Abstract
Peroxiredoxins are cysteine-dependent peroxidases that react with hydrogen peroxide, larger hydroperoxide substrates, and peroxynitrite. Protocols are provided to measure Prx activity with peroxide by 1) a coupled reaction with NADPH, thioredoxin reductase, and thioredoxin, 2) the direct monitoring of thioredoxin oxidation, 3) competition with horseradish peroxidase, and 4) peroxide consumption using the FOX assay.
Keywords: antioxidants, antioxidant enzymes, sulfenic acids, hydrogen peroxide, hydroperoxides, thiol peroxidase, Prx, PRDX, cysteine oxidation, redox catalysis, HRP, FOX
INTRODUCTION
Peroxiredoxin (Prx) enzymes (EC 1.11.1.15) are a family of cysteine-dependent peroxidases that react with hydrogen peroxide, aliphatic and aromatic hydroperoxide substrates, and peroxynitrite, (Hall, et al., 2010) as described in the preceding overview. These antioxidant enzymes are ubiquitous and highly expressed in many organisms, including plants, bacteria and animals where they can comprise up to 1% or more of cellular proteins (Winterbourn, 2008). It has been suggested that Prx proteins are responsible for the reduction of more than 90% of intracellular peroxide in humans (Cox, et al., 2010, Winterbourn, 2008). Prxs are critical components of the antioxidant defense systems of many bacteria (Parsonage, et al., 2008, Seaver and Imlay, 2001) and are important components in the plant chloroplast (Dietz, et al., 2006). In eukaryotes, Prxs have been implicated in the regulation of a variety of signaling processes including cell proliferation, differentiation, and cell death (Veal, et al., 2007). Because Prxs are induced by oxidative stress, highly expressed in various cancers, and linked to decreased sensitivity to radiation therapy (Zhang, et al., 2005), these proteins are considered possible prognostic or therapeutic targets (Kang, et al., 2005).
Interest in Prx enzymes has recently increased with the realization that these proteins react with a range of hydroperoxides with high catalytic rate constants on the order of ~107 M−1s−1 (Horta, et al., 2010, Manta, et al., 2009, Ogusucu, et al., 2007, Parsonage, et al., 2005, Peskin, et al., 2007). The importance of Prx proteins was slow to be recognized largely because early assays dramatically underestimated this rate for two reasons. First, in many cases, reduction of the Prx is rate limiting; only with the development of the assays described in this chapter (Basic Protocols II and III) was it possible to measure the true peroxide-dependent rate of Prxs (Ogusucu, et al., 2007, Parsonage, et al., 2005). Second, early assays often used hydrogen peroxide concentrations around 1 mM and many of the eukaryotic Prxs are inactivated under these conditions; the half-life of human Prx2 is just 20 seconds in the presence of 1 mM hydrogen peroxide and reductant (Rabilloud, et al., 2002, Yang, et al., 2002). These results highlight how important it is to carefully design and implement any kinetic assay with Prxs.
This chapter includes four protocols that have been used with some success to characterize one or more Prx. Basic Protocol I provides a general absorbance-based assay in which Prx activity is coupled to oxidation of NADPH via thioredoxin reductase (TrxR) and thioredoxin (Trx). Basic Protocol II is an adaptation of this assay, which allows for the measurement of a true peroxide-dependent rate and directly monitors the changes to Trx fluorescence as Trx becomes oxidized. Basic Protocol III measures the second order rate constant of Prx reacting with hydrogen peroxide by monitoring the ability of Prx to prevent peroxide from reacting with horseradish peroxidase. Basic protocol IV utilizes the ferrous oxidation-xylenol orange (FOX) assay which directly measures peroxide-dependent oxidation of Fe(II) to Fe(III).
All of the assays described here, with the exception of Alternate Protocol IVA, utilize purified protein. Physiologically, Prxs detoxify peroxides at the expense of either NADPH or NADH. In most cell types, hydrogen peroxide disappearance is also catalyzed by catalase and glutathione peroxidase making it difficult, if not impossible, to measure Prx activity with any specificity. Alternate Protocol IVA provides one example of an in vivo assay and avoids this difficulty by using the FOX assay to measure the disappearance of cumene hydroperoxide, which is a good substrate for most Prxs and not readily reduced by either catalase or glutathione peroxide. All of these assays, with the exception of Basic Protocol III, can be used with lipid hydroperoxides such as linoleic acid hydroperoxide (Baker, et al., 2001). Although the protocols presented can be adapted to utilize peroxynitrite, this substrate is treated in more detail elsewhere (Trujillo, et al., 2008).
STRATEGIC PLANNING
We are providing a starting set of assays and concentrations that have been used with success to measure Prx activity. However, because Prxs utilize a range of reductants and exhibit a wide range of rates, the most appropriate assay will need to be selected based on the characteristics of each individual protein (more information provided in critical parameters section), and assays will almost certainly have to be adapted to fit individual situations. It is also suggested that some time be spent prior to setting up the assays to identify some critical characteristics of the Prx. In particular, it will be helpful to identify the Prx subfamily, the genomic context of the Prx of interest, and the possible reductants present in the native environment; more details about these characteristics are provided in the Critical Parameters section.
BASIC PROTOCOL I: MEASUREMENT OF NADPH-DEPENDENT PEROXIREDOXIN ACTIVITY IN PRESENCE OF TRX, TRXR, AND PEROXIDE
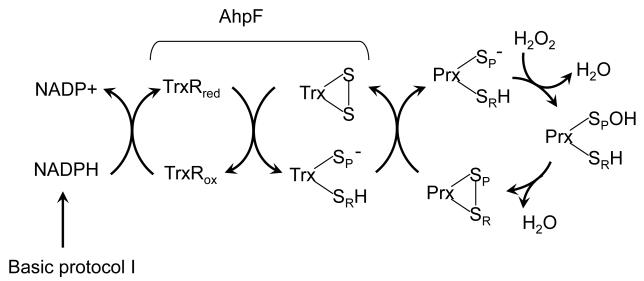
Although some Prxs are reduced physiologically by specialized disulfide reductases, Trx is the most common reductant. This protocol describes a general assay for Trx-linked Prx activity. Prx is assayed by coupling its activity to oxidation of NADPH via TrxR (Figure 1), as occurs physiologically. The reaction as described here is carried out in a conventional, thermostat-controlled spectrophotometer and can be used with a variety of peroxide substrates. For this assay, care must be taken to optimize the concentrations of the various components so that it is limited as little as possible by the reduction of Trx by TrxR and NADPH. The concentrations listed here have been used with success previously, but care must be taken to ensure that they are appropriate for each situation. Although it is preferable to utilize the Trx and TrxR system from the same organism as the Prx to be studied, we have found that E. coli Trx and TrxR often reduce Prxs from other bacterial species with similar kinetic parameters as their native Trx/TrxR system (Baker, et al., 2001, Parsonage, et al., 2010a).
Figure 1.
Determination of Prx activity in the presence of NADPH, Trx, and TrxR. The peroxidatic cysteine in Prx (-SP–) reacts with peroxide to form a cysteine sulfenic acid (Prx-SPOH) which, in 2-Cys Prxs, then forms a disulfide bond with the resolving cysteine (SRH). The Prx disulfide is reduced by Trx with electrons ultimately coming from NADPH through TrxR. As described in Basic Protocol 1, Prx turnover is monitored as a change in absorbance at 340 nm as NADPH becomes oxidized. For bacterial AhpC-like proteins, AhpF can replace both Trx and TrxR.
Materials
1 mL semimicro cuvettes
Reaction buffer (see recipe)
Purified E. coli Trx (500 μM diluted into Prx assay buffer
Purified E. coli TrxR (50 μM diluted into Prx assay buffer)
15 mM NADPH (dissolved in 10 mM Tris-SO4 pH 8.5)
10 mM Peroxide solution: hydrogen peroxide, cumene hydroperoxide or t-butyl hydroperoxide (see recipe)
Peroxiredoxin (50 μM diluted into Prx assay buffer)
Spectrophotometer with kinetic capabilities (required) and temperature regulation (preferred; i.e. with an attached waterbath).
Establish conditions for Prx-dependent assay
Turn on the spectrophotometer and set the waterbath to 25 °C; allow them to warm up and become stable (~ 30 min). Set the spectrophotometer to kinetics mode with a wavelength of 340 nm.
In a 1 ml quartz cuvette, mix Prx reaction buffer, 10 μl Trx, 10 μl TrxR, and various peroxide concentrations between 15 and 500 μM (100 μM peroxide is a good choice for initial assays). Add Prx assay buffer for a total of 980 μl. Place the cuvette in the spectrophotometer and allow for thermal equilibration (3-5 min). Final concentrations in the assay should be 5 μM Trx and 0.5 μM TrxR.
Add 10 μL of 15 mM NADPH. The final concentration is 150 μM and the A340 should be ~0.9. Mix the contents and begin recording the 340 nm absorbance. This value will provide the background rate of peroxide with Trx and TrxR. Monitor until a steady reading, or a linear decrease is established (~1 minute; make sure that no more than 10% of the NADPH is used up at this step).
- Add 10 μL of a 50 μM Prx solution to start the assay (final concentration of 0.5 μM). Monitor the change in absorbance at 340 nm for 1-2 min, ensuring that less than 10-20 % of the NADPH has been oxidized.In general, assays should be designed so that Trx levels are roughly 10-fold higher than Prx and TrxR concentrations to ensure that Prx peroxidase activity is rate limiting and not electron transfer to either Trx or Prx.
From the difference in rate before and after the addition of Prx, calculate the Prx-dependent rate of NADPH oxidation (ε340 = 6,220 M−1cm−1). Only measure the initial, linear portion of the curve.
Repeat steps 2-5 using 1 μM Prx. Test to ensure that the rate is approximately twice the rate with 0.5 μM Prx.
Repeat steps 2-5 using 0.25 μM Prx. Test to ensure that the rate is approximately half of the rate with 0.5 μM Prx.
If altering Prx concentrations (steps 6-7) does not change the activity proportionately, adjust the Trx and TrxR concentrations so that it does. If the assay is correctly set up, increasing the TrxR concentration should not increase the rate.
Measure Rates for Prx turnover with peroxide
- 9. Once the conditions have been established, repeat steps 2-5 using different peroxide concentrations between 15 and 500 μM. Fit the rates at each peroxide concentration to the Michaelis-Menten equation to find the apparent Km for peroxide under these conditions. Ideally, the final peroxide concentrations should range from one fifth to five-fold the Km for peroxide substrate. Determine the preliminary Km for peroxide and adjust the range of peroxide concentrations for all future measurements as appropriate.If peroxide concentrations below 15 μM are required, it may be useful to decrease the concentration of all assay components and measure the reaction for a longer period of time to ensure that ~ 10% of the NADPH is used during the course of the assay and the signal is large enough to be reliably measured. Alternatively, increased sensitivity may be obtained by measuring changes to NADPH fluorescence rather than absorbance. See (Baker, et al., 2001) for more details.
Measure Rates for Prx turnover with Trx
10. Repeat steps 2-5 using a different concentration of Trx. Ideally, the apparent Vmax should start to saturate as the Trx concentration increases. As the Trx concentration increases, the apparent Km for peroxide may also increase and it may require higher peroxide concentrations to saturate the reaction.
