR in the human brain neocortex illustrates the novel action of miR convergence that is further discussed below.
Altered Immune and Inflammatory Signaling in Alzheimer’s Disease—Novel Therapeutic Targets
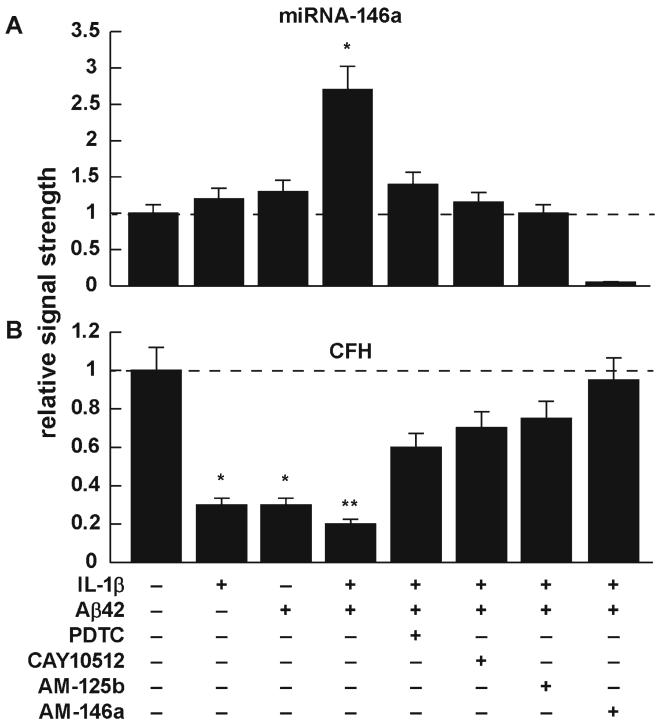
The pro-inflammatory transcription factor nuclear factor kappa B (NF-κB) regulates a battery of inducible miR (regulatory) and mRNA (protein encoding and structural) genes that are critical to innate and adaptive immunity, cell proliferation and inflammation during development and aging [6, 7, 22, 26, 36, 41-45]. These same miR and mRNA genes, essential for homeostatic regulation of brain and retinal cell function, are often mis-activated in neurological and retinal diseases such as AMD [41-43, 46]. One illustrative example of these regulatory actions is summarized in the data presented in bar graph format in Fig. 3. In these experiments, human neural (HNG) cells, a primary co-culture of human neurons and glia, were stressed with physiologically relevant amounts of IL-1β and Aβ42 peptides, and the effects on miR-146a abundance and CFH expression were examined in some detail [2, 6, 7]. The basal, homeostatic amounts of miR-146a and CFH are shown in the leftmost lanes (Fig. 3a, b). In Fig. 3a, miR-146a is induced 1.2-to 1.3-fold by IL-1β or Aβ42 peptides alone; in the presence of IL-1β and Aβ42 together, we see a synergistic effect with induction to 2.6-fold over control. The cooperative or synergistic effects of cytokines and beta-amyloid peptides have been previously demonstrated in a variety of human primary brain cell types [2, 6-8, 22, 23]. In the presence of the NF-kB inhibitor pyrollidine dithiocarbamate (PDTC; a chelator, antioxidant, and NFκB translocation inhibitor) or CAY10512 (a resveratrol (trans-3,5,4′-trihydroxystilbene) analog, polyphenolic antioxidant and inhibitor of NFκB-DNA binding), we see a significant quenching effect—this up-regulated miR-146a signal is also partially quenched in the presence of anti-sense to miR-125b (AM-125b); there is also a miR-125b recognition feature in the CFH mRNA 3′-UTR [22]. There is almost a completely quenched miR-146a signal in the presence of anti-sense to miR-146a (AM-146a). As might be expected, under equivalent treatment conditions CFH protein is down-regulated by IL-1β and/or Aβ42 peptides, thereby promoting an innate immune and inflammatory response, and this effect is reversed by the NF-kB inhibitors PDTC and CAY10512, or more effectively by AM-125b or AM-146a, thereby restoring CFH to basal homeostatic levels.
Fig. 3.
