Abstract
Posttranslational protein modification by the Small Ubiquitin-like MOdifiers (SUMO) is involved in many cellular functions including organization of nuclear structures and chromatin, transcriptional regulation, and nucleo-cytoplasmic transport. Both genetic and biochemical studies indicate that the SUMO modification pathway plays an important role in proper cell cycle control, especially in the normal progression of mitosis. DNA topoisomerase II has been shown to be modified by SUMO in budding yeast as well as in vertebrates. We have shown by biochemical analysis using the Xenopus egg extract (XEE) cell-free assay system that DNA topoisomerase IIα (Topo IIα) is modified by SUMO-2/3 on mitotic chromosomes in the early stages of mitosis. Inhibition of mitotic SUMOylation in the XEE assay system causes aberrant sister chromatid separation in anaphase and alters Topo IIα association with chromosomes.
Keywords: Topoisomerase II, SUMO, Xenopus, egg extracts, chromosomes
1. Introduction
Posttranslational modification by SUMO, Small Ubiquitin-like MOdifier, was discovered 10 years ago (1, 2). Since then it has become clear that SUMOylation is a major protein modification system with a broad impact on cellular functions. SUMO proteins and the mechanism of SUMOylation are highly conserved in eukaryotes. In vertebrates, there are multiple SUMO isoforms. SUMO-2 and SUMO-3 are closely related in their primary sequence (95% identical), whereas SUMO-1 is around 45% identical to either of the other two isoforms. All isoforms are around 50% identical to the SUMO homologue, Smt3, in budding yeast, which has a single SUMO gene in its genome (3). The biochemistry of modification by SUMO, SUMOylation, is similar to ubiquitinylation. The conjugation process requires three sequential enzymes, E1, E2, and E3, and conjugation can be reversed by specific protease activity (1–3). Because of highly active deconjugation enzymes, SUMO proteases, studying SUMOylation under physiological conditions has been one of the challenging topics in this field (4, 5).
DNA topoisomerase II (Topo II) is an essential enzyme, required for cell proliferation, which regulates chromosomal structure (6). Genetic studies indicate that Topo II has an essential function in both chromosome condensation and segregation during mitosis (7, 8). This genetic evidence was further confirmed in the Xenopus egg extract (XEE) cell-free assay system, where only the α form of topoisomerase II (Topo IIα) is present (9). Immunodepletion of Topo IIα from XEEs completely inhibits the assembly of condensed chromosomes either from sperm chromatin (9) or from chicken erythrocyte nuclei (10). Inhibition of Topo IIα in XEEs with a specific inhibitor prevents remodeling and condensation of chromosomes derived from sperm nuclei and also prevents the proper function of the condensin complex, which participates in the assembly of condensed chromosomes (9, 11). Moreover, inhibition of Topo II in XEE using VP-16 at the metaphase–anaphase transition compromises sister chromatid separation (12). These findings indicate the importance of Topo IIα to various process of mitosis and emphasize the benefits of XEEs in the study of Topo IIα.
Findings that Topo II is modified by SUMO in budding yeast revealed a novel mechanism of Topo II regulation on mitotic chromosomes (13, 14). Similarly, we have identified Topo IIα as major SUMO-modified protein on mitotic chromosomes in XEE (15). SUMOylation of Topo II can be observed in mammalian cells when they are treated with Topo II inhibitors (16), and Topo II inhibitors enhance SUMO-2/3 modification of Topo IIα in mitotic mammalian cells (17).
Utilizing XEEs, we have demonstrated cell cycle-dependent SUMOylation of Topo IIα. Interestingly, SUMOylation of Topo IIα utilizes exclusively SUMO-2/3 under physiological conditions not SUMO-1. SUMO-1 modification of Topo IIα, however, can be observed after addition of exogenous SUMO-1 into XEE (15). This result suggests that there is a precise mechanism for selection of SUMO paralogues under physiological conditions and for temporal regulation during the cell cycle. XEEs are an excellent model system for studying SUMOylation because of their highly synchronized and manipulable cell cycle progression and the simplicity of biochemical fractionation of this material (18, 19). This article includes detailed protocols for the production of mitotic chromosomes in XEE and for the analysis of Topo II SUMOylation in this context.
