SUMMARY
Streptococcus agalactiae is a genetically diverse organism; when typed by pulsed-field gel electrophoresis (PFGE), multiple types appear within a single serotype. We tested whether S. agalactiae PFGE types correspond to a specific serotype within individuals, and different individuals from the same geographic area. A total of 872 S. agalactiae isolates from 152 healthy individuals were classified by PFGE and capsular serotype. Serotype V was the most homogeneous (Simpson’s diversity index 0.54); and types III, II and Ib were mostly heterogeneous (Simpson’s diversity index ≥0.90). Within an individual, isolates with the same PFGE patterns had identical capsular types, but across individuals the same PFGE types sometimes occurred in different serotypes. Capsular type alone is insufficient to define epidemiological relatedness. Although PFGE types appear to be a valid surrogate for capsular typing of isolates from the same individual, it is not a valid surrogate for serotype in isolates from different individuals.
Keywords: Diversity, Group B Streptococcus
INTRODUCTION
Streptococcus agalactiae, also called Group B Streptococcus (GBS), is a facultative Gram-positive coccus distinguished from other types of Streptococcus by the presence of a type B-specific carbohydrate antigen known as the C substance [1, 2]. There are nine known serotypes, the distribution of which vary with geographic location, although serotypes Ia, II, III and V predominate in invasive disease [3]. S. agalactiae is a common colonist of the gastrointestinal and genitourinary tracts, and can be found in up to 40% of the female population and 30% of the male population [4, 5]. It was first noted in the 1970s as a leading cause of neonatal disease but over the last two decades the species has emerged as an invasive pathogen in pregnant women and the elderly [6, 7]. While the incidence of early-onset neonatal disease and infection in pregnant women has declined over the past decade due to prenatal screening, that of late-onset neonatal disease has remained stable, and the incidence of disease in the elderly has increased [6, 8–12].
S. agalactiae is a genetically diverse organism. When characterized using pulsed-field gel electrophoresis (PFGE) multiple PFGE types often share the same serotype [13, 14]. However, the level of genetic diversity within the various molecular capsular types, as characterized by differing PFGE types, remains unclear. Previous studies have used PFGE types as a surrogate for serotyping, presuming that isolates from a single individual with the same PFGE type have the same serotype [5, 15]; however, this presumption has been based on the examination of a small number of isolates [16]. Given the importance of correctly characterizing capsular types, both for understanding transmission dynamics and vaccine formulation, there is a need to validate the presumption that isolates from one individual with the same PFGE type have the same capsular type.
Over time, the proportion of S. agalactiae isolates that are non-typable using the Lancefield serotyping method has increased [10], suggesting that the use of PFGE to infer capsular type in different individuals may be desirable. However, it is noteworthy that one study reported a single mother–child pair of isolates that were identical by PFGE but with different serotypes [13]. The capsular serotype of a strain could potentially be altered by as little as the horizontal transfer of a single gene [17, 18] which might not result in a change in PFGE type. Therefore the reliability of using PFGE profiles as a predictor of capsular type for isolates from different individuals, even from within the same geographic area, warrants investigation.
We examined a large collection of S. agalactiae isolates from individual cases in order to describe the genetic diversity of these isolates and test the hypotheses that S. agalactiae PFGE types correspond to a specific serotype within an individual, and between different individuals from the same geographic area.
METHODS
Study protocol
As described previously [19, 20], 738 male and female University of Michigan students were recruited to provide specimens from throat and mouth, vagina, anal orifice, and urine samples. In total, 913 S. agalactiae isolates were collected over a 12-week period, a subset of which was from students who provided multiple samples over the study period. Isolates were serologically confirmed with the Slidex Strepto B kit (bioMérieux, USA).
PFGE
Of the 913 original isolates, 882 were typed using PFGE and patterns were analysed for relatedness by visual inspection and using the BioNumerics software (Applied Maths, Belgium) as described previously [5, 20]. A subset of the isolates either failed to yield restriction fragments with SmaI (13), or were not analysed with BioNumerics (6), or were not viable at the time of the present study (12).
Capsular typing using library on a slide hybridization
Sixteen of the isolates from the original collection were not viable. The remaining 897 isolates were grown in 9 ml Todd–Hewitt broth (THB) (BD, USA) at 37 °C in 5%CO2 for 36 h. Genomic DNA was extracted and purified using sonication and heat treatment as previously described [21]. The DNA from each isolate was arrayed in duplicate on Vivid Gene Array slides with nylon membranes (Pall, USA) using the VersArray ChipWriter Compact system (Bio- Rad, USA).