Effects of interleukin 1-beta (IL-1β) and amyloid beta peptide 42 (Aβ42) on human neuronal-glial (HNG) cell lines in primary culture; effects of NF-kB inhibitors and anti-miRs. inhibition of miR actions using (a) the NF-kB translocation inhibitors pyrollidine dithiocarbamate (PDTC) and the polyphenolic trans-stilbene resveratrol analog CAY10512 (10009536; Cayman Chemical, Ann Arbor, MI) and (b) anti-miRs (AMs, antagomirs) AM-125b and AM-146a. Details of the growth and cytokine IL-1β- and Aβ42-peptide-induced stress of HNG cells have been extensively described by our group [4-8, 12, 14, 20, 22, 23, 27, 32, 37-39]. PDTC and CAY10512 each showed significant reversal of both miR-125b- and miR-146a-induced effects. Qualitatively similar results have been observed in other stressed brain cell types and in Tg2576 and 5xFAD Tg-AD mice [2, 6-8, 22, 55]; N= 5; significance over controls; *p<0.05; **p<0.01; a dashed horizontal line at 1.0 indicates baseline (homeostatic) miR-146a (a) and CFH levels (b) in control HNG cells
What might the advantage be in using either an NF-κB inhibitor or a specific anti-sense miR (AM) approach therapeutically? NF-κB inhibitors might be expected to quench an entire family of NF-κB-regulated miR- and mRNA-encoding genes, thus acting as a broad-range control strategy [22, 43, 44]. Indeed, there is accumulating evidence that NF-κB-regulated genes encoding both miR and pro-inflammatory mRNAs are significantly up-regulated in AD and other human inflammatory diseases, when compared with non-NF-κB-regulated genes [2, 6, 7, 22, 47-53]. Lying at the other end of this “NF-κB-activation” spectrum is the use of specific anti-miR (antagomir; AM) strategies that may be useful to “fine tune” this integrated therapeutic approach. Indeed, several clinical strategies directed at the down-regulation of NF-κB responses in NF-κB-related diseases and based on these ideas have been recently described [43-45].
Convergence and Divergence of miR Actions
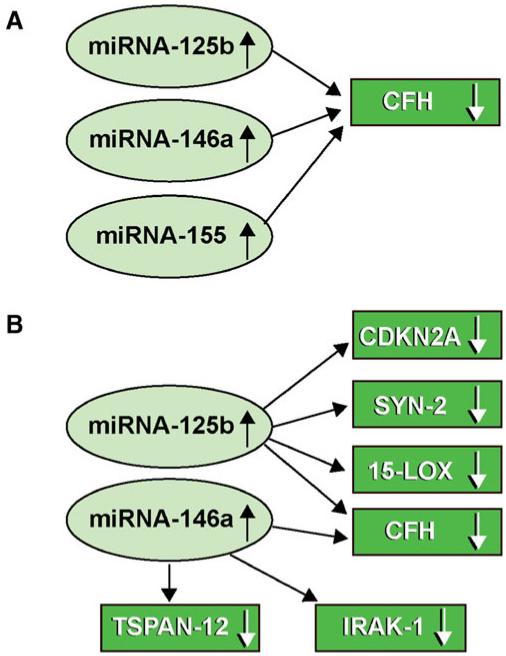
Recently, two major mechanistic ideas concerning brain miR function have been advanced. Firstly, Guo et al. demonstrated, using ribosome profiling, that mammalian miRs predominantly act to decrease target mRNA levels, and that changes in mRNA levels closely reflect the impact of miRs on gene expression, and indicate that destabilization of target mRNAs is the predominant reason for reduced gene expression and decreased protein output [3]. This has been specifically demonstrated in the human brain by observation of up-regulated miR-125b, miR-146a or miR-155 on the targeting and down-regulation of CFH mRNA and CFH expression in oxidation-stressed HNG cells, in AD brain and in other neurodegenerative conditions including human prion disease and Down’s syndrome [3-8, 22, 30-33, 40]. The second important recent idea is one of miR-action-convergence and miR action-divergence, the mechanisms of which are further illustrated in Fig. 4. Indeed, single miRs such as miR-125b and miR-146a (and miR-155 under special circumstances) appear to have the intrinsic capability to regulate multiple mRNA expression “nodes” within neurobiological and neuro-immunological pathways. Several of these mRNA targets are known to associate with neurodegenerative disease, and participate in complex positive or negative NF-κB-mediated feedback and signaling loops [6, 7, 53]. Interestingly, miR-146a and miR-155 recognition sites overlap within the CFH mRNA 3′-UTR, suggesting a developmental or evolutionary link between these two miRs in CFH expression regulation. It currently appears that either miR-146a or miR-155 may be used as alternate CFH mRNA abundance and CFH expression regulators in different human brain or retinal cells [2, 5, 22, 32, 37, 39, 40, 54, 55].