2. Materials
2.1. Preparation of CSF Extracts from Xenopus Eggs
MMR: 100 mM NaCl, 2 mM KCl, 1 mM MgSO4, 2 mM CaCl2, 0.1 mM EDTA, 5 mM HEPES, pH 7.8. (Prepare 10X concentrated and store at room temperature.)
Pregnant mare serum gonadotropin (PMSG, EMD/Calbiochem): Dissolve in water at 200 units/ml, store at −20°C.
Human chorionic gonadotropin (HCG, Sigma-Aldrich): Dissolve in water at 1000 units/ml, store at 4°C.
Dejellying solution: 2% w/v cysteine, free base (EMD/Calbiochem), dissolve in water, and adjust to pH 7.8 with NaOH.
CSF-XB: 100 mM KCl, 0.1 mM CaCl2, 2 mM MgCl2, 5 mM EGTA, 50 mM sucrose, and 10 mM HEPES, adjust to pH 7.7 with KOH.
Protease inhibitor (LPC) solution: Dissolve a mixture of leupeptin, pepstatin, and chymostatin (all from EMD/Calbiochem) at a final concentration of 20 mg/ml each in dimethyl sulfoxide (DMSO, Sigma-Aldrich). Store at −20°C in aliquots of 30 μl/tube.
Cytochalasin B (CyB) solution: Dissolve cytochalasin B (EMD/Calbiochem) at 10 mg/ml in DMSO. Store at −20°C in aliquots of 30 μl/tube.
50X Energy mixture: Dissolve in sterile water 375 mM phosphocreatine (Sigma-Aldrich), 50 mM ATP (Mg salt, Sigma-Aldrich), and 5 mM EGTA, pH 7.7. Adjust pH to ~7.0 and store at −80°C in aliquots of 100 μl/tube.
Calcium solution: 6 mM CaCl2, 50 mM KCl, and 2 mM MgCl2.
2.2. Preparation of Demembraned Sperm Nuclei
Buffer T: 15 mM PIPES, 15 mM NaCl, 80 mM KCl, 5 mM EDTA, 7 mM MgCl2, and 200 mM sucrose. Adjust pH to 7.4 with KOH.
Demembrane buffer: Buffer-T containing 0.05% lysophosphatidyl choline (Sigma-Aldrich) and 20 mM maltose (Sigma-Aldrich).
Washing buffer: Buffer-T containing 3% BSA.
Haemocytometer.
2.3. Chromosome Assembly and Isolation
CaCl2 solution: 6 mM CaCl2, 50 mM KCl, and 2 mM MgCl2.
Dilution buffer: 0.5X CSF-XB containing 18 mM β-glycerophosphate (Sigma-Aldrich), 0.25% (v/v) triton-X100 (Sigma-Aldrich), 1/2000 volume of LPC solution, 1/2000 volume of CyB solution, 0.4 μg/ml nocodazole (EMD/Calbiochem), and 0.2 μM okadaic acid.
Glycerol cushion: 0.5X CSF-XB containing 18 mM β-glycerophosphate (Sigma-Aldrich), 0.1% (v/v) triton-X100 (Sigma-Aldrich), and 30% (v/v) glycerol.
2 ml conical bottomed microcentrifuge tubes (Corning).
Fix solution: 0.3 ml of 37% formaldehyde, 0.1 ml of 10X MMR, 0.6 ml 70% glycerol, 1 μg/ml Hoechst 33342 (EMD/Calbiochem).
Standard SDS-PAGE sample buffer (3X): 187 mM Tris–HCl, pH 6.8, 6% (w/v) SDS, 30% (v/v) glycerol, 0.01 mg bromophenol blue, 10% (v/v) 2-mercaptoethanol. A stock solution should be prepared without 2-mercaptoethanol and stored at room temperature.
2.4. Analysis of SUMOylation of Topoisomerase II
Gradient SDS-PAGE gel: 8–16% Tris–glycine gel (Invitrogen).