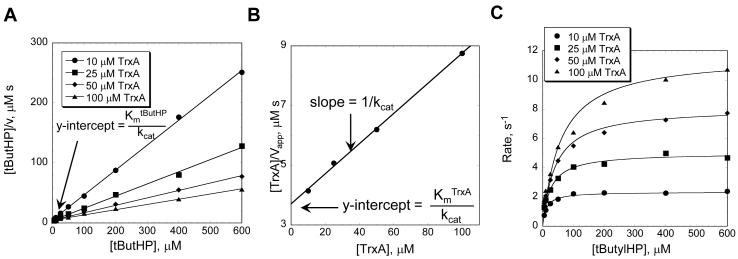
- 11. Plot the data using a linearized form of the Michaelis-Menten equation. We typically use a Hanes-Woolf plot which plots [peroxide] on the x-axis and [peroxide]/rate on the y-axis (Figure 2A) because this type of plot does not exaggerate the error at low substrate concentrations as is often observed with Lineweaver-Burk plots. A Hanes-Woolf plot should result in a series of straight lines, each one corresponding to a set of experiments carried out at one Trx concentration.The point of intersection of the lines indicates the appropriate kinetic mechanism to use for data analysis. When lines intersect on the y-axis, this suggests a substituted enzyme (ping-pong) mechanism, where the first substrate (i.e. peroxide) reacts and dissociates prior to binding of the second substrate (i.e. Trx), which reacts, dissociates, and returns the enzyme to its starting state. The Prx family members studied so far fit best to a ping-pong mechanism. When the lines of the Hanes-Woolf plot intersect to the left of the y-axis, this suggests the formation of a ternary complex, in which both substrates must bind prior to undergoing the chemical reaction. For more information about these different mechanisms see (Cornish-Bowden, 2004).
12. The reciprocal slope for each Trx concentration provides the apparent Vmax for each Trx concentration. Generate secondary Hanes–Woolf plots with [Trx] on the x-axis and [Trx]/Vmaxapp on the y-axis to obtain the true kcat and the true Km value for Trx (Figure 2B). As an alternative to steps 11-12, all of the data can be fit globally (Figure 2C) as described in Basic Protocol II, step 27. Dalziel analysis has also been used to obtain similar rate constants for Prx proteins; for details refer to: (Baker, et al., 2001, Jaeger, et al., 2004, Sayed and Williams, 2004).
Figure 2. Kinetic analysis to determine rate constants for Treponema pallidum AhpC with Escherichia coli Trx and t-butyl hydroperoxide (tButHP).
A The data for all substrate concentrations were plotted according to the Hanes-Woolf representation of the Michaelis-Menten equation to obtain the apparent Vmax for each Trx concentration. B The apparent Vmax values obtained for each Trx concentration were plotted against Trx concentration to obtain the true kcat and KmTrx values. The true kcat value obtained from plot B can then be used to calculate the KmtButHP using the y-intercept in plot A. C All of the data shown in plots A and B can be fit globally using the multi-function nonlinear regression capabilities of a computer program such as Sigmaplot (Systat Software Inc. San Jose, CA).
ALTERNATIVE PROTOCOL 1A: MEASURING PEROXIDASE ACTIVITY WITH AHPF AS REDUCTANT
Bacterial Prxs of the AhpC/Prx1 subfamily frequently have a dedicated flavoprotein reductant, AhpF; in these cases, the AhpF gene is found immediately downstream of the coding sequence for the AhpC it reduces. Functionally, AhpF is a fusion of a Trx-like N-terminal domain to a C-terminal TrxR-like domain (Poole, et al., 2000, Wood, et al., 2001). Unlike TrxR which prefers NADPH, AhpF typically utilizes NADH. AhpC activity with AhpF is assayed using a similar protocol to that described above except that 0.1 μM AhpF replaces both the Trx and TrxR and 150 μM NADH replaces NADPH in the assay. Starting conditions for the assay should utilize final concentrations of 0.1 μM AhpF, 0.5 μM AhpC and 100 μM peroxide in 50 mM potassium phosphate pH 7.0, 1 mM EDTA, 100 mM ammonium sulfate.
AhpF isolated from Salmonella typhimurium, Streptococcus mutans and Amphibacillus xylanus have significant NADH oxidase activity, as oxygen can readily react directly with the reduced FAD cofactor of these enzymes (Poole, 2005). It is important to measure this background rate prior to the addition of AhpC. The background rate should be subtracted from the measured rate obtained after Prx addition to calculate the final AhpC-dependent rate.
BASIC PROTOCOL II: MEASUREMENT OF PEROXIDE-DEPENDENT PEROXIREDOXIN ACTIVITY WITH EXCESS TRX (BY MONITORING TRX OXIDATION)
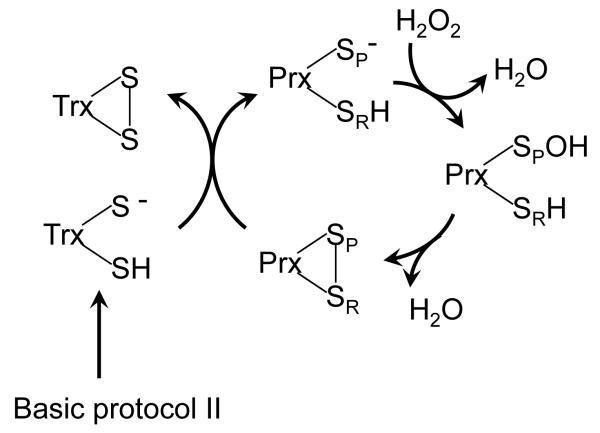
Although the assay described in Basic Protocol I is the most representative of the physiological activity of Prxs, it is frequently unable to distinguish the specificity for various peroxides because it is limited by the rate with which electrons can be passed from NADPH through TrxR and Trx to reduce the Prx (Parsonage, et al., 2010b, Poole and Ellis, 1996). The assay described in this section has been designed to overcome this limitation by using a high concentration of pre-reduced Trx (Figure 3). In many Trx proteins, at least one tryptophan (Trp) residue near the catalytic CXXC motif is appropriately oriented to act as a fluorescent reporter of the active site redox state (Trp31 in E. coli Trx) (Krause and Holmgren, 1991, Parsonage, et al., 2010b). Because of this, Trx oxidation can be monitored directly as a decrease in fluorescence.
Figure 3.
Determination of Prx activity by directly monitoring Trx oxidation. As described in Basic Protocol 2, Prx reacts with peroxide to form a sulfenic acid on the peroxidatic cysteine (SPOH) and, ultimately, a disulfide bond. The fluorescence of Trx decrease as it becomes oxidized and reduces the Prx disulfide.
Although it is theoretically possible to use this assay with a standard fluorimeter possessing kinetic capabilities, many Prxs have been shown to have a very low Km for peroxide (~1 μM in the case of S. typhimurium AhpC and hydrogen peroxide, (Parsonage, et al., 2005)). Due to the low concentrations of substrate used in these cases, it is very difficult if not impossible to observe the linear initial rate with the typical initial deadtime required for manual mixing. In order to overcome this problem, a stopped-flow spectrometer is typically used to follow the change in Trx fluorescence over the first 5 s after mixing with Prx and peroxide. This assay can be used with a variety of peroxide substrates.
Although many of the Trx proteins (including human and E. coli) are suitable for this assay, this is not always the case. For proteins such as AhpF and Grx, the conversion between disulfide and dithiol forms is spectrally silent or minimally detectable. In these cases, it is possible to engineer in a sensitive fluorescent reporter by substituting in a strategically-located Trp (Parsonage, et al., 2010b, Parsonage, et al., 2005).
Materials
Prx Reaction buffer (see recipe)
Pre-reduced, purified E. coli Trx (~5 mg/ml)
Purified peroxiredoxin (typically stored at ~10 mg/ml)
Syringe tip filters, 0.22 or 0.45 μm
Spectrophotometer capable of measuring UV absorbance
10 mM peroxide substrate: hydrogen peroxide, cumene hydroperoxide or t-butyl hydroperoxide (see recipe)
Stopped-flow spectrophotometer, capable of measuring fluorescence with excitation at 280 nm and emission monitored at >320 nm.
Reduce Trx
Pre-reduce ~10 mg of Trx using an approximately 20-fold molar excess of DTT and remove excess DTT using a PD-10 desalting column (see Support Protocol I).
Combine the peak fractions and filter them through a 0.22 μm syringe-tip filter. Determine the concentration of the reduced Trx, by measuring the 280 nm absorbance of a dilution (typically, 50 μl of Trx solution added to 600 μl of buffer). Calculate the Trx concentration using the ε280 (13,700 M−1 cm−1 for E. coli Trx) (Holmgren and Reichard, 1967).
Prepare a series of solutions containing Prx and reduced Trx
3. Prepare a ~25 μM solution of Prx protein in reaction buffer. If the extinction coefficient of the enzyme is known, use the diluted Prx from the cuvette to prepare the Prx plus Trx solutions. If the extinction coefficient is not known, perform a protein assay on the peroxiredoxin solution to determine the concentration and then prepare a suitable dilution in reaction buffer.
4. Prepare a series of solutions in Prx reaction buffer containing Prx and different Trx concentrations between 2.5 and 50 μM (for example, 2.5, 5, 10, 20, and 50 μM final concentration after mixing in the stopped-flow). A typical starting concentration for this solution would be 20.2 μM Trx and 0.2 μM Prx. The reduced Trx will reduce the Prx, leaving 20 μM reduced Trx and 0.2 μM reduced Prx as the starting concentrations. Stopped-flow spectrophotometers typically mix equal volumes of reactants, so this solution should be prepared at twice the final desired protein concentration. After mixing with peroxide substrate in the stopped-flow, the final concentration of protein will be 10 μM Trx and 0.1 μM Prx.
5. Filter the Trx/Prx mixtures through a 0.22 or 0.45 μm syringe-tip filter.
Prepare a series of peroxide dilutions
6. Make multiple peroxide solutions in a final volume of 15 ml Prx reaction buffer. All solutions should be prepared at twice the final desired peroxide concentration. A minimum of 3 ml is required for each set of reactions at one Trx concentration. Ideally, the final peroxide concentrations should range from one fifth to five-fold the Km for peroxide substrate. Since the Km for a given Prx is initially unknown, prepare concentrations from 1 to 500 μM (for example, 1, 2, 5, 10, 25, 50 100, 250 and 500 μM final concentrations). After examining the first set of results, determine the preliminary Km for peroxide and adjust the range of peroxide concentrations for all future measurements to appropriately cover the preliminary Km.
7. Filter all peroxide dilutions using a 0.22 or 0.45 μm syringe-tip filter.
Prepare stopped-flow spectrophotometer
You should be familiar with operation of the stopped-flow instrument before starting these experiments
8. Adjust the waterbath to give a temperature of 25 °C in the mixing chamber. Turn on the stopped-flow instrument and allow the lamp output, photomultiplier detector and temperature of the waterbath to stabilize for 1 hour.
9. Flush the instrument drive syringes, flow cell and tubing with reaction buffer. Set up the instrument to monitor fluorescence changes with excitation wavelength of 280 nm and with emission >320 nm.
Determine settings and fluorescence changes for each Trx concentration
10. Leave reaction buffer in one drive syringe and load one Prx/Trx solution in the other. Allow the solutions to equilibrate to the required temperature. This usually happens quickly (3-5 minutes) in most instruments since thermostated water surrounds the drive syringes and mixing chamber
11. Activate mixing between Prx/Trx and reaction buffer to obtain the starting fluorescence of reduced Trx in the mixing chamber.
12. Set the instrument sensitivity by adjusting the photomultiplier (PMT) voltage so that the fluorescent signal is 80 % of the maximum signal. This PMT voltage should be used for all reactions at the same Trx concentration. Monitor the signal for 10-20 s to observe any background rate. Usually this will be negligible as Trx is a stable protein. If the background rate is significant, it will have to be subtracted from all rates measured in the presence of peroxide.
13. Replace the reaction buffer in the drive syringe with a peroxide concentration that is greater than the Trx concentration; the peroxide concentration must be high enough to ensure that all the Trx is oxidized by the end of the reaction. A minimum of 3 ml volume should be used to load the drive syringe. Drive the reaction 2 times to ensure that the previous buffer has been fully flushed from the mixing chamber.