PCR amplification of probe sequences was performed as follows: DNA lysates were prepared from 3 ml S. agalactiae overnight culture in THB. Each reaction mixture (48 μl) included 1 μl of each primer (25 pmol), 1 μl DNA lysate, and 45 μl Platinum PCR SuperMix (Invitrogen, USA). The primers and PCR conditions used to amplify capsular types Ia, Ib, II, III, IV, V and VI–VIII genes were as described previously [12, 22], with the following modifications: 94 °C for 30 s, 49 °C (type V) or 55 °C (type III) for 30 s, and 72 °C for 25 s, for 35 cycles. PCR product (10 μl) was analysed by electrophoresis on a 1% agarose gel, stained with 0.02 μg ethidium bromide/ml, and visualized by UV transillumination.
PCR amplification products were purified using the QIAquick gel extraction kit and the QIAquick PCR purification kit (Qiagen Inc., USA) and fluorescein labelled with 2.5 μl of 1mM fluorescein-12-dCTP (PerkinElmer, USA) using a modification of the BioPrime DNA Labeling System (Invitrogen).
The probes were hybridized to genomic DNA arrayed on slides in duplicate using previously described methods [23] and hybridization signals were detected using a ArrayIt Microarray SpotWare Colorimetric Scanner (TeleChem International Inc., USA) and analysed with IconoClust software (Clondiag Chip Technologies, Germany). The signal intensity of the spots was adjusted for background and normalized to the signal intensity of a quantification probe which consisted of a mixture of S. agalactiae housekeeping genes (adhP, glcK, glnA, pheS). These values were used to account for differences in genomic DNA concentrations at different spots. Cut-offs for probe positive strains were established using previously published methods [24].
A hierarchical strategy for capsular-type confirmation was employed, such that at least one isolate with a unique PFGE pattern was assigned a capsular type via traditional dot-blot hybridization, in addition to the Library on a Slide dot-blot hybridization. Isolates with non-confirmatory capsular-type results in the Library on a Slide assay or that were discrepant with both traditional and Library on a Slide were confirmed by PCR and dot-blot hybridization methods using the primer sequences and methods described earlier [12, 22]; 892/897 isolates originally printed on the Library on a Slide were confirmed using the hierarchal typing strategy and were assigned a unique capsular-type designation.
RESULTS
Of the 892 isolates assigned a unique capsular type and 882 isolates assigned a PFGE pattern, a total of 872 S. agalactiae isolates from 152 individuals had both PFGE pattern data and a unique capsular-type assignment. These 872 isolates were used for the final analyses. Overall, an average of six isolates were available from each individual.
Capsular type
The capsular-type distribution of the S. agalactiae isolates was examined in the final collection. In order to account for the possibility that an individual was persistently colonized with the same organism over the course of the study, the capsular-type distribution of the collection was also examined by choosing one isolate from each unique PFGE pattern from a given individual (168). The capsular-type distributions were very similar whether analysing one isolate or all isolates per person, suggesting few individuals were colonized with more than one serotype (Fig. 1).
Fig. 1.
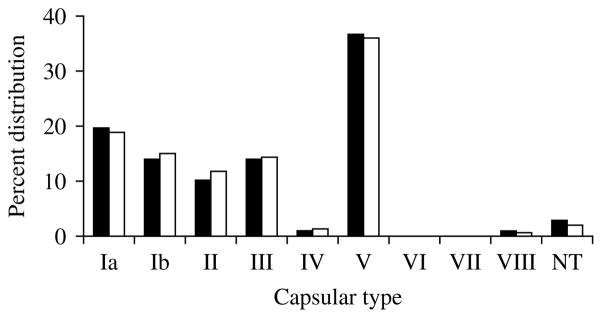
Capsular-type distribution of S. agalactiae isolates collected between January and April 2001, from healthy male and female University of Michigan undergraduates based on the total collection (■, n=872) and a subgroup collection of one isolate for each unique PFGE pattern from an individual (□, n=168). NT, Non-typable.
As reported previously, based on results of traditional dot-blot hybridization to determine capsular type [20], there was a predominance of type V isolates (322, 36·9%). Additionally the collection included type Ia (170, 19·4%), type III (124, 14·2%), type Ib (123, 14·1%), type II (88, 10·1%), type VIII (10, 1·1%) and type IV (8, 0·9%). Only 27 isolates from the entire collection were non-typable (3%) by all molecular capsular-typing strategies. Interestingly, while a majority of the isolates from a given individual had the same capsular type, nine (5·9%) of the 152 individuals were concurrently colonized with two different capsular types; an additional two individuals were colonized with two different capsular types over the course of the study.