Standard SDS-PAGE running buffer (10X): 250 mM Tris, 1.92 M glycine, 1% (w/v) SDS. Store at room temperature.
PVDF membrane: Immobilon-P transfer membrane (Millipore).
Transfer buffer (20X): 1 M Tris, 750 mM glycine, 0.4% (w/v) SDS. Store at room temperature. Prepare 1X solution with 7.5% (v/v) methanol.
PBS-T: PBS containing 0.1% (v/v) Tween 20 (Sigma-Aldrich).
Blocking solution: 5% w/v skimmed milk powder in PBS-T.
Anti-Topo IIα/β monoclonal antibody (clone AK5).
Stripping buffer: 62.5 mM Tris–HCl, pH 6.8, 1% SDS.
3. Methods
3.1. Preparation of CSF Extracts from Xenopus Eggs
There are several established protocols for obtaining frog egg extracts that retain mitotic activity. The method described here preserves meiosis II arrest through preservation of Cyto Static Factor (CSF) (18, 19). Extracts prepared in this manner are called CSF XEEs.
To induce oocyte maturation, the female frogs are primed by injecting 100 units of PMSG (0.5 ml/frog) at least 4 days before eggs are required. Primed frogs should be used within 2 weeks, or the quality of eggs will often become insufficient for use. The day before the egg collection, each female should be injected with 500 units of HCG (0.5 ml/frog) and placed in an individual bucket that contains 2 l of MMR.
Fifteen hours after the HCG injection, eggs are recovered from each bucket into individual glass beakers, then washed once with MMR. Discard as much MMR as possible by decantation, then add dejellying solution to dissolve the jelly coat of the eggs. The completion of dejellying can be monitored by observing the disappearance of the space between the eggs. Discard the dejellying solution and briefly rinse the eggs once more with dejellying solution (see Note 1).
Quickly rinse the eggs by decanting twice with about 200 ml of MMR. During this rinse step, damaged and poor-quality eggs can be removed using a Pasteur pipet. Exchange the MMR for CSF-XB and carefully remove poor-quality eggs. Eggs that show clear separation of animal (dark colored) and vegetal (light colored) hemispheres of approximately equal size are usually of sufficient quality for XEE preparations. Enlarged, small, and abnormal colored eggs should be removed. Rinse the eggs by decanting three times more with CSF-XB (see Note 2).
Prepare a 14-ml round-bottom polypropylene tube with 1.5 ml of CSF-XB containing 7.5 μl each of LPC and CyB solution. Transfer eggs using a large bore transfer pipette. After removing excess buffer, centrifuge the tube at 170g for 30 s to pack the eggs. Again remove excess buffer and leave just enough buffer to protect the eggs from contact with air (see Note 3).
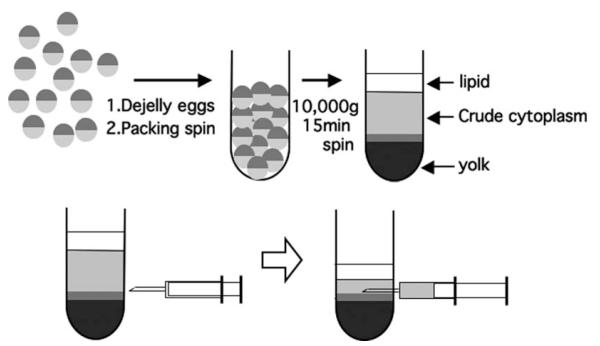
Centrifuge the packed eggs at 10,000g for 15 min at 16°C in a swinging bucket rotor (Beckman/Coulter JS13.1 or Sorval HB-6). After centrifugation, lysed eggs will be separated into three layers: lipids (bright yellow), crude cytoplasm (light brown or golden), and yolk (dark brown or gray) from top to bottom. There will be a thin layer of mitochondrial material between the cytoplasm and yolk, which is cloudier than the cytoplasm (see Fig. 17.1).