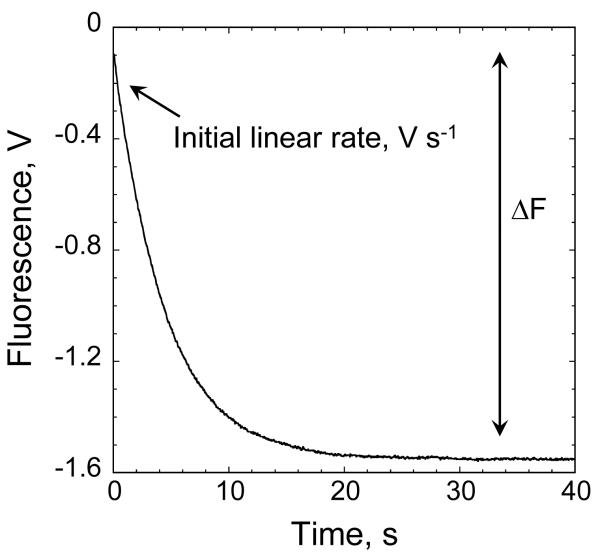
- 14. After allowing for thermal equilibration, set the timescale of the reaction to 20 s and start the instrument to mix enzymes and peroxide using a low sensitivity scale (the window showing the range of observed fluorescence changes should be zoomed out so that the whole reaction can be observed). Trx fluorescence will decrease as Trx becomes oxidized during the reaction of Prx with peroxide (Figure 4); when the reaction is complete, the fluorescence signal will remain constant. If the reaction is not complete by the end of 20 s, increase the observation time. Repeat the longest time course to confirm the magnitude of the total fluorescence change. Calculate the starting fluorescent signal by extrapolating the initial rate back to time zero. Record the value for the total fluorescence change (ΔF = starting fluorescence – final fluorescence).The total change in fluorescence during the reaction is important as this value is used to convert the change in fluorescence signal to a rate of substrate depletion (see step 25). This calibration run can be used for all further measurements at this Trx concentration and PMT voltage.
Figure 4. Thioredoxin fluorescence decreases during oxidation.
The total fluorescence change (ΔF) in the presence of Prx and excess peroxide is used to convert the rate of change in fluorescence in V s−1 into μM peroxide s−1 μM−1 Prx using the equation: Rate (μM peroxide s−1 μM−1 Prx) = rate (V s−1) · [Trx]/(ΔF x [Prx]), where [Trx] and [Prx] are the final concentrations of thioredoxin and Prx respectively.
Measure Rate of Prx reaction with peroxide
15. Adjust the measurement time to examine the initial 2-5 s of the reaction
16. Measure the fluorescence changes with the same peroxide concentration until you have at least three consistent traces. You should see an initial (albeit brief at low peroxide concentrations) linear phase for the decrease in fluorescence of the reaction mixture (Figure 4). The lowest peroxide concentrations may require more traces (up to 10-12) to obtain a trustworthy average. Use a more sensitive signal scale for these observations and zoom in on the initial signal, as only the first few seconds of the reaction need to be visible.
- 17. Discard the used peroxide solution from the drive syringe to minimize the risk of contamination by enzyme. Load the next concentration of peroxide into the drive syringe and allow 3-5 minutes for thermal equilibration.Do not choose the peroxide solutions in order from high concentration to low, but “randomize” the order. This will prevent the creation of a systematic error. If going directly from a high to a very low concentration, rinse the syringe with reaction buffer before adding the second peroxide solution to prevent carryover from the high peroxide solution.
- 18. While solutions are equilibrating, average the traces for the previous peroxide concentration. Use linear regression to calculate the initial rate (v); units should be V s−1. Record this number for each peroxide and Trx concentration.At low peroxide concentrations (< 1 μM) a decrease in fluorescence may only be observed after the individual traces have been averaged.
- 19. Repeat steps 16-18 until all the concentrations of peroxide have been used to calculate rates of reaction with one Prx/Trx solutionIt is useful to plot the calculated rates (V s−1) directly against the peroxide concentration as you perform the experiments. In this way, you can spot anomalous measurements, caused by incorrect dilution of substrate, or instrument artifact. Bubbles accidentally introduced into the stopped-flow instrument can give spurious rates, and hence should be avoided. Also, this plot will quickly reveal if the correct peroxide concentration range has been covered. The plot should indicate approach to saturation at the higher peroxide concentrations, while measurements at the lowest peroxide concentrations will be significantly slower (see Figure 2C).
Measure rate of Prx reaction with next Trx concentration
20. Once you have a satisfactory range of measurements, replace the Trx/Prx solution with one with the next Trx concentration.
- 21. For each new Trx concentration, remove the peroxide solution and rinse the syringe 2-3X with buffer. Adjust the photomultiplier voltage and determine the full fluorescence change during the reaction as described in steps 10-14.Adjusting the photomultiplier voltage for each Trx concentration has the advantage that the instrumental sensitivity is increased for the lower Trx concentrations, making small fluorescence changes easier to observe.
22. Measure the reaction rate with each peroxide concentration as described in steps 15-19.
23. Repeat for each Trx concentration.
Measure rate of Trx reaction with peroxide (optional)
24. If high peroxide concentrations (>1 mM) are needed, you will need to perform blank reactions in the absence of Prx, as there will be non-negligible rates of Trx reacting directly with the peroxide substrate. For these reactions, make solutions containing each concentration of Trx and no Prx. Use these solutions to perform steps 10-23.
Data analysis
- 25. Convert observed fluorescent change rates in V s−1 into the rate in μM peroxide s−1 μM−1 Prx using Equation 1:
Where [Trx] and [Prx] are the final micromolar concentrations of Trx and Prx respectively, and ΔF is the total Trx fluorescence change in the presence of excess peroxide (step 14) for the same Trx concentration as that being analyzed.(Equation 1) 26. Plot the data using a Hanes-Woolf plot as described in detail in Basic Protocol I, step 11. The Prx family members studied so far fit best to a ping-pong mechanism (also called a substituted-enzyme mechanism) and a the Hanes-Woolf plot should result in a series of straight lines, each one corresponding to a set of experiments carried out with varying peroxide concentrations at one Trx concentration.
- 27. All the data (different peroxide and Trx concentrations) can be fit to Equation 2 using the multi-function nonlinear regression capabilities of a computer program such as Sigmaplot (Systat Software Inc. San Jose, CA).
These global fits will simultaneously calculate the Km for Trx (KmTrx) and the Km for peroxide (KmROOH), the kcat, and the errors associated with these constants. See Figure 2C for an example of a global fit using data obtained with Treponema pallidum AhpC using E. coli Trx and t-butyl hydroperoxide.(Equation 2) Alternatively, these kcat and Km values can be obtained using primary and secondary plots as described in Basic Protocol I, steps 11-12 (Figure 2A-B). Dalziel analysis has also been used to obtain similar rate constants for Prx proteins; for details refer to: (Baker, et al., 2001, Jaeger, et al., 2004, Sayed and Williams, 2004).
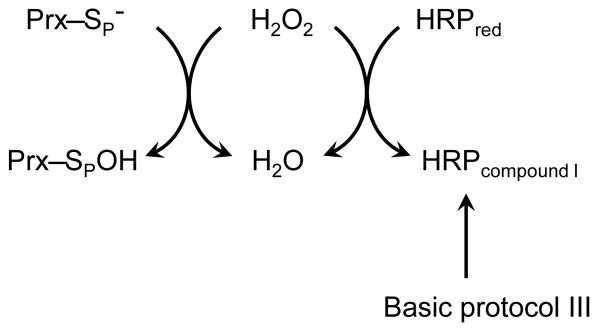
BASIC PROTOCOL III: MEASUREMENT OF SECOND ORDER RATE CONSTANT WITH HYDROGEN PEROXIDE USING HORSERADISH PEROXIDASE (HRP) COMPETITION ASSAY
This protocol describes an assay to determine the second order rate constant for Prx reduction of peroxide by monitoring the enzyme’s ability to compete with HRP (Figure 5). HRP reacts with hydrogen peroxide to form compound I with a second order rate constant of 1.8 × 107 M−1 s−1 over a wide range of pH (Dolman, et al., 1975). The formation of compound I can be measured spectroscopically as a decrease in absorbance at 403 nm (Δε403 = 5.4 × 104 M−1 cm−1) (Dolman, et al., 1975). When the hydrogen peroxide concentration is less than the HRP concentration, Prx activity can thus be detected as a function of the decreased formation of compound I (and thus decreased change in absorbance) as the Prx competes with HRP for the available peroxide. This assay monitors a partial turnover and provides a rate that is independent of the rate of reduction of the peroxidase – an advantage when the reductant is unknown or rate-limiting. Because HRP does not react efficiently with larger substrates such as cumene hydroperoxide and t-butyl hydroperoxide, this assay is only suitable to measure the rate with hydrogen peroxide. As the Prx rate constant decreases, greater concentrations of Prx are required to efficiently compete with HRP; at higher protein concentrations, the assay becomes more technically difficult. We have found this assay most useful to analyze peroxidases with rate constants greater than 105 M−1s−1. A similar assay using the much slower peroxidase, lignin peroxidase (6.5 × 105 M−1 s−1), has been used to measure the second order rate constant of Mycobacterium tuberculosis AhpE with hydrogen peroxide (Hugo, et al., 2009). HRP competition has also been used to measure Prx reactivity with peroxynitrite (Trujillo, et al., 2008).
Figure 5.
Determination of Prx activity by competition with horseradish peroxidase (HRP). As described in Basic Protocol 3, HRP is oxidized by H2O2 to form compound I, which can be monitored as a decrease in absorbance at 403 nm. As reduced Prx (Prx-SP–) reacts with H2O2 to form sulfenic acid (Prx-SPOH), less peroxide is available to oxidize HPR, resulting in a decrease in compound I formation.
Materials
10 mM hydrogen peroxide (see recipe)
Standard Prx buffer (see recipe)
Pre-reduced Prx (see Support protocol I)
HRP solution (see recipe)
96 well UV Half Area Plate (no lid required)
Multichannel Pipets (10 μl and 100-200 μl)
Create peroxide assay solution
Dilute 100 μl of a 10 mM hydrogen peroxide solution (see recipe) into 0.9 ml dH2O to make a 1 mM solution.
Add 180 μl of the 1 mM hydrogen peroxide to 3.82 ml dH2O in a clean Petri dish (100 mm) to make 4 ml of a 45 μM hydrogen peroxide solution.
Aliquot HRP and increasing concentrations of Prx into a 96 well plate
- 3. To a row of wells, add 15 μM HRP, 6-8 different concentrations of Prx, and enough standard buffer to make a final volume of 150 μl. All concentrations are the final values after mixing. Be sure to include a set of samples with no Prx present (0). For each Prx concentration, include 3-4 replicates. A typical layout for an assay is shown in Table 1. Carefully pipette up and down 3-4 times to mix, taking care to prevent the introduction of bubbles which interfere with good absorbance measurements. Existing bubbles can be popped using a clean 10 μl pipet tip.For Prx proteins with a kcat/Km for peroxide of ~107 M−1s−1 (for example, S. typhimurium AhpC (Parsonage, et al., 2005), Yeast TSA1 & 2 (Ogusucu, et al., 2007), and Xylella fastidiosa PrxQβ (Horta, et al., 2010)), suggested starting concentrations of Prx are: 0, 2, 4, 8, 12, and 16 μM. For Prxs with second order rate constants 105-106 M−1s−1, Prx concentrations will have to be increased. Concentrations of 0, 30, 60, 100, 150, and 200 μM are suggested for a protein with a second order rate constant of ~2 × 105 M−1s−1 (unpublished data and (Trujillo, et al., 2007)). As the Prx concentration increases, it becomes increasingly more difficult to prevent the formation of bubbles, and great care must be taken during pipetting and mixing.
