Abstract
Genotype imputation provides imputation of untyped SNPs that are present on a reference panel such as those from the HapMap Project. It is popular for increasing statistical power and comparing results across studies using different platforms. Imputation for African American populations is challenging because their LD blocks are shorter and also because no ideal reference panel is available due to admixture. In this paper, we evaluated three imputation strategies for African Americans. The intersection strategy used a combined panel consisting of SNPs polymorphic in both CEU and YRI. The union strategy used a panel consisting of SNPs polymorphic in either CEU or YRI. The merge strategy merged results from two separate imputations, one using CEU and the other using YRI. Because recent investigators are increasingly using the data from the 1000 Genomes (1KG) Project for genotype imputation, we evaluated both 1KG-based imputations and HapMap-based imputations. We used 23,707 SNPs from chromosomes 21 and 22 on Affymetrix SNP Array 6.0 genotyped for 1,075 HyperGEN African Americans. We found that 1KG-based imputations provided a substantially larger number of variants than HapMap-based imputations, about three times as many common variants and eight times as many rare and low frequency variants. This higher yield is expected because the 1KG panel includes more SNPs. Accuracy rates using 1KG data were slightly lower than those using HapMap data before filtering, but slightly higher after filtering. The union strategy provided the highest imputation yield with next highest accuracy. The intersection strategy provided the lowest imputation yield but the highest accuracy. The merge strategy provided the lowest imputation accuracy. We observed that SNPs polymorphic only in CEU had much lower accuracy, reducing the accuracy of the union strategy. Our findings suggest that 1KG-based imputations can facilitate discovery of significant associations for SNPs across the whole MAF spectrum. Because the 1KG Project is still underway, we expect that later versions will provide better imputation performance.
INTRODUCTION
Genotype imputation provides imputation of untyped single nucleotide polymorphisms (SNPs) that are present on a reference panel such as those from the HapMap Project. As reviewed by Marchini and Howie [2010], it is popular with many genome wide association (GWA) studies and meta-analyses for several reasons. First, imputation can boost the statistical power for finding causal SNPs that are not directly typed in GWA studies and can provide a higher resolution of associated regions. Second, it enables researchers to genotype fewer SNPs and then impute the desired untyped SNPs [Anderson et al., 2008]. Those imputed SNPs have been commonly used for many GWA studies [Hao et al., 2009]. For analysis of imputed SNPs, the expected allele count (also called the posterior mean or allele dosage) is commonly used to take account of imputation uncertainties and implemented in several programs such as BIMBAM [Servin and Stephens, 2007], MACH2DAT/MACH2QTL [Li et al., 2010], SNPTEST [Marchini et al., 2007], PLINK [Purcell et al., 2007] and ProbABEL [Aulchenko et al., 2010]. It was also shown to provide a good approximation to a Bayesian approach [Guan and Stephens, 2008]. Third, when multiple studies use different genotyping platforms, imputation using the same HapMap II reference panels produces an identical set of imputed SNPs in all studies regardless of the platform used. This makes comparison of results across studies readily possible [de Bakker et al., 2008].
Genotype imputation for African populations is challenging because their linkage disequilibrium (LD) blocks are shorter than in other populations. Phase II of the HapMap Project provides reference panels for African, European and Asian ancestry: Yoruba ethnic group in Africa (YRI), Utah residents with European ancestry (CEU), Han Chinese in Beijing (CHB) and Japanese in Tokyo (JPT) [The International HapMap Consortium, 2007]. Among 29 populations in the Human Genome Diversity Project, African populations were shown to have the worst imputation accuracy [Huang et al. 2009]. Imputation for admixed African American populations has an additional challenge because no ideal reference panel is available from the HapMap II project. The first strategy is to simply use the YRI reference panel [Fridley et al., 2010] because African American genomes are mostly of African ancestry, with their admixture proportions ranging from 77% to 93% [Parra et al., 1998]. The second strategy is the “cosmopolitan strategy” that combines the CEU and YRI panels to better account for European ancestry [Huang et al., 2010; Shriner et al., 2010]. Huang et al. [2009] showed that, for most populations, imputation using all four panels (CEU, YRI, CHB and JPT) provided accuracy almost as good as imputation using a specific reference panel. However, because their combined reference panel only included SNPs that were polymorphic in multiple panels, there were fewer SNPs, resulting in lower imputation yields. The third strategy is to merge imputed results from two separate imputations, once using the CEU panel and another using the YRI panel, to achieve higher imputation yields [Shriner et al., 2010].