Extract the crude cytoplasmic fraction by withdrawing it from the side of the tube with a 16-G needle attached to a syringe. Withdrawing the cytoplasm too rapidly will cause contamination by either the lipid or the mitochondrial fractions, and those contaminations may compromise the quality of the XEE (see Fig. 17.1) (see Note 4).
Place the CSF extracts on ice. Add 1/2000 volume each of LPC and CyB solution to the extracts and 1/50 volume of energy mixture. Gently mix the extracts by taping the tube and keep it on ice.
To obtain interphase extracts, take an aliquot of the CSF extract, and warm it up to room temperature. Add 1/10 volume of calcium solution to the CSF extracts and mix it rapidly but gently without forming bubbles. Incubate the extracts for 5–10 min at room temperature to complete the induction of interphase (see Note 5).
Fig. 17.1.
Xenopus egg extract (XEE) preparation.
3.2. Preparation of Demembraned Sperm Nuclei
This protocol is designed to isolate a large number of sperm nuclei from more than six male frogs at one time. For small-scale preparation, reduce the amount of buffers accordingly. All the centrifugation should be done with a swinging bucket rotor at room temperature.
Anesthetize male frogs by injection of tricaine methanesulfonate (100 mg/kg) and kill by double pithing.
Dissect to obtain the testes, which are located behind the intestinal tract. Carefully remove the connective tissue and blood vessels around the testes (see Note 6).
Rinse the testes with buffer-T twice, and put the testes in round-bottom 2-ml microcentrifuge tubes containing 1 ml of buffer-T (four testes/tube). Mince the testes with forceps in the tubes.
Centrifuge at 150g for 10–20 s and collect the supernatant that contains sperm. Re-extract the sperm from the pellet with 0.5 ml of buffer-T and re-centrifuge to obtain a second supernatant.
Combine the first and the second supernatants, then sediment the sperm by centrifugation at 1350g for 2 min. A white pellet with a pink/red colored bottom will be obtained. Carefully separate the white pellet with a 1 ml pipetman and transfer to a 14 ml round-bottom tube containing 10 ml of buffer-T.
Centrifuge again at 1350g for 2 min to sediment the sperm, and resuspend the pellet with 10 ml of 0.2X buffer-T. Quickly mix and then centrifuge again at 1350g for 2 min (see Note 7).
Resuspend the pellet in 1 ml of buffer-T and add 3 ml of demembrane buffer. Quickly mix, then incubate at room temperature for 7 min. There will be a large chunk of precipitate formed during the incubation. At the end of the incubation, transfer the supernatant to a new tube containing 10 ml of a washing buffer, then mix the transferred supernatant quickly by inverting the tube.
Centrifuge the tube at 300g for 10 min to sediment the demembraned sperm nuclei. Resuspend the pellet in 10 ml of washing buffer and centrifuge again.
Resuspend the pellet in 1 ml of buffer-T, then estimate the concentration of sperm using a haemocytometer. Adjust the concentration to ~300,000 nuclei/ml with buffer-T, and aliquot 20 μl/tube. Snap freeze the sperm in liquid nitrogen and store at −80°C (see Note 8).
3.3. Assembly and Isolation of Chromosomes or Interphase Chromatin
SUMOylated Topo II from mitotic chromosomes can be observed as slow migrating isoforms on SDS-PAGE gels. So far, we have not detected SUMOylated isoforms of Topo II isolated from interphase chromatin. Therefore, the isolation of interphase chromatin is included as a negative control for analysis of Topo II SUMOylation. Another negative control that can be employed utilizes a dominant negative mutant of Ubc9, a SUMO E2 enzyme, that has the following mutations: C93S and L97S. Addition of this mutant protein to XEEs prevents Topo II SUMOylation (note that this method is not described in detail herein).
Put 100 μl of CSF extract or interphase extract into 2-ml round-bottom microcentrifuge tubes and add demembraned sperm nuclei to a final concentration of 3000–6000 nuclei/μl. Mix the extracts gently by tapping (see Note 9).
Incubate the extracts at room temperature and at 10–15 min intervals, take 1 μl of the reaction and mix with 4 μl of fixation solution on a slide glass to observe the DNA morphology with a fluorescence microscope. Usually, after 30–40 min of incubation, assembly of mitotic chromosomes or interphase nuclei will be complete (see Note 10).