Determine the starting HRP absorbance
4. Set up a plate reader to monitor absorbance in endpoint mode. Select a fixed wavelength of 403 nm. In the case that a reference wavelength is requested, use 600 nm.
5. Measure absorbance for all wells at the same time.
Measure change in HRP absorbance after addition of hydrogen peroxide in the presence of Prx
6. Compound 1is unstable, therefore, the change in absorbance at 403 nm should be measured within 90 s of peroxide addition. Set the plate reader so that it only reads 6-8 wells of a single row.
7. Using a multichannel pipettor, add 10 μL of a 45 μM hydrogen peroxide solution (final concentration of hydrogen peroxide = 3 μM) to one set of 6-8 wells. Using a 200 μl multichannel pipettor set at 100 μl, pipet up and down three times to mix solutions (being careful to minimize the creation of bubbles).
8. Read absorbance at 403 nm on a plate reader.
9. Change settings on the plate reader to read the next set of wells. Repeat steps 6-8 until all samples have been read.
Calculate the Prx rate constant based upon competition with HRP
The extent of inhibition of HRP oxidation (ΔAmax-ΔAobs/ΔAobs) at each Prx concentration can be plotted against [Prx] in order to obtain the second-order rate constant for Prx (kPrx) as established by Equation 3 (Ogusucu, et al., 2007, Winterbourn, 1987).
| (Equation 3) |
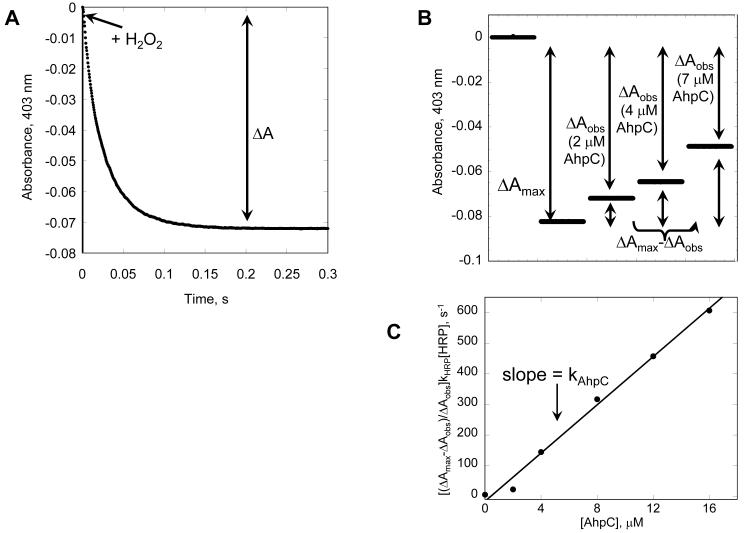
10. Determine the change in absorbance for each well by subtracting the absorbance after peroxide addition from the starting absorbance for the same well (Figure 6A). To obtain ΔAmax, calculate the average change in HRP absorbance in the absence of Prx (average all 0 samples).
11. The fraction of peroxide reacted with HRP is proportional to the observed change in absorbance (ΔAobs).
12. The fraction of peroxide reacted with Prx and not HRP is proportional to the difference between the observed absorbance change and the maximal absorbance change observed in the absence of Prx (ΔAmax-ΔAobs) (Figure 6B).
13. Calculate kHRP [HRP] [(ΔAmax-ΔAobs)/ΔAobs] for each well using a second order rate constant of 1.7 × 107 M−1 s−1 for the reaction of HRP with hydrogen peroxide and an HRP concentration of 7.5 μM (Dolman, et al., 1975).
14. Plot all values for kHRP [HRP] [(ΔAmax-ΔAobs)/ΔAobs] (y-axis) against Prx concentration (x-axis) and fit to a linear equation. The slope of the line gives the second order rate constant for Prx reacting with hydrogen peroxide (Figure 6C). As Prx concentrations increase, the percent of peroxide left to react with HRP approaches 0 and the slope of the line will start to flatten out, only the linear portion of the line should be used to calculate the Prx second order rate constant.
Figure 6. Prx inhibits the HRP reaction with hydrogen peroxide and can be used to measure the kPrx.
A HRP absorbance at 403 nm decreases as compound I is formed in the presence of hydrogen peroxide. B As Prx concentrations increase, the amount of compound I formed decreases due to competition for the available peroxide. The relative amounts of hydrogen peroxide reacting with HRP (ΔAobs) and Prx (ΔAmax - ΔAobs) can be used to calculate the kapp for each Prx concentration using equation 3. C The values for ((ΔAmax - ΔAobs)/ΔAobs) · kHRP[HRP] are plotted versus [Prx] and fit by linear regression. The slope of the line provides the kPrx for hydrogen peroxide. Shown is representative data for 7.5 μM HRP, 4 μM hydrogen peroxide and increasing concentrations of pre-reduced Salmonella typhimurium AhpC in 25 mM potassium phosphate, 1 mM EDTA, pH 7.0
ALTERNATE PROTOCOL IIIA: MEASUREMENT OF THE CATALYTIC CYSTEINE pKA USING THE HRP COMPETITION ASSAY
The HRP competition assay has been successfully used to determine the pKa of the catalytic cysteine for Prx proteins including S. typhimurium AhpC (Nelson, et al., 2008) yeast TSA1 and TSA2 (Ogusucu, et al., 2007), and X. fastidiosa PrxQβ (Horta, et al., 2010). For these experiments, the second order rate constants are obtained across a range of pH values, then the values for kPrx are plotted against pH and used to calculate a pKa for the Prx. More detailed procedures are provided in Nelson 2008 (Nelson, et al., 2008) and Ogusucu 2007(Ogusucu, et al., 2007). For this analysis, it is helpful that kHRP does not change across a wide range of pH values from 5-10. For pH <5, the kHRP in step 13 will need to be replaced with a corrected value that can be calculated for each pH using the equation provided by (Dunford, 1999), or determined directly by stopped-flow spectroscopic analysis.
BASIC PROTOCOL IV: MEASUREMENT OF PEROXIDASE ACTIVITY USING FOX ASSAY TO MEASURE PEROXIDE DISAPPEARANCE
The assay described in this section directly monitors decreasing peroxide concentrations (Figure 7) in the presence of Fe(II) and xylenol orange. Under acidic conditions, peroxides are able to oxidize Fe(II) to Fe(III), which forms a complex with xylenol orange [o-cresolsulfonphthalein-3,3-bis(methyliminodiacetic acid sodium salt)]; the production of this complex causes the generation of a blue-purple color with ε560 of 1.5 × 104 M−1 cm−1 (Wolff, 1994). This assay is suitable for hydrogen peroxide, cumene hydroperoxide, t-butyl hydroperoxide as well as lipid peroxides. This assay can be used to measure peroxide concentrations as low as 10 μM up to the mM range. Prx peroxidase activity can be measured under partial or single turnover conditions or under steady-state conditions in the presence of Trx and TrxR (or other appropriate physiological reductants) or using DTT as the reductant. This protocol uses DTT as a reductant; however we note that this chemical reductant can be much slower than the physiological reductant (Horta, et al., 2010) and at higher concentrations (~1 mM), DTT can directly react with the peroxide. While the FOX assay is useful when a reductant is unknown, in our hands it is also much noisier and less sensitive than the assays described in Basic Protocols I-III. This assay is most useful for peroxidases with rate constants less than 105 M−1s−1; for Prxs with rates on the order of 107 M−1s−1, the reaction is completed in a couple seconds and the Km tends to be lower than the detectable limit of the assay.
Figure 7.
Prx inhibits the HRP reaction with hydrogen peroxide and can be used to measure the kPrx. (A) HRP absorbance at 403 nm decreases as compound I is formed in the presence of hydrogen peroxide. (B) As Prx concentrations increase, the amount of compound I formed decreases due to competition for the available peroxide. The relative amounts of hydrogen peroxide reacting with HRP (1Aobs) and Prx (1Amax – 1Aobs) can be used to calculate the kapp for each Prx concentration using Equation 7.10.3. (C) The values for ((1Amax – 1Aobs)/1Aobs) ·kHRP[HRP] are plotted versus [Prx] and fit by linear regression. The slope of the line provides the kPrx for hydrogen peroxide. Shown are representative data for 7.5 μM HRP, 4 μM hydrogen peroxide, and increasing concentrations of prereduced Salmonella typhimurium AhpC in 25mMpotassium phosphate, 1mMEDTA, pH 7.0.
Materials
FOX Reagent A (see recipe)
FOX Reagent B (see recipe)
10 mM peroxide substrate: hydrogen peroxide, cumene hydroperoxide, or tert-butyl hydroperoxide
Pure Prx protein
100 mM DTT (see recipe)
Make FOX working reagent
Make enough FOX working reagent to have 1 ml for each sample. Mix 1 volume FOX reagent A with 100 volumes FOX reagent B. For example, for 20 samples, use 0.2 ml FOX Reagent A and 20 ml FOX Reagent B. Make fresh daily.
Make standard curve
2. Create a series of samples containing the peroxide of interest at known concentrations. At least 6 concentrations should be used between 10 μM and 200 μM peroxide. Standards should include controls containing no peroxide and 2-3 replicates for each standard concentration. Standard volumes should be the same as the volumes used for the samples.
Start Prx reaction
3. Set up peroxidase assays in microfuge tubes containing 5 μM Prx, 100 μM DTT (optional), and peroxide concentrations ranging from 10 to 200 μM. Under these conditions, DTT is used to reduce the Prx protein and allows for multiple turnovers. For all assays including DTT, be sure to include controls containing DTT and peroxide but no Prx. For single turnover reactions, the amount of Prx should be increased so that the amount of peroxide reduced in one turnover is sufficient to provide a reliably detectable change in peroxide (e.g. 100 μM peroxide to 10 μM pre-reduced Prx, see Support Protocol I.
Measure peroxide concentration
- 4. Quench each sample at the appropriate time by adding 50 μl of sample to 950 μl of FOX working reagent. This reagent is acidic and will stop the reaction since the protonated catalytic cysteine is unable to act as a nucleophile and reduce peroxides at low pH.The volume can be adjusted according to the amount of test sample and the volumes required for the spectrophotometer or plate reader to be used. To use a plate reader, we typically add 200 μl of FOX working reagent to 20 μl of sample in microfuge tubes and then transfer 200 μl of the developed solution to a 96-well plate. All volumes should be held constant between the samples and the standards.
5. Vortex each sample and incubate at room temperature for a minimum of 30 min to allow the absorbance to stabilize. Once developed, the absorbance is stable for up to 24 hours.
6. Read the absorbance at 560 nm of standards and samples. Plot the standard curve and use a linear fit to calculate the peroxide concentration for each sample.
7. Plot the peroxide concentration (y-axis) vs. time (x-axis) and find the initial rate by linear regression of the earliest points, which should fall on a straight line. The slope of the line is the rate (μM peroxide reduced s−1).
Notes: Sorbitol can be omitted from FOX Reagent B if the concentration of peroxide is high (<100 μM) because the role of sorbitol is to amplify the signal. With peroxide concentrations higher than 100 μM, the amount of assay added to the FOX working reagent can be decreased; just ensure that the points in the standard curve are treated the same way as the sample to be measured.
ALTERNATE PROTOCOL IVA: MEASUREMENT OF PEROXIDASE ACTIVITY IN VIVO USING FOX ASSAY
With some small adaptations, the FOX assay can be used with live bacteria to compare strains with Prx knockouts to the wild-type strain. It is also possible to compare mutant protein to the wild-type by over-expressing both. For these assays, we do not recommend using hydrogen peroxide as the substrate since multiple proteins use this substrate including catalase and glutathione peroxidase. In contrast, cumene hydroperoxide, t-butyl hydroperoxide and linoleic acid hydroperoxide are much more efficiently reduced by peroxiredoxins than by other peroxidases and have been used with some success to monitor intracellular Prx activity in Xanthomonas campestris (Klomsiri, et al., 2005, Vattanaviboon, et al., 2002).
