Abstract
Noroviruses are an important cause of non-bacterial epidemic gastroenteritis, but no specific antiviral therapies are available. We investigated the inhibitory effect of phosphorodiamidiate morpholino oligomers (PMOs) targeted against norovirus sequences. A panel of peptide-conjugated PMOs (PPMOs) specific for the murine norovirus (MNV) genome was developed, and two PPMO compounds directed against the first AUG of the ORF1 coding sequence near the 5′-end of the genome proved effective in inhibiting MNV replication in cells. A consensus PPMO (designated Noro 1.1), designed to target the corresponding region of several diverse human norovirus genotypes, decreased the efficiency of protein translation in a cell-free luciferase reporter assay and inhibited Norwalk virus protein expression in replicon-bearing cells. Our data suggest that PPMOs directed against the relatively conserved 5′-end of the norovirus genome may show broad antiviral activity against this genetically diverse group of viruses.
Keywords: Norovirus, Antisense, PMO, Antiviral
Introduction
Noroviruses are the most important cause of non-bacterial epidemic gastroenteritis in all age groups, and are the second most important cause of sporadic endemic diarrhea in infants and young children after rotaviruses (Glass et al., 2000, Green, 2007). Currently, there are no vaccines or specific antiviral therapies for the control of norovirus disease (Green, 2007). Noroviruses belong to the family Caliciviridae, which consists of four genera: Norovirus, Sapovirus, Vesivirus and Lagovirus. These 27–35 nm, non-enveloped viruses have single-stranded, positive-sense RNA genomes. Norovirus genomes are organized into three open reading frames (ORFs). The 5′ terminal ORF1 encodes a large polyprotein that is processed into mature nonstructural proteins (Green, 2007). ORF2 encodes the major capsid protein, VP1, and the 3′ terminal ORF3 encodes a minor structural protein, VP2.
The genus Norovirus is divided into five different genogroups, designated GI through GV. Genogroups I, II, and IV, which include human noroviruses, are further subdivided into several genotypes based on the similarities of the capsid (ORF2) gene (Zheng et al., 2006). Noroviruses have highly variable genomes (up to 43% nucleotide divergence between genogroups), with over 100 distinct viruses identified to date. The 5′-end sequence of the virus is one of the most conserved regions among norovirus genomes. Like other caliciviruses, the 5′-end of the norovirus genome shares sequence similarity with the 5′-end of the viral subgenomic RNA that maps near the beginning of ORF2 (Green, 2007).
Vaccine development has been impeded by the lack of a cell culture system for the propagation of human noroviruses. The absence of in vitro propagation techniques has been compensated for somewhat by the availability of recombinant virus-like particles (VLPs) produced by the expression of VP1 in the baculovirus expression system (Bertolotti-Ciarlet et al., 2003). Norovirus VLPs have served as antigens for the study of norovirus immunity, and as a vaccine candidate (Estes et al., 2000). An alternative approach to the study of norovirus RNA replication was established when a human norovirus replicon-bearing cell line was created by transfecting a plasmid containing most of the Norwalk virus genome into mammalian cell lines (Chang et al., 2006). These cell lines are capable of constitutively expressing the replicative enzymes and other nonstructural proteins, allowing the study of RNA replication and providing a platform for screening antiviral compounds.
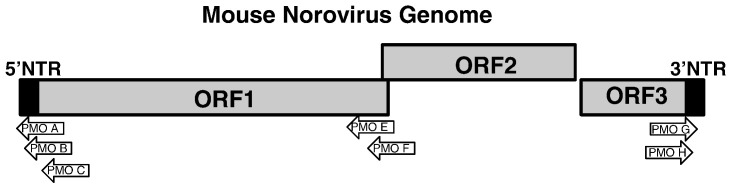
The recent discovery of mouse norovirus-1 (MNV-1) in immunodeficient mice led to the development of the first cell culture system for the noroviruses (Wobus et al.) MNV shares molecular properties with other noroviruses (Wobus, Thackray, and Virgin, 2006), although various MNV strains are less divergent in their genome sequence and therefore comprise a single genotype (Thackray et al., 2007). Analysis of MNV genomes identified the three open reading frames characteristic of noroviruses and vesiviruses (Fig. 1 ) (Wobus et al., 2006), and an additional ORF4 that overlaps ORF2 in a different reading frame (Thackray et al., 2007). ORF1 of MNV also encodes the nonstructural proteins while ORFs 2 and 3 encode the two proteins present in the viral capsid (Sosnovtsev et al., 2006). MNV-1 replication and dissemination in wild-type mice are limited by STAT-1 dependent interferons (Mumphrey et al., 2007). Thus, virus replication in lymph nodes and intestinal tissue in vivo does not result in clinical disease in mice. In vitro, MNV-1 grows efficiently in dendritic cells and macrophages (Chaudhry et al., 2007, Ward et al., 2007, Wobus et al., 2004), facilitating studies of viral gene expression, replication, cytopathology, and antiviral effector molecules. In addition, MNV is the only norovirus for which a reverse genetic system has been developed (Chaudhry et al., 2007, Ward et al., 2007).
Fig. 1.
Murine norovirus genome organization. The murine norovirus genome is shown organized into three different open reading frames, together with the 5′- and 3′-nontranslated regions (NTR). The murine norovirus PPMO targets are indicated in the figure (PMOs were synthesized as PPMOs). The direction of the arrow indicates the sense of the PPMO. Not shown is ORF4, which partially overlaps ORF2 (Thackray et al., 2007).
In this study, we investigated the use of phosphorodiamidiate morpholino oligomers (PMOs) as an inhibitor of norovirus replication. PMOs are similar to single-stranded DNA oligonucleotides in that they contain the same bases as DNA with a morpholine ring in place of a deoxyribose sugar and a neutral backbone (Iversen, 2001). PMOs are uncharged, water-soluble, and highly resistant to degradation in the host. PMOs bind to mRNA by the Watson–Crick base-pairing, and can reduce gene expression by the steric blockade of mRNA (Ghosh et al., 2000, Stein et al., 1997, Summerton, 1999). To improve uptake into cells, PMO can be conjugated to an arginine-rich cell-penetrating peptide to produce peptide-PMO (PPMO) (Abes et al., 2006, Deas et al., 2007, Moulton et al., 2004).
PMOs have been reported as effective inhibitors against a number of viruses including hepatitis B and C viruses, influenza virus, picornavirus (FMDV), coronavirus (mouse hepatitis and SARS), calicivirus and West Nile virus (Burrer et al., 2007, Deas et al., 2007, Ge et al., 2006, Heintges et al., 2001, Neuman et al., 2005, Smith et al., 2002, Vagnozzi et al., 2007, Wang et al., 2001, Yang et al., 1999). The titers of feline calicivirus (FCV), a vesivirus, were significantly reduced in cell culture by treatment with PMOs targeted to the upstream region of ORF1 (Stein et al., 2001). Furthermore, this PMO was reported to be effective in treating FCV disease in kittens (Smith et al., 2008). Because the corresponding 5′-end sequence is relatively conserved within the noroviruses, we hypothesized that this region might also function as a target for inhibition of norovirus replication. We designed a panel of PPMOs directed against selected regions of the murine norovirus genome and in addition, developed a PPMO targeted to a 5′-end consensus sequence representing several circulating human norovirus genotypes. We used recently available assays to test the ability of the PPMOs to inhibit viral protein synthesis and replication in their corresponding homologous systems. Our study identified a target site near the 5′-end of the norovirus genome that may prove useful for the inhibition of viral translation and replication by specific PPMOs. The application of PMOs to basic studies of norovirus replication in vitro and in animal models might lead to the evaluation of this class of chemical compounds as antiviral drugs for the prevention and treatment of norovirus disease in humans.
Results
Effect of mouse norovirus PPMOs in cell culture
Prior to our main objective of identifying PPMOs that effectively inhibited MNV-1 replication, it was necessary to first demonstrate that PPMO uptake into RAW 264.7 macrophages was dose responsive, and to determine the maximum dose of each PPMO that was nontoxic to these cells. To fulfill the former, we used flow cytometry to quantitatively measure the uptake of an FITC-labeled PPMO. Cells incubated with 5, 10 20, 40, or 80 μM FITC-PPMO were 1%, 5%, 15% 38%, 63%, and 88% positive for fluorescence, respectively. Although two structural differences, the addition of an FITC label and the 3′ hydroxyl group, prevent a direct comparison with the test PPMOs, these results demonstrate an efficient and dose-responsive uptake of peptide-conjugated PMOs into RAW264.7 macrophages.
