Abstract
Epigenetic events establish a particular gene expression signature for each cell type during differentiation and fertilization. Disruption of these epigenetic programs in response to environmental stimuli during prenatal exposure dysregulates the fetal epigenome, potentially impacting susceptibility to disease later in life (the fetal basis of adult disease). Maternal dietary modifications during gestation and lactation play a pivotal role in the period of fetal (re)programming. Recently, many studies have demonstrated the impact of maternal nutrition on the fetal epigenome. In this review, we discuss the complex interplay between various environmental factors and epigenetic mechanisms which have been found to affect the human and animal models. We summarize here the impact of various dietary phytochemicals capable of modulating the epigenome in diverse human cancers and childhood cancer with potential environmental etiology through maternal consumption during pregnancy and lactation and discuss other dietary agents that are still untested as to their effectiveness in transplacental studies. Recent developments discussed here enhance our understanding of how chemopreventive agents act, their potential to impact the prenatal epigenome and to identify dietary interventions that can be beneficial in treating and preventing disease.
Keywords: fetal epigenome, transplacental, environmental pollutants, dietary phytochemicals, cancer chemoprevention
Introduction
Epigenetics is broadly defined as the field of research which studies changes in gene expression without underlying changes in DNA sequence. Epigenetic changes mediate global alterations in DNA methylation, posttranslational modifications of histones, chromatin remodeling enzymes and microRNA (miRNA) that produce widespread changes in gene expression and can play a fundamental role in tumor development. Epigenetic regulation in multicellular organisms produce a particular signature, which changes during differentiation or fertilization and for each cell type during development. Disruption of these epigenetic signatures, by internal factors and environmental stimuli, can result in hypomethylation, hypermethylation of gene promoter enriched CpG islands (CGIs), histone modifications, deregulated miRNA profiles and result in altered epigenetic programming that promotes genomic instability. These epigenetic alterations do not cause changes in the underlying DNA sequence and, moreover, most are reversible and provide promising therapies to restore “correct” epigenetically control to neoplastic genomes. A number of dietary factors have been investigated as effective chemopreventive agents that can target the epigenome and potentially reduce the risk of developing various chronic diseases.
Epigenetic alterations play an important role in the risk of developing cancer, the leading cause of death worldwide. Epigenetic modifications have the potential to be inherited and thereby impact the health of future generations. Despite modern advances in adjuvant, surgical, chemo- and pharmacotherapies, cancer is still a challenge and also a burden on a world-wide scale. The WHO estimates that the number of cancer deaths will increase by 45% over the next decade with 12 million newly diagnosed cancer cases per year by 2030. Cancer is the second leading cause of death in children, after accidents, in the United States, among which leukemia and lymphomas are the most common and account for about 31% of all cancers in children.1,2 The etiology of malignant tumors of childhood is largely unknown, but several epidemiological studies conclude that childhood cancers are attributed, in part, to maternal exposure to environmental factors, e.g. polycyclic aromatic hydrocarbons (PAHs).3-6 Research seems to indicate that plant-based dietary agents, given in maternal diet, have an ability to target the epigenome, particulary DNA methylation, and provide protection to the developing fetus which carries into adulthood.7 Excellent reviews on the epigenome, as a therapeutic target, have been published.8-10 In this review, we discuss the possible environmental etiology of childhood cancer with a focus on chemopreventive strategies of supplementation of maternal diet with phytochemicals. We will also consider other dietary agents that are still untested in the context of transplacental studies and that target the epigenome of the fetus for reducing risk and mortality and highlight potential new directions for the field of cancer prevention and research treatment.
Epigenetic Mechanisms
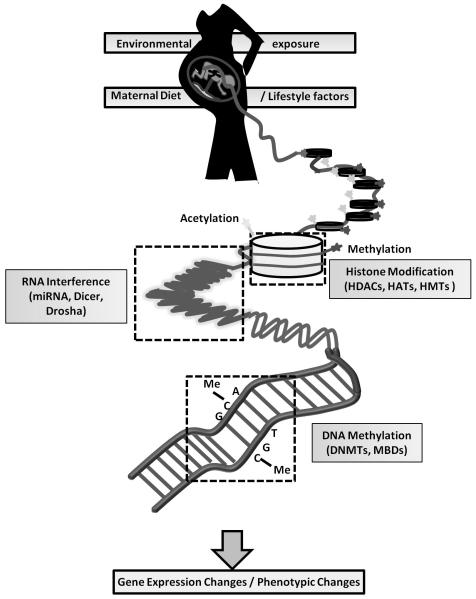
Maternal nutrition and environmental exposure induce changes in the epigenome of the developing fetus. The most widely recognized epigenetic regulatory mechanisms include DNA methylation, histone modifications and small interfering RNAs (siRNAs) (Fig.1). Diet, lifestyle and other environmental factors have been shown to induce epigenetic alterations and thereby effect changes in cell phenotype and gene expression that leads to an altered risk of various diseases including cancer.11 DNA methylation, predominantly at 5′-CpG-3′, regulates various developmental processes and is thought to be a major factor in differentiation and embryogenesis. CpG-dense sequences, known as CGIs are around 300-3000 bp in length and are randomly distributed at or near the transcription start site of many genes and overlap with the promoter region of 50-60% of total human genes, including most house-keeping genes.
Fig.1. Impact of maternal diet on fetal epigenetic mechanisms.
Maternal dietary, lifestyle and environmental factors have an important impact on fetal epigenetic mechanisms (DNA methylation, histone modifications and RNA interference), regulating the epigenome, which in turn contributes to phenotypic and genotypic change.
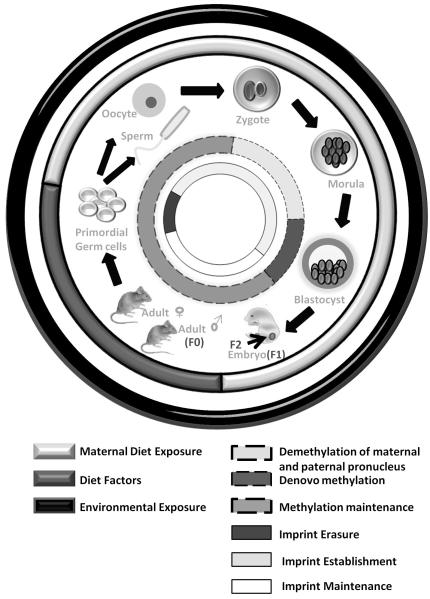
In mammals, the first phase of epigenetic reprogramming occurs in primordial germ cells (PGCs). In mice, after embryonic day 7.25 (E7.25), PGCs migrate into the genital ridges and start to proliferate and differentiate. Genome-wide demethylation is thought to occur on E12.5. At this time, highly methylated PGCs undergo complete demethylation. This phase of demethylation coincides with erasure and reestablishment of imprinted differentially methylated regions (DMRs). The second phase of methylation programming occurs between fertilization and formation of the blastocyst. After fertilization, active demethylation occurs in the paternal pronucleus followed by a step-wise decline in methylation until the morula stage, known as passive demethylation. Then de novo methylation occurs in the inner cell mass of the blastocyst and the new DNA methylation pattern is established and copied through successive cell divisions.12,13 Environmental exposures and maternal diet are significant factors for at least three generations, the F0 female, the F1 embryo and the F2 germline (Fig. 2).
Fig. 2. Epigenetic consequences of fetal programming.
In mammals, epigenetic reprogramming occurs in two phases. Primordial Germ Cells (PGCs), derived from epiblast, start to migrate to the germinal ridge after E7.25 and proliferate and differentiate. Highly methylated PGCs undergo loss of methylation after the arrival of PGCs in the genital ridge. This first phase of demethylation also coincides with erasure and reestablishment of imprinted differentially methylated regions (DMRs). The second round of reprogramming occurs between fertilization and blastocyst formation. After fertilization, active demethylation of paternal pronucleus occurs in the zygote followed by the step-wise decline in methylation until the morula stage which is known as passive demethylation. De novo methylation occurs at the blastocyst stage and the new DNA methylation pattern are established and copied through successive cell divisions. Exposure of the pregnant female to environmental toxicant affects the F1 generation embryo and F2 generation germline.
DNA methylation is essential for embryonic development and cell differentiation however, in normal cells CpG islands in promoter regions are primarily hypomethylated, with the exception of imprinted genes and those islands located on the inactive X-chromosome. Methylation of CpG islands in the promoter recruits methyl-CpG binding domain proteins (MBDs) that interact with histone deacetylases (HDACs). This complex network between MBDs and chromatin remodeling factors also bring about chromatin condensation and thus make promoter regions inaccessible to various transcription factors. Several studies have clearly revealed that DNA hypermethylation stably alters gene expression and is predominantly associated with gene silencing.
The importance of CpG island methylation in cancer has been well recognized. To date, inadequate data exist on methylation of genes in childhood cancer. There is some evidence in the literature emphasizing the importance of maternal diet.7,14-16 Maternal diet during pregnancy, placental insufficiency, psychosocial characteristics and other factors such as stress, anxiety and fatigue induce changes in the epigenome of the embryo.17 Experimental studies have been conducted in animal models by feeding pregnant mice with genistein which results in shifting the coat color from yellow to brown due to increased methylation of the Agouti viable yellow (Avy) intracisternal A particle (IAP). The expression of the agouti gene protects the offspring from obesity later in life.7 Sandovici and Sapienza18 also demonstrated in rats that the transcription factor Hnf4a, required for pancreatic β-cell differentiation and glucose homeostasis, is epigenetically regulated by maternal diet. Prenatal exposure to maternal smoking and to other persistent environmental agents also modifies the epigenetic machinery and was associated with subsequent adult diseases.17
Gene methylation patterns are frequently dysregulated and affect the clinical outcome of patients with ALL (Acute Lymphoblastic Leukemia). Many reports have been published on methylation status of tumor suppressor genes (e.g., p73, p16, p15, p14, RASSF6, RASSF10 and E-cadherin), DNA repair (O6MGMT), cyclin dependent kinase (CDK) inhibitors, Cdkn2a (p16INK4a), Cdkn2b (p15INK4b), Cdkn2c (p18INK4c), p21 and DAP-kinase in childhood ALL19-21 and a broad range of other cancers such as colon, kidney, liver, pancreas, stomach and thyroid. Dunwell et al.22 studied the pathogenesis of ALL and identified cancer-specific novel methylation markers in childhood leukemia that could be used for prognosis and potentially therapy. The authors found methylation hotspots on chromosome 3 by NotI microarray in cells of T cell-lineage (T-ALL) and B cell-lineage leukemia (B-ALL). Direct readouts of genome-wide methylation studies can be used to identify epigenetic predictors of malignant transformation.
Genomic imprinting is one phenomenon for control of expression of maternal and paternal alleles. Few studies have provided information about dysregulation of imprinted genes in development of leukemia. Promoter CpG methylation of multiple genes on the inactive X chromosome is responsible for silencing the inactive maternal allele. The active maternal allele has also been shown to be silenced by promoter hypermethylation in ALL and Burkitt’s lymphoma.23 Doherty and colleagues24 identified another possible mechanism of imprinting, disruption in the early embryo by environmental influences. They found aberrant expression of H19, a gene for long non-coding RNA (ncRNA), in the mouse preimplantation embryos when cultured in Whitten’s medium. Lane and Gardner25 showed that addition of ammonium to the culture of mouse zygotes in the blastocyst stage elevated H19 expression. Loss of imprinting and aberrant DNA methylation of the IGF-2 (insulin growth factor-2) locus has been associated with various tumors such as Wilms tumor and chronic myelogenous leukemias26, ovarian27 and nasopharyngeal carcinoma.28 Kuerbitz et al.29 have observed complete loss of the imprinted neuronatin (NNAT) gene in 69% of acute lymphoid and myeloid leukemias. Maternal nutrient depreviation during pregnancy has also been documented in the context of imprinted genes. Gallou-Kabani et al.30 observed sex- specific epigenetic alterations within CpGs in offspring of dams fed high fat diets during pregnancy. Prenatal exposure to maternal low protein and folic acid diets infuences expression of H19 and Igf2 in liver of day 0 rat male offspring.31
miRNAs are small endogenous non-coding RNAs that are believed to control the translation or stability of mRNAs and play an important role in development, cell proliferation, differentiation and apoptosis. An increasing body of evidence has shown that deregulated miRNA expression is involved in many cancer types. However, in childhood cancer, little is known about the molecular alterations of these functional non-coding sequences. A recent study provided a more comprehensive view on medulloblastoma, a solid tumor of childhood, by identifying somatic mutations of miRNA genes. Most interestingly, 16% of the medulloblastoma patients had inactivating mutations in histone-lysine N-methyltransferase genes (MLL2 and MLL3).32 It should be particularly informative to evaluate the methylation status of CGIs of these genes and to study their loss of activity. Mi et al.33 sheds light on the molecular mechanisms involved in leukemogenesis by genome-wide miRNA analysis using a bead-based flow cytometric method and by using large-scale genome-wide miRNA expression profiling in which expression signatures of 27 miRNAs were differentially expressed between ALL and acute myeloblastic leukemia (AML).