Fig. 4.
Integrated actions of up-regulated miRs and down-regulated mRNA abundance for AD-relevant gene expression. (a) miR action— convergence: multiple miRs down-regulate a single mRNA target; miR-155 has also been recently implicated in the regulation of CFH expression [22, 40]; miR-146a and miR-155 have overlapping targets in the CFH mRNA 3′-UTR [19]; see also Fig. 2; (b) miR action— divergence: single miRs have multiple mRNA targets. The integrated signaling actions of only a few miRs (miR-125b, miR-146a, and miR-155) can explain many of the pathogenic features of AD including glial cell proliferation (CDKN2A), synaptogenesis (SYN-2), neurotrophism (15-LOX), altered cytokine signaling (IRAK-1; with compensatory IRAK-2 up-regulation) and non-homeostatic activation of innate immunity and inflammatory signaling (CFH) [2, 6, 7, 14, 16, 17, 23, 26, 55]. Interestingly, the expression of miR-125b, miR-146a and miR-155 are under transcriptional regulatory control by the pro-inflammatory transcription factor NF-kB (p50/p65 subunit) [22, 41-46, 56]
It will be highly informative to further analyze the participation of NF-κB with other pro-inflammatory transcription factors, chromatin-mediated mechanisms, and other epigenetic influences on specific miR-mRNA deactivation pathways to further understand their surprisingly dynamic interactive roles, and their contribution to the neurogenetics of brain and retinal cell aging, and age-related neuropathologies such as AD and AMD. Lastly, the ideas of miR convergence and divergence further underscore the possibility of an important human brain miR regulatory network that through only a relatively small number of miRs could have large genetic impact and potential to affect many mRNAs and their expression in both health and degenerative disease.
Summary
Human neurodegenerative brain disorders such as AD appear to be associated in part with a disruption in an incompletely understood innate immune and chronic inflammatory response. Immune- and stress-induced transcription factors such as NF-κB have been shown to play determinant roles in the regulation of pathology-related miRs and their mRNA targets involved in the AD process [4-8, 40, 51-53, 56]. The current studies focused on up-regulated miRs, as down-regulated miRs may be, in part, a consequence of their relatively short observed half-life in vitro, and their uncontrolled and rapid degradation in vivo, especially in post-mortem human brain tissues [8, 22]. Moreover, it has been recently shown that the primary mode of up-regulated miR action is to down-regulate target mRNA levels, such as in the miR-146a-CFH mRNA 3′-UTR pairing [3-9]. It is remarkable that only as few as two or three significantly up-regulated brain miRs—miR-125b, miR-146a, and miR-155—may contribute to so many of the observed deficits in AD including increased glial cell proliferation, altered synaptogenesis, deficits in neurotrophism, altered cytokine signaling, and non-homeostatic activation of innate immunity and inflammatory signaling (CFH) (Fig. 4). At this time we cannot exclude the idea that other brain-relevant miRs or sncRNAs are involved in the mis-regulation of mRNAs and pathogenic gene expression regulation in the AD brain. For example, the miR-146a and miR-155 recognition sites in the CFH mRNA 3′-UTR overlap, and the chromosome21-encoded miR-155 appears to have specific effects on CFH down-regulation in Down’s syndrome, a congenital human neurological disorder that shows a remarkable AD-like neuropathology with age [40]. Indeed, expansion of our understanding of the human brain’s reliance on a selective subset of NF-κB-regulated miRs to shape the brain’s gene expression patterns may redirect our therapeutic perspectives to novel treatment strategies for AD that have not yet been considered.