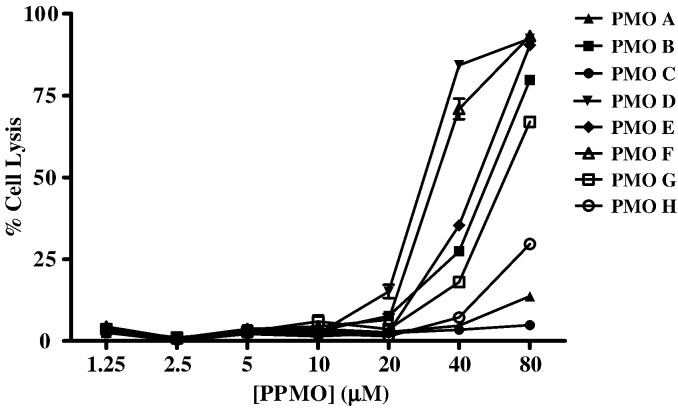
We subsequently identified the maximal nontoxic dose of each MNV-1 PPMO in cytotoxicity assays. All the PPMOs designed for this study (Table 1 and Fig. 1) were nontoxic up to a concentration of 20 μM (Fig. 2 ). Higher concentrations of most PPMOs induced cytotoxicity, with the exception of PPMOs A and C, which remained nontoxic even at concentrations of 80 μM.
Table 1.
PPMO compounds tested aligned to corresponding target sequence
| PPMO (Target)a | Sequenceb (5′-3′) | Length | Target |
|---|---|---|---|
| Human norovirusc | 19TGACGCCATCATCATTCAC1 | ORF1, antisense | |
| Noro 1.1 | CGACGCCATCATCATTCAC | 19 nt | 5′-end of NV genome |
| Murine norovirusd | 32CGCAGAAGATGGCGTTGCCATCCTCATTTCAC1 | ORF1, antisense | |
| PPMO A | GGCGTTGCCATCCTCATTTCAC | 22 nt | 5′-end of MNV genome |
| PPMO B | CGTTGCCATCCTCATTTCAC | 20 nt | 5′-end of MNV genome |
| PPMO C | CGCAGAAGATGGCGTTGCCATCCTC | 25 nt | 5′-end of MNV genome |
| PPMO D | GGCGTTGCCTTCTAGAATTCAC | 22 nt | 5′-end (no AUG) |
| Murine noroviruse | 5064CATCCTCATTCACAAAGACTGCTG5044 | ORF2, antisense | |
| PPMO E | CTCATTCACAAAGACTGCTG | 20 nt | ORF2 of MNV genome |
| PPMO F | CATCCTCATTCACAAAGACTG | 21 nt | ORF2 of MNV genome |
| Murine noroviruse | 7353TCTTTTCTTTGTGGTAGTTAGATGC7377 | 3′-end of ORF3, sense | |
| PPMO G | TTTCTTTGTGGTAGTTAGATGC | 22 nt | 3′-end of MNV genome |
| PPMO H | TCTTTTCTTTGTGGTAGTTAG | 21 nt | 3′-end of MNV genome |
| FITC-PPMO | FITC-GCATAATTCATAACTTAAC | 19 nt | N/Af |
All PMOs synthesized as peptide-PMO (PPMO).
AUG (start codon) is underlined.
Based on prototype Hu/NoV/GI.1/Norwalk virus/1968/US, minus strand.
Based on prototype Mu/NoV/GV/MNV1/2002/US, minus strand.
Based on prototype Mu/NoV/GV/MNV1/2002/US, positive strand.
N/A: not available.
Fig. 2.
The toxicity of MNV-1 PPMOs in RAW264.7 cells. RAW264.7 cells were incubated with various concentrations of each PPMO, ranging from 1.25 to 80 μM (per ml), as indicated, in replicates of 8. Untreated cells served as controls. Following treatment, the viable, adherent cells were fixed and stained with crystal violet. The methanol-dissolved stain was then quantitated on an ELISA plate reader at an absorbance of 570 nm. The average absorbance of the 8 replicates was converted to % cell lysis by the following formula: (untreated − PMO-treated / untreated) × 100. PMO toxicity is expressed as the average percent cell lysis for measurements at each concentration, with error bars indicating standard deviations for quadruplicate measurements.
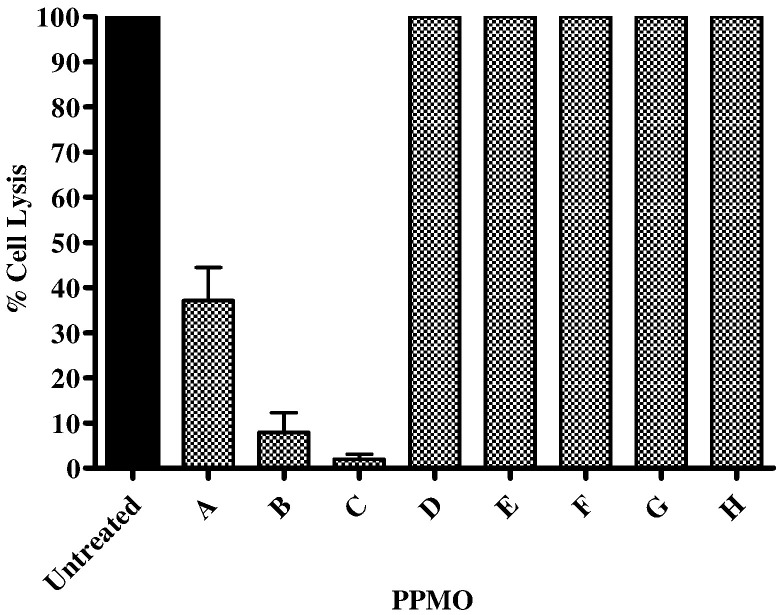
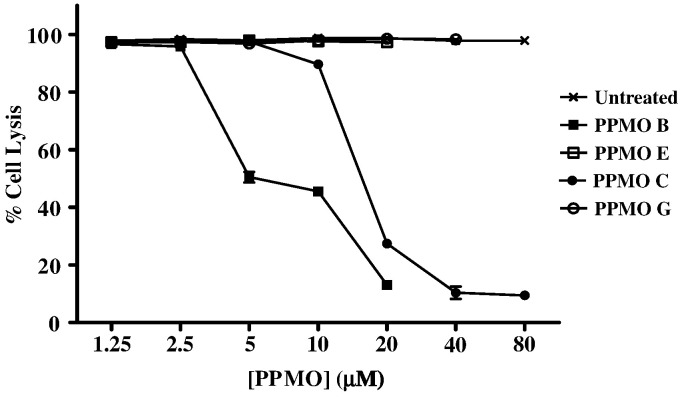
The antiviral effect of each PPMO at its maximum nontoxic concentration (see Fig. 2) was then tested (Fig. 3 ). Two of the PPMOs targeting the 5′-end of MNV-1 ORF1, PPMOs B and C, inhibited virus replication by at least 90%. The other PPMO targeting the 5′ terminus AUG of MNV-1, PPMO A, was significantly less inhibitory. As expected, PPMO D presented no inhibitory effect since the sequence fragment complimentary to the AUG had been replaced in this compound. None of the compounds targeting ORF2, or the 3′-end demonstrated antiviral activity (Fig. 3, MNV PPMOE through H).
Fig. 3.
PPMOs B and C inhibit MNV-1 replication in RAW264.7 cells. Each PPMO was tested for its ability to inhibit MNV-1 replication in a PMO inhibition assay using the highest concentration of the PPMO found to be nontoxic in RAW264.7 cells (PPMOs A: 80 μM, PPMO B, D–G: 20 μM; PPMO C: 80 μM; PPMO H: 40 μM). Cells were treated for 4 h with one of twelve PPMOs, then infected with 2 × 104 PFU (per well) of MNV-1. Three days later, the cells were fixed, stained, and the methanol-dissolved stain was quantitated at an absorbance of 570 nm. For each PPMO assayed, controls included PPMO-treated cells (no virus) and virus-infected cells with no PPMO (untreated). Inhibition of MNV-1 replication is expressed as the average percent cell lysis for each PMO, with error bars indicating standard deviations for quadruplicate measurements.
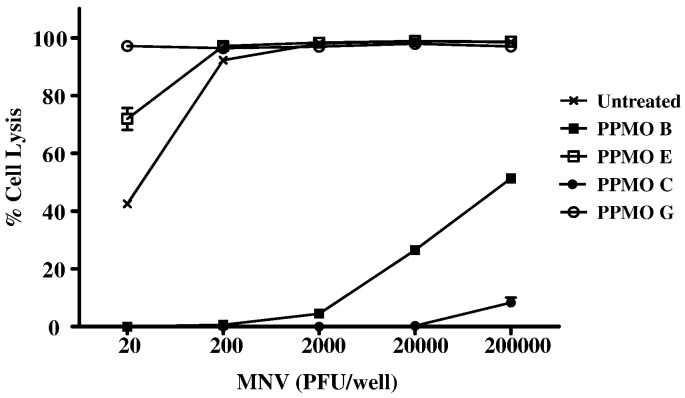
Focusing on MNV-1 PPMOs B and C, we next determined the maximal dose of MNV-1 inhibited by the PPMOs (Fig. 4 ). For this screen, each antiviral compound was tested at its maximal nontoxic concentration (20 μM for PPMO B; 80 μM for PPMO C) against increasing concentrations of MNV-1. To control for sequence specificity, we used compounds E and G as negative controls. These controls, with no demonstrable antiviral activity, were selected based on their similarities to PPMO B or C in nucleotide length and in the presence of a 5′ P007-(pip-PDA) tag and 3′ acetyl group. PPMO C (80 μM) inhibited replication of up to 2 × 105 PFU virus. PPMO B (20 μM) completely inhibit replication of 2 × 103 PFU of MNV-1. The inhibition was sequence-specific, as the control compounds were not effective in preventing virus replication (Fig. 4).