A nucleosome consists of small amounts of DNA (200 bp) wrapped around a histone core of four proteins (H2A, H2B, H3 and H4), and it’s conformation regulates transcription. The tight association of histone proteins to the DNA result in chromatin condensation and a series of posttranslational modifications includes methylation, acetylation, phosphorylation, biotinylation, sumoylation and ubiquitination, among which histone acetylation and methylation are the major players in epigenetic regulation. Modification of amino acid residues by acetylation and methylation in core histones H3 and H4 is also a common hallmark of human cancer.
Prenatal Exposure to Environmental Pollutants
Exposure to inhaled carcinogens, such as PAHs, tobacco-specific N-nitrosamines, aromatic amines, dietary factors such as alcohol, aflatoxin B1 (AFB1) and high fat or cholesterol often result in pre-malignant changes including hyperplasia and metaplasia that precede malignancy. Througout the in utero development of the fetus, the placenta is of utmost importance to ensure proper growth and development. Although much effort has been focused on gene-environment interactions, epigenetic alteration in response to in utero nutritional and environmental factors also deregulates histone modifications, microRNAs and alters the CpG methylation thus playing an important role in disease susceptibility. Observations of how the environment influences epigenetic mechanisms has been well demonstrated in animals34,35 and plants.36,37 There is a growing body of evidence that alteration of the epigenome is associated with prenatal exposure to dietary and environmental factors. Exposure to dietary and environmental compounds can impact the epigenome of future generations. It is assumed that during fetal development, pollutants that can cross the placenta impact fetal target organs by altering the expression of genes. The question we face is: what is known about transplacental and translactation exposure of these dietary phytochemicals to the fetus/neonate? Many studies have shown that dietary phytochemicals also cross the placenta such as genistein7 and quercetin.38 But are these concentrations high enough to impact DNMT or HDAC activity? In this review we focus on how food and environmental factors function as epigenetic modifiers that may lead to alterations in the epigenome resulting in increased disease risk in offspring later in life.
Alcohol
Prenatal exposure to maternal alcohol consumption has been reported to be associated with an increased risk of various development disorders collectively known as Fetal Alcohol Spectrum Disorder (FASD).39 Research suggests that alcohol consumption results in altered recruitment of transcriptional machinery and abnormal gene expression associated with various cancers. Many studies in rodent models have shown that transplacental ethanol exposure alters gene expression during development. The mouse model of gestational ethanol exposure resulted in the transcriptional silencing of an epigenetically sensitive allele, Avy, in the offspring and also postnatal growth restriction in the siblings of Avy mice.40 Previous work in rats indicates that alcohol exposure in utero increases the risk of mammary tumorigenesis in female offspring by inducing changes in mammary gland morphology and gene expression.41
Cytochrome P450 2E1 (CYP2E1), plays a major role in the detoxification of various xenobiotics including alcohol. Jones et al.42 observed no CYP2E1 expression in fetal tissues (lung, liver, kidney and placenta) collected at the termination of pregnancy from mothers that had consumed alcohol. The authors also documented methylation of the 3′ region of the CYP2E1 gene in fetal liver. However, Tong et al.43 found that chronic gestational exposure of rat dams to 18% and 37% ethanol significantly reduced the expression of alcohol dehydrogenase (ADH), aldehyde dehydrogenase 2 (ALDH2) and catalase and significantly increased CYP2E1 expression in rat pup cerebellar tissue. Higher expression of CYP2E1 in fetal tissues could lead to an increased formation of DNA and protein adducts and also to an increased mutational burden in the fetus. Infante-Rivard et al.44 observed a protective effect for leukemia in offspring by maternal alcohol consumption during pregnancy and suggested that the reduced risk is due to the potential chemopreventive effects of flavonoids present in red wine and beer. Chronic paternal alcohol use has been correlated with reduced DNA methyltransferase (DNMT) expression in sperm and also linked to imprinting disturbances which may have an effect on fetal development.45 Another study by Guo et al.46 found that fetal alcohol exposure affects the CREB binding protein (CBP) which is critical for the activation of neuronal gene expression and normal brain development.
Drugs and Pesticides
The effects of prenatal drug use, such as cocaine, opiates (heroin and methadone), amphetamine and methamphetamine, phencyclidine hydrochloride (PCP), permethrin, insect repellent N,N-diethyl-meta-toluamide (DEET) and marijuana, can have many detrimental effects on development of the offspring. Various experimental studies have used animal models to test the effects of several other toxic chemicals at early life exposure, e.g., the hormonally active substances, diethylstilbestrol, tributyl tin, bisphenol A (BPA) and genistein produce enhanced susceptibility to reproductive abnormalities, metabolic disorders including obesity and diabetes, and cancer through epigenetic mechanisms. The first trimester of the pregnancy is considered as the most critical period to exposures of environmentally induced epigenetic transgenerational inheritance. Manikkam et al.47 investigated the effect of various environmental mixtures including pesticides (permethrin and DEET) on F1-F3 generations and identified unique epigenetic signature (363 DMRs in pesticides exposure group) in the F3 generation. Robinson et al.48 reported that prenatal exposure from maternal use of marijuana is highly associated with monocytic leukaemia in children under 5 years of age. Recently, Desaulniers et al.49 revealed that high doses of polychlorinated biphenyls (PCBs) and a mixture of organochlorine pesticides, methylmercury chloride and PCBs, given to the pregnant rats from gestational day 1 (GD1) until postnatal day 21, reduces the global genome DNA methylation in hepatic tissue and also reduces the level of S-adenosylmethionine. The data were also validated by pyrosequencing which showed that the high dose group had a reduction in promoter methylation of the tumor suppressor gene p16INK4a.
Metals
Numerous experimental studies have shown that transplacental exposure to toxic metals may alter the epigenome and thus effect gene expression and fetal growth mechanisms. Toxic metals are widely dispersed in the environment from numerous sources and remain a major human health risk. Cadmium is classified as a human carcinogen by the International Agency for Research on Cancer (IARC) 50. In rodent models, exposure to cadmium produces a variety of abnormalities in development of the offspring.51,52 Ronco et al.53 demonstrated that cadmium exposure affects cortisol production and changes expression of 11β-hydroxysteroid dehydrogenase type 2 (11β-HSD2), an important enzyme in protecting the fetus from elevated level of maternal glucocorticoids, by DNA methylation of the promoter region. These results suggest that accumulation of cadmium in the placenta alters gene expression related to fetal development. Prenatal exposure to methylmercury from a high fish diet is related to delayed developmental milestones and neurological abnormalities in the Seychelles child development study.54 Lead exposure produces a wide range of health effects in children and also appears to be related to prenatal and postnatal exposures.55 Studies in rodents revealed that exposure to lead during brain development elevated the expression of amyloid precursor protein (APP) and its proteolytic product, β-amyloid (Aβ), involved in Alzheimer’s disease pathogenesis in later life.56 Wu et al.57 reported the upregulation of Alzheimer’s disease-related genes and also reduced expression of DNMT activity in monkeys exposed to lead as infants. Pilsner and colleagues58 examined the impact of lead stored in mother’s bone on fetal development by analyzing DNA methylation in umbilical cord blood samples of two important repititive elements, Alu and LINE-1, which constitute about 50% of the human genome and are highly methylated in normal tissues. DNA methylation of Alu and LINE-1 decreased as the level of lead increased in maternal bone suggesting how prenatal exposure influences gene programming. However, more research is required to confirm these early findings and to investigate whether early-life exposure influences epigenome-wide or gene-specific DNA methylation. Exposure to arsenic in relation to epigenetic changes can also lead to various human diseases such as pulmonary diseases, cardiovascular diseases, diabetes and obesity as well as cancer of the liver, bladder, lung and skin (reviewed in 59 and 60). Studies have shown an association between these diseases in terms of in utero exposure to arsenic. Arsenic accumulation in the placenta might have toxic effects on fetal development. Fetal exposure to arsenic has been shown to cause skin cancer in a mouse model and also tended to increase the number of cancer stem cells.61 These results clearly indicate that attempts should be made to develop preventive approaches that target stem cells as these are a critical target in transplacental carcinogenesis. In utero arsenic exposure results in significant reduction in global methylation levels at GC-rich regions and causes aberrant transcription and gene silencing related to insulin like growth factors and CYP enzymes in liver tissue of new-born mice.62
PAHs (Polycyclic Aromatic Hydrocarbons)
PAHs are ubiquitous environmental pollutants and are released into the air during burning of coal, oil, wood, gasoline, and tobacco and from residential uses such as heating and cooking.63 Humans are exposed to some PAHs through diet also, especially, overcooked, grilled, smoked food and vegetable oils. PAHs are thought to require metabolic activation to exert their tumorigenic effect.64 Preliminary evidence suggests transplacental exposure to PAHs affects the fetal epigenome and is a risk factor for various diseases65,66 that may potentially result in cancer. Accumulation of PAHs in the placenta poses a toxicological threat to the growing fetus. In a small cohort of 56 pregnant women, methylation of the acyl-CoA synthetase long chain family member 3 (ACSL3) gene, expressed in both lung and thymus tissue, was associated with childhood asthma. The most striking difference seen was the increased methylation of the ASCL3 gene promoter in 81% of asthmatic children born to mothers with high PAH exposure, compared to the 23% of the children born to mothers with low PAH exposure.67 In another study of a New York City (NYC) cohort, prenatal exposure to PAHs was associated with altered global methylation in umblical cord white blood cells (UCWBC) which suggest that maternal airborne PAH exposure may influence the degree of methylation.68 Previous studies on animal models by Andersen et al.69 and Miller70 have shown that exposure of the dam during third trimester to the PAH, 3-methylcholanthrene, produced lung and liver tumor in her offspring. Our laboratory has also demonstrated that exposure of pregnant mice to a single dose of DBC (dibenzo[def,p]chrysene), a PAH, during third trimester results in offspring mortality due to an aggressive T-cell lymphoblastic leukemia. The 10 months survivors had a 100% incidence of pulmonary tumors and 70% of the males had liver lesions.71 These studies indicate that the fetal genome and epigenome are sensitive to PAH exposure and thus this stage of development is of importance in assessing the efficiency of chemopreventive agents.
Bisphenol A
BPA is a monomer used primarily in the manufacture of polycarbonate plastics (used in food and drink containers, dental materials) and epoxy resins (commonly used for lining and protective packaging such as toys, food and drink cans, bottle tops and water supply pipes). Due to extensive use of BPA, human exposure to the chemical is increasing, according to the Center for Disease Control and Prevention (CDC).72 Studies have detected73,74 BPA in urine in 95% of adult human samples, suggesting human exposure to BPA is widespread. The CDC studies have shown that children have higher body burdens of BPA than do adolescents and adults. BPA exposure in the womb can also exert an adverse effect on development of mammary gland, the immune system, brain and male and female reproductive tracts. Pre- and perinatal exposure to BPA in rodents is associated with increased body weight, reduced sperm concentration, altered pituitary and reproductive functions and also to the formation of breast and prostate cancer. 75 BPA exposure can result in altered phenotype through an epigenetic mechanism. Dolinoy et al.15 examined the shift in coat color from yellow to brown in Avy mouse offspring after maternal exposure to BPA and also documented the association of this phenotypic change with DNA hypomethylation in the IAP of the Agouti gene. Bromer et al.75 further demonstrated that in utero exposure to BPA leads to aberrant methylation in the Hoxa10 gene, responsible for embryo viability and regulation of hematopoietic lineage commitment. Neonatal exposure at low doses of BPA induces hypomethylation in the phosphodiesterase type 4 variant 4 (PDE4D4) gene in prostate tissue and increased susceptibility of prostate cancer with aging.76 These findings help in understanding the mechanisms of epigenomic dysregulation during pre- and neonatal exposure to BPA in a critical period of development.
Epigenetic targeted therapy
Several therapeutic drugs and dietary phytochemicals are under consideration that target the epigenome, identify epigenomic biomarkers and have promising results in cancer prevention. Recent advances in molecular diagnostics have highlighted DNA methylation machinery, histone modifications and miRNAs as highly promising targets for epigenetic therapy. However, there are many as-yet undiscovered molecules that play a role in the cross-talk between different epigenetic pathways. Unlike conventional chemotherapy, the goal of epigenetic treatments are not to kill the cancer cells directly but to restore the expression and function of epigenetically silenced genes.