Materials
Bacterial expression vector containing Prx gene
Bacterial growth media (LB or other media of choice)
FOX Reagent A (see recipe)
FOX Reagent B (see recipe)
10 mM peroxide substrate: cumene hydroperoxide, or t-butyl hydroperoxide
Transform bacterial strain with Prx expression plasmid
1. Transform the desired bacterial strain with an expression plasmid containing the coding region for the Prx of choice using standard methods. At the same time, transform a different sample of the same strain with an empty vector to provide a negative control. Plate cells using an appropriate antibiotic for the vector to select for cells containing the plasmid.
2. Take cells from each plate and inoculate tubes containing 5 ml bacterial growth media with either the Prx vector or the control vector. Incubate overnight with shaking at 37 °C.
Prepare FOX working reagent
3. Make enough FOX working reagent to have 1 ml for each sample. Mix 1 volume FOX reagent A with 100 volumes FOX reagent B. For example, for 20 samples, use 0.2 ml FOX Reagent A and 20 ml FOX Reagent B. Make fresh daily.
Treat cells with peroxide
4. Measure absorbance of overnight cultures at 600 nm. Add enough of each overnight culture to separate flasks containing 20 ml growth medium to give a final A600 of 0.1.
5. Grow samples for 4 hours (should still be in log phase growth) with shaking at 37 °C and measure A600. Adjust absorbance of each sample to 0.5 using fresh media.
6. Add cumene hydroperoxide, t-butyl hydroperoxide, or linoleic acid hydroperoxide to a final concentration of 200 μM.
7. For cumene hydroperoxide (dissolved in DMSO) or linoleic acid hydroperoxide (dissolved in methanol), be sure to include a control where the cells are treated with an equivalent volume of the solvent and no peroxide.
Create standard curve
8. To create a standard curve, add different concentrations of peroxide to 0.5 ml fresh media to yield final peroxide concentrations ranging from 10 – 200 μM.
Measure residual hydroperoxide concentrations
9. At 5 min intervals, remove 1 ml of the culture and centrifuge for 3 min at 10,000 × g.
10. Add 100 μl of the cleared supernatant to 900 μl of FOX working reagent and incubate for at least 30 min at room temperature.
11. Read the absorbance at 560 nm of standard curve and samples. Plot the standard curve and calculate peroxide concentration in samples as described in Section IV, 5-8.
SUPPORT PROTOCOL I: PRE-REDUCTION OF PRX OR TRX PROTEINS
Some of the protocols described in this chapter require at least one of the proteins to be fully reduced at the start of each assay. Prx proteins are commonly oxidized after prolonged exposure to air. In our hands, various Prxs and Trx will remain fully reduced for up to 6 hours on ice after removal of DTT; however, auto-oxidation may be a problem with other proteins and situations. In this case, reduced proteins can be stored in the presence of bead-linked TCEP (Immobilized TCEP Disulfide Reducing Gel from Thermo Scientific). In this case, the sample is centrifuged immediately prior to each assay and protein removed from the top of the beads.
Materials
100 mM DTT
Purified Prx or Trx protein (~10 mg/ml)
Suitable buffer for desired assay (Prx Reaction buffer or Standard Prx buffer)
PD-10 column
- Thaw purified protein. Add 1/10 volume DTT from a 100 mM stock to the protein solution (i.e. for 1 ml protein, add 0.1 ml DTT). To ensure complete reduction, incubate Prx for 1 hour at room temperature. Alternatively, incubate overnight at 4 °C.It is desirable to have at least a 20 fold molar excess of DTT over protein. If the protein is more concentrated than 10 mg/ml, the final DTT concentration should be increased accordingly.
Remove DTT using a PD-10 desalting column (GE Healthcare). First, equilibrate the column with 25 ml of buffer. Load protein. As much as 2.5 ml of protein may be loaded on the column, but better separation is achieved with smaller volumes. After protein has completely loaded onto the column, add buffer to make a total of 2.5 ml. Add another 0.5 ml buffer and collect the resulting eluate in a microcentrifuge tube. When no more eluate flows from the column, add another 0.5 ml buffer to the column, collect the resulting eluate in a new microcentrifuge tube, and repeat for a total of 10 fractions.
- Measure the 280 nm absorbance of each aliquot in a spectrophotometer. Combine those fractions containing the peak of the A280 absorbance.DTT will start to elute towards the end of the peak so only include the main portion of the peak and do not include the tail end.
Determine the concentration of the combined fractions. When an extinction coefficient is known, it is preferable to measure absorbance, which can be converted to concentration using Beer’s law; A=ε280cl where c = concentration (M) and l = pathlength (cm). Otherwise, a protein assay such as the BCA Assay (Pierce) can be used.
In our hands, reduced Prxs and Trxs have been successfully stored on ice for 6 hours without a loss of reduced cysteine. This time may vary for other proteins and in other locations. The number of reduced cysteines remaining in the original sample can be determined at the end of the assay by measuring the number of cysteines available to react with DTNB (see Support Protocol II).
Note: This support protocol will typically result in a >2-fold dilution of protein from its original concentration. If higher concentrations of pre-reduced Prx are required for the subsequent assay, Prxs should be concentrated in the presence of DTT prior to removal of this reductant.
SUPPORT PROTOCOL II: MEASUREMENT OF THIOL CONTENT BY REACTION WITH DTNB
In the presence of free thiols, 5,5′-dithiobis(2-nitrobenzoate) (DTNB, also referred to as Ellman’s reagent) undergoes a disulfide rearrangement to produce a mixed disulfide between the target thiol and 2-nitro-5-thiobenzoate (TNB) and release a free TNB molecule. This reaction is stoichiometric with one molecule of free TNB produced per molecule of free thiol. The absorbance of free TNB at 412 nm is used to calculate the original thiol concentration. The protocol here employs denaturing conditions to allow for accessibility to all protein thiols.
Materials
50 mM Tris-HCl, 4 M GuHCl, 0.5 mM EDTA, pH 8.0
30 mM 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB) in DMSO
Pre-reduced protein with DTT removed (see Support Protocol I)
Set up a spectrophotometer to read absorbance at 412 nm. Add enough 50 mM Tris-HCl, 4 M GuHCl, 0.5 mM EDTA, pH 8.0 to a cuvette to make up a final volume of 1 ml after addition of DTNB and Prx.
Add 5 μl DTNB, mix and measure absorbance at 412 nm after 2-5 minutes to determine spontaneous TNB production.
Add Prx and measure absorbance at 412 nm. Subtract out background production measured in step 2 and calculate the number of reduced thiols using ε412 = 14,150 M−1cm−1.
Notes: DTT will reduce DTNB and should be removed prior to the assay.
Sulfenic acids react with TNB to form a mixed disulfide, resulting in a decrease in absorbance at 412 nm (Poole and Ellis, 2002, Poole, 2008) and, thus, interfere with the correct quantification of protein thiols. Pre-reduction with DTT as described in Support Protocol I will reduce sulfenic acids. Alternately, sulfenic acids can be blocked prior to DTNB analysis (Poole and Ellis, 2002, Poole, 2008).
SUPPORT PROTOCOL III: MEASUREMENT OF PEROXIDE CONCENTRATION
While hydrogen peroxide concentrations can be measured using its absorbance at 240 nm (ε240 = 43.6 M−1cm−1) or monitoring HRP oxidation (Poole and Ellis, 2002), these methods are not suitable for other peroxides. The method described here can quickly be used to measure an accurate concentration for all peroxide substrates under very similar conditions as those used for subsequent Prx assays.
Materials
1 mL semimicro cuvettes
Reaction buffer (see recipe)
Purified E. coli Trx (500 μM diluted into Prx assay buffer)
Purified E. coli TrxR (50 μM diluted into Prx assay buffer)
Peroxiredoxin (50 μM diluted into Prx assay buffer)
15 mM NADPH (dissolved in 10 mM Tris-SO4 pH 8.5)
10 mM Peroxide solution to be measured: cumene hydroperoxide or t-butyl hydroperoxide (see recipe)
Spectrophotometer
Turn on the spectrophotometer and allow it to warm up and become stable (~ 30 min). Set the spectrophotometer to kinetics mode with a wavelength of 340 nm.
In a 1 ml quartz cuvette, mix Prx reaction buffer, 10 μl Trx, 10 μl TrxR, and 10 μL of a 50 μM Prx solution. Final concentrations in the assay should be 5 μM Trx, 0.5 μM Prx, and 0.5 μM TrxR.
Add 10 μL of 15 mM NADPH. The final concentration is 150 μM and the A340 should be ~0.9. Mix the contents and begin recording the 340 nm absorbance. This value will provide the background activity in the absence of peroxide (~1 minute; make sure that no more than 10% of the NADPH is used up at this step).
- Add 10 μl of peroxide solution to start the assay (100 μM peroxide final concentration). Monitor the change in absorbance at 340 nm until the absorbance remains constant or returns to the rate before the peroxide addition, indicating that all of the peroxide has been reduced.Ensure that some NADPH remains, so that the extent of the reaction is limited by the peroxide concentration.
Calculate the difference in absorbance before and after the addition of peroxide and convert this value to μM NADPH consumed using ε340 = 6,220 M−1cm−1; this is equal to the amount of peroxide added to the reaction.
REAGENTS AND SOLUTIONS
Purified peroxiredoxin protein
Purified Prx proteins are typically stored ~10 mg/ml and can be stored at −80 or −20 °C (without freeze thaw cycles) for an indefinite period of time.
Standard Prx buffer
25 mM potassium phosphate, pH 7.0, 1 mM EDTA
Prx Reaction buffer
25 mM potassium phosphate, pH 7.0, 1 mM EDTA, 100 mM ammonium sulfate
100 mM 1,4-Dithio-DL-threitol (DTT)
154.2 g/mole made in dH2O. Aliquots of 100 mM DTT (200 μl each) can be stored at −80 °C for up to one year. Once thawed, use each aliquot within 10 hours and do not refreeze.
10 mM hydrogen peroxide
Add approximately 456 μl of a 30% hydrogen peroxide solution to dH2O to make 100 ml total. The concentration of the 30% hydrogen peroxide solution should be confirmed by measuring the 240 nm absorbance of a 1000-fold dilution (perform a 50-fold dilution followed by a 20-fold dilution) and using an ε240 of 43.6 M−1cm−1. Make the 10 mM peroxide solution fresh daily.
10 mM cumene hydroperoxide
Dilute cumene hydroperoxide into DMSO to make a 100 mM stock (for an 80% purity solution, this will be a ~60-fold dilution). Add 100 μl of the 100 mM stock to 0.9 ml standard Prx buffer to make a 10 mM solution. Confirm the concentration of the 10 mM solution using Support Protocol III. Make the 10 mM peroxide solution fresh daily. For all assays using this substrate, control reactions should be done to make sure the solvent does not affect Prx activity. This can best be done by measuring hydrogen peroxide-dependent activity with and without DMSO present in the assay at the largest final concentration required for the desired set of cumene hydroperoxide assays.
10 mM t-butyl hydroperoxide
Dilute stock solution into Prx Reaction buffer to make a 10 mM solution. Confirm the concentration of the 10 mM solution using Support Protocol III. Make the 10 mM peroxide solution fresh daily.