Fig. 4.
Effect of MNV-1 load on the ability of PPMO B and PPMO C to inhibit virus replication in RAW264.7 cells. To determine the highest dose of MNV-1 for which PPMO B and PPMO C can effectively inhibit viral replication in RAW264.7 cells, the PPMO inhibition assay was conducted with serial 10-fold dilutions of MNV-1, ranging from 20 PFU/well up to 2 × 105 PFU/well, as indicated. Cells were treated with 20 μM (per ml) of PPMO B or 80 μM (per ml) of PPMO C (see Fig. 3), and then infected with 101, 102, 103, 104, or 105 PFU of MNV-1. The negative control for PPMO B is PPMO E, and the negative control for PPMO C is PPMO G. The untreated group represents cells that were incubated with DMEM only (no PPMO) and then infected with one of the five MNV-1 doses. Each MNV-1 dose for each PPMO was done in 8 replicates, and the average was used in the following formula to determine % cell lysis: ([PPMO-treated] − [PPMO-treated and virus-infected] / [PPMO-treated]) × 100. Error bars indicate standard deviations for quadruplicate measurements.
In order to compare the potency of PPMOs B and C, we determined the IC50 and IC90 for each compound, using 2 × 104 PFU/well MNV-1, based on the above results. As shown in Fig. 5 , PPMO B was 50% effective at a dose of 5 μM, and 90% effective in inhibiting MNV-1 replication at 20 μM. Higher concentrations of PPMO C were required to inhibit MNV-1 replication. At a concentration of 20 μM, this compound was 70% effective in inhibiting viral replication, and was 90% effective at 40 μM. Therefore, each compound effectively inhibited MNV-1 replication at a dose well below its level of cellular toxicity.
Fig. 5.
Effect of concentration on the ability of PPMO B and PPMO C to inhibit MNV-1 replication in RAW264.7 cells. To determine the lowest effective concentration of PPMO B and PPMO C capable of inhibiting MNV-1 replication in RAW264.7 cells, the PPMO inhibition assay was conducted with various concentrations of PPMOs, ranging from 1.25 to 80 μM (per ml), as indicated. The untreated group represents cells that were incubated with DMEM only (no PPMO) and then infected with MNV-1 (2 × 104 PFU/well). The negative control for PPMO B is PPMO E, and the negative control for PPMO C is PPMO G. Each concentration of each PPMO was tested in 8 replicates, and the average of these replicates was used in the following formula for % cell lysis: ([PMO-treated] − [PMO-treated and virus-infected] / [PMO-treated]) × 100. Error bars indicate standard deviations for quadruplicate measurements.
Effect of Noro 1.1 PPMO on circulating human noroviruses
A relatively limited number of 5′-end sequences were available for circulating noroviruses associated with acute illness, so we first analyzed the 5′-end sequences of four norovirus strains, KL724 (GII.4), CN3050 (GII.4), CN2753 (GII.12), and NR2210 (GII.12), that were collected during an epidemiologic survey of hospitalized infants in the U.S. (Zintz et al., 2005). A comparison of these new 5′-end sequences with those available in the GenBank database showed marked conservation with known sequences. However, noroviruses CN2753 and CN3050 represented unique 5′-end sequences. The alignment (Table 2 ) was used to design a PPMO designated Noro 1.1, that would target a consensus 5′-end sequence representing the predominant circulating genetic clusters of noroviruses (Zintz et al., 2005).
Table 2.
Variation among naturally occurring and laboratory-generated norovirus 5′ ORF1 sequences used as targets for PPMO Noro 1.1
| Norovirus ORF1 target name (nt)a | Variant sequence | Outcome of in vitro assayb |
|---|---|---|
| Consensus sequence | GUGAAUGAUGAUGGCGUCG | |
| PPMO Noro 1.1 [19 mer] | CACTTACTACTACCGCAGC-5′ | |
| Hu/NoV/GI.1/Norwalk virus/1968/US (1) | ––––––––––––––––––A | Inhibition |
| Hu/NoV/GI.2/Southampton/1991/UK (0) | ––––––––––––––––––– | Inhibition |
| Hu/NoV/GI.6/WUG1/2000/JP (1) | –T––––––––––––––––– | Inhibition |
| Hu/No.V/GII.4/Lordsdale/1993/UK (2) | ––––––––A–––––––––U | Inhibition |
| Hu/NoV/GII.12/CN2753/1998/USA (2) | ––––––––G–––––––––U | Inhibition |
| Hu/NoV/GII.4/CN3050/1998/USA (3) | ––––––––A–––U–––––U | Inhibition |
| Laboratory variant 1 (4) | A–––––––A–––U–––––U | Inhibition |
| Laboratory variant 2 (5) | A-A–––––A–––U–––––U | Inhibition |
| Laboratory variant 3 (6) | A-A–––––A–––A––––AU | No inhibition |
| Bo/NoV/GIII.1/Jena/1998/DEU (6) | ––––––––A––CUUU-A–– | No inhibition |
Number of mismatches in nucleotide sequence when compared to Noro 1.1 PPMO sequence. Laboratory variant sequences were designed to minimize number of hydrogen bonds at “hot spots” observed among circulating norovirus and/or at termini of the PMO–target interaction.
Inhibition defined as > 60% reduction of protein synthesis, as defined by luciferase signal.
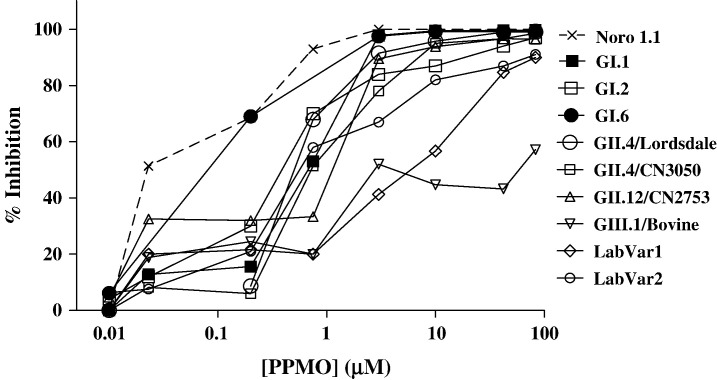
Based on the results presented above with MNV-1 and from previous data in the vesivirus cell culture system (Stein et al., 2001), we examined whether the 5′-end region of the human norovirus genome might serve as an effective target for inhibition by a PMO. The 5′-end sequences of the CN2753 and CN3050 viruses, as well as those from representative prototype noroviruses indicated in Table 2, were engineered as part of a 40 nt leader sequence into a luciferase reporter plasmid, designated pCiNeo-Luc (Fig. 6A). In addition to the naturally occurring 5′-end sequences, we also designed three constructs (designated laboratory variants 1–3) that incorporated varying combinations of the nucleotides that showed evidence for variation among circulating norovirus strains, in order to test the specificity of the PPMO. The 5′-end of the Jena GIII bovine norovirus was also included in the analysis. We examined whether the PPMO Noro 1.1 showed an inhibitory effect on translation of the luciferase protein under control of each of the 5′-end leader sequences (Fig. 6B). The Noro 1.1 compound was added in a range of concentrations (0.01 to 83 μM) to the translation reaction, and the percent inhibition was calculated by comparing the luciferase expression in each reaction with a nontreated control. Fig. 7 shows the percent inhibition as measured by luciferase expression levels in this cell-free system. PPMO Noro 1.1 at a concentration of 3 μM inhibited at least 80% of the luciferase expression from a reporter RNA containing an identical complementary target sequence and in addition, inhibited all other human norovirus ORF1 constructs. The bovine norovirus target never achieved more than 57% inhibition by Noro 1.1 in the range of concentrations tested. Of interest, Noro 1.1 inhibited translation of two artificial laboratory variant targets, LabVar 1 and 2, with 4 and 5 nucleotide changes, respectively, although less efficiently. The 5′-end of the GIII bovine norovirus genome (which contains 6 nucleotide changes from the precise Noro 1.1 target sequence), was the only naturally occurring viral target for which the PPMO failed to inhibit at least 80% of the luciferase translation, even with concentrations greater than 80 μM. Finally, Noro 1.1 completely failed to inhibit translation of the third laboratory variant, which contained 6 nucleotide differences (LabVar3, data not shown).