Gene silencing can be reactivated by using inhibitors of DNMTs which include nucleoside and non-nucleoside inhibitors. Many studies have shown great potential in treatment of a variety of cancers, particularly prostate, breast, colorectal, lung and leukemias with nucleoside inhibitors like 5-azacytidine (Vidaza), 5-aza-2′-deoxycytidine (Dacogen), 5-fluorouracil (Adrucil), arabinofuranosyl-5-azacytosine (Fazarabine), 1-β-D-ribofuranosyl-2(1H)-pyrimidinone (Zebularine) and 2′,3′-dideoxycytidine (Zalcitabine).77 5-Azacytidine and 5-aza-2′-deoxycytidine are FDA-approved drugs for the treatment of Myelodysplastic Syndrome (MDS). These agents arrest the release of DNMTs during replication and thus result in reduction in the level of DNA methylation. Sequential administration of demethylating drugs reactivates epigenetically silenced tumor suppressor genes and thus the reversible nature of DNMT inhibitors makes an attractive tool for epigenetic therapy. However, at higher concentration, these drugs induce chromosomal aberrations and contribute to cytotoxicity due to their incorporation into both DNA and RNA. Despite exciting results from nucleoside inhibitors, which have raised hopes for cancer therapy and especially for MDS, the clinical applications of these drugs are currently limited due to their toxicity. Non-nucleoside inhibitors have drawn much attention in recent years as they cause less toxicity and are not incorporated into DNA but still reactivate transcriptionally silenced genes by demethylation of the promoter region. These include procaine, procainamide, N-acetyl-procainamide, mitoxantrone, hydralazine, disulfiram78, RG-108, antisense oligodeoxynucleotides and natural compounds such as epigallocatechin-3-gallate (EGCG), psammaplins and polyphenols. The most promising ones are procaine and procainamide which inhibit DNMT activity by direct binding to CpG rich sequences. For clinical applications, the challenge is characterizing the role of DNMT inhibitors in combination with HDAC inhibitors and assessing the long-term efficacy and safety of these agents. HDAC inhibitors (HDACi) are emerging as a new class of therapeutic agents with promising applications for the treatment of cancer and non-cancer diseases. Histone acetylases (HATs) and HDACs play a major role in the regulation of gene expression.79 HDAC inhibitors induce cell cycle arrest and cellular differentiation, increase the expression of apoptotic genes and downregulate the expression of angiogenic markers such as vascular endothelial growth factor (VEGF) and the gene products that participate in angiogenesis and metastasis.80,81 An imbalance between these enzymes leads to deregulation of transcriptional regulators and results in chromatin condensation (Fig. 3). Various HDACis have been advanced for the treatment of different carcinomas in in vitro and in vivo models. Belinostat and Panobinostat (small hydroxamic acid HDACis), Mocetinostat and Entinostat (benzamide HDAC inhibitors) are being investigated for the treatment of solid tumors82,83 and haematological malignancy84 either alone or in combination with other agents. Vorinostat, also known as SAHA, is the first anticancer agent receiving FDA approval for the treatment of T-cell lymphoma85; and it has been used in clinical trials in patients for the treatment of solid tumors including ovarian cancer86 and non-small cell lung cancer (NSCLC).87 Romidepsin, a natural product derived from the bacteria Chromobacterium violaceum, is FDA-approved for treatment of cutaneous T-cell lymphoma (CTCL).
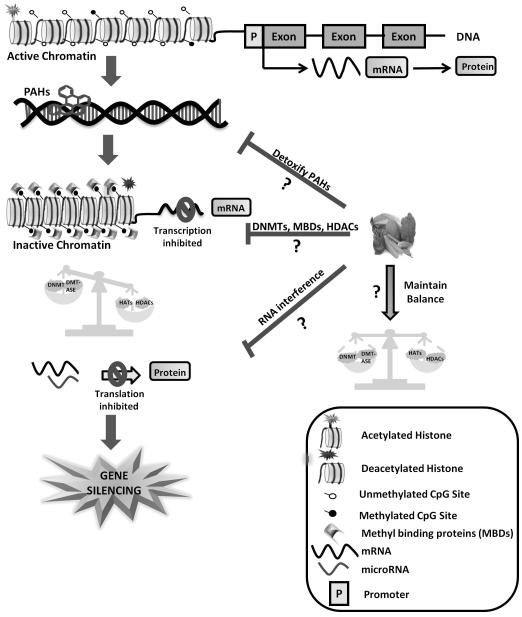
Fig. 3. Dietary phytochemicals impacting three epigenetic codes.
Schematic view showing possible effects of epigenetic changes of gene silencing during carcinogenesis. Epigenetic processes are important in regulating gene activity and are susceptible to epigenetic dysregulation by environmental toxicants. This dysregulation involves the aberrant methylation of CpG sites which in turn recruits CpG binding protein (methyl binding protein) and HDACs and changes the chromatin structure. miRNA interference leads to inhibition of mRNA translation. Strategies to reverse the epigenetic changes by the combination of dietary components, could be a chemopreventive approach to control tumorigenesis.
miRNAs have also been implicated in gene regulation, in addition to DNA methylation and histone modification, and have recently gained attention as potential therapeutic targets. Several studies have explored therapeutic strategies for these deregulated tiny non-coding RNAs in cancer. Epigenetic therapy based on DNMT and HDAC inhibitors could be used to reactivate epigenetically silenced miRNA. The advantage of using miRNA therapy is that individual miRNA reexpression targets multiple genes/pathways. For example, miR-29 targets not only DNMT3a and DNMT3b but also DNMT1. Restoration of functional miR-29 family into a lung cancer cell line reduced cytosine methylation in the promoter region of tumor suppressor genes and reactivated the epigenetically silenced genes.88
Although experimental studies have shown DNMT and HDAC inhibitors are effective at low doses, thus offsetting toxicities associated with them, there’s still a lot more to be improved. One major concern about DNMT inhibitors is the return of an aberrant DNA methylation pattern once the treatment period terminates.89 Secondly, HDAC inhibitors-induced DNA damage in normal cells also.90 Other concerns are that DNMT and HDAC inhibitors can potentially activate oncogenes due to lack of specificity.91 Further studies are needed to characterize the epigenetic regulation of miRNAs by these chromatin modifying drugs. Thus there is a need for the development of safe epigenetic chemopreventive agents in terms of long term protection and for future generations.
Dietary Agents and Cancer Chemoprevention
Several dietary factors are well known for their chemopreventive effects, among which dietary phytochemicals constitute a distinct group with anti-cancer activity. Phytochemicals are nonessential nutrients and can play a major role in disease prevention. Many phytochemicals, such as polyphenols, flavanoids, allyl sulfides and carotenoids, exhibit strong antioxidant and anticarcinogenic activities.92
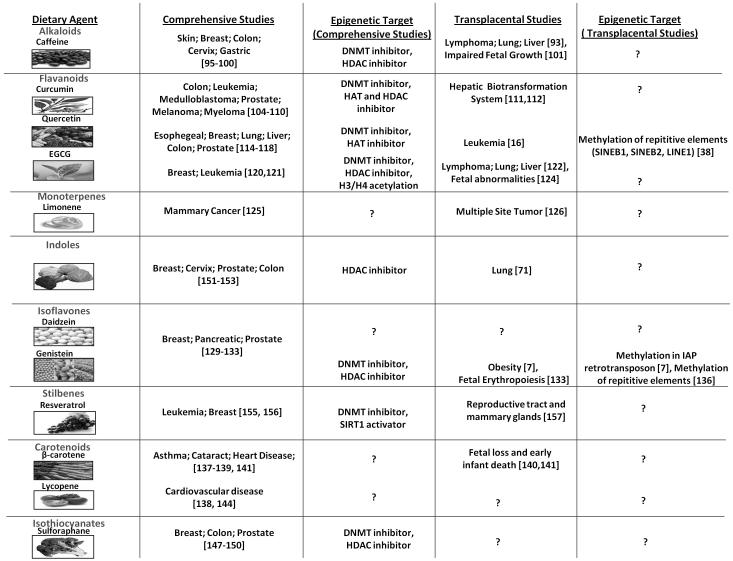
Phytochemicals, such as saponins, damage cancer cells by blocking cell replication and by stimulating cancer-fighting enzymes. There is a need to identify novel and promising approaches that use either pharmaceutical or natural agents that can suppress or prevent the premalignant lesions from developing or becoming invasive cancer. Epigenetic therapy by dietary phytochemicals is an emerging area of cancer research that could be an effective and valuable approach in fighting cancer at the level of gene expression. Studies conducted to date have shown significant effects from many dietary agents that target epigenetic pathways in preventing cancer and still many more agents are in the pipeline. The present article will summarize the epigenetic chemopreventive role of existing dietary phytochemicals in various human cancers and in the transplacental model developed by our research group.71,93,94 We discussed some of the early findings that in utero exposure to carcinogens may contribute to the development of chronic disease including cancer. A key question arises as to whether the exposure to dietary phytochemicals during lactation and pregnancy could modify the fetal epigenome providing protection to the offspring. Figure 4 gives a summary of dietary phytochemicals used in various human cancers and in transplacental studies.
Fig. 4. Various dietary phytochemicals with chemical structure and role in general cancers and transplacental models.
This figure depicts major dietary phytochemicals with epigenetic targets having effect on various cancers and in transplacental studies. Dietary agents which are still untested for epigenetic targets are shown with question mark (?). For illustrative purpose only, these images were taken from different websites.
Alkaloids
Alkaloids are naturally occurring compounds, derived from bacteria, fungi, plant sources and animals, and contain one or more basic nitrogen atoms. They possess antitumorigenic properties as well as antiaging and antiviral possibilities. Examples are: colchicine, piperine, quinine, emetine, brucine, caffeine and coniine. Among these, caffeic acid (3,4-dihydroxy-cinnamic acid) and chlorogenic acid (3-(3,4-dihydroxycinnamoyl)quinate), are naturally occurring dietary polyphenolic phytochemicals, widely distributed in many plants including coffee beans, that also act as antioxidant and carcinogenic inhibitors and are studied as DNA demethylating agents.95 Suppression by caffeic acid is more effective than chlorogenic acid.Caffeic acid may have utility in chemoprevention against skin cancer.96 Caffeic acid and chlorogenic acid inhibited DNA methylation through the formation of S-adenosyl-L-homocysteine (SAH).97 Interestingly, caffeic acid also revealed an ability to reduce HDAC activity in HT-29 human colon carcinoma98 and HeLa cells.99
Recently, Han et al.100 showed that caffeic acid 3,4-dihydroxy phenylethyl ester (CADPE) also has tumor-shrinking properties. The authors of this study observed that CADPE suppresses matrix metalloproteinase-9 (MMP-9) activity by inhibiting phosphorylation of FAK, MEK and ERK1/2 kinases in gastric carcinoma. Castro et al.93 developed a mouse model of transplacental induction of lymphoma, lung and liver cancer by the PAH, dibenzo[a,l]pyrene (DBP), and observed significant protection against T-cell lymphoma mortality when dams were administered caffeine in their drinking water. However, Bakker et al.101 studied the effect of caffeine intake in 7346 pregnant women from early pregnancy and found that high caffeine intake (>6 units/d) during pregnancy is associated with impaired fetal growth. Cytochrome P450 1A2 (CYP1A2), an enzyme involved in caffeine metabolism, is absent in the placenta and fetus. Caffeine and its metabolites can easily pass through placental barrier and adversely influence fetal programming.102 This helps in understanding how maternal diet during critical periods of development can have life-long consequences.
The risk to the fetus from gestational caffeine exposure remains the most immediate concern but the available literature suggests no harmful effects in babies born to mothers consuming moderate amounts of caffeine.103 However, the underlying mechanism of the adverse effect of high caffeine intake in offspring during early development remains to be fully established.
Flavanoids
Curcumin
Curcumin, a component of the perennial herbaceous plant Curcuma longa and also a caffeic acid derivative, has been shown to reverse epigenetic alterations that play a role in development and progression of tumors. Recent evidence has suggested that curcumin is able to activate genes which were silenced by hypermethylation in colon cancer104 and also induce global hypomethylation in leukemia.105
Studies provide evidence that curcumin suppresses HDAC activity both in vitro and in vivo cancer models. Lee et al.106 reported that curcumin supressed HDAC4 activity, increased tubulin acetylation and induced apoptosis and cell cycle arrest in medulloblastoma cells. These authors have also shown an antitumor effect of curcumin in a xenograft model, which suggests that it has chemopreventive potential for medulloblastoma.