Add four volumes of ice-cold dilution buffer to the reactions and mix rapidly by pipeting or inverting the tube.
Layer the diluted samples onto an ice-cold glycerol cushion in 2-ml conical-bottomed microcentrifuge tubes. Centrifuge with a swing bucket rotor at 10,000g for 5 min at 4°C.
Aspirate off the supernatant and resuspend the pellet in 500 μl of ice-cold glycerol cushion. Centrifuge again as above (step 4) then aspirate off the supernatant.
Add to the pellet 1X SDS-PAGE sample buffer (an equal volume to the original reaction). Mix by vortexing and heat the sample at ~100°C for 5 min. After heating, vortex the sample vigorously for 30 s and then reheat the sample for 5 min more. This process helps to disrupt DNA to make better separation in the SDS-PAGE gel. Centrifuge the boiled samples at 10,000g for 15 min at room temperature to pellet debris in the samples (see Note 11).
3.4. Analysis of SUMOylation of Topoisomerase II
To analyze the different molecular weight isoforms of Topo II and SUMOylated Topo II, we prefer to use a gradient SDS-PAGE gel. The gel electrophoresis and Western blotting conditions that are specific to this analysis are as follows:
Load 10–15 μl of the SDS-PAGE samples into each well of a pre-made gradient SDS-PAGE gel (8–16% or 4–20%).
Run the SDS-PAGE gel with 1X standard SDS-PAGE running buffer at 20–25 mA/gel for 2–3 h, until the bromophenol blue in the sample reaches the bottom of the gel.
Transfer the protein onto PVDF membrane with a semi-dry transfer apparatus and 1X transfer buffer at 45 mA/gel (8 × 7 cm gels) for 1.5 h.
Block the membrane with shaking in a blocking solution for 1 h at room temperature.
Probe the membrane with an anti-Topo IIα/β monoclonal antibody to detect Xenopus Topo IIα (see Note 12). The primary antibody should be diluted in blocking solution at 1/1000 and the membrane incubated with shaking overnight at 4°C.
Wash the membrane three times for 10 min each with shaking in PBS-T at room temperature.
Probe the membrane with the secondary antibody diluted in blocking solution at 1/2000–1/5000 with shaking for 1 h at room temperature.
Wash the membrane three times for 10 min each with shaking in PBS-T at room temperature.
Develop the membrane according to the secondary antibody conjugate that was employed (e.g., using ECL for a peroxidase-conjugated secondary antibody) and expose to X-ray film. A typical result of this analysis is shown in Fig. 17.2.
If the membrane needs to be stripped and re-probed with another antibody, strip the membrane by incubating it in stripping buffer at 55°C for 20 min. Next, wash the membrane briefly with water and then with PBS-T. Block the membrane with blocking buffer for 1 h and then proceed to the primary antibody step.
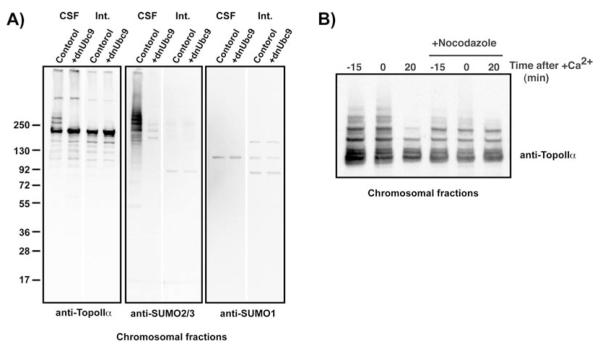
Fig. 17.2. Detection of SUMOylated Topo IIα on mitotic chromosomes.