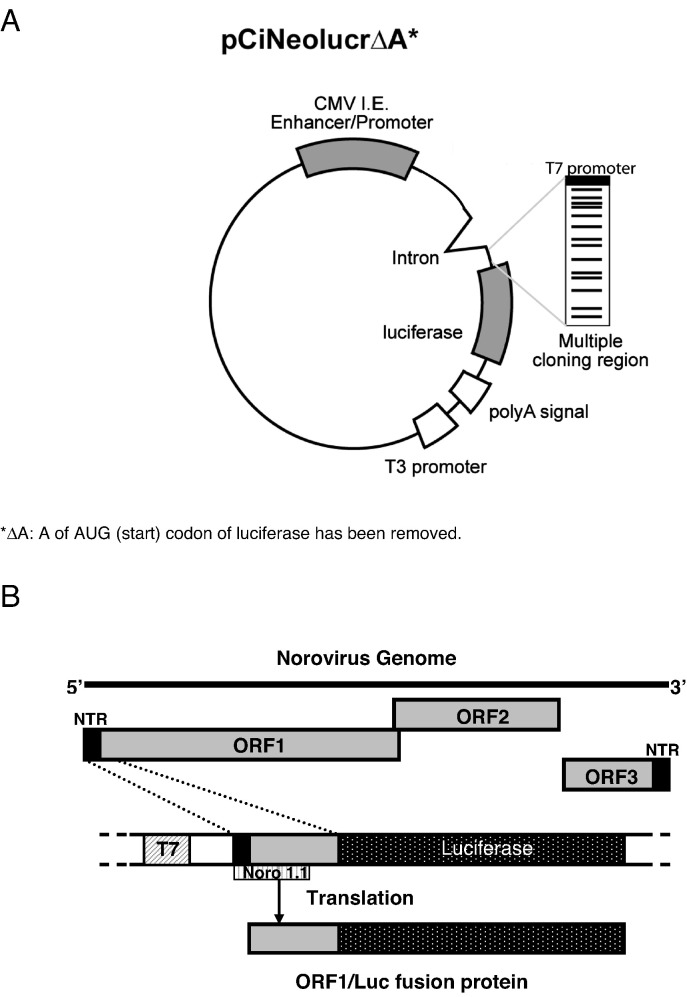
Fig. 6.
Cell-free transcription and translation assay. (A) Plasmid map of pCiNeo containing the luciferase gene. (B) Schematic representation of the norovirus genome and the transcription and translation experiment. The predicted target of Noro 1.1 is shown.
Fig. 7.
Protein translation inhibition by Noro 1.1 PPMO measured by a luciferase reporter assay. The inhibitory activity of Noro 1.1 against several targets (indicated in the graph) containing sequence variations is shown. Only targets that resulted in some degree of luciferase translation inhibition are plotted compared to the bovine strain which represents no inhibition of luciferase activity. These dose–response curves are representative of at least 2 independent repetitions of the protein translation inhibition experiment for each target, carried out by different investigators in independent laboratories.
Effect of Noro 1.1 PMO in cell culture
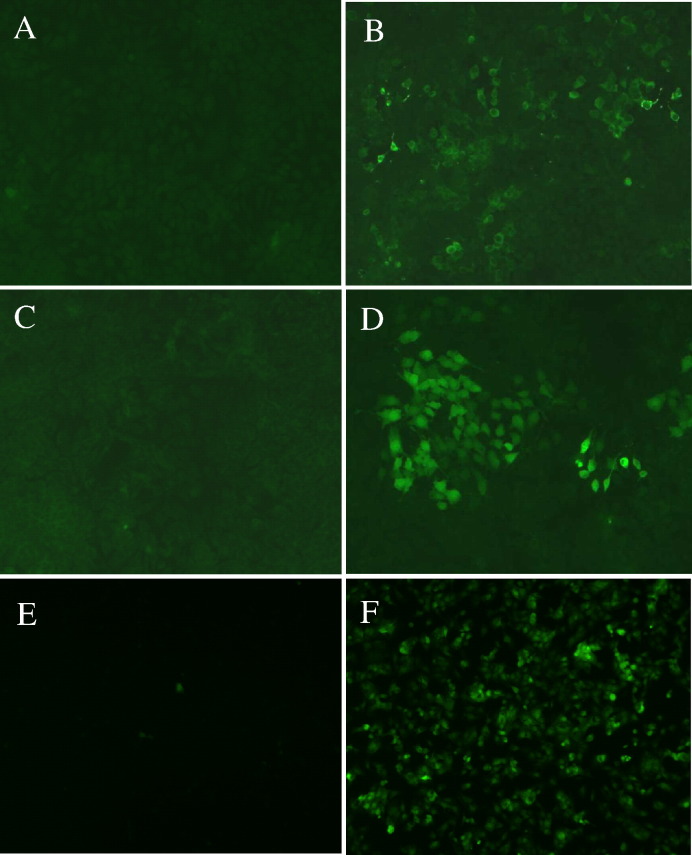
In order to further test the inhibitory effect of Noro 1.1 in a cellular environment, a replicon-bearing cell line expressing Norwalk virus proteins (HG23) was utilized. Approximately 50% of the HG23 cells expressed Norfolk viral proteins (as observed by immunofluorescence detecting the viral capsid protein). This fact, together with the finding that peptide-conjugated PMOs are not efficiently taken up by the HG23 cells, led us to choose electroporation as an efficient means to deliver Noro 1.1 to a majority of these cells. We tested the proportion of cells incorporating an FITC-labeled Noro 1.1 PMO following electroporation in order to examine the efficiency of the transfection method (Fig. 8F). The majority of cells in the monolayer showed a positive immunofluorescence signal compared to the cells receiving the FITC-Noro 1.1 PMO without electroporation (Fig. 8E). Because the replicon cell line was constitutively expressing viral proteins, pre-treatment with PMO was not feasible. For this reason, the highest nontoxic concentration (16 μM) was selected for further experiments. Noro 1.1 PPMO (16 μM) was introduced into cells by electroporation, and the cells were incubated at 37 °C for 5 days. An immunofluorescence assay was then performed with antisera specific for Norwalk virus proteins VP1 and the RNA-dependent RNA polymerase (RdRP) to examine viral protein synthesis. The level of VP1 expression was visibly reduced in the presence of PMO, compared to nontreated control cells (Figs. 8B and D). The expression of the norovirus RdRP protein was also visibly reduced in PMO-treated replicon cells when compared to nontreated cells (Figs. 8A and B). These results are consistent with reduced translation of the viral proteins by Noro 1.1.
Fig. 8.
Reduced norovirus protein translation by Noro 1.1 in a replicon-bearing cell line detected by immunofluorescence with polyclonal antibodies. Norovirus protease-polymerase detection with (A) and without (B) PMO. Norovirus main capsid protein detection with (C) and without (D) PMO. Fluorescein conjugated Noro 1.1 transfection efficiency in replicon-bearing cell line with (F) and without (E) PMO.
Discussion
Human noroviruses are antigenically diverse viruses that cause an acute illness of short duration but with prominent gastrointestinal clinical manifestations (Green, 2007). The presence of norovirus-specific serum antibodies does not correlate with resistance to subsequent norovirus infection and illness (Lindesmith et al., 2003, Parrino et al., 1977), complicating the impetus toward the development of an efficacious vaccine. Thus, novel approaches to controlling the infection are needed. However, in their design, consideration must be given to the fact these viruses are also genetically diverse, with multiple genotypes of noroviruses co-circulating worldwide. In 2006, Zheng et al. proposed a genomic norovirus classification system and a strain nomenclature that divide noroviruses into genogroups I to V, with each genogroup further divided into genetic clusters (or genotypes) (Zheng et al., 2006). The uncorrected pairwise distances between genogroups are unusually large among RNA viruses, ranging from 43% to 58%. Genogroups are then further subdivided into 8 and 17 clusters for NoVGI and NoVGII respectively (Zheng et al., 2006). This scenario makes the selection of an appropriate vaccine candidate, protective correlates aside, even more challenging.
It is in this context that antisense drugs represent viable alternatives for prevention as well as treatment of the disease. Norovirus diarrhea can be a life-threatening and incapacitating disease, especially in elderly or immuno-compromised patients and in addition, appears to be important in severe diarrhea of infants and young children, being second to the rotaviruses in this regard. Its important role in outbreaks of non-bacterial gastroenteritis is well known (Green, 2007). Antisense treatment, if shown to be effective clinically, might therefore be a promising option for various groups of patients, those with or without difficulties in mounting an effective immune response.