Diversity within capsular type
Fifty-two PFGE patterns were identified in the 872 isolates. To explore genetic diversity we chose one representative isolate for each PFGE pattern from each individual, resulting in a total of 168 isolates with both a PFGE type and a capsular type. Based on Simpson’s diversity index, which indicates the probability of randomly choosing two isolates of a given capsular type with a different PFGE type [25, 26], capsular types III, II and Ib exhibited the greatest heterogeneity; while capsular type V isolates were the most homogeneous (Table 1).
Table 1.
Simpson’s diversity index of PFGE within capsular serotypes of S. agalactiae
| No. of isolates | Simpson’s diversity index | 95% CI | |
|---|---|---|---|
| Ia | 32 | 0·72 | 0·546–0·896 |
| Ib | 25 | 0·90 | 0·832–0·961 |
| II | 20 | 0·93 | 0·901–0·973 |
| III | 24 | 0·94 | 0·908–0· ·982 |
| V | 61 | 0·54 | 0·391–0·691 |
| All | 168 | 0·92 | 0·893–0·952 |
PFGE type within individuals
Fourteen of the 152 individuals (9·2%) carried isolates with more than one PFGE type. In three of these individuals, all isolates, which included an average of seven isolates per person, were of the same capsular type. In the remaining 11 individuals, one had three isolates with unique PFGE patterns, representing two distinct capsular types, nine persons had two PFGE types representing two distinct capsular types (Fig. 2), and one individual had 11 isolates representing two PFGE types and two serotypes. Two of the isolates sharing PFGE types had different serotypes. There was a 2·0% probability of an individual having more than one unique PFGE type but representing a single capsular type and a 6·6%probability of an individual having more than one unique PFGE type each representing a distinct capsular type. We observed one individual (1/152) where both these phenomena occurred (0·7%). Therefore, within a given individual, while differing PFGE patterns potentially represented identical capsular types, the probability that all isolates with a unique PFGE pattern represent the same capsular type was 99%, which may reflect technical rather than true biological variability.
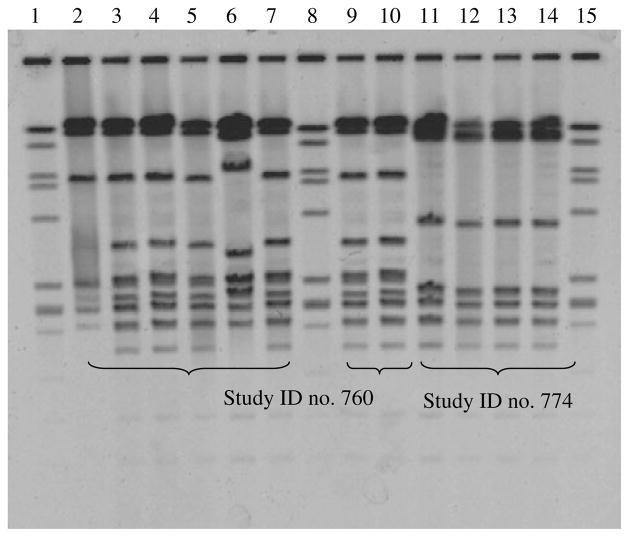
Fig. 2.
Representative PFGE pattern of 12 colonizing S. agalactiae isolates recovered from two healthy individuals: Group B Streptococcus recovered from throat, rectal and urine samples of study participant no. 760 are shown in lanes 2–7 and 9–10, and from study participant no. 774 in lanes 11–14. Lanes 2–5, 7 and 9–10 from participant no. 760 were capsular type III, while lane 6 was capsular type II. Lanes 11–14 from participant no. 774 were capsular type V. Lanes 1, 8, 15 are control strain A909.
PFGE type in individuals
When examining the distribution of capsular types and PFGE patterns across individuals, 10/52 (19·2%) identified PFGE patterns occurred in more than one capsular type. Seven PFGE patterns included two capsular types, while three PFGE patterns included three capsular types. Based on these results, there could be between an 8·3% and 15·4% error rate in capsular-type assignment if PFGE was used as a surrogate for capsular typing in these individuals from similar geographic areas.