(A) Chromosomal fractions were isolated either from CSF (indicated as CSF) or interphase (indicated as Int.) egg extracts in the presence or absence of dnUbc9. The chromosomal fractions were analyzed by immunoblotting with the indicated antibodies. On mitotic chromosomes (CSF), Topo IIα shows molecular weight shifts, which corresponds to the protein detected with the anti-SUMO2/3 antibody. (B) CSF extracts were incubated with 8000 sperm nuclei/ml in the presence or absence of nocodazole. After 45 min of incubation (time 0), CaCl2 was added to the reaction to drive the reactions into interphase. Chromosomal fractions were isolated at the indicated time points, and then isolated chromosomes were analyzed by immunoblotting with the anti-Topo II antibody. When the mitotic checkpoint was activated by the inclusion of nocodazole, this prevented exit from mitosis (20) and de-SUMOylation of Topo II induced by Ca2+.
Acknowledgments
I would like to thank to Mary Dasso for critical reading and her support. This work was supported by NIH/NCRR, CCET-COBRE (P20 RR015563), and is currently supported by NIH/NIGMS (GM080278).
4. Notes
We use a 500–600 ml glass beaker for dejellying and washing the eggs. This size of beaker is large enough to mix the eggs gently and to observe the morphology of the eggs. Adjust the size of the beaker according to the volume of eggs you have.
Usually, low-quality eggs accumulate on the top and center of the eggs after you mix by rotating the beaker horizontally. If you observe more than 30% of the eggs are not of sufficient quality, they should not be used for XEE preparation.
Do not drop the eggs from the top of the tube. Place the tip of a transfer pipette in the buffer at the bottom and slowly put the eggs down into the buffer. Try not to put excess buffer in the tube with the eggs.
The color of the extracts will vary depending on the pigmentation of the eggs. Contamination by mitochondrial material will damage the extracts and make them no longer useful for cell cycle analysis. Use the appropriate size of syringe depending on the volume of the cytoplasmic fraction.
Mix CaCl2 as quickly as possible. Mixing thoroughly is required to complete the release from CSF arrest.
Rolling the dissected testes on a paper towel is one of the easiest ways to remove extra tissue attached to the testes.
This wash step under hypotonic conditions is very effective when you are trying to isolate a large quantity of sperm from a large number of testes. After this wash step, all of the contaminating red blood cells will be disrupted and other contaminating somatic cells will tend to form an aggregate in the next step.
If you observed aggregated sperm, you can remove the aggregates by centrifuging the sperm at 150g for 10 s. The concentration of the sperm can be lower or higher depending on the application. For a morphological assay, a lower concentration of the stock sperm will be desired.
Assembled chromosomes tend to sink to the bottom of the tube. Using a round-bottom tube makes mixing the large volume of the reaction easier. When a larger reaction volume, more than 200 μl, is required, we use 5- or 14-ml round-bottom test tubes.
Condensed chromosomes and round nuclear morphology will be observed after 30 min of incubation with sperm [e.g., (11)].
Chromatin samples often separate poorly on SDS-PAGE gels. Occasionally, both Topo II and SUMO show smeary signals upon western blotting. We have found that high concentrations of salt, above 300 mM, and vigorous mixing by vortexing help to avoid such unclear separation on SDS-PAGE gels.
We are currently using both anti-SUMO-2/3 and anti-Topo IIα antibodies that have been generated in our laboratory. We tested several anti-Topo II antibodies (15), but some of them are not available currently. Our antibodies were made against the full-length SUMO-2/3 and the C-terminal fragment of Topo IIα.