A murine norovirus was recently described, providing the first isolate in the genus that can be propagated in cell culture (Wobus et al., 2004). This model offers an opportunity to assess PMO activity in vivo, with a well-characterized host species. The results observed with mouse norovirus PPMOs tested in cell culture showed that two PPMOs targeting the MNV-1 5′ AUG translational start site of ORF1 inhibited MNV-1 replication. These data are consistent with previous PMO studies that proposed translation inhibition as a major mechanism of action (Ghosh et al., 2000, Stein et al., 1997, Summerton, 1999). The failure to translate the nonstructural proteins encoded in ORF1 of the MNV genome would result in the inability of the virus to replicate its RNA genome. However, it is not yet understood why certain specific PPMOs do not inhibit the virus growth (e.g. PPMOs directed against ORF2), and we hypothesize that additional mechanisms might be involved in the reduction of MNV infectivity beyond the inhibition of translation. The PPMOs directed against the 5′-end of the virus might be alternatively interfering with the RNA secondary structure that has been proposed to be essential for viral RNA replication (Simmonds et al., 2008). The disruption of this specific structure by mutagenesis of the MNV genome has been reported to decrease virus titers by 15 to 20 fold (Simmonds et al., 2008). The observation that PPMO A was less effective on virus replication than PPMOs B and C was unexpected. Its sequence completely overlaps that of PPMO B, with two additional 5′ guanine nucleotides and its length is intermediate (22 nt) between PPMO B (20 nt) and PPMO C (25 nt). Comparative studies of the mechanism of action of the PPMOs in this study could provide additional insight into the design of an optimal antisense drug.
Noro 1.1 showed a broad range of inhibition by affecting the protein translation of genetically diverse targets. Noro 1.1 PPMO treatment was associated with in vitro inhibition of translation (≥ 80% inhibition with ≤ 3 μM PMO) of all norovirus targets with up to 3 nucleotide mismatches. Therefore, based on the variability of naturally circulating 5′-end norovirus sequences found in this study and additional sequences submitted to GenBank since the design of this compound (which only show 3 mismatches or less compared to Noro 1.1, Table 3 ), Noro 1.1 has the potential to inhibit translation of every human norovirus genotype described so far. However, Noro 1.1 would likely not inhibit certain animal noroviruses such as MNV-1 (12 mismatches) and bovine norovirus (6 mismatches). Noro 1.1 was a successful inhibitor of translation of the two GII.4 variants tested, achieving 100% inhibition in both cases. This is an important property of Noro 1.1 since GII.4 is the most commonly detected norovirus genotype worldwide (Bull et al., 2006, Kirkwood, 2004). These results suggest that the efficacy of a single PPMO compound targeted to the conserved 5′-end may not be affected by the marked genetic variability of circulating noroviruses in other regions of the genome.
Table 3.
Variation among 5′ ORF1 sequences of each norovirus genogroup and/or genotype available in the GenBank database
| Norovirus ORF1 target name (nt)a | Variant sequence | Outcome (predicted) of in vitro assay |
|---|---|---|
| PPMO Noro 1.1 [19 mer] | CACTTACTACTACCGCAGC-5′ | |
| Hu/NoV/GI.1/Norwalk virus/1968/US (1) | GUGAAUGAUGAUGGCGUCA | Inhibition |
| Hu/NoV/GI.2/Southampton/1991/UK (0) | ––––––––––––––––––– | Inhibition |
| Hu/NoV/GI.4/Chiba 407/1987/JP (0) | ––––––––––––––––––– | (Inhibition) |
| Hu/NoV/GI.5/SzUG1/1997/JP (0) | ––––––––––––––––––– | (Inhibition) |
| Hu/NoV/GI.6/WUG1/2000/JP (1) | –T––––––––––––––––– | Inhibition |
| Hu/NoV/GI ?/Otofuke/1979/JPb (1) | ––––––––––––––––––A | (Inhibition) |
| Hu/NoV/GII.1/Hawaii/1971/USA (2) | ––––––––A–––––––––U | (Inhibition) |
| Hu/NoV/GII.2/Snow Mountain/1976/USA (2) | ––––––––A–––––––––U | (Inhibition) |
| Hu/NoV/GII.3/SU201/1997/JP (2) | ––––––––A–––––––––U | (Inhibition) |
| Hu/No.V/GII.4/Lordsdale/1993/UK (2) | ––––––––A–––––––––U | Inhibition |
| Hu/NoV/GII.4/CN3050/1998/USA (3) | ––––––––A–––U–––––U | Inhibition |
| Hu/NoV/GII.6/SU17/1997/JP (2) | ––––––––A–––––––––U | (Inhibition) |
| Hu/NoV/GII.8/SU25/1997/JP (2) | ––––––––A–––––––––U | (Inhibition) |
| Hu/NoV/GII.10/Mc37/2000/THA (2) | ––––––––A–––––––––U | (Inhibition) |
| Hu/NoV/GII.12/Hiroshima/1999/JP (2) | ––––––––A–––––––––U | (Inhibition) |
| Hu/NoV/GII.12/CN2753/1998/USA (2) | ––––––––G–––––––––U | Inhibition |
| Hu/NoV/GII.16/Neustrelitz260/2000/DE (2) | ––––––––A–––––––––U | (Inhibition) |
| Hu/NoV/GII.?/Guangzhou/2001/CHN (2) | ––––––––A–––––––––U | (Inhibition) |
| Bo/NoV/GIII.1/Jena/1998/DEU (6) | ––––––––A––CUUU-A–– | No inhibition |
| Mu/NoV/GV/MNV-1/2002/USA (12) | –––––ATGA–GAT–GCAAC | (No inhibition) |
| Bo/NoV/BEC/NB/80/US (13) | ––––T–T–ATTATAGAGAG | (No inhibition) |
Number of mismatches in nucleotide sequence when compared to Noro 1.1 PPMO sequence.
Unclassified strain.
Noro 1.1 was also shown to be effective in our in vitro reporter assay well below the toxic concentrations of PPMOs described in cell culture as well as in in vivo studies (Smith et al., 2008, Vagnozzi et al., 2007). Only 3 μM of Noro 1.1 was needed to inhibit 80% or more of the total protein translation of representative circulating norovirus variants, as measured in a luciferase reporter assay system. Greater concentrations were needed to achieve similar inhibition rates only when testing artificially diverse “laboratory variants” or bovine norovirus. In any case, the PPMO IC50 value was usually between 150 and 300 nM, comparable to previously reported values (Patel et al., 2008). In addition, the murine norovirus PPMO experiments showed that 20 μM of compound, a concentration similar to that previously reported in PMO studies, is capable of inhibiting the replication of a wide range of virus titers, showing an antisense effect even when 2 × 105 PFU of MNV were in the presence of an anti 5′-end PPMO (PPMO C) (Burrer et al., 2007, Ge et al., 2006, Patel et al., 2008).
The inhibition of protein translation by Noro 1.1 was further confirmed in the experiments carried out in cell culture. Noro 1.1 effectively inhibited norovirus protein translation in HG23 cells as evidenced by immunofluorescence. This effect was observed even after a 5-day incubation period with Noro 1.1 at a concentration of 16 μM demonstrating the stability of the compound when incubated at 37 °C for long periods of time. This stability could potentially reduce the number of doses needed for successful treatment. The HG23 clone expressing the Norwalk virus replicon constitutively expresses Norwalk virus RNA and proteins in all cells. It has been used as a screen for antiviral agents, such as interferon alpha (Chang and George, 2007). However, treatment of HG23 with as high as 20 units of interferon alpha, a concentration ten times the ED50, significantly reduced levels of polymerase protein, yet approximately 100 copies of RNA genome per cell were retained. With this antiviral agent, reductions in viral polymerase levels did not directly correlate with a reduction in viral RNA. We, too, found a significant reduction in Norwalk proteins after treatment of HG23 with Noro 1.1, yet did not see a significant reduction in RNA with this dose of PMO (data not shown). Likely, viral polymerase at levels below the limit of detection of IF remained, and as seen with IFNa, a reduction in viral RNA may require higher concentrations of the compound. How this observation relates to a natural infection awaits cell culture methods for propagating human noroviruses. Indirect evidence that MNV-1 PPMOs B and C effectively inhibited viral genome replication in the context of virus infection was demonstrated by the fact that infection of control-treated RAW264.7 cells with an m.o.i. as low as 0.0001 (20 PFU/2 × 105 cells) produced 100% CPE within 3 days, while PPMO B or C treatment significantly inhibited CPE resulting from 1000 times this dose (m.o.i. of 0.1) during this time.
A recent study using PPMOs to inhibit the replication of feline calicivirus (FCV), a norovirus-related virus, showed that the antiviral effect previously observed in vitro could be translated into a clinical application. The antiviral strategy used was similar, testing a FCV-PMO that masked the first AUG of the virus genome. This compound was initially shown to reduce FCV viral titers, in a dose-dependent manner, by 70% when 20 μM of the PMO was present in cell culture, which is comparable to that used in our in vitro experiments. The FCV-PMO was then tested in vivo, administered subcutaneously to kittens during an outbreak of FCV disease, and 80% of treated kittens (47/59) survived severe FCV infection, compared with 10% of non-treated kittens (3/31) (Smith et al., 2008). The dosage used to achieve these results in vivo was estimated to be approximately 1 μM (2 mg/kg). This study implicates that the current PPMO doses used with murine and human noroviruses “in vitro” might be translated into a successful clinical application. The use of such an antiviral compound could represent a huge advantage to prevent and control gastroenteritis outbreaks. While the duration of the clinical symptoms caused by norovirus is usually short and self-limited, shedding of the virus in stool commonly lasts for up to a month, which greatly increases the possibility of transmission of the disease (Green, 2007). Thus, the administration of an antiviral to sick patients and their contacts might prevent the spread of the disease, especially in sensitive settings such as hospital wards, military ships, and nursing homes where it might also prevent death of elderly patients.