DISCUSSION
DNA sequencing of various strains of S. agalactiae has demonstrated the genetic diversity of the species and isolates of different serotypes have been noted to share more sequence similarities than isolates of the same serotype [27, 28]. Tettelin et al. suggest that the level of S. agalactiae genetic diversity and evolutionary patterns are not adequately reflected with serotype classification [27]; thus the level of heterogeneity within serotype classification could vary widely. We describe the heterogeneity of S. agalactiae and the impact of this heterogeneity on scientific inference by examining the PFGE patterns and capsular types of 872 isolates from 152 individuals. S. agalactiae is highly heterogeneous but the heterogeneity varies by capsular type, with the greatest heterogeneity occurring in capsular types III, Ib and II. Within an individual, isolates with the same PFGE patterns had identical capsular types, while across individuals PFGE types appeared in more than one capsular type. Therefore, capsular type alone was not sufficient to characterize epidemiological relatedness. Although PFGE types appear to be a valid surrogate for capsular typing of isolates from the same individual, it is not a valid surrogate for serotype in isolates from different individuals.
The heterogeneity we observed is consistent with previous reports describing much smaller collections [14, 29], and as observed in these earlier studies, capsular type V was the most homogeneous [14, 30]. These studies did not report a diversity index score and so we cannot make a direct comparison with our study findings. Interestingly, the relative homogeneity of type V isolates is consistent with the emergence of a predominant capsular type V clonal lineage [31].
Assuming that a common PFGE type represented the same capsular type across individuals would have resulted in an error rate in capsular-type assignment of as much as 15%. Thus, using 2–3 band differences in PFGE pattern might not be sufficiently stringent for outbreak investigations [32], as geographically and temporally linked isolates with distinct serotypes shared almost identical PFGE patterns, providing a strong potential for linking epidemiologically unrelated strains and obscuring the role and routes of transmission in colonization and disease.
Multiple researchers have demonstrated that S. agalactiae isolates sharing identical multilocus sequence types (MLST) can possibly represent differing capsular types [18, 33, 34]. Jones et al. described the increased potential of S. agalactiae sequence type 17 to cause invasive disease within neonates when compared to other sequence tyoes independent of capsular type [35]. Given that S. agalactiae capsule is a key determinant of virulence and vaccine development is based on capsular type, it is important to recognize that MLST typing is likely to encounter similar problems as PFGE typing if used as a surrogate for capsular typing.
Our study collection was large, enabling us to characterize the genetic diversity more precisely than had been done previously. Furthermore, we quantified the degree of diversity using Simpson’s diversity index. The consistent effect across a large number of isolates strongly supports the hypothesis that PFGE typing distinguishes between capsular types isolated from a single individual. Using the hierarchical capsular- typing strategy we employed of dot-blot hybridization and PCR, only 27 isolates (3%) of our entire collection could not be assigned a capsular type, consistent with previous reports [3, 36]. The non-typable isolates might contain a defective cps operon not detected using a genotype-based strategy. It is also possible that these few non-typable isolates represent the newly described serotype IX, although the few reported serotype IX isolates were collected from a geographic region distant from our study population [37]. An additional potential limitation of our study is the omission of 20 isolates (2·2%) which had corresponding serotype data but could not be digested and characterized by PFGE. It is possible that exclusion of these isolates could have resulted in a decreased Simpson’s diversity index for some of the capsular types. However, given the small fraction of undigested isolates, it is unlikely that their inclusion would significantly alter the relative heterogeneity of capsular types.
In summary, by PFGE, S. agalactiae is a genetically diverse organism, with diversity varying within capsular types. Neither S. agalactiae capsular-type nor PFGE-type designation alone appear sufficiently discriminatory to determine the relatedness of S. agalactiae isolates, given that either typing method alone can link epidemiologically unrelated isolates or fail to identify epidemiologically related isolates. Thus, a combined typing strategy such as PFGE or MLST with capsular typing should be considered to more adequately understand the epidemiology of S. agalactiae.
Acknowledgments
We gratefully acknowledge Shannon Manning and Sandra McCoy for their work on typing the S. agalactiae isolates by PFGE. We also acknowledge the work of Sheng Li, Elizabeth Levin, Sara McNamara, Anne Styka, Suhael Momin and Caitlin Murphy who assisted with DNA isolation. This work was supported, in part, by R01 AI051675 (B.F.) and T32 HD007513 from the National Institutes of Health.
Footnotes
DECLARATION OF INTEREST
None.