Most genotype imputations have used the reference panels from the HapMap II Project. However, recent investigators are increasingly using the reference panels from the 1000 Genomes (1KG) Project for genotype imputation, detecting stronger associations with SNPs that are not available in the HapMap data [see Sanna et al., 2010, and Ellinghaus et al., 2010, using MACH; Liu et al., 2010, and Padmanabhan et al., 2010, using IMPUTE]. The major advantage of imputations based on the 1KG data, instead of the HapMap data, is the ability to impute a much larger number of variants. The 1KG Project aims to discover most low frequency and common variants across the genome and most rare variants in gene regions [The 1000 Genomes Project Consortium, 2010]. Therefore, 1KG-based imputations should provide many more variants that are rare and low frequency than HapMap-based imputations. However, there are concerns about 1KG-based imputations. The HapMap data were based on direct genotyping of previously discovered SNPs and have been thoroughly scrutinized, whereas currently available 1KG data were based on low-depth whole-genome sequencing data and, hence, are expected to be of lower quality. Furthermore, there have not been many evaluations of 1KG-based imputations [Fridley et al., 2010; The 1000 Genomes Project Consortium, 2010; Li et al., 2011].
In this paper, we had two main objectives. The first objective was to evaluate three imputation strategies that have been used for African Americans. The second objective was to evaluate both 1KG-based imputations and HapMap-based imputations. In particular, we investigated whether the 1KG data provided imputation of a much larger number of rare and low frequency variants. We used 23,707 SNPs from chromosomes 21 and 22 on Affymetrix SNP Array 6.0. Because Affymetrix SNP Array 6.0 contained a relatively small number of rare variants, all rare variants were masked to evaluate imputation performance of rare variants.
MATERIALS AND METHODS
STUDY SAMPLE
In this paper, we used 23,707 SNPs from chromosomes 21 and 22 genotyped for 1,083 African Americans in the Hypertension Genetic Epidemiology Network (HyperGEN). The study recruited African American and Caucasian participants at five field centers to investigate the genetic causes of hypertension and related conditions [Williams et al., 2000]. Study participants were one of three types: 1) individuals in a hypertensive sibship with at least two siblings diagnosed with hypertension; 2) random subjects, who were age-matched with hypertensive sibs; or 3) unmedicated adult offspring of one of the hypertensive siblings. The study obtained informed consent from participants and approval from the appropriate institutional review boards. Most of the African Americans were genotyped on Affymetrix SNP Array 6.0; about 10% were genotyped on Affymetrix SNP Array 5.0. In this paper, we used only those genotyped on Affymetrix SNP Array 6.0. We removed control samples, corrected sample mix-ups and pedigree errors. We removed monomorphic SNPs and SNPs with missing rate >5% or Hardy-Weinberg p-value <10−6 and removed any genotypes with a non-Mendelian pattern of inheritance.
IMPUTATION STRATEGIES
To impute untyped markers in African Americans, we considered three strategies. The first strategy, denoted by INT for intersection, used a combined panel consisting of SNPs polymorphic in both CEU and YRI. The second strategy, denoted by UNI for union, used a combined panel consisting of SNPs polymorphic in either CEU or YRI. The third strategy, denoted by MER for merge, used results from two separate imputations, one using CEU and the other using YRI. Our MER strategy used YRI-based imputations for SNPs polymorphic in YRI and otherwise used CEU-based imputations, because the HyperGEN African Americans had 80.7% African ancestry on average [Zhu et al., 2005]. Therefore, the MER strategy corresponded to adding imputed results of CEU-specific SNPs (that were polymorphic only in CEU) to imputed results using YRI.
To evaluate imputation performance using HapMap and 1KG data, we used haplotypes for CEU and YRI populations from phase II of the HapMap Project (release 22, build 36) and pilot 1 of the 1KG Project (released in June 2010). The CEU panel consisted of 120 haplotypes of the same 60 individuals in both HapMap and 1KG data. The YRI panel consisted of 120 haplotypes in the HapMap data and 118 haplotypes in the 1KG data. The UNI strategy used the combined panel consisting of 240 and 238 haplotypes in the HapMap and 1KG data, respectively. The INT strategy used the same number of haplotypes as in the UNI strategy with a smaller number of SNPs (shown in Table I).
Table I.