Linoleic acid hydroperoxide
This is prepared by enzymatic oxidation of linoleic acid with soybean lipoxygenase as described by Evans et al (Evans, et al., 1998). Using this method, small volumes of linoleic acid hydroperoxide are prepared in methanol and the concentration is determined by the absorbance at 234 nm (ε234 = 25,000 M−1cm−1). Make the hydroperoxide solution fresh daily. For all assays using this substrate, control reactions should be done to make sure the solvent does not affect Prx activity. This can best be done by measuring hydrogen peroxide dependent activity with and without methanol present in the assay in the largest final concentration required for the desired set of linoleic acid hydroperoxide assays.
HRP solution
Type VI horseradish peroxidase can be purchased from Sigma Aldrich (P8375). Dissolve HRP in Standard Prx buffer to an approximate concentration of 0.5 mM. The final HRP concentration should be confirmed by measuring A403 of the stock solution and using an ε403 of 1.02×105 M−1cm−1 (Dolman 1975 Dunford 1999). Once dissolved, aliquots of HRP can be stored at −80 °C for up to one month. Multiple freeze-thaw cycles should be avoided.
FOX Reagent A
25 mM ammonium ferrous sulfate in 2.5 M sulfuric acid (H2SO4). The iron salt must be dissolved directly in acid because ferrous ions are prone to autoxidation at physiological pH. This solution can be stored for several weeks at 4 °C.
FOX Reagent B
100 mM sorbitol and 125 μM xylenol orange. This solution can be stored for 1 week at 4 °C.
Commentary
Background Information
Peroxiredoxin enzymes contain an absolutely conserved active site cysteine, referred to as the peroxidatic cysteine (CP), which reacts with peroxide to form a cysteine sulfenic acid (R-SOH) and releases water or the corresponding alcohol {Hall, 2010 #349}. In contrast to the peroxide reduction reaction common to all Prxs, regeneration of the reduced, active form of these enzymes varies. In many Prxs, a second cysteine, referred to as the resolving cysteine (CR), attacks the R-SOH to form a water molecule and a disulfide bond in the protein. In the typical 2-Cys Prxs, arguably the earliest Prx group to be recognized, the CR is found in the C-terminus of the partner subunit, yielding an intersubunit disulfide bond upon oxidation. As more examples of Prxs were discovered, the CR participating in the recycling process was recognized to reside in other positions, where it generally forms an intrasubunit disulfide bond (atypical 2-Cys Prxs). In 1-Cys Prxs, there is no CR and a thiol from another protein or small molecule presumably takes the place of this residue in the recycling process. While the CR shows up in different locations within the structure, formation of a disulfide bond with the CP always involves localized unfolding of the structure around both cysteines (Hall, et al., 2010). Prxs are then typically reduced by a thiol-containing disulfide reductase system that utilizes a CXXC-containing oxidoreductase domain or protein [e.g., Trx (Trx), AhpF, AhpD or glutaredoxin (Grx)] as the direct reductant which is in turn reduced by a flavin-containing, NAD(P)H-dependent disulfide reductase component (Poole, 2005). The mechanism for reduction of the CP in some of the 1-Cys Prx proteins is still unclear. Other features which vary across Prxs include their peroxide specificity and susceptibility towards inactivation by high peroxide concentrations, oligomeric state, and subcellular localization.
More recent bioinformatic and structural analysis has resulted in the identification of 6 subfamilies designated as AhpC/Prx1, Prx6, BCP/PrxQ, Tpx, Prx5, and AhpE. Prx proteins are widely distributed and subfamily distributions for Prxs are frequently not correlated with their phylogenetic distribution. For example, T. pallidum contains one highly expressed Prx similar to the prototypical AhpC protein from S. typhimurium. In contrast, Treponema denticola has only one Prx, but this protein is more similar to the Homo sapiens Prx6 protein (Parsonage, et al., 2010a). Humans express 6 different Prx proteins from three different subfamilies (Knoops, et al., 2007), while Escherichia coli contain 3 different Prxs from three different subfamilies (Baker and Poole, 2003). Much remains to be learned about why multiple Prx proteins are needed in a given organism although mice with knockouts to either Prx1 or Prx2 (which are both found in the cytoplasm and share high sequence and structural similarity) have been shown to exhibit different defects (Lee, et al., 2003, Neumann, et al., 2003).
Critical Parameters
Because of differences among different Prx proteins, some prior research will have to be done to determine how best to assay Prx activity. Important assay components that vary greatly depending on the Prx to be measured include selection of the peroxide substrate, substrate concentration, and Prx reductant. Many of these choices are aided by some preliminary work to identify the genetic context, subfamily, and which of the previously studied Prxs is most similar to the Prx of interest.
Prx subfamily
Within a single Prx subfamily, there is frequently still variation as far as substrate specificity, sensitivity to hyperoxidation, and mechanism of disulfide formation; however, identification of the correct subfamily is still a very useful way to identify the most appropriate starting assay parameters for a given Prx. More information about the Prx subfamilies can be found it the previous chapter in this volume, (Nelson, et al., 2011), and (Hall, et al., 2010). An online database tool is also available that provides a putative subfamily assignment for over 6,000 Prx sequences found in GenBank(nr) (http://www.csb.wfu.edu/prex/) (Soito, et al., 2010).
Reductants
Most Prxs are reduced in the cell by a thiol-containing disulfide reductase system typically composed of a CXXC-containing oxidoreductase domain or protein (e.g., Trx) as the direct reductant working in concert with a flavin-containing disulfide reductase component (e.g. TrxR) which is reduced by NAD(P)H (Poole, 2005). Although Trx is the most common reductant, some Prxs utilize another disulfide reductase such as AhpF, AhpD or glutaredoxin (Grx). AhpF contains an N-terminal domain that interacts with AhpC and a C-terminal TrxR–like flavin containing domain. For those Prxs that use AhpF as reductant as well as those linked to AhpD (all bacterial typical 2-Cys proteins belonging to the AhpC/Prx1 subfamily), the ahpF or ahpD coding sequence is located immediately downstream of the ahpC sequence (Bryk, et al., 2002, Poole, 2005). In the absence of data suggesting otherwise, Trx is the best starting choice as a general Prx reductant and protocols using Trx are provided in Basic Protocols I and II (Trx can also be used as a reductant for the assay described in Section IV after leaving out DTT). In the NADPH-linked assay described in Basic Protocol I, the rate of turnover is frequently limited by the ability of Prx to be reduced by Trx. This limitation has been dealt with to some degree in Basic Protocol II in which a large excess of reduced Trx is added and monitored directly. In all the protocols described, AhpF can replace Trx and TrxR when appropriate (details provided in Alternate Protocol IA and the introduction to Basic Protocol II).
In the absence of a resolving Cys, 1-Cys members of the BCP subfamily have been shown to be reduced by Trx or Grx (Clarke, et al., 2010, Rouhier, et al., 2004). Further information about how to best perform a Grx-linked Prx assay can be found in references (Parsonage 2010) and (Rouhier, et al., 2004). The mechanism for reduction of the CP in 1-Cys members of the Prx6 subfamily is still unclear. Trx is able to reduce Saccharomyces cerevisiae Ahp1p (Pedrajas, et al., 2000), but not human PrxVI (Kang, et al., 1998). Other potential reductants include GSTπ and glutathione (Manevich, et al., 2004), glutaredoxin (Pedrajas, et al., 2010), and ascorbate (Monteiro, et al., 2007). The assays described in Basic Protocol III, Basic Protocol IV, and Alternate Protocol IVA can be used to measure rates of Prx oxidation by peroxide in the absence of any information about the reductant.
Peroxide substrates
Peroxide concentrations should be chosen carefully as some (mostly eukaryotic) members of the AhpC/Prx1 subfamily are hyperoxidized at high peroxide concentration to form sulfinic (−SPO2H) and sulfonic (−SPO3H) acids. These species are not reducible by Trx or DTT, although in eukaryotes the sulfinic acid form of some Prxs can be rescued by an ATP-dependent repair enzyme called sulfiredoxin (Lowther and Haynes, epub Dec. 17, 2010).
Members of the AhpC/Prx1 subfamily have been shown to react very efficiently with hydrogen peroxide (~107 M−1s−1). S. typhimurium AhpC prefers hydrogen peroxide to more bulky peroxides such as cumene hydroperoxide and t-butyl hydroperoxide (Parsonage, et al., 2008).
E. coli Tpx (in contrast to AhpC) is more specific for cumene hydroperoxide (a bulky, hydrophobic substrate) than for hydrogen peroxide (Km of 9 μM compared with 1.7 mM, respectively) (Baker and Poole, 2003). Other Tpx proteins have also been shown to react with alkyl hydroperoxides suggesting that members of the Tpx subfamily are important for lipid hydroperoxide removal in bacteria (Cha, et al., 2004, Jaeger, et al., 2004).
Human PrxV is somewhat less reactive with hydrogen peroxide than PrxI and PrxII, but is very efficient with organic hydroperoxides and peroxynitrite (Trujillo, et al., 2007).
Less information is available for the BCP subfamily. The current data suggest that many of the enzymes in this subfamily react with similar specificities with a variety of peroxides including hydrogen peroxide, cumene hydroperxoide, t-butyl hydroperoxide, and linoleic acid hydroperoxide (Horta, et al., 2010, Jeong, et al., 2000).
Troubleshooting
Inactivation of Prxs by peroxide
While eukaryotic Prx1 and 2 are more prone to over-oxidation than many of the bacterial members of the same subfamily, most Prxs can be hyperoxidized and thus inactivated at high peroxide concentrations. Inactivation is best observed when the assays described in Basic Protocol I are allowed to proceed through multiple turnovers until NADPH and/or hydrogen peroxide consumption is complete. Under conditions where Prx is inactivated, the decrease in NADPH quickly becomes non-linear in the absence of peroxide or NADPH exhaustion. Turnover appears to be a critical component for inactivation to occur (Baker and Poole, 2003). Example data showing peroxide-dependent inactivation of Prxs can be found for human Prx1 (Yang, et al., 2002), S. typhimurium AhpC (Wood, et al., 2003) and E. coli Tpx (Baker and Poole, 2003). Because of this, peroxide concentrations should be selected to minimize the occurrence of hyperoxidation. Starting concentrations around 100 μM are generally safe, but this should be confirmed experimentally.
In some cases, Prxs may become inactivated during purification. It is possible to confirm the presence of a full complement of reducible cysteines using a DTNB assay (see Support Protocol II). This is particularly a problem for proteins in which the resolving cysteine has been mutated. If a Prx is inactivated during purification, the inclusion of 1 mM DTT in all buffers used for purification can generally circumvent the problem.
Non-saturating kinetics
Some Prxs appear not to follow Michaelis-Menten kinetics and show no saturation with peroxide and/or Trx (Baker, et al., 2001, Sayed and Williams, 2004). This might suggest that the Km for the non-saturable substrate is so high that the rate is linearly proportional to substrate concentration under technically feasible conditions. In this case, kinetic parameters can be calculated using the integrated Dalziel rate equation to calculate kinetic parameters analogous to maximum velocity and Km for peroxide (Forstrom, et al., 1979).
It should be noted that an N-terminally His-tagged T. pallidum AhpC exhibited non-saturable kinetics with Trx. Without the tag, this protein interacted in a saturable manner with Trx and also had a lower Km and higher Vmax with various peroxides than the tagged protein (Parsonage, et al., 2010a). Oligomerization and activity of human PrxIII was also altered by the presence of a tag (Cao, et al., 2007). We suggest purifying Prxs without a tag or, at the very least, cleaving the tag prior to kinetic analysis.
Anticipated Results
Although it is expected that kinetic analysis of various Prx proteins will reveal the presence of a peroxidase activity under some conditions, there is almost certainly going to be considerable variability in the details of this activity among different Prx family members. Of the Prxs measured to date, second order rate constants for the reaction with peroxide have varied between 104 (Hugo, et al., 2009) and 108 M−1s−1 (Manta, et al., 2009). Even among members of the same Prx subfamily in the same organism, peroxide substrate specificity and sensitivity toward hyperoxidation can vary substantially (Cox, et al., 2009, Sayed and Williams, 2004).