There is mounting evidence that curcumin may regulate expression of various genes which, in turn, can influences various cancer-associated pathways, e.g. PI3K/Akt, apoptosis, STAT3, NF-kB, MAPK and Wnt signalling pathways107-109, and may also regulate miRNAs. Due to its low oral bioavailability, a concentration of 10μM may not be acheivable in vivo. Interestingly, miRNA expression profiling in curcumin treated pancreatic cancer cells at a concentration of 10 μM/L revealed 11 upregulated and 18 downregulated miRNAs, which in turn, regulates many genes critical for tumorigenesis.110
Neonatal exposure during lactation to curcumin modulated the hepatic detoxifying machinery in lactating dams and F1 generation by inducing the expression of glutathione-S-transferase (GST), cytochrome b5, and CYP as well as acid soluble sulfhydryls 111,112.
Quercetin
Quercetin, a flavanoid found in large amounts in apple skin, onion, tea and red wine, has been studied extensively as a chemotherapeutic model for various cancers. The mutagenicity of quercetin has been known for a long time. Recently, this flavanoid has also drawn greater attention for its chemopreventive properties due to its antioxidant and antitumor potential. Quercetin induces growth arrest by regulating cell cycle and apoptotic genes in human esophageal squamous cell carcinoma113, breast114, lung115 and liver116 cancer cells. Tan et al.117 found reversal of hypermethylation and reactivation of the p16INK4a gene in a quercetin-treated human colon RKO cancer cell line. Quercetin also mediates decreases in histone H3 acetylation in prostate cancer, resulting in reduced expression of survivin, a member of a family of inhibitors of apoptosis.118 These results suggested an interference of quercetin with methylation patterns and histone alterations that contributes to the initiation and progression of human cancers.
Maternal diet containing quercetin significantly enhanced expression of Cyp1a1, Cyp1b1, Nqo1 and Ugt1a6 in mouse fetal liver tissues at GD14.5 which also persisted into adulthood in a tissue- and gender-dependent manner.119 Prenatal quercetin exposure also produced hypomethylation of repetitive elements (SINEB1) in liver of adult female mice. Vanhees et al.16 have also shown that transplacental exposure to quercetin introduces double-stranded breaks into DNA and also causes rearrangement of the mixed-lineage leukemia (MLL) gene in humanized transgenic mice (homozygous/heterozygous expressing human ATM mutation, Atm-ΔSRI). There are still some unanswered questions regarding ideal dosage and whether quercetin is a safe supplement in cancer therapy.
(−) Epigallocatechin-3-gallate
EGCG, a green-tea-derived polyphenol derived from the Camellia sinensis plant, has potential clinical applications for preventing various diseases and as a chemotherapeutic agent in several cancers. EGCG has been reported to efficiently block the growth of cancer cell lines, and so is a potential tool for cancer prevention. Berletch et al.120 observed decreases in promoter methylation in human telomerase reverse transcriptase (hTERT), increased histone H3Lys9 acetylation in MCF-7 breast cancer cells and HL60 promyelocytic leukemia cells with EGCG treatment. Reduced DNMT1, DNMT3a and DNMT3b expression, histone deacetylase activity and increased acetylated activity of H3 and H4 was observed in EGCG-treated in vitro and in vivo models of skin cancer.121
Little research has been undertaken on the impact of green tea and EGCG on the fetus. The transplacental chemoprevention for lymphoma by green tea in our murine model appeared to be due to caffeine although decaffeinated green tea and EGCG alone decreased lung tumor multiplicity by 40%.93 It has also been shown that maternal tea intake, not caffeine, during the preconceptual period is associated with developmental neural tube defects122. Some maternal toxicity was also seen after the administration of >91% pure EGCG to pregnant rats during organogenesis, however, there was no evidence of embryotoxicity123. Park et al.124 studied the effect of green tea extract on fetal skeletal abnormalities in rats induced by cyclophosphamide. Pregnant rats were given 100 mg/kg green tea extract by gavage and 11 mg/kg cyclophosphamide by intraperitoneal injection. Administration of cyclophosphamide along with green tea extract caused body weight loss and fetal abnormalities. Furthermore, repeated green tea treatment resulted in increased expression of CYP2B and reduced the expression of CYP3A. The practical consequence of this observation suggests that repeated intake of green tea results in fetal deformity by altering the expression of CYP2B and CYP3B genes. These initial studies raise important question/concern(s) of whether epigenetic mechanisms contribute substantially to toxicity.
Monoterpenes
Limonene
Monoterpenes are naturally occurring compounds that contain many plant essential oils. Limonene (also known as d-limonene) from orange peel; perillyl alcohol from lavender and peppermint have been widely used in cancer chemoprevention. Elegbede et al.125 found that rats treated with 7,12-dimethylbenz[a]anthracene (DMBA), along with 5% orange peel oil, had a significantly lower risk of developing mammary cancer than with DMBA alone. DMBA transplacental and translactation carcinogenesis models, developed by Rao and Hashima126, observed reduction in multiple-site tumor incidence in the F1 generation. The chemopreventive action of the food seasoning spice mixture garam masala (Indian spice), containing monoterpenes significantly reduced the tumor incidence in F1 progeny from 62% to 19% when dams were fed garam masala during GD13-19 and to 10% when fed during GD15-17. A similar trend in tumor incidence at multiple sites in F1 progeny was also seen in a translactational carcinogenesis model when garam masala was given to lactating mice at concentrations of 10 and 30 mg/day during the first 15 days of lactation. The mechanism by which limonene has chemopreventive effect is still unclear. This transplacental and translactational chemopreventive model has been shown to effect F1 generation, but the transmission across multiple generations has not been thoroughly studied. Reports on transgenerational inheritance in humans are also very limited. While unanswered questions still remain, evidence points to a window of susceptibility early in life when the developing fetus is particularly vulnerable to environmental influences. Whether there is any connection between epigenetic and genetic regulation by limonene has not been well established. More detailed mechanistic studies are required to assess the potential usefulness of monoterpenes in cancer chemoprevention.
Isoflavones
Daidzein and Genistein
Isoflavones from soybeans, daidzein and genistein, show tremendous potential as anticancer agents. These naturally occurring compounds, widely distributed in many soy products and certain herbs such as red clover and have attracted much attention recently due to their antioxidant and antitumor properties in prostate, breast, colon, pancreas and cervix.127
Isoflavones, from soybeans, exhibit estrogenic and antiestrogenic effects and have provided beneficial effects among postmenopausal women.128 Daidzein conversion by intestinal bacteria to equol, another isoflavone, has been shown to be more potent than estradiol in inducing the expression of the estrogen-responsive pS2 gene and in effecting cellular proliferation in MCF7 breast cancer cells.129 Guo et al.130 highlighted the antiproliferative effects of daidzein on estrogen positive (MiaPaCa-2) and estrogen negative (PANC-1) pancreatic cancer cells. Effects of daidzein and genistein were also reported by Singh-Gupta et al.131 in mice bearing a PC-3-derived prostate tumor; daidzein inhibited metastasis to lymph nodes and act as a radiosensitizer.
Fang et al.132 found a strong effect of genistein on reversal of epigenetic silencing of RARβ and an additional role in inhibiting DNMT and HDAC activity as compared to daidzein and Biochanin A in androgen-responsive human prostate carcinoma (LNCaP) and PC-3 prostate cells. The demethylating effect of daidzein and genistein was also observed on BRCA1, EPHB2 and GSTP1 genes in human prostate cancer DU-145 and the PC-3 cell lines.133
Diadzein and genistein can cross the placenta and have been found in breast milk and animal studies have shown protection against cancer when given during early stages of development.134,135 Supplementing the maternal diet with genistein during gestation alters the coat color of heterozygous Avy/a offspring toward pseudoagouti due to increased expression of the Agouti gene.7 Hypermethylation of repetitive elements was observed during prenatal exposure of genistein which also effects fetal erythropoiesis.136 All of these results suggest that genistein, daidzein and other isoflavones may produce additive or even synergistic effects in inhibiting cancer growth.
Carotenoids
β-carotene
Carotenoids are yellow/orange/red pigments in fruits, vegetables and whole grains. β-Carotene, the main source of vitamin A and its derivative retinoic acid, has been reported to have beneficial effects in patients with asthma, cataracts and heart disease137-139 and it also has been linked to reduced prostate cancer risk. The effect of maternal β-carotene supplementation, during pregnancy and lactation, in the hope of improving fetal loss and infant survival, was assessed in 43,559 women in Nepal. Compared with control, β-carotene supplementation did not have any discernable effect in reducing fetal loss and early infant death140. Evidence of such an effect has also been reported in Bangladesh, where the weekly use of vitamin A or β-carotene did not reduce fetal loss or infant mortality141. It remains to be seen whether β-carotene, in combination with other vitamins and dietary components can improve fetal and infant survival and has the potential for epigenetic reprogramming.
Lycopene
Lycopene, a carotenoid, has been found to possess antioxidant and antitumor properties. The highest source of lycopene in the typical diet is tomato, although other food sources such as guava, apricot, watermelon and papaya also provide lycopene but in smaller amounts.142 King-Batoon et al.143 have shown that lycopene at 2 μM alters GSTP1, RARβ and HIN-1 gene expression in non-cancer fibrocystic breast cells by demethylating the promoter region. The results of some epidemiological studies suggest that higher intake of lycopene reduces the risk of cancer144 and cardiovascular diseases.138
The lycopene content in the maternal diet has also been proven to prevent inadequate growth of the fetus145, but the underlying mechanism is still unclear.
Isothiocyanates
Sulforaphane
Isothiocyanates, such as phenethylisothiocyanate (PEITC) and sulforaphane (SFN), have been shown to prevent tumorigenesis in in vitro and in vivo models.146,147 The major source of isothiocyanates, are cruciferous vegetables such as broccoli, Brussels sprouts, kale, radish, cabbage, watercress and turnip. Several lines of evidence provide a strong case for SFN-mediating chemoprotection through various cellular (apoptosis and cell cycle arrest) and epigenetic mechanisms. Recently, the ability of SFN to inhibit DNMT activity has been discovered in breast cancer stem cells.148 Our laboratory has also found that SFN treatments in human T-cell acute lymphoblastic (T-ALL) cells significantly reduce DNMT1 and DNMT3b activity (unpublished data) which supports other studies providing key evidence for SFN impacting epigenetics. The effect of SFN on HDAC activity has also been shown in cancer models.146,147 Sulforaphane has the ability to inhibit breast cancer stem cells and it also plays a role in downregulating the Wnt signaling pathway.148 Meeran et al.149 also tested this compound as a demethylating agent. Although research on SFN as a chemopreventive agent is very promising for many cancers, determining the degree of chemoprotection by SFN for the fetus during in utero and lactational exposure to carcinogens is still a challenging task.
Indoles
Indole-3-Carbinol (I3C) and Diindolylmethane (DIM)
I3C is a common dietary constituent present in almost all cruciferous vegetables. I3C is known to induce estrogen metabolism and has been demonstrated to reduce the risk of breast150 and cervical151 cancers as well as having a significant effect in reducing prostate tumors152 in vitro and in vivo. I3C is converted in the gut into a variety of condensation products.153 DIM, a phytonutrient also found in cruciferous vegetables, is derived from digestion of I3C and is also known to possess cancer chemopreventive and chemotherapeutic properties. I3C and DIM supplementation increase the activity of certain detoxification enzymes and facilitates elimination of carcinogens.152 Gene expression profiling studies have revealed that I3C and DIM regulate the transcription of genes involved in cell cycle regulation, cell proliferation, signal transduction and other cellular processes in prostate cancer cells.152 Our lab has also examined the risk/benefit of these chemoprotective phytochemicals in the transplacental mouse carcinogenesis model employing the PAH dibenzo[def,p]chrysene (DBC, also known as dibenzo[a,l]pyrene). Maternal diet supplementation with I3C from GD9 till weaning significantly reduced lung cancer mortality in offspring to 10 months of age71, which suggests that offspring born to mothers consuming chemoprotective phytochemicals exhibit significant protection against PAH-dependent cancer. Recently, Li et al. examined the effect of DIM on HDAC activity in colon cancer cells and observed the proteasome–mediated degradation of HDAC 1, 2, 3 and 8 with increased protein expression of p21 and p27 genes in HT-29 cells following DIM treatment. Growing evidence suggests that isothiocyanates and cruciferous vegetables provide an avenue for epigenetic therapy to fight cancer.