Acknowledgments
These studies were presented in part at the 11th annual Alzheimer’s Association International Conference on Alzheimer’s disease (AAICAD 11) conference in Versailles, France, 16–20 July 2011; thanks are extended to the families, physicians, and researchers who contributed to the murine and human brain bank tissue and total RNA resources used in these studies and especially to Drs. R. I. Carp, C. Chen, S. Gettner, E. Head, W. Poon, T. Saing, and Jian Zhang at donor institutions. Thanks are also extended to S. Bhattacharjee, B.M. Jones, K.P Pfefferle, and D. Guillot for expert technical assistance. Some of the brain tissues used in this study was provided by the Institute for Memory Impairments and Neurological Disorders and the University of California at Irvine Alzheimer’s Disease Research Center (UCI-ADRC); funding for the UCI-ADRC was provided by NIH/NIA grant P50 AG16573. Research on the structure and function of NF-κB and miR expression in the Lukiw laboratory were supported through Translational Research Initiative (TRI) Grants from LSU Health Sciences Center New Orleans (WJL), an Alzheimer Association Investigator-Initiated Research Grant IIRG-09-131729 (WJL), and NIH NIA grants AG18031 and AG038834 (WJL). The content of this manuscript is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute on Aging or the National Institutes of Health.
References
- 1.Liang WS, Dunckley T, Beach TG, Grover A, Mastroeni D, Ramsey K, Caselli RJ, Kukull WA, McKeel D, Morris JC, Hulette CM, Schmechel D, Reiman EM, Rogers J, Stephan DA. Altered neuronal gene expression in brain regions differentially affected by Alzheimer’s disease: a reference data set. Physiol Genomics. 2008;33:240–256. doi: 10.1152/physiolgenomics.00242.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li YY, Cui JG, Dua P, Pogue AI, Bhattacharjee S, Lukiw WJ. Differential expression of miRNA-146a-regulated inflammatory genes in human primary neural, astroglial and microglial cells. Neurosci Lett. 2011;499:109–113. doi: 10.1016/j.neulet.2011.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lukiw WJ. Micro-RNA speciation in fetal, adult and Alzheimer’s disease hippocampus. Neuroreport. 2007;18:297–300. doi: 10.1097/WNR.0b013e3280148e8b. [DOI] [PubMed] [Google Scholar]
- 5.Lukiw WJ, Pogue AI. Induction of specific micro RNA (miRNA) species by ROS-generating metal sulfates in primary human brain cells. J Inorg Biochem. 2007;101:1265–1269. doi: 10.1016/j.jinorgbio.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lukiw WJ, Zhao Y, Cui JG. A NF-κB-sensitive miRNA-146a-mediated inflammatory circuit in AD and in stressed human brain cells. J Biol Chem. 2008;283:31315–31322. doi: 10.1074/jbc.M805371200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cui JG, Li YY, Zhao Y, Bhattacharjee S, Lukiw WJ. Differential regulation of interleukin-1 receptor-associated kinase-1 (IRAK-1) and IRAK-2 by miRNA-146a and NF-kB in stressed human astroglial cells and in Alzheimer’s disease. J Biol Chem. 2010;285:38951–38960. doi: 10.1074/jbc.M110.178848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sethi P, Lukiw WJ. Micro-RNA abundance and stability in human brain: specific alterations in Alzheimer’s disease temporal lobe neocortex. Neurosci Lett. 2009;459:100–104. doi: 10.1016/j.neulet.2009.04.052. [DOI] [PubMed] [Google Scholar]
- 9.Deangelis MM, Silveira AC, Carr EA, Kim IK. Genetics of age-related macular degeneration: current concepts, future directions. Semin Ophthalmol. 2011;26:77–93. doi: 10.3109/08820538.2011.577129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loring JF, Wen X, Lee JM, Seilhamer J, Somogyi R. A gene expression profile of Alzheimer’s disease. DNA Cell Biol. 2001;20:683–695. doi: 10.1089/10445490152717541. [DOI] [PubMed] [Google Scholar]