Time considerations
For all these protocols, the time required to accomplish the assays is secondary to the time required to obtain the appropriate purified proteins. We also suggested that sufficient time be allotted for multiple repeats of all the assays to ensure the reproducibility of the data prior to publication.
All solutions can be prepared the day before unless stated otherwise.
For all assays, data analysis will take some time, but is often done on a subsequent day to performing the assays. All datasets should be analyzed prior to performing the next replicate to ensure that the assay does not need to be modified.
Basic Protocol I and Alternate Protocol IA
Using a Prx with typical activity, these assays can be carried out in a half-day. An enzyme displaying low activity would need longer assays, and hence more time is required to complete the set of experiments.
Basic Protocol II
A complete bi substrate kinetic analysis using this procedure can be completed in one day. Assume that between 2-4 hours will be spent preparing the solutions and the stopped flow instrument for the assays. Incubating the Trx with DTT overnight at 4 °C shortens the preparation time required the day of the assays. It is frequently preferable to only use one Trx concentration the first day to provide time for optimization of the correct range of peroxide concentrations and to familiarize oneself with the assays.
Basic Protocol III
Allow 2-3 hours to reduce the Prx and to prepare protein and peroxide solutions. Incubating the Prx with DTT overnight at 4 °C shortens the preparation time required the day of the assays. The time required to aliquot samples in the plate and read the results will vary depending on the number of samples to be assayed. Allow 1 hour total for 24 samples and ~2 hours to prepare and ~1.5 hours to read two 96 well plates.
Basic Protocol IV
FOX working reagent should be prepared the day of the assay; other solutions can be prepared before. Under these conditions, Prxs should utilize all of the peroxide within the first 1 – 10 minutes. Although each sample must be incubated at least 30 minutes with the working reagent before reading the absorbance, the final product is stable, allowing samples quenched at various times to all be read together.
ACKNOWLEDGEMENTS
This work was supported by funding from the National Institutes of Health [F32 GM074537 to K.J.N. and RO1 GM050389 to L.B.P]. We thank Dr. Chananat Klomsiri for her advice in preparing this manuscript.
Footnotes
Internet Resources http://www.csb.wfu.edu/prex/: PREX is a searchable database containing > 6,000 Prx protein sequences unambiguously classified into one of six distinct subclasses. Subfamily classifications use information around the active sites of structurally characterized subfamily members to search for sequences with conserved functionally-relevant motifs (Nelson, et al., 2011, Soito, et al., 2010).
LITERATURE CITED
- Baker LM, Raudonikiene A, Hoffman PS, Poole LB. Essential thioredoxin-dependent peroxiredoxin system from Helicobacter pylori: genetic and kinetic characterization. J. Bacteriol. 2001;183:1961–1973. doi: 10.1128/JB.183.6.1961-1973.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker LM, Poole LB. Catalytic mechanism of thiol peroxidase from Escherichia coli. Sulfenic acid formation and overoxidation of essential CYS61. J. Biol. Chem. 2003;278:9203–9211. doi: 10.1074/jbc.M209888200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryk R, Lima CD, Erdjument-Bromage H, Tempst P, Nathan C. Metabolic enzymes of mycobacteria linked to antioxidant defense by a thioredoxin-like protein. Science. 2002;295:1073–1077. doi: 10.1126/science.1067798. [DOI] [PubMed] [Google Scholar]
- Cao Z, Bhella D, Lindsay JG. Reconstitution of the mitochondrial PrxIII antioxidant defence pathway: general properties and factors affecting PrxIII activity and oligomeric state. J. Mol. Biol. 2007;372:1022–1033. doi: 10.1016/j.jmb.2007.07.018. [DOI] [PubMed] [Google Scholar]
- Cha MK, Kim WC, Lim CJ, Kim K, Kim IH. Escherichia coli periplasmic thiol peroxidase acts as lipid hydroperoxide peroxidase and the principal antioxidative function during anaerobic growth. J. Biol. Chem. 2004;279:8769–8778. doi: 10.1074/jbc.M312388200. [DOI] [PubMed] [Google Scholar]
- Clarke DJ, Ortega XP, Mackay CL, Valvano MA, Govan JR, Campopiano DJ, Langridge-Smith P, Brown AR. Subdivision of the bacterioferritin comigratory protein family of bacterial peroxiredoxins based on catalytic activity. Biochemistry. 2010;49:1319–1330. doi: 10.1021/bi901703m. [DOI] [PubMed] [Google Scholar]
- Cornish-Bowden A. Fundamentals of Enzyme Kinetics. Portland Press Ltd.; London: 2004. [Google Scholar]
- Cox AG, Pearson AG, Pullar JM, Jonsson TJ, Lowther WT, Winterbourn CC, Hampton MB. Mitochondrial peroxiredoxin 3 is more resilient to hyperoxidation than cytoplasmic peroxiredoxins. Biochem. J. 2009;421:51–58. doi: 10.1042/BJ20090242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox AG, Winterbourn CC, Hampton MB. Mitochondrial peroxiredoxin involvement in antioxidant defence and redox signalling. Biochem. J. 2010;425:313–325. doi: 10.1042/BJ20091541. [DOI] [PubMed] [Google Scholar]
- Dietz KJ, Jacob S, Oelze ML, Laxa M, Tognetti V, de Miranda SM, Baier M, Finkemeier I. The function of peroxiredoxins in plant organelle redox metabolism. J. Exp. Bot. 2006;57:1697–1709. doi: 10.1093/jxb/erj160. [DOI] [PubMed] [Google Scholar]
- Dolman D, Newell GA, Thurlow MD. A kinetic study of the reaction of horseradish peroxidase with hydrogen peroxide. Can. J. Biochem. 1975;53:495–501. doi: 10.1139/o75-069. [DOI] [PubMed] [Google Scholar]
- Dunford HB. Spectroscopy of horseradish peroxidase. I. Optical, resonance raman, magnetic circular dichroism, x-ray absorption, and diffraction. In: Dunford HB, editor. Heme peroxidases. Wiley; New York: 1999. pp. 135–174. [Google Scholar]
- Evans MV, Turton HE, Grant CM, Dawes IW. Toxicity of linoleic acid hydroperoxide to Saccharomyces cerevisiae: involvement of a respiration-related process for maximal sensitivity and adaptive response. J. Bacteriol. 1998;180:483–490. doi: 10.1128/jb.180.3.483-490.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forstrom JW, Stults FH, Tappel AL. Rat liver cytosolic glutathione peroxidase: reactivity with linoleic acid hydroperoxide and cumene hydroperoxide. Arch. Biochem. Biophys. 1979;193:51–55. doi: 10.1016/0003-9861(79)90007-9. [DOI] [PubMed] [Google Scholar]
- Hall A, Nelson KJ, Poole LB, Karplus PA. Structure-based insights into the catalytic power and conformational dexterity of peroxiredoxins. Antioxid. Redox Signal. 2010;402:194–209. doi: 10.1089/ars.2010.3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmgren A, Reichard P. Thioredoxin 2: cleavage with cyanogen bromide. Eur. J. Biochem. 1967;2:187–196. doi: 10.1111/j.1432-1033.1967.tb00125.x. [DOI] [PubMed] [Google Scholar]
- Horta BB, de Oliveira MA, Discola KF, Cussiol JR, Netto LE. Structural and biochemical characterization of peroxiredoxin Qbeta from Xylella fastidiosa: catalytic mechanism and high reactivity. J. Biol. Chem. 2010;285:16051–16065. doi: 10.1074/jbc.M109.094839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugo M, Turell L, Manta B, Botti H, Monteiro G, Netto LE, Alvarez B, Radi R, Trujillo M. Thiol and sulfenic acid oxidation of AhpE, the one-cysteine peroxiredoxin from Mycobacterium tuberculosis: kinetics, acidity constants, and conformational dynamics. Biochemistry. 2009;48:9416–9426. doi: 10.1021/bi901221s. [DOI] [PubMed] [Google Scholar]
- Jaeger T, Budde H, Flohe L, Menge U, Singh M, Trujillo M, Radi R. Multiple thioredoxin-mediated routes to detoxify hydroperoxides in Mycobacterium tuberculosis. Arch. Biochem. Biophys. 2004;423:182–191. doi: 10.1016/j.abb.2003.11.021. [DOI] [PubMed] [Google Scholar]
- Jeong W, Cha MK, Kim IH. Thioredoxin-dependent hydroperoxide peroxidase activity of bacterioferritin comigratory protein (BCP) as a new member of the thiol-specific antioxidant protein (TSA)/Alkyl hydroperoxide peroxidase C (AhpC) family. J. Biol. Chem. 2000;275:2924–2930. doi: 10.1074/jbc.275.4.2924. [DOI] [PubMed] [Google Scholar]
- Kang SW, Baines IC, Rhee SG. Characterization of a mammalian peroxiredoxin that contains one conserved cysteine. J. Biol. Chem. 1998;273:6303–6311. doi: 10.1074/jbc.273.11.6303. [DOI] [PubMed] [Google Scholar]
- Kang SW, Rhee SG, Chang TS, Jeong W, Choi MH. 2-Cys peroxiredoxin function in intracellular signal transduction: therapeutic implications. Trends Mol. Med. 2005;11:571–578. doi: 10.1016/j.molmed.2005.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karplus PA, Hall A. Structural Survey of the Peroxiredoxins. In: Flohé L, Harris JR, editors. Peroxiredoxin Systems. Springer; New York: 2007. pp. 41–60. [DOI] [PubMed] [Google Scholar]
- Klomsiri C, Panmanee W, Dharmsthiti S, Vattanaviboon P, Mongkolsuk S. Novel roles of ohrR-ohr in Xanthomonas sensing, metabolism, and physiological adaptive response to lipid hydroperoxide. J. Bacteriol. 2005;187:3277–3281. doi: 10.1128/JB.187.9.3277-3281.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoops B, Loumaye E, Van Der Eecken V. Evolution of the peroxiredoxins. In: Flohé L, Harris JR, editors. Peroxiredoxin Systems. Springer; New York: 2007. pp. 27–40. [DOI] [PubMed] [Google Scholar]
- Krause G, Holmgren A. Substitution of the conserved tryptophan 31 in Escherichia coli thioredoxin by site-directed mutagenesis and structure-function analysis. J. Biol. Chem. 1991;266:4056–4066. [PubMed] [Google Scholar]
- Lee TH, Kim SU, Yu SL, Kim SH, Park do S, Moon HB, Dho SH, Kwon KS, Kwon HJ, Han YH, Jeong S, Kang SW, Shin HS, Lee KK, Rhee SG, Yu DY. Peroxiredoxin II is essential for sustaining life span of erythrocytes in mice. Blood. 2003;101:5033–5038. doi: 10.1182/blood-2002-08-2548. [DOI] [PubMed] [Google Scholar]