Future studies include testing PPMO compounds in the MNV mouse model. It will also be important to investigate whether escape variants might arise under the selective pressure of a PPMO. Further testing of PPMOs in animal models will address important questions regarding the application of norovirus-specific PPMO drugs to the prevention and control of disease in humans.
Materials and methods
Cells
The macrophage cell line RAW264.7 (ATCC TIB-71) was used to propagate MNV-1. The cells were cultured in Dulbecco's modified Eagle's medium (DMEM) with 4 mM l-glutamine, 1.5 g/L sodium bicarbonate, and 10% fetal bovine serum. The norovirus replicon cell line, HG23, expressing the Norwalk virus RNA replicon, was derived from the human liver cell line, Huh-7 (Chang et al., 2006). The replicon cells were maintained in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum and 0.25 mg/ml of neomycin.
Viruses and cells
Murine norovirus strain MNV-1.CW1 (Wobus et al., 2004) was provided by Herbert W. Virgin (Washington University School of Medicine, St. Louis, MO). The virus was received as a titered stock (4 × 107 PFU/ml) and used directly in the assays in this study. The RAW 264.7 cells were obtained from American Type Culture Collection (Manassas, VA) and maintained in Dulbecco's Modified Essential Medium (DMEM) with 10% fetal bovine serum and antibiotics (penicillin, 250 units/ml and streptomycin 250 μg/ml, complete medium).
PMO sequences
NeuGene® morpholino oligomers (PMOs) were synthesized at AVI BioPharma Inc (Corvallis, OR). The PMOs were constructed with an arginine-rich peptide (CytoPorter™) conjugated to the 5′-end to facilitate cellular uptake, and an acetyl group at the 3′-end (PPMO). Table 1 shows the PPMO compounds tested in this study. Eight different PPMOs were generated to target the 5′-end of ORF 1 or ORF2, or the 3′-end of the MNV-1 genome (see Fig. 1). An additional sequence-nonspecific FITC-labeled PPMO was generated to quantitate cellular uptake and intracellular stability of the compounds. A single PPMO was designed to target the 5′-end of human noroviruses ORF 1 (Noro 1.1, see Table 1). Noro 1.1 was also synthesized by Gene Tools (Philomath, OR) as a nonconjugated PMO and a fluorescein-conjugated PMO for use in the norovirus replicon cell culture studies. All PPMOs and PMOs were resuspended in sterile distilled water and further diluted in DMEM to the indicated concentrations.
Cellular uptake of a FITC-labeled PPMO
A 19-residue, peptide-conjugated PMO with an additional 5′-fluorescein (FITC) tag was used to quantitate the uptake of PPMOs in RAW264.7 cells. The cells were seeded at 2 × 106 cells per well of a 6-well plate and incubated overnight at 37 °C. The following day, the cells were rinsed twice with DMEM, and treated with 2.5, 5, 10, 20, 40, or 80 μM of FITC-PPMO for 4 h at 37 °C. The PPMO was removed, the cells were rinsed twice with plain DMEM, and the cells were incubated in complete medium overnight at 37 °C. The following day, the cells were harvested, washed with BD Pharmingen stain buffer (BD Biosciences, San Diego, CA), resuspended in 0.5 ml of stain buffer, and passed through a 40 μm cell strainer. The samples were then acquired on a flow cytometer (FACStar Plus, Becton Dickinson) to detect FITC staining.
Mouse norovirus PPMO toxicity assay
A toxicity assay was developed in order to examine whether treatment of RAW264.7 cells with MNV-1-specific PPMOs caused nonspecific cytotoxic effects. RAW264.7 cells (2 × 105 cells/well) were seeded into flat-bottom 96-well plates. The following day, the cells were rinsed twice with plain DMEM and incubated for 4 h with varying concentrations of PPMO diluted in plain DMEM. The concentrations of each PPMO tested (in replicates of eight) were 1.25, 2.5, 5, 10, 20, 40, and 80 μM. Following incubation, the cells were rinsed twice with DMEM, complete medium was added, and the plates were incubated at 37 °C for 24 h. The following day, the plates were rinsed twice with warm PBS, and the cells were fixed with 10% buffered formalin (Fisher Scientific, Fairlawn, NJ) overnight. The plates were then rinsed with tap water and dried overnight. Following staining of the cell monolayer with 0.4% crystal violet, the plates were rinsed extensively and left to dry at room temperature. The following day, 200 μl of 100% methanol was added per well, allowed to incubate at room temperature for 1 h, and the methanol-dissolved stain was transferred to the wells of a clean 96-well plate. The plates were then read on an ELISA plate reader (EL340, Bio-Tek Instruments) at an absorbance of 570 nm. The average absorbance of the 8 replicates was converted to percent cell lysis by the following formula: % cell lysis = untreated] − [PMO-treated] / [untreated] × 100.
Mouse norovirus PPMO inhibition assay
Each PPMO was tested for its ability to inhibit MNV-1 replication in a PPMO inhibition assay using the highest concentration of the PPMO found to be nontoxic in RAW264.7 cells (see above). Briefly, 2 × 105 RAW264.7 cells/well were seeded into flat-bottom 96-well plates. The following day, the cells were rinsed twice with plain DMEM, and the PPMO was added at the desired concentration to eight replicate wells. Following treatment with PPMO for 4 h, the cells were rinsed twice with plain DMEM, and 50 μl of complete medium containing 2 × 104 PFU of MNV-1 was added to each well and allowed to incubate for 2 h at 37 °C. The inoculum was removed, and following one rinse with complete medium, 200 μl of complete media was added per well and the plates were incubated for 3 days at 37 °C. Controls included PPMO-treated cells without virus and virus-infected cells with no PPMO (untreated). The plates were rinsed twice with PBS, fixed with 10% buffered formalin, stained with crystal violet, and the intensity of the methanol-dissolved stain was quantitated as above. The average absorbance at 570 nm of the 8 replicates was converted to percent cell lysis using the following formula: % cell lysis = ([PMO-treated] − [PMO-treated and virus-infected] / [PMO-treated]) × 100.
Virus dose–response in PPMO inhibition assay
The PPMO inhibition assay described above was carried out to determine the highest dose of MNV-1 for which PPMO B and PPMO C could effectively inhibit viral replication. Serial 10-fold dilutions of MNV-1 (ranging from 20 PFU/well up to 2 × 105 PFU/well, as indicated) were used to infect RAW264.7 cells. Briefly, cells were treated with 20 μM of PPMO B or 80 μM of PPMO C, and then infected (in replicates of eight) with one of the five doses of MNV-1. PPMO E and PPMO G were used as negative controls. The untreated group represents cells that were incubated with DMEM only (no PPMO) and then infected with one of the five MNV-1 doses. Percent cell lysis was quantitated with the following formula: % cell lysis (for each virus dose) = ([PMO-treated] − [PMO-treated and virus-infected] / [PMO-treated]) × 100.
PPMO dose–response in PPMO inhibition assay
A PPMO inhibition assay was carried out to determine the lowest effective concentration of PPMO B and PPMO C capable of inhibiting MNV-1 replication in RAW264.7 cells. The virus concentration remained constant (2 × 104 PFU MNV-1 per well) while various concentrations of PPMO B or C, ranging from 1.25 to 80 μM (in replicates of eight) were used. The negative controls were PPMOs E and G. Percent cell lysis was quantitated using the following formula: % cell lysis (for each PMO dose) = ([PMO-treated] − [PMO-treated and virus-infected] / [PMO-treated]) × 100.
Consensus sequence analysis of the 5′-end region of human norovirus genomes
In order to design a PMO targeted to human noroviruses, available 5′-end genome sequences were aligned and examined for conserved regions. In addition, we selected representative circulating norovirus specimens from an epidemiological study that was conducted at three hospital sites across the U.S. from November 1997 to December 1999 (Zintz et al., 2005) for partial genomic sequence analysis. Stool samples were collected within 24 h of admission, frozen and sent to the Center for Pediatric Research (Norfolk, Virginia) where they were stored at −70 °C until screening by RT-PCR. One-hundred thirty-five children with norovirus illness were identified and portions of the norovirus genome were sequenced from each infecting strain to confirm the norovirus diagnosis and examine genetic diversity. Norovirus variants (NR2210, KL724, CN2753, CN3050) representing the most divergent phylogenetic clusters in the collection, were selected for 5′-end sequence analysis. Briefly, a 5′-end RACE kit (Roche Applied Science, Indianapolis, IN) was used according to the manufacturer's protocols, employing primers deduced from related viruses belonging to the same genotype as the variant (primer sequences available upon request).