- 11.Colangelo V, Schurr J, Ball MJ, Pelaez RP, Lukiw WJ. Gene expression profiling of 12633 genes in Alzheimer hippocampal CA1: transcription and neurotrophic factor down-regulation and up-regulation of apoptotic and pro-inflammatory signaling. J Neurosci Res. 2002;70:462–473. doi: 10.1002/jnr.10351. [DOI] [PubMed] [Google Scholar]
- 12.Lukiw WJ. Gene expression profiling in fetal, aged, and Alzheimer hippocampus: a continuum of stress-related signaling. Neurochem Res. 2004;29:1287–1297. doi: 10.1023/b:nere.0000023615.89699.63. [DOI] [PubMed] [Google Scholar]
- 13.Lee JC, Simonyi A, Sun AY, Sun GY. Phospholipase A2 and neural membrane dynamics: implications for Alzheimer’s disease. J Neurochem. 2011;116:813–819. doi: 10.1111/j.1471-4159.2010.07033.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lukiw WJ, Bazan NG. Inflammatory, apoptotic, and survival gene signaling in Alzheimer’s disease. A review on the bioactivity of neuroprotectin D1 and apoptosis. Mol Neurobiol. 2010;42:10–16. doi: 10.1007/s12035-010-8126-4. [DOI] [PubMed] [Google Scholar]
- 15.Rapoport SI. In vivo approaches and rationale for quantifying kinetics and imaging brain lipid metabolic pathways. Prostaglandins Other Lipid Mediat. 2005;77:185–196. doi: 10.1016/j.prostaglandins.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 16.Valtorta F, Pozzi D, Benfenati F, Fornasiero EF. The synapsins: multitask modulators of neuronal development. Dev Biol. 2011;22:378–386. doi: 10.1016/j.semcdb.2011.07.008. [DOI] [PubMed] [Google Scholar]
- 17.Evergren E, Benfenati F, Shupliakov O. The synapsin cycle: a view from the synaptic endocytic zone. J Neurosci Res. 2007;85:2648–2656. doi: 10.1002/jnr.21176. [DOI] [PubMed] [Google Scholar]
- 18.Griffiths MR, Neal JW, Fontaine M, Das T, Gasque P. Complement factor H protects against experimental autoimmune encephalomyelitis. J Immunol. 2009;18:4368–4377. doi: 10.4049/jimmunol.0800205. [DOI] [PubMed] [Google Scholar]
- 19.Song F, Poljak A, Smythe GA, Sachdev P. Plasma biomarkers for mild cognitive impairment and Alzheimer’s disease. Brain Res Rev. 2009;61:69–80. doi: 10.1016/j.brainresrev.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 20.Alexander JJ, Quigg RJ. The simple design of complement factor H: Looks can be deceiving. Mol Immunol. 2007;44:123–132. doi: 10.1016/j.molimm.2006.07.287. [DOI] [PubMed] [Google Scholar]
- 21.Griffin WS. Inflammation and neurodegenerative diseases. Am J Clin Nutr. 2006;83:470–474. doi: 10.1093/ajcn/83.2.470S. [DOI] [PubMed] [Google Scholar]
- 22.Lukiw WJ. NF-κB-regulated micro RNAs (miRNAs) in primary human brain cells. Exp Neurol. 2011 doi: 10.1016/j.expneurol.2011.11.022. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lukiw WJ, Cui JG, Marcheselli VL, Bodker M, Botkjaer A, Gotlinger K, Serhan CN, Bazan NG. A role for docosahexaenoic cid-derived neuroprotectin D1 in neural cell survival and Alzheimer disease. J Clin Invest. 2005;115:2774–2783. doi: 10.1172/JCI25420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hanke ML, Kielian T. Toll-like receptors in health and disease in the brain: mechanisms and therapeutic potential. Clin Sci (Lond) 2011;121:367–387. doi: 10.1042/CS20110164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 26.Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS. Non-coding RNAs: regulators of disease. J Pathol. 2010;220:126–139. doi: 10.1002/path.2638. [DOI] [PubMed] [Google Scholar]
- 27.Lukiw WJ, Handley P, Wong L, McLachlan DRC. BC200 and other small RNAs RNA in normal human neocortex, non-Alzheimer dementia (NAD), and senile dementia of the Alzheimer type (AD) Neurochem Res. 1992;17:591–597. doi: 10.1007/BF00968788. [DOI] [PubMed] [Google Scholar]