References
- 1.Edwards MS, Baker CJ. Group B streptococcal infections. In: Remington JS, Klein JO, editors. Infectious Diseases of the Fetus and Newborn Infant. Philadelphia: W. B. Saunders; 2000. pp. 1091–1156. [Google Scholar]
- 2.Manning SD. Molecular epidemiology of Streptococcus agalactiae (group B streptococcus) Frontiers in Bioscience. 2003;8:1–18. doi: 10.2741/985. [DOI] [PubMed] [Google Scholar]
- 3.Harrison LH, et al. Serotype distribution of invasive group B streptococcal isolates in Maryland: implications for vaccine formulation. Maryland Emerging Infections Program. Journal of Infectious Diseases. 1998;177:998–1002. doi: 10.1086/515260. [DOI] [PubMed] [Google Scholar]
- 4.Committee on Infectious Diseases, American Academy of Pediatrics. Group B streptococcal infections. In: Pickering LK, editor. Red Book: 2006 Report of the Committee on Infectious Diseases. 27. Elk Grove Village: American Academy of Pediatrics; 2006. pp. 620–627. [Google Scholar]
- 5.Bliss SJ, et al. Group B streptococcus colonization in male and nonpregnant female university students: a cross-sectional prevalence study. Clinical Infectious Diseases. 2002;34:184–190. doi: 10.1086/338258. [DOI] [PubMed] [Google Scholar]
- 6.Schrag SJ, et al. Group B streptococcal disease in the era of intrapartum antibiotic prophylaxis. New England Journal of Medicine. 2000;342:15–20. doi: 10.1056/NEJM200001063420103. [DOI] [PubMed] [Google Scholar]
- 7.Schuchat A. Group B streptococcus. Lancet. 1999;353:51–56. doi: 10.1016/S0140-6736(98)07128-1. [DOI] [PubMed] [Google Scholar]
- 8.Blancas D, et al. Group B streptococcal disease in nonpregnant adults: incidence, clinical characteristics, and outcome. European Journal of Clinical Microbiology and Infectious Disease. 2004;23:168–173. doi: 10.1007/s10096-003-1098-9. [DOI] [PubMed] [Google Scholar]
- 9.Rothrock G, et al. Decreasing incidence of perinatal Group B streptococcal disease – United States, 1993– 1995. Centers for Disease Control and Prevention. Morbidity and Mortality Weekly Report. 1997;46:473–477. [PubMed] [Google Scholar]
- 10.Farley MM. Group B streptococcal disease in nonpregnant adults. Clinical Infectious Diseases. 2001;33:556–561. doi: 10.1086/322696. [DOI] [PubMed] [Google Scholar]
- 11.Schrag SJ, Schuchat A. Easing the burden: characterizing the disease burden of neonatal group B streptococcal disease to motivate prevention. Clinical Infectious Diseases. 2004;38:1209–1211. doi: 10.1086/382889. [DOI] [PubMed] [Google Scholar]
- 12.Borchardt SM, et al. Comparison of DNA dot blot hybridization and Lancefield capillary precipitin methods for group B streptococcal capsular typing. Journal of Clinical Microbiology. 2004;42:146–150. doi: 10.1128/JCM.42.1.146-150.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benson KD, et al. Pulsed-field fingerprinting of vaginal group B streptococcus in pregnancy. Obstetrics and Gynecology. 2002;100:545–551. doi: 10.1016/s0029-7844(02)02139-7. [DOI] [PubMed] [Google Scholar]
- 14.Skjaervold NK, Bergh K, Bevanger L. Distribution of PFGE types of invasive Norwegian group B streptococci in relation to serotypes. Indian Journal of Medical Research. 2004;119:201–204. [PubMed] [Google Scholar]
- 15.Manning SD, et al. Determinants of co-colonization with Group B streptococcus among heterosexual college couples. Epidemiology. 2002;13:533–539. doi: 10.1097/00001648-200209000-00008. [DOI] [PubMed] [Google Scholar]
- 16.Gordillo ME, et al. Typing of group B streptococci: comparison of pulsed-field gel electrophoresis and conventional electrophoresis. Journal of Clinical Microbiology. 1993;31:1430–1434. doi: 10.1128/jcm.31.6.1430-1434.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chaffin DO, et al. The serotype of type Ia and III group B streptococci is determined by the polymerase gene within the polycistronic capsule operon. Journal of Bacteriology. 2000;182:4466–4477. doi: 10.1128/jb.182.16.4466-4477.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luan SL, et al. Multilocus sequence typing of Swedish invasive group B streptococcus isolates indicates a neonatally associated genetic lineage and capsule switching. Journal of Clinical Microbiology. 2005;43:3727–3733. doi: 10.1128/JCM.43.8.3727-3733.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Manning SD, et al. Prevalence of group B streptococcus colonization and potential for transmission by casual contact in healthy young men and women. Clinical Infectious Diseases. 2004;39:380–388. doi: 10.1086/422321. [DOI] [PubMed] [Google Scholar]