Number of SNPs from chromosomes 21 and 22 across the MAF spectrum in HyperGEN African Americans genotyped on Affymetrix SNP Array 6.0, reference panels and three strategies from HapMap II (HMII) and 1000 Genomes (1KG) Projects. Total number of SNPs are red boldfaced.
| Rare | Low Freq | Common | Total | |
|---|---|---|---|---|
| HyperGEN | 294 | 2,317 | 21,096 | 23,707 |
| HMII-YRI | 2,644 | 9,283 | 64,878 | 76,805 |
| HMII-CEU | 2,701 | 7,611 | 57,366 | 67,678 |
| HMII-INT | 82 | 2,493 | 55,522 | 58,097 |
| HMII-UNI | 1,400 | 6,903 | 70,553 | 78,856 |
| HMII-MER | 3,457 | 11,383 | 71,546 | 86,386 |
| 1KG-YRI | 571 | 69,744 | 197,352 | 267,667 |
| 1KG-CEU | 336 | 41,227 | 149,202 | 190,765 |
| 1KG-INT | 0 | 4,760 | 123,334 | 128,094 |
| 1KG-UNI | 13,405 | 62,188 | 254,745 | 330,338 |
| 1KG-MER | 830 | 97,159 | 232,349 | 330,338 |
Rare variants with MAF ≤ 0.01; Low frequency variants with 0.01 < MAF ≤ 0.05; Common variants with MAF > 0.05
INT refers the intersection strategy; UNI refers the union strategy; MER refers the merge strategy
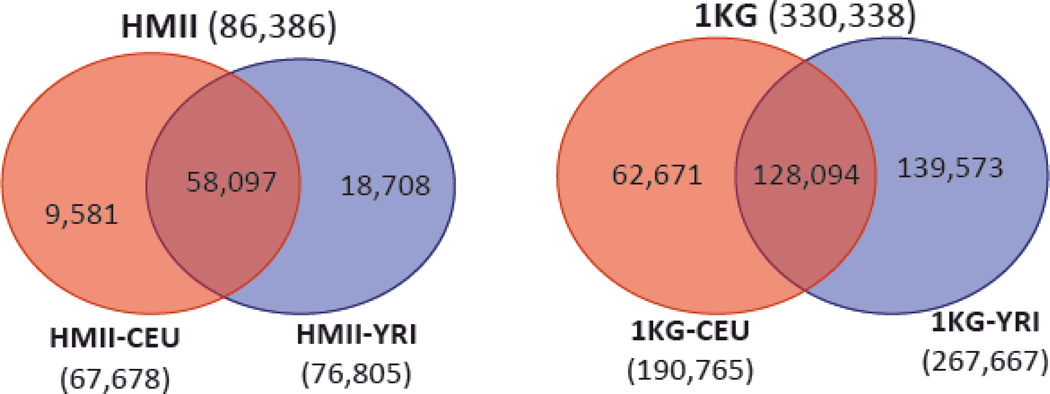
Figure 1 shows Venn diagrams of SNPs from chromosomes 21 and 22 in CEU and YRI for both projects. In the HapMap data, 67% of SNPs were in both CEU and YRI, whereas only 39% of SNPs were in CEU and YRI in the 1KG data. This made a larger difference in the number of SNPs in the INT and UNI strategies for the 1KG data (Table I). In the HapMap data, some SNPs were polymorphic in either CEU or YRI but not genotyped in the other panel, and these were excluded from the UNI strategy. Therefore, in the HapMap data, the MER strategy contained more SNPs than the UNI strategy (86K vs. 79K SNPs, Table I). In the 1KG data, 234 SNPs did not have the same alleles in both panels and were excluded from all three strategies.
Figure 1.
Venn diagrams showing the number of SNPs from chromosomes 21 and 22 in the CEU and YRI reference panels from the HapMap II (HMII) and 1000 Genomes (1KG) Projects.