Stilbenes
Resveratrol
Resveratrol (3,5,4′-trihydroxystilbene) is a potent dietary antioxidant compound present in grapes, peanuts, berries and red wine. Resveratrol possesses promising cancer chemopreventive properties against the three major carcinogenesis stages, initiation, promotion and progression, by inducing phase II drug metabolizing enzymes and inhibiting cyclooxygenases and peroxidases. Resveratrol induces human promyelocytic leukemia cell differentiation.154 Dietary administration of all-trans retinoic acid (ATRA), vitamin D3 and resveratrol alone, or in combination, to MCF-7 and MDA-MB-231 breast cancer cells was associated with reduced PTEN promoter methylation due to DNMT downregulation and p21 upregulation which shows the promise of plant-based dietary agents for cancer prevention.155 Maternal exposure to a wide variety of endocrine disruptors including resveratrol showed transient effects on the reproductive tracts and mammary glands in offsprings.156
Conclusion
Epigenetic mechanisms function as an interface between environmental factors, dietary factors and the genome, and are effective in altering cellular machinery during early embryonal, neonatal development, onset of puberty and old age. The early development stage is highly sensitive to combinations of genetic and environmental insults since the DNA replication rate is high and establishment of the DNA methylation pattern occurs during embryogenesis. With respect to long-term effects on human health and for further generations, epigenetic dysregulation is a hallmark of various diseases and cancer. Various dietary and lifestyle factors and environmental variation experienced by parents, before mating, may have an influence on offspring behavior and development, inherited by germline cells. Recent findings indicate that alterations in the in utero environment promote a permanent change in the gamete epigenome and may have an impact on the transcriptomes of developing organs which perpetuates in successive generations. For instance, BPA exposure in rats during gestation (F0 generation) alters expression of testicular steroid receptor coregulators in F1, F2 and F3 generations.157 Vinclozolin exposure leads to an altered DNA methylation pattern of imprinted genes in subsequent generations158 and also increases infertility by reducing sperm count which lasts for four generations.159 If these environmental agents can reprogram early development and induce transgenerational diseases then protective phytochemicals given in maternal diet may be able to prevent future generations from diseases and cancer. Experimental animal studies have proven that maternal diet is an important factor in shaping the epigenome of her offspring.
The exciting field of nutritional epigenomics is particularly concerned with how nutrients affect the epigenome and potential as a chemotherapy medication for prevention and treatment of various diseases including cancer. Although individual dietary components targeting DNA methylation, HDAC activity and miRNAs discussed in this review article, have chemopreventive potential, the effect of a combination of dietary components, which is still at early stage, could be a promising strategy for chemoprevention and therapy (Fig. 3).
Despite significant advances, little is known about the mechanism of action of cancer protection by many phytochemicals and many questions remain to be answered such as:
Although, these dietary phytochemicals are considered to be safe, what dosages/concentrations are likely to be effective in humans and by what mechanism(s)?
In transplacental studies, what is the critical time window in in utero exposure and during certain growth phases?
What is the bioavailability of dietary supplements?
Nanomaterials encapsulation technology enhances bioavailability and offers significant promise for chemoprevention. Recently, a few studies attempted nano therapies in human cancer cell lines by encapsulating various dietary phytochemicals such as curcumin, resveratrol, EGCG and genistein (reviewed in 160) in nanoparticles to circumvent the issues of poor bioavailability with dietary agents.
Reliable high throughput approaches such as methylation microarrays, transcriptomics and proteomics profiling to identify diet modulated signatures, which might serve as surrogate biomarkers, have implications for cancer chemoprevention and can also be used in transplacental fetal therapy.
Acknowledgements
We apologize to authors whose contributions we may have overlooked.
Funding: This work was supported by National Institute of Health (NIH) grant P01 CA90890.
Abbreviations
- ALL
Acute Lymphoblastic Leukemia
- Avy
Agouti viable yellow
- BPA
Bisphenol A
- CYP2E1
Cytochrome P450 2E1
- DIM
3,3′-Diindolylmethane
- DNMT
DNA methytransferase
- EGCG
Epigallocatechin-3-gallate
- GD1
Gestational Day1
- I3C
Indole-3-carbinol
- miRNA
microRNA
- PAHs
Polycyclic Aromatic Hydrocarbons
- SFN
Sulforaphane
Footnotes
Conflict of Interest Statement: The authors declare no conflict of interest.
References
- 1.Howlader N NA, Krapcho M, Neyman N, Aminou R, et al. SEER Cancer Statistics Review, 1975-2008. National Cancer Institute; Bethesda, MD: 2011. [Google Scholar]
- 2.Society AC. Cancer Facts and Figures 2012. Atlanta, GA: 2012. [Google Scholar]
- 3.Costas K, Knorr RS, Condon SK. A case-control study of childhood leukemia in Woburn, Massachusetts: the relationship between leukemia incidence and exposure to public drinking water. Sci. Total Environ. 2002 Dec 2;300(1-3):23–35. doi: 10.1016/s0048-9697(02)00169-9. [DOI] [PubMed] [Google Scholar]
- 4.Infante-Rivard C, Labuda D, Krajinovic M, Sinnett D. Risk of childhood leukemia associated with exposure to pesticides and with gene polymorphisms. Epidemiology. 1999 Sep;10(5):481–487. [PubMed] [Google Scholar]
- 5.Shu XO, Gao YT, Brinton LA, et al. A population-based case-control study of childhood leukemia in Shanghai. Cancer. 1988 Aug 1;62(3):635–644. doi: 10.1002/1097-0142(19880801)62:3<635::aid-cncr2820620332>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 6.Shu XO, Stewart P, Wen WQ, et al. Parental occupational exposure to hydrocarbons and risk of acute lymphocytic leukemia in offspring. Cancer Epidemiol. Biomarkers Prev. 1999 Sep;8(9):783–791. [PubMed] [Google Scholar]
- 7.Dolinoy DC, Weidman JR, Waterland RA, Jirtle RL. Maternal genistein alters coat color and protects Avy mouse offspring from obesity by modifying the fetal epigenome. Environ Health Perspect. 2006 Apr;114(4):567–572. doi: 10.1289/ehp.8700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Link A, Balaguer F, Goel A. Cancer chemoprevention by dietary polyphenols: promising role for epigenetics. Biochem. Pharmacol. 2010 Dec 15;80(12):1771–1792. doi: 10.1016/j.bcp.2010.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meeran SM, Ahmed A, Tollefsbol TO. Epigenetic targets of bioactive dietary components for cancer prevention and therapy. Clin Epigenetics. 2010 Dec 1;1(3-4):101–116. doi: 10.1007/s13148-010-0011-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perera F, Herbstman J. Prenatal environmental exposures, epigenetics, and disease. Reprod. Toxicol. 2011 Apr;31(3):363–373. doi: 10.1016/j.reprotox.2010.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herceg Z. Epigenetics and cancer: towards an evaluation of the impact of environmental and dietary factors. Mutagenesis. 2007 Mar;22(2):91–103. doi: 10.1093/mutage/gel068. [DOI] [PubMed] [Google Scholar]
- 12.Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001 Aug 10;293(5532):1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- 13.Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat Rev Genet. 2002 Sep;3(9):662–673. doi: 10.1038/nrg887. [DOI] [PubMed] [Google Scholar]
- 14.Jensen CD, Block G, Buffler P, Ma X, Selvin S, Month S. Maternal dietary risk factors in childhood acute lymphoblastic leukemia (United States) Cancer Causes Control. 2004 Aug;15(6):559–570. doi: 10.1023/B:CACO.0000036161.98734.17. [DOI] [PubMed] [Google Scholar]
- 15.Dolinoy DC, Huang D, Jirtle RL. Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. Proc Natl Acad Sci U S A. 2007 Aug 7;104(32):13056–13061. doi: 10.1073/pnas.0703739104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vanhees K, de Bock L, Godschalk RWL, van Schooten FJ, van Doorn-Khosrovani SBV. Prenatal Exposure to Flavonoids: Implication for Cancer Risk. Toxicological Sciences. 2011 Mar;120(1):59–67. doi: 10.1093/toxsci/kfq388. [DOI] [PubMed] [Google Scholar]
- 17.Burdge GC, Lillycrop KA. Nutrition, epigenetics, and developmental plasticity: implications for understanding human disease. Annu Rev Nutr. 2010 Aug 21;30:315–339. doi: 10.1146/annurev.nutr.012809.104751. [DOI] [PubMed] [Google Scholar]
- 18.Sandovici I, Sapienza C. PRDM9 sticks its zinc fingers into recombination hotspots and between species. F1000 Biol Rep. 2010:2. doi: 10.3410/B2-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gutierrez MI, Siraj AK, Bhargava M, et al. Concurrent methylation of multiple genes in childhood ALL: Correlation with phenotype and molecular subgroup. Leukemia. 2003 Sep;17(9):1845–1850. doi: 10.1038/sj.leu.2403060. [DOI] [PubMed] [Google Scholar]
- 20.Takeuchi S, Matsushita M, Zimmermann M, et al. Clinical significance of aberrant DNA methylation in childhood acute lymphoblastic leukemia. Leuk. Res. 2011 Oct;35(10):1345–1349. doi: 10.1016/j.leukres.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hesson LB, Dunwell TL, Cooper WN, et al. The novel RASSF6 and RASSF10 candidate tumour suppressor genes are frequently epigenetically inactivated in childhood leukaemias. Mol Cancer. 2009;8:42. doi: 10.1186/1476-4598-8-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dunwell TL, Hesson LB, Pavlova T, et al. Epigenetic analysis of childhood acute lymphoblastic leukemia. Epigenetics. 2009 Apr 1;4(3):185–193. doi: 10.4161/epi.4.3.8752. [DOI] [PubMed] [Google Scholar]
- 23.Corn PG, Kuerbitz SJ, van Noesel MM, et al. Transcriptional silencing of the p73 gene in acute lymphoblastic leukemia and Burkitt’s lymphoma is associated with 5′ CpG island methylation. Cancer Res. 1999 Jul 15;59(14):3352–3356. [PubMed] [Google Scholar]
- 24.Doherty AS, Mann MR, Tremblay KD, Bartolomei MS, Schultz RM. Differential effects of culture on imprinted H19 expression in the preimplantation mouse embryo. Biol Reprod. 2000 Jun;62(6):1526–1535. doi: 10.1095/biolreprod62.6.1526. [DOI] [PubMed] [Google Scholar]
- 25.Lane M, Gardner DK. Ammonium induces aberrant blastocyst differentiation, metabolism, pH regulation, gene expression and subsequently alters fetal development in the mouse. Biol Reprod. 2003 Oct;69(4):1109–1117. doi: 10.1095/biolreprod.103.018093. [DOI] [PubMed] [Google Scholar]
- 26.Randhawa GS, Cui H, Barletta JA, et al. Loss of imprinting in disease progression in chronic myelogenous leukemia. Blood. 1998 May 1;91(9):3144–3147. [PubMed] [Google Scholar]
- 27.Murphy SK, Huang Z, Wen Y, et al. Frequent IGF2/H19 domain epigenetic alterations and elevated IGF2 expression in epithelial ovarian cancer. Mol Cancer Res. 2006 Apr;4(4):283–292. doi: 10.1158/1541-7786.MCR-05-0138. [DOI] [PubMed] [Google Scholar]
- 28.Ng A, Tang JP, Goh CH, Hui KM. Regulation of the H19 imprinting gene expression in human nasopharyngeal carcinoma by methylation. Int J Cancer. 2003 Mar 20;104(2):179–187. doi: 10.1002/ijc.10926. [DOI] [PubMed] [Google Scholar]