- 28.Perron MP, Provost P. Protein components of the micro-RNA pathway and human diseases. Methods Mol Biol. 2009;487(1):369–385. doi: 10.1007/978-1-60327-547-7_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsitsiou E, Lindsay MA. microRNAs and the immune response. Curr Opin Pharmacol. 2009;9(4):514–520. doi: 10.1016/j.coph.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lukiw WJ, Cui JG, Yuan LY, Bhattacharjee PS, Corkern M, Clement C, et al. Acyclovir or Aβ42 peptides attenuate HSV-1-induced miRNA-146a levels in human primary brain cells. Neuroreport. 2010;21:922–927. doi: 10.1097/WNR.0b013e32833da51a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Madathil SK, Nelson PT, Saatman KE, Wilfred BR. Micro-RNAs in CNS injury: potential roles and therapeutic implications. Bioessays. 2011;33:21–26. doi: 10.1002/bies.201000069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pogue AI, Li YY, Cui JG, Zhao Y, Kruck TPA, Percy ME, et al. Characterization of an NF-kB-regulated miRNA-146a in metal-sulfate-stressed human brain cells. J Inorg Biochem. 2009;11:156–164. doi: 10.1016/j.jinorgbio.2009.05.012. [DOI] [PubMed] [Google Scholar]
- 33.Burmistrova OA, Goltsov AY, Abramova LI, Kaleda VG, Orlova VA, Rogaev EI. MicroRNA in schizophrenia: genetic and expression analysis of miR-130b (22q11) Biochemistry (Mosc) 2007;72:578–582. doi: 10.1134/s0006297907050161. [DOI] [PubMed] [Google Scholar]
- 34.Wang K, Zhang S, Weber J, Baxter D, Galas DJ. Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res. 2010;38:7248–7259. doi: 10.1093/nar/gkq601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, Mitchell PS, Bennett CF, Pogosova-Agadjanyan EL, Stirewalt DL, Tait JF, Tewari M. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci USA. 2011;108:5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-κB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lukiw WJ, Pogue AI. Induction of specific micro RNA (miRNA) species by ROS-generating metal sulfates in primary human brain cells. J Inorg Biochem. 2007;101:1265–1269. doi: 10.1016/j.jinorgbio.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hill JM, Zhao Y, Clement C, Neumann DM, Lukiw WJ. HSV-1 infection of human brain cells induces miRNA-146a & Alzheimer-type inflammatory signaling. Neuroreport. 2009;20:1500–1505. doi: 10.1097/WNR.0b013e3283329c05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao Y, Cui JG, Lukiw WJ. Natural secretory products of human neural and microvessel endothelial cells: implications in pathogenic “spreading” and Alzheimer’s disease. Mol Neurobiol. 2006;34:181–192. doi: 10.1385/MN:34:3:181. [DOI] [PubMed] [Google Scholar]
- 40.Li YY, Alexandrov PN, Pogue AI, Zhao Y, Bhattacharjee S, Lukiw WJ. miRNA-155 up-regulation and complement factor H (CFH) deficits in Down’s Syndrome. Neuroreport. 2012 doi: 10.1097/WNR.0b013e32834f4eb4. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma X, Becker Buscaglia LE, Barker JR, Li Y. MicroRNAs in NF-kB signaling. J Mol Cell Biol. 2011;3:159–166. doi: 10.1093/jmcb/mjr007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sarnico I, Lanzillotta A, Benarese M, Alghisi M, Baiguera C, Battistin L, Spano P, Pizzi M. NF-kB dimers in the regulation of neuronal survival. Int Rev Neurobiol. 2009;85:351–362. doi: 10.1016/S0074-7742(09)85024-1. [DOI] [PubMed] [Google Scholar]
- 43.Cai D. NF-kB-mediated metabolic inflammation in peripheral tissues versus central nervous system. Cell Cycle. 2009;8:2542–2548. doi: 10.4161/cc.8.16.9386. [DOI] [PubMed] [Google Scholar]
- 44.Ruland J. Return to homeostasis: down-regulation of NF-κB responses. Nat Immunol. 2011;12:709–714. doi: 10.1038/ni.2055. [DOI] [PubMed] [Google Scholar]
- 45.Suzuki J, Ogawa M, Muto S, Itai A, Isobe M, Hirata Y, Nagai R. Novel IkB kinase inhibitors for treatment of nuclear factor-kB-related diseases. Expert Opin Investig Drugs. 2011;20:395–405. doi: 10.1517/13543784.2011.559162. [DOI] [PubMed] [Google Scholar]