Luciferase reporter assay
The ability of the PPMO Noro 1.1 to target a panel of diverse 5′-end sequences was evaluated in an in vitro reporter assay. Briefly, each reporter plasmid was engineered to include a 5′-end sequence of interest (summarized in Table 2) as a translational “leader” sequence (approximately 40 nt in length) fused in frame immediately upstream of the firefly luciferase gene in which the first A of the AUG codon was removed. The leader sequences were synthesized as oligonucleotides with engineered restriction enzyme sites at the 5′- and 3′-ends to facilitate cloning into the NheI and SalI sites of the expression plasmid, pCiNeo (Promega, Madison, WI). The expression cassette in the vector was organized as follows: CMV promoter–intervening intron–T7 promoter–leader sequence–luciferase gene–poly-A signal (Fig. 6A). Each construct was verified by sequence analysis. The resulting plasmid DNA was digested with a restriction enzyme (NotI), purified, and RNA was transcribed from the plasmid with T7 polymerase (Megascript, Ambion, Austin, TX). The transcribed RNA was purified and run in an agarose gel to verify quality and size. In addition, the RNA was quantitated by UV spectroscopy and 6 nM solutions were prepared for use in translation assays. The translation reaction was carried out in a rabbit reticulocyte lysate (Promega, Madison, WI). To assess the effect of the Noro 1.1 PPMO on translation, a reaction included the following: 3 μl PPMO (final concentration 0.01-83 μM), 3 μl RNA (final concentration 1 nM), 1 μl amino acid mix, and 11 μl nuclease-treated rabbit reticulocyte lysate. This reaction was incubated for 75 min at 37 °C and terminated by chilling on ice. Ten microliters of this reaction was then added to 50 μl of luciferase assay reagent (Promega, Madison, WI), mixed gently 5 times and incubated for 30 s at room temperature, after which light emission was measured (Dynex MLX luminometer, Dynex, Chantilly, VA). Controls for this experiment included (i) a nonspecific PMO directed against ORF3 of vesiviruses (Stein et al., 2001), (ii) non-treated (no PMO), and (iii) a specimen without RNA. The Noro 1.1 PPMO was assayed within a range of concentrations that were effective and nontoxic in previous cell culture studies. Each assay was performed at least twice and constructs were analyzed by two independent investigators in two separate laboratories to verify reproducibility. Percent inhibition was determined by comparing the reduction in the firefly luciferase signal of different concentrations of a specific PPMO with the signal obtained without PPMO in the well.
PMO inhibition of norovirus in a replicon cell line
The cloned cell line HG23 expressing self-replicating norovirus RNA (norovirus replicon) was used to test Noro 1.1 PMO activity in cell culture (Chang et al., 2006). First, the degree of incorporation of PMO into the replicon-bearing cell line was assayed using a fluorescein conjugated Noro 1.1 PMO (FITC-Noro 1.1). In order to determine the best electroporation conditions for the HG23 cell line, an optimization kit was utilized to test several electroporation solutions combined with different nucleofection programs (Nucleofector, Amaxa, Gaithersburg, MD). Electroporation solution “L” combined with the nucleofection program “A020” was selected as the best compromise between cell viability and PMO incorporation. A range of concentrations of FITC-Noro 1.1 (1 to 16 μM) was electroporated (Nucleofector, Amaxa, Gaithersburg, MD) into the replicon cell line, and cell monolayers were observed 24 h later for fluorescence to determine the toxicity and optimal concentration of PMO. Based on this experiment, 16 μM (maximum concentration and nontoxic) of the Noro 1.1 PMO was electroporated into 2 × 106 replicon-bearing cells, which were then seeded into 100 mm cell culture plates. The monolayers were incubated for 5 days, and then fixed with ice-cold methanol for analysis by immunofluorescence with antibodies raised in guinea pigs against the Norwalk virus VP1 and polymerase proteins as previously described (Chang et al., 2006). In order to assess the effect of such Noro 1.1 treatment on the levels of RNA in the replicon-bearing cells, the RNA present in treated and untreated samples was quantified using a single-step quantitative real-time RT-PCR. One Taq-Man probe and two primers that specifically amplified the GI genotype (the norovirus genotype present in the replicon cell line) were utilized (Liu and Moe, 2006). Briefly, 5 μl of RNA extracted from 2 × 106 cells/ml using a Qiagen RNA extraction kit (Qiagen, Valencia, CA) was added to an RT-PCR mix (Brilliant II q RT-PCR core reagent kit, Stratagene, La Jolla, CA). The reaction conditions were as follows: 45 °C for 30 min, 95 °C for 10 min, followed by 45 cycles of 95 °C for 30 s, 50 °C for 1 min and 72 °C for 30 s in an ABI 7900 HT real time detector (Applied Biosystems, Foster City, CA). An appropriate standard curve was included in each run.
Acknowledgments
We acknowledge the generosity of Dr. Skip Virgin in providing MNV-1 stocks. The authors would like to thank Dr. Mary Staat and Dr. Parvin Azimi for their work in the calicivirus epidemiologic study. This work was supported by grant 02820-4 from the Thrasher Research Fund, Salt Lake City, UT, awarded to K.B, D.O.M, and A.E.C. This research was also supported [in part] by the Intramural Research Program of the NIH, NIAID.
References
- Abes S., Moulton H.M., Clair P., Prevot P., Youngblood D.S., Wu R.P., Iversen P.L., Lebleu B. Vectorization of morpholino oligomers by the (R-Ahx-R)4 peptide allows efficient splicing correction in the absence of endosomolytic agents. J. Control. Release. 2006;116(3):304–313. doi: 10.1016/j.jconrel.2006.09.011. [DOI] [PubMed] [Google Scholar]
- Bertolotti-Ciarlet A., Crawford S.E., Hutson A.M., Estes M.K. The 3¢ end of Norwalk virus mRNA contains determinants that regulate the expression and stability of the viral capsid protein VP1: a novel function for the VP2 protein. J. Virol. 2003;77(21):11603–11615. doi: 10.1128/JVI.77.21.11603-11615.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull R.A., Tu E.T., McIver C.J., Rawlinson W.D., White P.A. Emergence of a new norovirus genotype II.4 variant associated with global outbreaks of gastroenteritis. J. Clin. Microbiol. 2006;44(2):327–333. doi: 10.1128/JCM.44.2.327-333.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrer R., Neuman B.W., Ting J.P., Stein D.A., Moulton H.M., Iversen P.L., Kuhn P., Buchmeier M.J. Antiviral effects of antisense morpholino oligomers in murine coronavirus infection models. J. Virol. 2007;81(11):5637–5648. doi: 10.1128/JVI.02360-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang K.O., George D.W. Interferons and ribavirin effectively inhibit Norwalk virus replication in replicon-bearing cells. J. Virol. 2007;81(22):12111–12118. doi: 10.1128/JVI.00560-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang K.O., Sosnovtsev S.V., Belliot G., King A.D., Green K.Y. Stable expression of a Norwalk virus RNA replicon in a human hepatoma cell line. Virology. 2006;353(2):463–473. doi: 10.1016/j.virol.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Chaudhry Y., Skinner M.A., Goodfellow I.G. Recovery of genetically defined murine norovirus in tissue culture by using a fowlpox virus expressing T7 RNA polymerase. J. Gen. Virol. 2007;88(Pt 8):2091–2100. doi: 10.1099/vir.0.82940-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deas T.S., Bennett C.J., Jones S.A., Tilgner M., Ren P., Behr M.J., Stein D.A., Iversen P.L., Kramer L.D., Bernard K.A., Shi P.Y. In vitro resistance selection and in vivo efficacy of morpholino oligomers against West Nile virus. Antimicrob. Agents. Chemother. 2007;51(7):2470–2482. doi: 10.1128/AAC.00069-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estes M.K., Ball J.M., Guerrero R.A., Opekun A.R., Gilger M.A., Pacheco S.S., Graham D.Y. Norwalk virus vaccines: challenges and progress. J. Infect. Dis. 2000;181(Suppl 2):S367–373. doi: 10.1086/315579. [DOI] [PubMed] [Google Scholar]
- Ge Q., Pastey M., Kobasa D., Puthavathana P., Lupfer C., Bestwick R.K., Iversen P.L., Chen J., Stein D.A. Inhibition of multiple subtypes of influenza A virus in cell cultures with morpholino oligomers. Antimicrob. Agents. Chemother. 2006;50(11):3724–3733. doi: 10.1128/AAC.00644-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh C., Stein D., Weller D., Iversen P. Evaluation of antisense mechanisms of action. Methods. Enzymol. 2000;313:135–143. doi: 10.1016/s0076-6879(00)13008-3. [DOI] [PubMed] [Google Scholar]
- Glass R.I., Noel J., Ando T., Fankhauser R., Belliot G., Mounts A., Parashar U.D., Bresee J.S., Monroe S.S. The epidemiology of enteric caliciviruses from humans: a reassessment using new diagnostics. J. Infect. Dis. 2000;181(Suppl 2):S254–261. doi: 10.1086/315588. [DOI] [PubMed] [Google Scholar]
- Green K.Y. Caliciviridae: the Noroviruses. In: Knipe D.M., editor. Fifth ed. Vol. I. Lippincott Williams and Wilkins, a Wolters Kluwer Business; Philadelphia: 2007. pp. 949–979. (Fields Virology). 2 vols. [Google Scholar]
- Heintges T., Encke J., zu Putlitz J., Wands J.R. Inhibition of hepatitis C virus NS3 function by antisense oligodeoxynucleotides and protease inhibitor. J. Med. Virol. 2001;65(4):671–680. doi: 10.1002/jmv.2089. [DOI] [PubMed] [Google Scholar]
- Iversen P.L. Antisense Drug Technology. Marcel Dekker; New York: 2001. Phosphorodiamidate morpholino oligomers; pp. 375–389. [Google Scholar]
- Kirkwood C. Viral gastroenteritis in Europe: a new norovirus variant? Lancet. 2004;363(9410):671–672. doi: 10.1016/S0140-6736(04)15661-4. [DOI] [PubMed] [Google Scholar]
- Lindesmith L., Moe C., Marionneau S., Ruvoen N., Jiang X., Lindblad L., Stewart P., LePendu J., Baric R. Human susceptibility and resistance to Norwalk virus infection. Nat. Med. 2003;9(5):548–553. doi: 10.1038/nm860. [DOI] [PubMed] [Google Scholar]
- Liu, P., Moe, C., 2006. Real-time RT-PCR detection of Norwalk virus.