- 29.Kuerbitz SJ, Pahys J, Wilson A, Compitello N, Gray TA. Hypermethylation of the imprinted NNAT locus occurs frequently in pediatric acute leukemia. Carcinogenesis. 2002 Apr;23(4):559–564. doi: 10.1093/carcin/23.4.559. [DOI] [PubMed] [Google Scholar]
- 30.Gallou-Kabani C, Gabory A, Tost J, et al. Sex- and diet-specific changes of imprinted gene expression and DNA methylation in mouse placenta under a high-fat diet. PLoS One. 2010;5(12):e14398. doi: 10.1371/journal.pone.0014398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gong L, Pan YX, Chen H. Gestational low protein diet in the rat mediates Igf2 gene expression in male offspring via altered hepatic DNA methylation. Epigenetics. 2010 Oct 1;5(7):619–626. doi: 10.4161/epi.5.7.12882. [DOI] [PubMed] [Google Scholar]
- 32.Parsons DW, Li M, Zhang X, et al. The genetic landscape of the childhood cancer medulloblastoma. Science. 2011 Jan 28;331(6016):435–439. doi: 10.1126/science.1198056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mi S, Lu J, Sun M, et al. MicroRNA expression signatures accurately discriminate acute lymphoblastic leukemia from acute myeloid leukemia. Proc Natl Acad Sci U S A. 2007 Dec 11;104(50):19971–19976. doi: 10.1073/pnas.0709313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Michaud EJ, van Vugt MJ, Bultman SJ, Sweet HO, Davisson MT, Woychik RP. Differential expression of a new dominant agouti allele (Aiapy) is correlated with methylation state and is influenced by parental lineage. Genes Dev. 1994 Jun 15;8(12):1463–1472. doi: 10.1101/gad.8.12.1463. [DOI] [PubMed] [Google Scholar]
- 35.Millar SE, Miller MW, Stevens ME, Barsh GS. Expression and transgenic studies of the mouse agouti gene provide insight into the mechanisms by which mammalian coat color patterns are generated. Development. 1995 Oct;121(10):3223–3232. doi: 10.1242/dev.121.10.3223. [DOI] [PubMed] [Google Scholar]
- 36.Sheldon CC, Rouse DT, Finnegan EJ, Peacock WJ, Dennis ES. The molecular basis of vernalization: the central role of FLOWERING LOCUS C (FLC) Proc Natl Acad Sci U S A. 2000 Mar 28;97(7):3753–3758. doi: 10.1073/pnas.060023597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feil R. Environmental and nutritional effects on the epigenetic regulation of genes. Mutat Res. 2006 Aug 30;600(1-2):46–57. doi: 10.1016/j.mrfmmm.2006.05.029. [DOI] [PubMed] [Google Scholar]
- 38.Vanhees K, van Schooten FJ, Moonen EJ, Maas LM, van Waalwijk van Doorn-Khosrovani SB, Godschalk RW. Maternal intake of quercetin during gestation alters ex vivo benzo[a]pyrene metabolism and DNA adduct formation in adult offspring. Mutagenesis. 2012 Jul;27(4):445–451. doi: 10.1093/mutage/ges002. [DOI] [PubMed] [Google Scholar]
- 39.Hoyme HE, May PA, Kalberg WO, et al. A practical clinical approach to diagnosis of fetal alcohol spectrum disorders: clarification of the 1996 institute of medicine criteria. Pediatrics. 2005 Jan;115(1):39–47. doi: 10.1542/peds.2004-0259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaminen-Ahola N, Ahola A, Maga M, et al. Maternal ethanol consumption alters the epigenotype and the phenotype of offspring in a mouse model. PLoS Genet. 2010 Jan;6(1):e1000811. doi: 10.1371/journal.pgen.1000811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hilakivi-Clarke L, Cabanes A, de Assis S, et al. In utero alcohol exposure increases mammary tumorigenesis in rats. Br J Cancer. 2004 Jun 1;90(11):2225–2231. doi: 10.1038/sj.bjc.6601793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jones SM, Boobis AR, Moore GE, Stanier PM. Expression of CYP2E1 during human fetal development: methylation of the CYP2E1 gene in human fetal and adult liver samples. Biochem. Pharmacol. 1992 Apr 15;43(8):1876–1879. doi: 10.1016/0006-2952(92)90726-y. [DOI] [PubMed] [Google Scholar]
- 43.Tong M LL, Nguyen QG, Chen WC, Spaisman A, Monte SM. Acetaldehyde-Mediated Neurotoxicity: Relevance to Fetal Alcohol SpectrumDisorders. Oxidative Medicine and Cellular Longevity. 2011:1–13. [Google Scholar]
- 44.Infante-Rivard C, Krajinovic M, Labuda D, Sinnett D. Childhood acute lymphoblastic leukemia associated with parental alcohol consumption and polymorphisms of carcinogen-metabolizing genes. Epidemiology. 2002 May;13(3):277–281. doi: 10.1097/00001648-200205000-00007. [DOI] [PubMed] [Google Scholar]
- 45.Bielawski DM, Zaher FM, Svinarich DM, Abel EL. Paternal alcohol exposure affects sperm cytosine methyltransferase messenger RNA levels. Alcohol Clin Exp Res. 2002 Mar;26(3):347–351. [PubMed] [Google Scholar]
- 46.Guo W, Crossey EL, Zhang L, et al. Alcohol exposure decreases CREB binding protein expression and histone acetylation in the developing cerebellum. PLoS One. 2011;6(5):e19351. doi: 10.1371/journal.pone.0019351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Manikkam M, Guerrero-Bosagna C, Tracey R, Haque MM, Skinner MK. Transgenerational actions of environmental compounds on reproductive disease and identification of epigenetic biomarkers of ancestral exposures. PLoS One. 2012;7(2):e31901. doi: 10.1371/journal.pone.0031901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robison LL, Buckley JD, Daigle AE, et al. Maternal drug use and risk of childhood nonlymphoblastic leukemia among offspring. An epidemiologic investigation implicating marijuana (a report from the Childrens Cancer Study Group) Cancer. 1989 May 15;63(10):1904–1911. [PubMed] [Google Scholar]
- 49.Desaulniers D, Xiao GH, Lian H, et al. Effects of mixtures of polychlorinated biphenyls, methylmercury, and organochlorine pesticides on hepatic DNA methylation in prepubertal female Sprague-Dawley rats. Int J Toxicol. 2009 Jul-Aug;28(4):294–307. doi: 10.1177/1091581809337918. [DOI] [PubMed] [Google Scholar]
- 50.Cadmium and cadmium compounds IARC Monogr Eval Carcinog Risks Hum. 1993;58:119–237. [PMC free article] [PubMed] [Google Scholar]
- 51.Holt D, Webb M. Teratogenicity of ionic cadmium in the Wistar rat. Arch Toxicol. 1987 Apr;59(6):443–447. doi: 10.1007/BF00316212. [DOI] [PubMed] [Google Scholar]
- 52.Soukupova D, Dostal M. Developmental toxicity of cadmium in mice. I. Embryotoxic effects. Funct Dev Morphol. 1991;1(2):3–9. [PubMed] [Google Scholar]
- 53.Ronco AM, Llaguno E, Epunan MJ, Llanos MN. Effect of cadmium on cortisol production and 11beta-hydroxysteroid dehydrogenase 2 expression by cultured human choriocarcinoma cells (JEG-3) Toxicol In Vitro. 2010 Sep;24(6):1532–1537. doi: 10.1016/j.tiv.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 54.Myers GJ, Davidson PW, Shamlaye CF, et al. Effects of prenatal methylmercury exposure from a high fish diet on developmental milestones in the Seychelles Child Development Study. Neurotoxicology. 1997;18(3):819–829. [PubMed] [Google Scholar]
- 55.Mushak P, Davis JM, Crocetti AF, Grant LD. Prenatal and postnatal effects of low-level lead exposure: integrated summary of a report to the U.S. Congress on childhood lead poisoning. Environ. Res. 1989 Oct;50(1):11–36. doi: 10.1016/s0013-9351(89)80046-5. [DOI] [PubMed] [Google Scholar]
- 56.Basha MR, Wei W, Bakheet SA, et al. The fetal basis of amyloidogenesis: exposure to lead and latent overexpression of amyloid precursor protein and beta-amyloid in the aging brain. J Neurosci. 2005 Jan 26;25(4):823–829. doi: 10.1523/JNEUROSCI.4335-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu J, Basha MR, Brock B, et al. Alzheimer’s disease (AD)-like pathology in aged monkeys after infantile exposure to environmental metal lead (Pb): evidence for a developmental origin and environmental link for AD. J Neurosci. 2008 Jan 2;28(1):3–9. doi: 10.1523/JNEUROSCI.4405-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pilsner JR, Hu H, Ettinger A, et al. Influence of prenatal lead exposure on genomic methylation of cord blood DNA. Environ Health Perspect. 2009 Sep;117(9):1466–1471. doi: 10.1289/ehp.0800497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Martinez VD, Vucic EA, Becker-Santos DD, Gil L, Lam WL. Arsenic exposure and the induction of human cancers. J Toxicol. 2011;2011:431287. doi: 10.1155/2011/431287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reichard JF, Puga A. Effects of arsenic exposure on DNA methylation and epigenetic gene regulation. Epigenomics. 2010 Feb;2(1):87–104. doi: 10.2217/epi.09.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Waalkes MP, Liu J, Germolec DR, et al. Arsenic exposure in utero exacerbates skin cancer response in adulthood with contemporaneous distortion of tumor stem cell dynamics. Cancer Res. 2008 Oct 15;68(20):8278–8285. doi: 10.1158/0008-5472.CAN-08-2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xie Y, Liu J, Benbrahim-Tallaa L, et al. Aberrant DNA methylation and gene expression in livers of newborn mice transplacentally exposed to a hepatocarcinogenic dose of inorganic arsenic. Toxicology. 2007 Jul 1;236(1-2):7–15. doi: 10.1016/j.tox.2007.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu G, Niu Z, Van Niekerk D, Xue J, Zheng L. Polycyclic aromatic hydrocarbons (PAHs) from coal combustion: emissions, analysis, and toxicology. Rev Environ Contam Toxicol. 2008;192:1–28. doi: 10.1007/978-0-387-71724-1_1. [DOI] [PubMed] [Google Scholar]
- 64.Bostrom CE, Gerde P, Hanberg A, et al. Cancer risk assessment, indicators, and guidelines for polycyclic aromatic hydrocarbons in the ambient air. Environ Health Perspect. 2002 Jun;110(Suppl 3):451–488. doi: 10.1289/ehp.110-1241197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Miller RL, Garfinkel R, Horton M, et al. Polycyclic aromatic hydrocarbons, environmental tobacco smoke, and respiratory symptoms in an inner-city birth cohort. Chest. 2004 Oct;126(4):1071–1078. doi: 10.1378/chest.126.4.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Miller RL, Ho SM. Environmental epigenetics and asthma: current concepts and call for studies. Am. J. Respir. Crit. Care Med. 2008 Mar 15;177(6):567–573. doi: 10.1164/rccm.200710-1511PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Perera F, Tang WY, Herbstman J, et al. Relation of DNA methylation of 5′-CpG island of ACSL3 to transplacental exposure to airborne polycyclic aromatic hydrocarbons and childhood asthma. PLoS One. 2009;4(2):e4488. doi: 10.1371/journal.pone.0004488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Herbstman JB, Tang D, Zhu D, et al. Prenatal exposure to polycyclic aromatic hydrocarbons, benzo[a]pyrene-DNA adducts, and genomic DNA methylation in cord blood. Environ Health Perspect. 2012 May;120(5):733–738. doi: 10.1289/ehp.1104056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Anderson LM, Diwan BA, Fear NT, Roman E. Critical windows of exposure for children’s health: cancer in human epidemiological studies and neoplasms in experimental animal models. Environ Health Perspect. 2000 Jun;108(Suppl 3):573–594. doi: 10.1289/ehp.00108s3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Miller MS. Transplacental lung carcinogenesis: a pharmacogenetic mouse model for the modulatory role of cytochrome P450 1A1 on lung cancer initiation. Chem Res Toxicol. 1994 Jul-Aug;7(4):471–481. doi: 10.1021/tx00040a001. [DOI] [PubMed] [Google Scholar]
- 71.Yu Z, Mahadevan B, Lohr CV, et al. Indole-3-carbinol in the maternal diet provides chemoprotection for the fetus against transplacental carcinogenesis by the polycyclic aromatic hydrocarbon dibenzo[a,l]pyrene. Carcinogenesis. 2006 Oct;27(10):2116–2123. doi: 10.1093/carcin/bgl072. [DOI] [PubMed] [Google Scholar]
- 72.www.cdc.gov.