- 46.Meffert MK, Baltimore D. Physiological functions for brain NF-kB. Trends Neurosci. 2005;28:37–43. doi: 10.1016/j.tins.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 47.Lukiw WJ, LeBlanc HJ, Carver LA, McLachlan DR, Bazan NG. Run-on gene transcription in human neocortical nuclei. Inhibition by nanomolar aluminum and implications for neurodegenerative disease. J Mol Neurosci. 1998;11:67–78. doi: 10.1385/JMN:11:1:67. [DOI] [PubMed] [Google Scholar]
- 48.Lukiw WJ, Percy ME, Kruck TP. Nanomolar aluminum induces pro-inflammatory and pro-apoptotic gene expression in human brain cells in primary culture. J Inorg Biochem. 2005;99:1895–1898. doi: 10.1016/j.jinorgbio.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 49.Hill JM, Lukiw WJ, Gebhardt BM, Higaki S, Loutsch JM, Myles ME, Thompson HW, Kwon BS, Bazan NG, Kaufman HE. Gene expression analyzed by microarrays in HSV-1 latent mouse trigeminal ganglion following heat stress. Virus Genes. 2001;23:273–280. doi: 10.1023/a:1012517221937. [DOI] [PubMed] [Google Scholar]
- 50.Cui JG, Kuroda H, Chandrasekharan NV, Pelaez RP, Simmons DL, Bazan NG, Lukiw WJ. Cyclooxygenase-3 gene expression in Alzheimer hippocampus and in stressed human neural cells. Neurochem Res. 2004;29:1731–1737. doi: 10.1023/b:nere.0000035809.70905.8a. [DOI] [PubMed] [Google Scholar]
- 51.Coolen M, Bally-Cuif L. Micro RNAs in brain development and physiology. Curr Opin Neurobiol. 2009;19:461–470. doi: 10.1016/j.conb.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 52.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 53.Ma X, Becker Buscaglia LE, Barker JR, Li YY. Micro-RNAs in NF-kB signaling. J Mol Cell Biol. 2011;3:159–166. doi: 10.1093/jmcb/mjr007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pogue AI, Cui JG, Li YY, Zhao Y, Culicchia F, Lukiw WJ. Micro RNA-125b (miRNA-125b) function in astrogliosis and glial cell proliferation. Neurosci Lett. 2010;476:18–22. doi: 10.1016/j.neulet.2010.03.054. [DOI] [PubMed] [Google Scholar]
- 55.Alexandrov PN, Pogue A, Bhattacharjee S, Lukiw WJ. Retinal amyloid peptides and complement factor H (CFH) in transgenic models of Alzheimer’s disease. Neuroreport. 2011;22:623–627. doi: 10.1097/WNR.0b013e3283497334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lukiw WJ, Bazan NG. Strong nuclear factor-kB-DNA binding parallels cyclooxygenase-2 gene transcription in aging and in sporadic Alzheimer’s disease superior temporal lobe neocortex. J Neurosci Res. 1998;53:583–592. doi: 10.1002/(SICI)1097-4547(19980901)53:5<583::AID-JNR8>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 57.Palacios-Pelaez R, Lukiw WJ, Bazan NG. Omega-3 essential fatty acids modulate initiation and progression of neurodegenerative disease. Mol Neurobiol. 2010;41:367–374. doi: 10.1007/s12035-010-8139-z. [DOI] [PubMed] [Google Scholar]
- 58.Flannery S, Bowie AG. The interleukin-1 receptor-associated kinases: critical regulators of innate immune signalling. Biochem Pharmacol. 2010;80:1981–1991. doi: 10.1016/j.bcp.2010.06.020. [DOI] [PubMed] [Google Scholar]
- 59.Gan L, Li L. Regulations and roles of the interleukin-1 receptor associated kinases (IRAKs) in innate and adaptive immunity. Immunol Res. 2006;35:295–302. doi: 10.1385/IR:35:3:295. [DOI] [PubMed] [Google Scholar]
- 60.Xu D, Sharma C, Hemler ME. Tetraspanin12 regulates ADAM10-dependent cleavage of amyloid precursor protein. FASEB J. 2009;23:3674–3681. doi: 10.1096/fj.09-133462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yáñez-Mó M, Sánchez-Madrid F, Cabañas C. Membrane proteases and tetraspanins. Biochem Soc Trans. 2011;39:541–546. doi: 10.1042/BST0390541. [DOI] [PubMed] [Google Scholar]