- Moulton H.M., Nelson M.H., Hatlevig S.A., Reddy M.T., Iversen P.L. Cellular uptake of antisense morpholino oligomers conjugated to arginine-rich peptides. Bioconjug. Chem. 2004;15(2):290–299. doi: 10.1021/bc034221g. [DOI] [PubMed] [Google Scholar]
- Mumphrey S.M., Changotra H., Moore T.N., Heimann-Nichols E.R., Wobus C.E., Reilly M.J., Moghadamfalahi M., Shukla D., Karst S.M. Murine norovirus 1 infection is associated with histopathological changes in immunocompetent hosts, but clinical disease is prevented by STAT1-dependent interferon responses. J. Virol. 2007;81(7):3251–3263. doi: 10.1128/JVI.02096-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman B.W., Stein D.A., Kroeker A.D., Churchill M.J., Kim A.M., Kuhn P., Dawson P., Moulton H.M., Bestwick R.K., Iversen P.L., Buchmeier M.J. Inhibition, escape, and attenuated growth of severe acute respiratory syndrome coronavirus treated with antisense morpholino oligomers. J. Virol. 2005;79(15):9665–9676. doi: 10.1128/JVI.79.15.9665-9676.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrino T.A., Schreiber D.S., Trier J.S., Kapikian A.Z., Blacklow N.R. Clinical immunity in acute gastroenteritis caused by Norwalk agent. N. Engl. J. Med. 1977;297(2):86–89. doi: 10.1056/NEJM197707142970204. [DOI] [PubMed] [Google Scholar]
- Patel D., Opriessnig T., Stein D.A., Halbur P.G., Meng X.J., Iversen P.L., Zhang Y.J. Peptide-conjugated morpholino oligomers inhibit porcine reproductive and respiratory syndrome virus replication. Antiviral. Res. 2008;77(2):95–107. doi: 10.1016/j.antiviral.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmonds P., Karakasiliotis I., Bailey D., Chaudhry Y., Evans D.J., Goodfellow I.G. Bioinformatic and functional analysis of RNA secondary structure elements among different genera of human and animal caliciviruses. Nucleic. Acids. Res. 2008 doi: 10.1093/nar/gkn096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A.W., Matson D.O., Stein D.A., Skilling D.E., Kroeker A.D., Berke T., Iversen P.L. Antisense treatment of Caliciviridae: an emerging disease agent of animals and humans. Curr. Opin. Mol. Ther. 2002;4(2):177–184. [PubMed] [Google Scholar]
- Smith A.W., Iversen P.L., O'Hanley P.D., Skilling D.E., Christensen J.R., Weaver S.S., Longley K., Stone M.A., Poet S.E., Matson D.O. Virus-specific antiviral treatment for controlling severe and fatal outbreaks of feline calicivirus infection. Am. J. Vet. Res. 2008;69(1):23–32. doi: 10.2460/ajvr.69.1.23. [DOI] [PubMed] [Google Scholar]
- Sosnovtsev S.V., Belliot G., Chang K.O., Prikhodko V.G., Thackray L.B., Wobus C.E., Karst S.M., Virgin H.W., Green K.Y. Cleavage map and proteolytic processing of the murine norovirus nonstructural polyprotein in infected cells. J. Virol. 2006;80(16):7816–7831. doi: 10.1128/JVI.00532-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein D., Foster E., Huang S.B., Weller D., Summerton J. A specificity comparison of four antisense types: morpholino, 2¢-O-methyl RNA, DNA, and phosphorothioate DNA. Antisense. Nucleic. Acid. Drug. Dev. 1997;7(3):151–157. doi: 10.1089/oli.1.1997.7.151. [DOI] [PubMed] [Google Scholar]
- Stein D.A., Skilling D.E., Iversen P.L., Smith A.W. Inhibition of Vesivirus infections in mammalian tissue culture with antisense morpholino oligomers. Antisense. Nucleic. Acid. Drug. Dev. 2001;11(5):317–325. doi: 10.1089/108729001753231696. [DOI] [PubMed] [Google Scholar]
- Summerton J. Morpholino antisense oligomers: the case for an RNase H-independent structural type. Biochim. Biophys. Acta. 1999;1489(1):141–158. doi: 10.1016/s0167-4781(99)00150-5. [DOI] [PubMed] [Google Scholar]
- Thackray L.B., Wobus C.E., Chachu K.A., Liu B., Alegre E.R., Henderson K.S., Kelley S.T., Virgin H.W.T. Murine noroviruses comprising a single genogroup exhibit biological diversity despite limited sequence divergence. J. Virol. 2007;81(19):10460–10473. doi: 10.1128/JVI.00783-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagnozzi A., Stein D.A., Iversen P.L., Rieder E. Inhibition of foot-and-mouth disease virus infections in cell cultures with antisense morpholino oligomers. J. Virol. 2007;81(21):11669–11680. doi: 10.1128/JVI.00557-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A., Cheung P.K., Zhang H., Carthy C.M., Bohunek L., Wilson J.E., McManus B.M., Yang D. Specific inhibition of coxsackievirus B3 translation and replication by phosphorothioate antisense oligodeoxynucleotides. Antimicrob. Agents. Chemother. 2001;45(4):1043–1052. doi: 10.1128/AAC.45.4.1043-1052.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward V.K., McCormick C.J., Clarke I.N., Salim O., Wobus C.E., Thackray L.B., Virgin H.W.T., Lambden P.R. Recovery of infectious murine norovirus using pol II-driven expression of full-length cDNA. Proc. Natl. Acad. Sci. U. S. A. 2007;104(26):11050–11055. doi: 10.1073/pnas.0700336104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wobus C.E., Karst S.M., Thackray L.B., Chang K.O., Sosnovtsev S.V., Belliot G., Krug A., Mackenzie J.M., Green K.Y., Virgin H.W. Replication of Norovirus in cell culture reveals a tropism for dendritic cells and macrophages. PLoS. Biol. 2004;2(12):e432. doi: 10.1371/journal.pbio.0020432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wobus C.E., Thackray L.B., Virgin H.W.T. Murine norovirus: a model system to study norovirus biology and pathogenesis. J. Virol. 2006;80(11):5104–5112. doi: 10.1128/JVI.02346-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Yao J., Deng L. Inhibitory effect of phosphorothioae oligodeoxynucleotides on HBV replication and synthesis of antigen in vitro. Zhonghua. Yi. Xue. Za. Zhi. 1999;79(11):857–859. [PubMed] [Google Scholar]
- Zheng D.P., Ando T., Fankhauser R.L., Beard R.S., Glass R.I., Monroe S.S. Norovirus classification and proposed strain nomenclature. Virology. 2006;346(2):312–323. doi: 10.1016/j.virol.2005.11.015. [DOI] [PubMed] [Google Scholar]
- Zintz C., Bok K., Parada E., Barnes-Eley M., Berke T., Staat M.A., Azimi P., Jiang X., Matson D.O. Prevalence and genetic characterization of caliciviruses among children hospitalized for acute gastroenteritis in the United States. Infect. Genet. Evol. 2005;5(3):281–290. doi: 10.1016/j.meegid.2004.06.010. [DOI] [PubMed] [Google Scholar]