- 73.Calafat AM, Ye X, Wong LY, Reidy JA, Needham LL. Exposure of the U.S. population to bisphenol A and 4-tertiary-octylphenol: 2003-2004. Environ Health Perspect. 2008 Jan;116(1):39–44. doi: 10.1289/ehp.10753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ouchi K, Watanabe S. Measurement of bisphenol A in human urine using liquid chromatography with multi-channel coulometric electrochemical detection. J Chromatogr B Analyt Technol Biomed Life Sci. 2002 Nov 25;780(2):365–370. doi: 10.1016/s1570-0232(02)00547-0. [DOI] [PubMed] [Google Scholar]
- 75.Bromer JG, Zhou Y, Taylor MB, Doherty L, Taylor HS. Bisphenol-A exposure in utero leads to epigenetic alterations in the developmental programming of uterine estrogen response. FASEB J. 2010 Jul;24(7):2273–2280. doi: 10.1096/fj.09-140533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Prins GS, Tang WY, Belmonte J, Ho SM. Perinatal exposure to oestradiol and bisphenol A alters the prostate epigenome and increases susceptibility to carcinogenesis. Basic Clin Pharmacol Toxicol. 2008 Feb;102(2):134–138. doi: 10.1111/j.1742-7843.2007.00166.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Komashko VM, Farnham PJ. 5-azacytidine treatment reorganizes genomic histone modification patterns. Epigenetics. 2010 Apr 4;5(3) doi: 10.4161/epi.5.3.11409. [DOI] [PubMed] [Google Scholar]
- 78.Lin J, Haffner MC, Zhang Y, et al. Disulfiram is a DNA demethylating agent and inhibits prostate cancer cell growth. Prostate. 2011 Mar 1;71(4):333–343. doi: 10.1002/pros.21247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ropero S, Esteller M. The role of histone deacetylases (HDACs) in human cancer. Mol Oncol. 2007 Jun;1(1):19–25. doi: 10.1016/j.molonc.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Deroanne CF, Bonjean K, Servotte S, et al. Histone deacetylases inhibitors as anti-angiogenic agents altering vascular endothelial growth factor signaling. Oncogene. 2002 Jan 17;21(3):427–436. doi: 10.1038/sj.onc.1205108. [DOI] [PubMed] [Google Scholar]
- 81.Bolden JE, Peart MJ, Johnstone RW. Anticancer activities of histone deacetylase inhibitors. Nat Rev Drug Discov. 2006 Sep;5(9):769–784. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- 82.Qian X, Ara G, Mills E, LaRochelle WJ, Lichenstein HS, Jeffers M. Activity of the histone deacetylase inhibitor belinostat (PXD101) in preclinical models of prostate cancer. Int J Cancer. 2008 Mar 15;122(6):1400–1410. doi: 10.1002/ijc.23243. [DOI] [PubMed] [Google Scholar]
- 83.Marks PA, Xu WS. Histone deacetylase inhibitors: Potential in cancer therapy. J Cell Biochem. 2009 Jul 1;107(4):600–608. doi: 10.1002/jcb.22185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gimsing P, Hansen M, Knudsen LM, et al. A phase I clinical trial of the histone deacetylase inhibitor belinostat in patients with advanced hematological neoplasia. Eur J Haematol. 2008 Sep;81(3):170–176. doi: 10.1111/j.1600-0609.2008.01102.x. [DOI] [PubMed] [Google Scholar]
- 85.Marks PA, Breslow R. Dimethyl sulfoxide to vorinostat: development of this histone deacetylase inhibitor as an anticancer drug. Nat Biotechnol. 2007 Jan;25(1):84–90. doi: 10.1038/nbt1272. [DOI] [PubMed] [Google Scholar]
- 86.Reid T, Valone F, Lipera W, et al. Phase II trial of the histone deacetylase inhibitor pivaloyloxymethyl butyrate (Pivanex, AN-9) in advanced non-small cell lung cancer. Lung Cancer. 2004 Sep;45(3):381–386. doi: 10.1016/j.lungcan.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 87.Traynor AM, Dubey S, Eickhoff JC, et al. Vorinostat (NSC# 701852) in patients with relapsed non-small cell lung cancer: a Wisconsin Oncology Network phase II study. J Thorac Oncol. 2009 Apr;4(4):522–526. doi: 10.1097/jto.0b013e3181952478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fabbri M, Garzon R, Cimmino A, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci U S A. 2007 Oct 2;104(40):15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Costello JF, Plass C. Methylation matters. J. Med. Genet. 2001 May;38(5):285–303. doi: 10.1136/jmg.38.5.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Thurn KT, Thomas S, Moore A, Munster PN. Rational therapeutic combinations with histone deacetylase inhibitors for the treatment of cancer. Future Oncol. 2011 Feb;7(2):263–283. doi: 10.2217/fon.11.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sato N, Maitra A, Fukushima N, et al. Frequent hypomethylation of multiple genes overexpressed in pancreatic ductal adenocarcinoma. Cancer Res. 2003 Jul 15;63(14):4158–4166. [PubMed] [Google Scholar]
- 92.Craig WJ. Health-promoting properties of common herbs. Am J Clin Nutr. 1999 Sep;70(3 Suppl):491S–499S. doi: 10.1093/ajcn/70.3.491s. [DOI] [PubMed] [Google Scholar]
- 93.Castro DJ, Yu Z, Lohr CV, et al. Chemoprevention of dibenzo[a,l]pyrene transplacental carcinogenesis in mice born to mothers administered green tea: primary role of caffeine. Carcinogenesis. 2008 Aug;29(8):1581–1586. doi: 10.1093/carcin/bgm237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shorey LE, Castro DJ, Baird WM, et al. Transplacental carcinogenesis with dibenzo[def,p]chrysene (DBC): timing of maternal exposures determines target tissue response in offspring. Cancer Lett. 2012 Apr 1;317(1):49–55. doi: 10.1016/j.canlet.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lee WJ, Zhu BT. Inhibition of DNA methylation by caffeic acid and chlorogenic acid, two common catechol-containing coffee polyphenols. Carcinogenesis. 2006 Feb;27(2):269–277. doi: 10.1093/carcin/bgi206. [DOI] [PubMed] [Google Scholar]
- 96.Kang NJ, Lee KW, Shin BJ, et al. Caffeic acid, a phenolic phytochemical in coffee, directly inhibits Fyn kinase activity and UVB-induced COX-2 expression. Carcinogenesis. 2009 Feb;30(2):321–330. doi: 10.1093/carcin/bgn282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lee WJ, Shim JY, Zhu BT. Mechanisms for the inhibition of DNA methyltransferases by tea catechins and bioflavonoids. Mol Pharmacol. 2005 Oct;68(4):1018–1030. doi: 10.1124/mol.104.008367. [DOI] [PubMed] [Google Scholar]
- 98.Waldecker M, Kautenburger T, Daumann H, Busch C, Schrenk D. Inhibition of histone-deacetylase activity by short-chain fatty acids and some polyphenol metabolites formed in the colon. Journal of Nutritional Biochemistry. 2008 Sep;19(9):587–593. doi: 10.1016/j.jnutbio.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 99.Bora-Tatar G, Dayangac-Erden D, Demir AS, Dalkara S, Yelekci K, Erdem-Yurter H. Molecular modifications on carboxylic acid derivatives as potent histone deacetylase inhibitors: Activity and docking studies. Bioorganic & Medicinal Chemistry. 2009 Jul 15;17(14):5219–5228. doi: 10.1016/j.bmc.2009.05.042. [DOI] [PubMed] [Google Scholar]
- 100.Han H, Du B, Pan X, et al. CADPE inhibits PMA-stimulated gastric carcinoma cell invasion and matrix metalloproteinase-9 expression by FAK/MEK/ERK-mediated AP-1 activation. Mol Cancer Res. 2010 Nov;8(11):1477–1488. doi: 10.1158/1541-7786.MCR-10-0114. [DOI] [PubMed] [Google Scholar]
- 101.Bakker R, Steegers EA, Obradov A, Raat H, Hofman A, Jaddoe VW. Maternal caffeine intake from coffee and tea, fetal growth, and the risks of adverse birth outcomes: the Generation R Study. Am J Clin Nutr. 2010 Jun;91(6):1691–1698. doi: 10.3945/ajcn.2009.28792. [DOI] [PubMed] [Google Scholar]
- 102.Xu D, Zhang B, Liang G, et al. Caffeine-induced activated glucocorticoid metabolism in the hippocampus causes hypothalamic-pituitary-adrenal axis inhibition in fetal rats. PLoS One. 2012;7(9):e44497. doi: 10.1371/journal.pone.0044497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bracken MB, Triche EW, Belanger K, Hellenbrand K, Leaderer BP. Association of maternal caffeine consumption with decrements in fetal growth. Am J Epidemiol. 2003 Mar 1;157(5):456–466. doi: 10.1093/aje/kwf220. [DOI] [PubMed] [Google Scholar]
- 104.Link FB A, Shen Y, Jose Lozano J, Leung HE, Boland CR, Goel A. M1182 Novel Evidence for Curcumin-Induced DNA Methylation Changes in Colon Cancer Cells. Gastroenterology. 2010;138(5, Supplement 1):S–349. [Google Scholar]
- 105.Liu Z, Xie Z, Jones W, et al. Curcumin is a potent DNA hypomethylation agent. Bioorganic & Medicinal Chemistry Letters. 2009 Feb 1;19(3):706–709. doi: 10.1016/j.bmcl.2008.12.041. [DOI] [PubMed] [Google Scholar]
- 106.Lee SJ, Krauthauser C, Maduskuie V, Fawcett PT, Olson JM. Rajasekaran SA. Curcumin-induced HDAC inhibition and attenuation of medulloblastoma growth in vitro and in vivo. BMC Cancer. 2011;11:144. doi: 10.1186/1471-2407-11-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Teiten MH, Gaascht F, Cronauer M, Henry E, Dicato M, Diederich M. Anti-proliferative potential of curcumin in androgen-dependent prostate cancer cells occurs through modulation of the Wingless signaling pathway. International Journal of Oncology. 2011 Mar;38(3):603–611. doi: 10.3892/ijo.2011.905. [DOI] [PubMed] [Google Scholar]
- 108.Zheng M, Ekmekcioglu S, Walch ET, Tang CH, Grimm EA. Inhibition of nuclear factor-kappaB and nitric oxide by curcumin induces G2/M cell cycle arrest and apoptosis in human melanoma cells. Melanoma Res. 2004 Jun;14(3):165–171. doi: 10.1097/01.cmr.0000129374.76399.19. [DOI] [PubMed] [Google Scholar]
- 109.Sarkar FH, Li Y. Cell signaling pathways altered by natural chemopreventive agents. Mutat Res. 2004 Nov 2;555(1-2):53–64. doi: 10.1016/j.mrfmmm.2004.04.015. [DOI] [PubMed] [Google Scholar]
- 110.Sun M, Estrov Z, Ji Y, Coombes KR, Harris DH, Kurzrock R. Curcumin (diferuloylmethane) alters the expression profiles of microRNAs in human pancreatic cancer cells. Mol Cancer Ther. 2008 Mar;7(3):464–473. doi: 10.1158/1535-7163.MCT-07-2272. [DOI] [PubMed] [Google Scholar]
- 111.Singh A, Singh SP, Bamezai R. Postnatal modulation of hepatic biotransformation system enzymes via translactational exposure of F1 mouse pups to turmeric and curcumin. Cancer Lett. 1995 Sep 4;96(1):87–93. doi: 10.1016/0304-3835(95)03909-g. [DOI] [PubMed] [Google Scholar]
- 112.Singh A, Singh SP, Bamezai R. Effect of arecoline on the curcumin-modulated hepatic biotransformation system enzymes in lactating mice and translactationally exposed F1 pups. Nutr Cancer. 1996;25(1):101–110. doi: 10.1080/01635589609514431. [DOI] [PubMed] [Google Scholar]
- 113.Zhang Q, Zhao XH, Wang ZJ. Cytotoxicity of flavones and flavonols to a human esophageal squamous cell carcinoma cell line (KYSE-510) by induction of G2/M arrest and apoptosis. Toxicol In Vitro. 2009 Aug;23(5):797–807. doi: 10.1016/j.tiv.2009.04.007. [DOI] [PubMed] [Google Scholar]
- 114.Jeong JH, An JY, Kwon YT, Rhee JG, Lee YJ. Effects of low dose quercetin: cancer cell-specific inhibition of cell cycle progression. J Cell Biochem. 2009 Jan 1;106(1):73–82. doi: 10.1002/jcb.21977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yang JH, Hsia TC, Kuo HM, et al. Inhibition of lung cancer cell growth by quercetin glucuronides via G(2)/M arrest and induction of apoptosis. Drug Metabolism and Disposition. 2006 Feb;34(2):296–304. doi: 10.1124/dmd.105.005280. [DOI] [PubMed] [Google Scholar]
- 116.Mu C, Jia P, Yan Z, Liu X, Li X, Liu H. Quercetin induces cell cycle G1 arrest through elevating Cdk inhibitors p21 and p27 in human hepatoma cell line (HepG2) Methods Find Exp Clin Pharmacol. 2007 Apr;29(3):179–183. doi: 10.1358/mf.2007.29.3.1092095. [DOI] [PubMed] [Google Scholar]
- 117.Tan SN, Wang C, Lu CL, et al. Quercetin Is Able to Demethylate the p16INK4a Gene Promoter. Chemotherapy. 2009;55(1):6–10. doi: 10.1159/000166383. [DOI] [PubMed] [Google Scholar]