- Lowther WT, Haynes AC. Reduction of Cysteine Sulfinic Acid in Eukaryotic, Typical 2-Cys Peroxiredoxins by Sulfiredoxin. Antioxid. Redox Signal. doi: 10.1089/ars.2010.3564. epub Dec. 17, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manevich Y, Feinstein SI, Fisher AB. Activation of the antioxidant enzyme 1-CYS peroxiredoxin requires glutathionylation mediated by heterodimerization with pi GST. Proc. Natl. Acad. Sci. U. S. A. 2004;101:3780–3785. doi: 10.1073/pnas.0400181101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manta B, Hugo M, Ortiz C, Ferrer-Sueta G, Trujillo M, Denicola A. The peroxidase and peroxynitrite reductase activity of human erythrocyte peroxiredoxin 2. Arch. Biochem. Biophys. 2009;484:146–154. doi: 10.1016/j.abb.2008.11.017. [DOI] [PubMed] [Google Scholar]
- Monteiro G, Horta BB, Pimenta DC, Augusto O, Netto LE. Reduction of 1-Cys peroxiredoxins by ascorbate changes the thiol-specific antioxidant paradigm, revealing another function of vitamin C. Proc. Natl. Acad. Sci. U. S. A. 2007;104:4886–4891. doi: 10.1073/pnas.0700481104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson KJ, Parsonage D, Hall A, Karplus PA, Poole LB. Cysteine pK(a) values for the bacterial peroxiredoxin AhpC. Biochemistry. 2008;47:12860–12868. doi: 10.1021/bi801718d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson KJ, Knutson ST, Soito L, Klomsiri C, Poole LB, Fetrow JS. Analysis of the peroxiredoxin family: using active site structure and sequence information for global classification and residue analysis. Proteins. 2011;79:947–964. doi: 10.1002/prot.22936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann CA, Krause DS, Carman CV, Das S, Dubey DP, Abraham JL, Bronson RT, Fujiwara Y, Orkin SH, Van Etten RA. Essential role for the peroxiredoxin Prdx1 in erythrocyte antioxidant defence and tumour suppression. Nature. 2003;424:561–565. doi: 10.1038/nature01819. [DOI] [PubMed] [Google Scholar]
- Ogusucu R, Rettori D, Munhoz DC, Soares Netto LE, Augusto O. Reactions of yeast thioredoxin peroxidases I and II with hydrogen peroxide and peroxynitrite: Rate constants by competitive kinetics. Free Radic. Biol. Med. 2007;42:326–334. doi: 10.1016/j.freeradbiomed.2006.10.042. [DOI] [PubMed] [Google Scholar]
- Parsonage D, Youngblood DS, Sarma GN, Wood ZA, Karplus PA, Poole LB. Analysis of the link between enzymatic activity and oligomeric state in AhpC, a bacterial peroxiredoxin. Biochemistry. 2005;44:10583–10592. doi: 10.1021/bi050448i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsonage D, Karplus PA, Poole LB. Substrate specificity and redox potential of AhpC, a bacterial peroxiredoxin. Proc. Natl. Acad. Sci. U. S. A. 2008;105:8209–8214. doi: 10.1073/pnas.0708308105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsonage D, Desrosiers DC, Hazlett KR, Sun Y, Nelson KJ, Cox DL, Radolf JD, Poole LB. Broad specificity AhpC-like peroxiredoxin and its thioredoxin reductant in the sparse antioxidant defense system of Treponema pallidum. Proc. Natl. Acad. Sci. U. S. A. 2010a;107:6240–6245. doi: 10.1073/pnas.0910057107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsonage D, Reeves SA, Karplus PA, Poole LB. Engineering of fluorescent reporters into redox domains to monitor electron transfers. Meth. Enzymol. 2010b;474:1–21. doi: 10.1016/S0076-6879(10)74001-5. [DOI] [PubMed] [Google Scholar]
- Pedrajas JR, Miranda-Vizuete A, Javanmardy N, Gustafsson JA, Spyrou G. Mitochondria of Saccharomyces cerevisiae contain one-conserved cysteine type peroxiredoxin with thioredoxin peroxidase activity. J. Biol. Chem. 2000;275:16296–16301. doi: 10.1074/jbc.275.21.16296. [DOI] [PubMed] [Google Scholar]
- Pedrajas JR, Padilla CA, McDonagh B, Barcena JA. Glutaredoxin participates in the reduction of peroxides by the mitochondrial 1-CYS peroxiredoxin in Saccharomyces cerevisiae. Antioxid. Redox Signal. 2010;13:249–258. doi: 10.1089/ars.2009.2950. [DOI] [PubMed] [Google Scholar]
- Peskin AV, Low FM, Paton LN, Maghzal GJ, Hampton MB, Winterbourn CC. The high reactivity of peroxiredoxin 2 with H2O2 is not reflected in its reaction with other oxidants and thiol reagents. J. Biol. Chem. 2007;282:11885–11892. doi: 10.1074/jbc.M700339200. [DOI] [PubMed] [Google Scholar]
- Poole LB, Ellis HR. Flavin-dependent alkyl hydroperoxide reductase from Salmonella typhimurium. 1. Purification and enzymatic activities of overexpressed AhpF and AhpC proteins. Biochemistry. 1996;35:56–64. doi: 10.1021/bi951887s. [DOI] [PubMed] [Google Scholar]
- Poole LB, Godzik A, Nayeem A, Schmitt JD. AhpF can be dissected into two functional units: tandem repeats of two thioredoxin-like folds in the N-terminus mediate electron transfer from the thioredoxin reductase-like C-terminus to AhpC. Biochemistry. 2000;39:6602–6615. doi: 10.1021/bi000405w. [DOI] [PubMed] [Google Scholar]
- Poole LB, Ellis HR. Identification of cysteine sulfenic acid in AhpC of alkyl hydroperoxide reductase. Methods in enzymology. 2002;348:122–136. doi: 10.1016/s0076-6879(02)48632-6. [DOI] [PubMed] [Google Scholar]
- Poole LB. Bacterial defenses against oxidants: mechanistic features of cysteine-based peroxidases and their flavoprotein reductases. Arch. Biochem. Biophys. 2005;433:240–254. doi: 10.1016/j.abb.2004.09.006. [DOI] [PubMed] [Google Scholar]
- Poole LB. The Catalytic Mechanism of Peroxiredoxins. In: Flohé L, Harris JR, editors. Peroxiredoxin Systems. Springer; New York: 2007. pp. 61–81. [Google Scholar]
- Poole LB. Maines Mahin D., editor. Measurement of protein sulfenic acid content. Current protocols in toxicology / editorial board. 2008. [et al Chapter 17: Unit17 12. [DOI] [PMC free article] [PubMed]
- Rabilloud T, Heller M, Gasnier F, Luche S, Rey C, Aebersold R, Benahmed M, Louisot P, Lunardi J. Proteomics analysis of cellular response to oxidative stress. Evidence for in vivo overoxidation of peroxiredoxins at their active site. J. Biol. Chem. 2002;277:19396–19401. doi: 10.1074/jbc.M106585200. [DOI] [PubMed] [Google Scholar]
- Rouhier N, Gelhaye E, Gualberto JM, Jordy MN, De Fay E, Hirasawa M, Duplessis S, Lemaire SD, Frey P, Martin F, Manieri W, Knaff DB, Jacquot JP. Poplar peroxiredoxin Q. A thioredoxin-linked chloroplast antioxidant functional in pathogen defense. Plant Physiol. 2004;134:1027–1038. doi: 10.1104/pp.103.035865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayed AA, Williams DL. Biochemical characterization of 2-Cys peroxiredoxins from Schistosoma mansoni. J. Biol. Chem. 2004;279:26159–26166. doi: 10.1074/jbc.M401748200. [DOI] [PubMed] [Google Scholar]
- Seaver LC, Imlay JA. Alkyl hydroperoxide reductase is the primary scavenger of endogenous hydrogen peroxide in Escherichia coli. J. Bacteriol. 2001;183:7173–7181. doi: 10.1128/JB.183.24.7173-7181.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soito L, Williamson C, Knutson ST, Fetrow JS, Poole LB, Nelson KJ. PREX: PeroxiRedoxin classification indEX, a database of subfamily assignments across the diverse peroxiredoxin family. Nucleic Acids Res. 2010;39:D332–337. doi: 10.1093/nar/gkq1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo M, Clippe A, Manta B, Ferrer-Sueta G, Smeets A, Declercq JP, Knoops B, Radi R. Pre-steady state kinetic characterization of human peroxiredoxin 5: taking advantage of Trp84 fluorescence increase upon oxidation. Arch. Biochem. Biophys. 2007;467:95–106. doi: 10.1016/j.abb.2007.08.008. [DOI] [PubMed] [Google Scholar]
- Trujillo M, Ferrer-Sueta G, Radi R. Kinetic studies on peroxynitrite reduction by peroxiredoxins. Meth. Enzymol. 2008;441:173–196. doi: 10.1016/S0076-6879(08)01210-X. [DOI] [PubMed] [Google Scholar]
- Vattanaviboon P, Whangsuk W, Panmanee W, Klomsiri C, Dharmsthiti S, Mongkolsuk S. Evaluation of the roles that alkyl hydroperoxide reductase and Ohr play in organic peroxide-induced gene expression and protection against organic peroxides in Xanthomonas campestris. Biochem. Biophys. Res. Commun. 2002;299:177–182. doi: 10.1016/s0006-291x(02)02602-5. [DOI] [PubMed] [Google Scholar]
- Veal EA, Day AM, Morgan BA. Hydrogen peroxide sensing and signaling. Mol. Cell. 2007;26:1–14. doi: 10.1016/j.molcel.2007.03.016. [DOI] [PubMed] [Google Scholar]
- Winterbourn CC. The ability of scavengers to distinguish OH· production in the iron-catalyzed Haber-Weiss reaction: comparison of four assays for OH·. Free Radic. Biol. Med. 1987;3:33–39. doi: 10.1016/0891-5849(87)90037-2. [DOI] [PubMed] [Google Scholar]
- Winterbourn CC. Reconciling the chemistry and biology of reactive oxygen species. Nat. Chem. Biol. 2008;4:278–286. doi: 10.1038/nchembio.85. [DOI] [PubMed] [Google Scholar]
- Wolff SP. Ferrous ion oxidation in presence of ferric ion indicator xylenol orange for measurement of hydroperoxides. Meth. Enzymol. 1994;233:182–189. [Google Scholar]
- Wood ZA, Poole LB, Karplus PA. Structure of intact AhpF reveals a mirrored thioredoxin-like active site and implies large domain rotations during catalysis. Biochemistry. 2001;40:3900–3911. doi: 10.1021/bi002765p. [DOI] [PubMed] [Google Scholar]
- Wood ZA, Poole LB, Karplus PA. Peroxiredoxin evolution and the regulation of hydrogen peroxide signaling. Science. 2003;300:650–653. doi: 10.1126/science.1080405. [DOI] [PubMed] [Google Scholar]
- Yang KS, Kang SW, Woo HA, Hwang SC, Chae HZ, Kim K, Rhee SG. Inactivation of human peroxiredoxin I during catalysis as the result of the oxidation of the catalytic site cysteine to cysteine-sulfinic acid. J. Biol. Chem. 2002;277:38029–38036. doi: 10.1074/jbc.M206626200. [DOI] [PubMed] [Google Scholar]
- Zhang B, Su Y, Ai G, Wang Y, Wang T, Wang F. Involvement of peroxiredoxin I in protecting cells from radiation-induced death. J. Radiat. Res. (Tokyo) 2005;46:305–312. doi: 10.1269/jrr.46.305. [DOI] [PubMed] [Google Scholar]
Key References
- 1.Poole LB, Hall A, Nelson KJ. Overview – Peroxiredoxins in oxidant defense and redox regulation. doi: 10.1002/0471140856.tx0709s49. chapter submitted for this issue. An overview of Prx catalytic cycles, classification, and biological roles.
- 2.Fourquet S, Huang ME, D’Autreaux B, Toledano MB. The dual functions of thiol-based peroxidases in H2O2 scavenging and signaling. Antioxid. Redox Signal. 2008;10:1565–1576. doi: 10.1089/ars.2008.2049. Review of biochemical and kinetic properties of peroxiredoxins and the related family of glutathione peroxidases.
- 3.Hall A, Nelson KJ, Poole LB, Karplus PA. Structure-based insights into the catalytic power and conformational dexterity of peroxiredoxins. Antioxid. Redox Signal. 2010;402:194–209. doi: 10.1089/ars.2010.3624. Review of Prx function and subfamily with an emphasis on structural characteristics