References
- 1.Hay RT. SUMO: a history of modification. Mol Cell. 2005;18:1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 2.Johnson ES. Protein modification by sumo. Annu Rev Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- 3.Dasso M. Emerging roles of the SUMO pathway in mitosis. Cell Div. 2008;3:5. doi: 10.1186/1747-1028-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mukhopadhyay D, Dasso M. Modification in reverse: the SUMO proteases. Trends Biochem Sci. 2007;32:286–295. doi: 10.1016/j.tibs.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 5.Hay RT. SUMO-specific proteases: a twist in the tail. Trends Cell Biol. 2007;17:370–376. doi: 10.1016/j.tcb.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 6.Yanagida M. Basic mechanism of eukaryotic chromosome segregation. Philos Trans R Soc Lond B Biol Sci. 2005;360:609–621. doi: 10.1098/rstb.2004.1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holm C, Goto T, Wang JC, Botstein D. DNA topoisomerase II is required at the time of mitosis in yeast. Cell. 1985;41:553–563. doi: 10.1016/s0092-8674(85)80028-3. [DOI] [PubMed] [Google Scholar]
- 8.Uemura T, Yanagida M. Isolation of type I and II DNA topoisomerase mutants from fission yeast: single and double mutants show different phenotypes in cell growth and chromatin organization. EMBO J. 1984;3:1737–1744. doi: 10.1002/j.1460-2075.1984.tb02040.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirano T, Mitchison TJ. Topoisomerase II does not play a scaffolding role in the organization of mitotic chromosomes assembled in Xenopus egg extracts. J Cell Biol. 1993;120:601–612. doi: 10.1083/jcb.120.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adachi Y, Luke M, Laemmli UK. Chromosome assembly in vitro: topoisomerase II is required for conden-sation. Cell. 1991;64:137–148. doi: 10.1016/0092-8674(91)90215-k. [DOI] [PubMed] [Google Scholar]
- 11.Cuvier O, Hirano T. A role of topoisomerase II in linking DNA replication to chromosome condensation. J Cell Biol. 2003;160:645–655. doi: 10.1083/jcb.200209023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shamu CE, Murray AW. Sister chromatid separation in frog egg extracts requires DNA topoisomerase II activity during anaphase. J Cell Biol. 1992;117:921–934. doi: 10.1083/jcb.117.5.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takahashi Y, Yong-Gonzalez V, Kikuchi Y, Strunnikov A. SIZ1/SIZ2 control of chromosome transmission fidelity is mediated by the sumoylation of topoisomerase II. Genetics. 2006;172:783–794. doi: 10.1534/genetics.105.047167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bachant J, Alcasabas A, Blat Y, Kleckner N, Elledge SJ. The SUMO-1 isopeptidase Smt4 is linked to centromeric cohesion through SUMO-1 modification of DNA topoisomerase II. Mol Cell. 2002;9:1169–1182. doi: 10.1016/s1097-2765(02)00543-9. [DOI] [PubMed] [Google Scholar]
- 15.Azuma Y, Arnaoutov A, Dasso M. SUMO-2/3 regulates topoisomerase II in mitosis. J Cell Biol. 2003;163:477–487. doi: 10.1083/jcb.200304088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mao Y, Desai SD, Liu LF. SUMO-1 conjugation to human DNA topoisomerase II isozymes. J Biol Chem. 2000;275:26066–26073. doi: 10.1074/jbc.M001831200. [DOI] [PubMed] [Google Scholar]
- 17.Agostinho M, Santos V, Ferreira F, Costa R, Cardoso J, Pinheiro I, Rino J, Jaffray E, Hay RT, Ferreira J. Conjugation of human topoisomerase 2 alpha with small ubiquitin-like modifiers 2/3 in response to topoisomerase inhibitors: cell cycle stage and chromosome domain specificity. Cancer Res. 2008;68:2409–2418. doi: 10.1158/0008-5472.CAN-07-2092. [DOI] [PubMed] [Google Scholar]
- 18.Desai A, Murray A, Mitchison TJ, Walczak CE. The use of Xenopus egg extracts to study mitotic spindle assembly and function in vitro. Methods Cell Biol. 1999;61:385–412. doi: 10.1016/s0091-679x(08)61991-3. [DOI] [PubMed] [Google Scholar]
- 19.Kornbluth S, Yang J, Powers M. Analysis of the cell cycle using Xenopus egg extracts. In: Yamada KM, editor. Current Protocols in Cell Biology. John Wiley & Sons, Inc.; New York: 2001. pp. 11.11.11–11.11.13. [DOI] [PubMed] [Google Scholar]
- 20.Chen RH, Murray A. Characterization of spindle assembly checkpoint in Xenopus egg extracts. Methods Enzymol. 1997;283:572–584. doi: 10.1016/s0076-6879(97)83045-5. [DOI] [PubMed] [Google Scholar]