- 118.Kim YH, Lee DH, Jeong JH, Guo ZS, Lee YJ. Quercetin augments TRAIL-induced apoptotic death: Involvement of the ERK signal transduction pathway. Biochemical Pharmacology. 2008 May 15;75(10):1946–1958. doi: 10.1016/j.bcp.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Vanhees K, van Schooten FJ, Moonen EJ, Maas LM, Barjesteh van Waalwijk van Doorn-Khosrovani S, Godschalk RW. Maternal intake of quercetin during gestation alters ex vivo benzo[a]pyrene metabolism and DNA adduct formation in adult offspring. Mutagenesis. 2012 Feb 14; doi: 10.1093/mutage/ges002. [DOI] [PubMed] [Google Scholar]
- 120.Berletch JB, Liu C, Love WK, Andrews LG, Katiyar SK, Tollefsbol TO. Epigenetic and genetic mechanisms contribute to telomerase inhibition by EGCG. Journal of Cellular Biochemistry. 2008 Feb 1;103(2):509–519. doi: 10.1002/jcb.21417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Nandakumar V, Vaid M, Katiyar SK. (-)-Epigallocatechin-3-gallate reactivates silenced tumor suppressor genes, Cip1/p21 and p16INK4a, by reducing DNA methylation and increasing histones acetylation in human skin cancer cells. Carcinogenesis. 2011 Apr;32(4):537–544. doi: 10.1093/carcin/bgq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Correa A, Stolley A, Liu Y. Prenatal tea consumption and risks of anencephaly and spina bifida. Ann Epidemiol. 2000 Oct 1;10(7):476–477. doi: 10.1016/s1047-2797(00)00144-7. [DOI] [PubMed] [Google Scholar]
- 123.Isbrucker RA, Edwards JA, Wolz E, Davidovich A, Bausch J. Safety studies on epigallocatechin gallate (EGCG) preparations. Part 3: teratogenicity and reproductive toxicity studies in rats. Food Chem Toxicol. 2006 May;44(5):651–661. doi: 10.1016/j.fct.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 124.Park D, Jeon JH, Shin S, et al. Green tea extract increases cyclophosphamide-induced teratogenesis by modulating the expression of cytochrome P-450 mRNA. Reprod Toxicol. 2009 Jan;27(1):79–84. doi: 10.1016/j.reprotox.2008.11.058. [DOI] [PubMed] [Google Scholar]
- 125.Elegbede JA, Elson CE, Qureshi A, Tanner MA, Gould MN. Inhibition of DMBA-induced mammary cancer by the monoterpene d-limonene. Carcinogenesis. 1984 May;5(5):661–664. doi: 10.1093/carcin/5.5.661. [DOI] [PubMed] [Google Scholar]
- 126.Rao AR, Hashim S. Chemopreventive action of oriental food-seasoning spices mixture Garam masala on DMBA-induced transplacental and translactational carcinogenesis in mice. Nutr Cancer. 1995;23(1):91–101. doi: 10.1080/01635589509514365. [DOI] [PubMed] [Google Scholar]
- 127.Sarkar FH, Li Y. Soy isoflavones and cancer prevention. Cancer Invest. 2003;21(5):744–757. doi: 10.1081/cnv-120023773. [DOI] [PubMed] [Google Scholar]
- 128.Messina M, Wu AH. Perspectives on the soy-breast cancer relation. Am J Clin Nutr. 2009 May;89(5):1673S–1679S. doi: 10.3945/ajcn.2009.26736V. [DOI] [PubMed] [Google Scholar]
- 129.Sathyamoorthy N, Wang TT. Differential effects of dietary phyto-oestrogens daidzein and equol on human breast cancer MCF-7 cells. Eur J Cancer. 1997 Dec;33(14):2384–2389. doi: 10.1016/s0959-8049(97)00303-1. [DOI] [PubMed] [Google Scholar]
- 130.Guo JM, Xiao BX, Dai DJ, Liu Q, Ma HH. Effects of daidzein on estrogen-receptor-positive and negative pancreatic cancer cells in vitro. World J Gastroenterol. 2004 Mar 15;10(6):860–863. doi: 10.3748/wjg.v10.i6.860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Singh-Gupta V, Zhang H, Yunker CK, et al. Daidzein effect on hormone refractory prostate cancer in vitro and in vivo compared to genistein and soy extract: potentiation of radiotherapy. Pharm Res. 2010 Jun;27(6):1115–1127. doi: 10.1007/s11095-010-0107-9. [DOI] [PubMed] [Google Scholar]
- 132.Fang MZ, Chen DP, Sun Y, Jin Z, Christman JK, Yang CS. Reversal of hypermethylation and reactivation of p16(INK4a) RAR beta, and MGMT genes by genistein and other isoflavones from soy. Clinical Cancer Research. 2005 Oct 1;11(19):7033–7041. doi: 10.1158/1078-0432.CCR-05-0406. [DOI] [PubMed] [Google Scholar]
- 133.Adjakly M, Bosviel R, Rabiau N, et al. DNA methylation and soy phytoestrogens: quantitative study in DU-145 and PC-3 human prostate cancer cell lines. Epigenomics. 2011 Dec;3(6):795–803. doi: 10.2217/epi.11.103. [DOI] [PubMed] [Google Scholar]
- 134.Todaka E, Sakurai K, Fukata H, et al. Fetal exposure to phytoestrogens--the difference in phytoestrogen status between mother and fetus. Environ Res. 2005 Oct;99(2):195–203. doi: 10.1016/j.envres.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 135.Nagata C, Iwasa S, Shiraki M, et al. Associations among maternal soy intake, isoflavone levels in urine and blood samples, and maternal and umbilical hormone concentrations (Japan) Cancer Causes Control. 2006 Nov;17(9):1107–1113. doi: 10.1007/s10552-006-0044-4. [DOI] [PubMed] [Google Scholar]
- 136.Vanhees K, Coort S, Ruijters EJ, Godschalk RW, van Schooten FJ, Barjesteh van Waalwijk van Doorn-Khosrovani S. Epigenetics: prenatal exposure to genistein leaves a permanent signature on the hematopoietic lineage. FASEB J. 2011 Feb;25(2):797–807. doi: 10.1096/fj.10-172155. [DOI] [PubMed] [Google Scholar]
- 137.Rimm EB, Stampfer MJ, Ascherio A, Giovannucci E, Colditz GA, Willett WC. Vitamin E consumption and the risk of coronary heart disease in men. N Engl J Med. 1993 May 20;328(20):1450–1456. doi: 10.1056/NEJM199305203282004. [DOI] [PubMed] [Google Scholar]
- 138.Gaziano JM, Manson JE, Branch LG, Colditz GA, Willett WC, Buring JE. A prospective study of consumption of carotenoids in fruits and vegetables and decreased cardiovascular mortality in the elderly. Ann Epidemiol. 1995 Jul;5(4):255–260. doi: 10.1016/1047-2797(94)00090-g. [DOI] [PubMed] [Google Scholar]
- 139.Sahyoun NR, Jacques PF, Russell RM. Carotenoids, vitamins C and E, and mortality in an elderly population. Am J Epidemiol. 1996 Sep 1;144(5):501–511. doi: 10.1093/oxfordjournals.aje.a008957. [DOI] [PubMed] [Google Scholar]
- 140.Katz J, West KP, Jr., Khatry SK, et al. Maternal low-dose vitamin A or beta-carotene supplementation has no effect on fetal loss and early infant mortality: a randomized cluster trial in Nepal. Am J Clin Nutr. 2000 Jun;71(6):1570–1576. doi: 10.1093/ajcn/71.6.1570. [DOI] [PubMed] [Google Scholar]
- 141.West KP, Jr., Christian P, Labrique AB, et al. Effects of vitamin A or beta carotene supplementation on pregnancy-related mortality and infant mortality in rural Bangladesh: a cluster randomized trial. JAMA. 2011 May 18;305(19):1986–1995. doi: 10.1001/jama.2011.656. [DOI] [PubMed] [Google Scholar]
- 142.Chalabi N, Le Corre L, Maurizis JC, Bignon YJ, Bernard-Gallon DJ. The effects of lycopene on the proliferation of human breast cells and BRCA1 and BRCA2 gene expression. Eur J Cancer. 2004 Jul;40(11):1768–1775. doi: 10.1016/j.ejca.2004.03.028. [DOI] [PubMed] [Google Scholar]
- 143.King-Batoon A, Leszczynska JM, Klein CB. Modulation of gene methylation by genistein or lycopene in breast cancer cells. Environ Mol Mutagen. 2008 Jan;49(1):36–45. doi: 10.1002/em.20363. [DOI] [PubMed] [Google Scholar]
- 144.Giovannucci E. A review of epidemiologic studies of tomatoes, lycopene, and prostate cancer. Exp Biol Med (Maywood) 2002 Nov;227(10):852–859. doi: 10.1177/153537020222701003. [DOI] [PubMed] [Google Scholar]
- 145.Sharma JB, Kumar A, Malhotra M, Arora R, Prasad S, Batra S. Effect of lycopene on pre-eclampsia and intra-uterine growth retardation in primigravidas. Int J Gynaecol Obstet. 2003 Jun;81(3):257–262. doi: 10.1016/s0020-7292(03)00040-7. [DOI] [PubMed] [Google Scholar]
- 146.Myzak MC, Dashwood WM, Orner GA, Ho E, Dashwood RH. Sulforaphane inhibits histone deacetylase in vivo and suppresses tumorigenesis in Apc-minus mice. FASEB J. 2006 Mar;20(3):506–508. doi: 10.1096/fj.05-4785fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Myzak MC, Tong P, Dashwood WM, Dashwood RH, Ho E. Sulforaphane retards the growth of human PC-3 xenografts and inhibits HDAC activity in human subjects. Exp Biol Med (Maywood) 2007 Feb;232(2):227–234. [PMC free article] [PubMed] [Google Scholar]
- 148.Li Y, Zhang T, Korkaya H, et al. Sulforaphane, a dietary component of broccoli/broccoli sprouts, inhibits breast cancer stem cells. Clin Cancer Res. 2010 May 1;16(9):2580–2590. doi: 10.1158/1078-0432.CCR-09-2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Meeran SM, Patel SN, Tollefsbol TO. Sulforaphane causes epigenetic repression of hTERT expression in human breast cancer cell lines. PLoS One. 2010;5(7):e11457. doi: 10.1371/journal.pone.0011457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Grubbs CJ, Steele VE, Casebolt T, et al. Chemoprevention of chemically-induced mammary carcinogenesis by indole-3-carbinol. Anticancer Res. 1995 May-Jun;15(3):709–716. [PubMed] [Google Scholar]
- 151.Jin L, Qi M, Chen DZ, et al. Indole-3-carbinol prevents cervical cancer in human papilloma virus type 16 (HPV16) transgenic mice. Cancer Res. 1999 Aug 15;59(16):3991–3997. [PubMed] [Google Scholar]
- 152.Li Y, Li X, Sarkar FH. Gene expression profiles of I3C- and DIM-treated PC3 human prostate cancer cells determined by cDNA microarray analysis. J Nutr. 2003 Apr;133(4):1011–1019. doi: 10.1093/jn/133.4.1011. [DOI] [PubMed] [Google Scholar]
- 153.Aggarwal BB, Ichikawa H, Garodia P, et al. From traditional Ayurvedic medicine to modern medicine: identification of therapeutic targets for suppression of inflammation and cancer. Expert Opin Ther Targets. 2006 Feb;10(1):87–118. doi: 10.1517/14728222.10.1.87. [DOI] [PubMed] [Google Scholar]
- 154.Jang M, Cai L, Udeani GO, et al. Cancer chemopreventive activity of resveratrol, a natural product derived from grapes. Science. 1997 Jan 10;275(5297):218–220. doi: 10.1126/science.275.5297.218. [DOI] [PubMed] [Google Scholar]
- 155.Stefanska B, Salame P, Bednarek A, Fabianowska-Majewska K. Comparative effects of retinoic acid, vitamin D and resveratrol alone and in combination with adenosine analogues on methylation and expression of phosphatase and tensin homologue tumour suppressor gene in breast cancer cells. Br J Nutr. 2011 Aug 1;:1–10. doi: 10.1017/S0007114511003631. [DOI] [PubMed] [Google Scholar]
- 156.Nikaido Y, Yoshizawa K, Danbara N, et al. Effects of maternal xenoestrogen exposure on development of the reproductive tract and mammary gland in female CD-1 mouse offspring. Reprod. Toxicol. 2004 Aug-Sep;18(6):803–811. doi: 10.1016/j.reprotox.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 157.Salian S, Doshi T, Vanage G. Impairment in protein expression profile of testicular steroid receptor coregulators in male rat offspring perinatally exposed to Bisphenol A. Life Sci. 2009 Jul 3;85(1-2):11–18. doi: 10.1016/j.lfs.2009.04.005. [DOI] [PubMed] [Google Scholar]
- 158.Stouder C, Paoloni-Giacobino A. Transgenerational effects of the endocrine disruptor vinclozolin on the methylation pattern of imprinted genes in the mouse sperm. Reproduction. 2010 Feb;139(2):373–379. doi: 10.1530/REP-09-0340. [DOI] [PubMed] [Google Scholar]
- 159.Anway MD, Leathers C, Skinner MK. Endocrine disruptor vinclozolin induced epigenetic transgenerational adult-onset disease. Endocrinology. 2006 Dec;147(12):5515–5523. doi: 10.1210/en.2006-0640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Irfana Muqbil AM, Sarkar Fazlul H., Mohammad Ramzi M., Azmi Asfar S. Progress in Nanotechnology Based Approaches to Enhance the Potential of Chemopreventive Agents. Cancer. 2011;3:428–445. doi: 10.3390/cancers3010428. [DOI] [PMC free article] [PubMed] [Google Scholar]