- 20.Foxman B, et al. Incidence and duration of group B Streptococcus by serotype among male and female college students living in a single dormitory. American Journal of Epidemiology. 2006;163:544–551. doi: 10.1093/aje/kwj075. [DOI] [PubMed] [Google Scholar]
- 21.Zhang L, et al. Bacterial genomic DNA isolation using sonication for microarray analysis. BioTechniques. 2005;39:640–644. doi: 10.2144/000112038. [DOI] [PubMed] [Google Scholar]
- 22.Wen L, et al. Use of a serotype-specificDNAmicroarray for identification of group B streptococcus (Streptococcus agalactiae) Journal of Clinical Microbiology. 2006;44:1447–1452. doi: 10.1128/JCM.44.4.1447-1452.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang L, et al. Library on a slide for bacterial comparative genomics. BMC Microbiology. 2004;4:1–7. doi: 10.1186/1471-2180-4-12. Published online: 22 March 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang L, et al. Optimization of a fluorescent-based phosphor imaging dot blot DNA hybridization assay to assess E. coli virulence gene profiles. Journal of Microbiological Methods. 2001;44:225–233. doi: 10.1016/s0167-7012(01)00222-6. [DOI] [PubMed] [Google Scholar]
- 25.Hunter PR, Gaston MA. Numerical index of the discriminatory ability of typing systems: an application of Simpson’s index of diversity. Journal of Clinical Microbiology. 1988;26:2465–2466. doi: 10.1128/jcm.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grundmann H, Hori S, Tanner G. Determining confidence intervals when measuring genetic diversity and the discriminatory abilities of typing methods for microorganisms. Journal of Clinical Microbiology. 2001;39:4190–4192. doi: 10.1128/JCM.39.11.4190-4192.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tettelin H, et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial ‘pan-genome’. Proceedings of the National Academy of Sciences USA. 2005;102:13950– 13955. doi: 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tettelin H, et al. Complete genome sequence and comparative genomic analysis of an emerging human pathogen, serotype V Streptococcus agalactiae. Proceedings of the National Academy of Sciences USA. 2002;99:12391–12396. doi: 10.1073/pnas.182380799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perez-Ruiz M, et al. Genetic diversity of Streptococcus agalactiae strains colonizing the same pregnant woman. Epidemiology and Infection. 2004;132:375–378. doi: 10.1017/s0950268803001845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Savoia D, et al. Streptococcus agalactiae in pregnant women: phenotypic and genotypic characters. Journal of Infection. 2008;56:120–125. doi: 10.1016/j.jinf.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 31.Elliott JA, Farmer KD, Facklam RR. Sudden increase in isolation of group B streptococci, serotype V, is not due to emergence of a new pulsed-field gel electrophoresis type. Journal of Clinical Microbiology. 1998;36:2115– 2116. doi: 10.1128/jcm.36.7.2115-2116.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tenover FC, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. Journal of Clinical Microbiology. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davies HD, et al. Multilocus sequence typing of serotype III group B streptococcus and correlation with pathogenic potential. Journal of Infectious Diseases. 2004;189:1097–1102. doi: 10.1086/382087. [DOI] [PubMed] [Google Scholar]
- 34.Jones N, et al. Multilocus sequence typing system for group B streptococcus. Journal of Clinical Microbiology. 2003;41:2530–2536. doi: 10.1128/JCM.41.6.2530-2536.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jones N, et al. Enhanced invasiveness of bovine-derived neonatal sequence type 17 group B streptococcus is independent of capsular serotype. Clinical Infectious Diseases. 2006;42:915–924. doi: 10.1086/500324. [DOI] [PubMed] [Google Scholar]
- 36.Hickman ME, et al. Changing epidemiology of group B streptococcal colonization. Pediatrics. 1999;104:203– 209. doi: 10.1542/peds.104.2.203. [DOI] [PubMed] [Google Scholar]
- 37.Slotved HC, et al. Serotype IX, a proposed new Streptococcus agalactiae serotype. Journal of Clinical Microbiology. 2007;45:2929–2936. doi: 10.1128/JCM.00117-07. [DOI] [PMC free article] [PubMed] [Google Scholar]