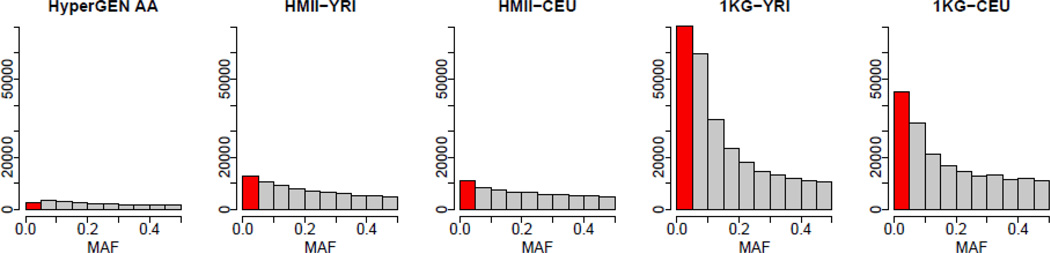
The 1KG data contained a much larger number of variants than the HapMap data across the minor allele frequency (MAF) spectrum (Figure 2). In both HapMap and 1KG data, YRI contained a larger number of rare (MAF ≤ 0.01) and low frequency (0.01 < MAF ≤ 0.05) variants than CEU. Table I shows the number of these rare and low frequency variants for all strategies. There were fewer rare variants in the 1KG data. This reflects that low-depth sequencing data may not be appropriate for detecting rare variants. It also reflects that stronger quality controls had been applied to these data than in the HapMap data, which were direct genotype calls of previously discovered SNPs. Histograms of MAF across the genome were similar to histograms in Figure 2.
Figure 2.
Histograms of minor allele frequencies (MAF) of SNPs from chromosomes 21 and 22 in reference panels from YRI and CEU populations in HapMap II (HMII) and 1000 Genomes (1KG) Projects. Histogram of MAF of SNPs in HyperGEN African Americans genotyped on Affymetrix SNP Array 6.0 is also shown for comparison. Rare and low frequency variants are in the red bin. Histograms of MAF across the genome were similar.
IMPUTATION USING MACH
Several programs are available for genotype imputations such as IMPUTE [Marchini et al., 2007;], MACH [Scott et al., 2007; Li and Abecasis 2006; Li et al., 2010], BIMBAM [Servin and Stephens, 2007] and BEAGE [Browning and Browning, 2007]. In this paper, we used MACH because MACH and IMPUTE, the two leading programs, have been shown to provide the most accurate results across various settings [Pei et al., 2008, Nothnagel et al., 2009]. Before imputation, negative strands of GWA SNPs were flipped with PLINK [Purcell et al., 2007] based on the most recent annotation file provided by Affymetrix.
Following the developers’ recommendation, we used a two-step procedure for running MACH (version 1.0.16) for each reference panel. In the first step, we used 50 rounds of iterations, 188 unrelated HyperGEN subjects and each reference panel to estimate model parameters: crossover rates between adjacent SNPs, which control breakpoints in haplotypes shared between HyperGEN data and the reference panel, and an error rate, which allows discrepancies between HyperGEN data and the panel. In the second step, using these estimated parameters, imputations were performed for all HyperGEN subjects. Imputations were performed separately by chromosomes.
IMPUTATION YIELD AND ACCURACY
To evaluate imputation performance, we used imputation yield and accuracy for each imputed data set. We applied a filtering rule that removed monomorphic SNPs and SNPs with MACH’s quality measure Rsq < 0.3. This follows MACH developers’ recommendation and is current practice for numerous imputed data sets that were used in published GWAS and meta-analyses. We defined imputation yield as the number of filtered SNPs (that remained after filter).
We measured imputation accuracy with dosage Rsq: the squared correlation between the true genotype and continuous-valued imputed genotype dosage for each masked SNP. Concordance rates between true and imputed genotype calls are often very high for rare and low frequency SNPs and it is hard to compare accuracy across SNPs with different MAFs. However, the dosage Rsq is not confounded by MAF and can be used to compare accuracy for rare and common SNPs. We computed imputation accuracy for each strategy using the mean of dosage Rsq values at filtered SNPs. To evaluate the effect of the filtering rule, we also computed accuracy using the mean dosage Rsq values at all imputed SNPs.
To evaluate imputation accuracy, we masked the genotype data of the HyperGEN GWA study at three levels: 5%, 50% and 80%. These masked SNPs were removed and imputation was performed using the remaining SNPs. Then the imputed results for these masked SNPs were compared with their actual genotype data. To evaluate accuracy of rare variants, all rare variants (shown in Table I) were masked. In addition, the 5% masked data removed a randomly selected 5% of the SNPs, the 50% masked data systematically removed every other SNP and the 80% masked data also systematically removed 4 in every 5 SNPs. The 5%, 50% and 80% masked data removed 1,468 SNPs, 11,636 SNPs, and 18,430 SNPs, respectively. The 50% masked data roughly corresponded to the coverage of Affymetrix SNP Array 5.0. We considered 80% masking because we wanted to evaluate imputation performance for less desirable conditions.
RESULTS
IMPUTATION YIELD
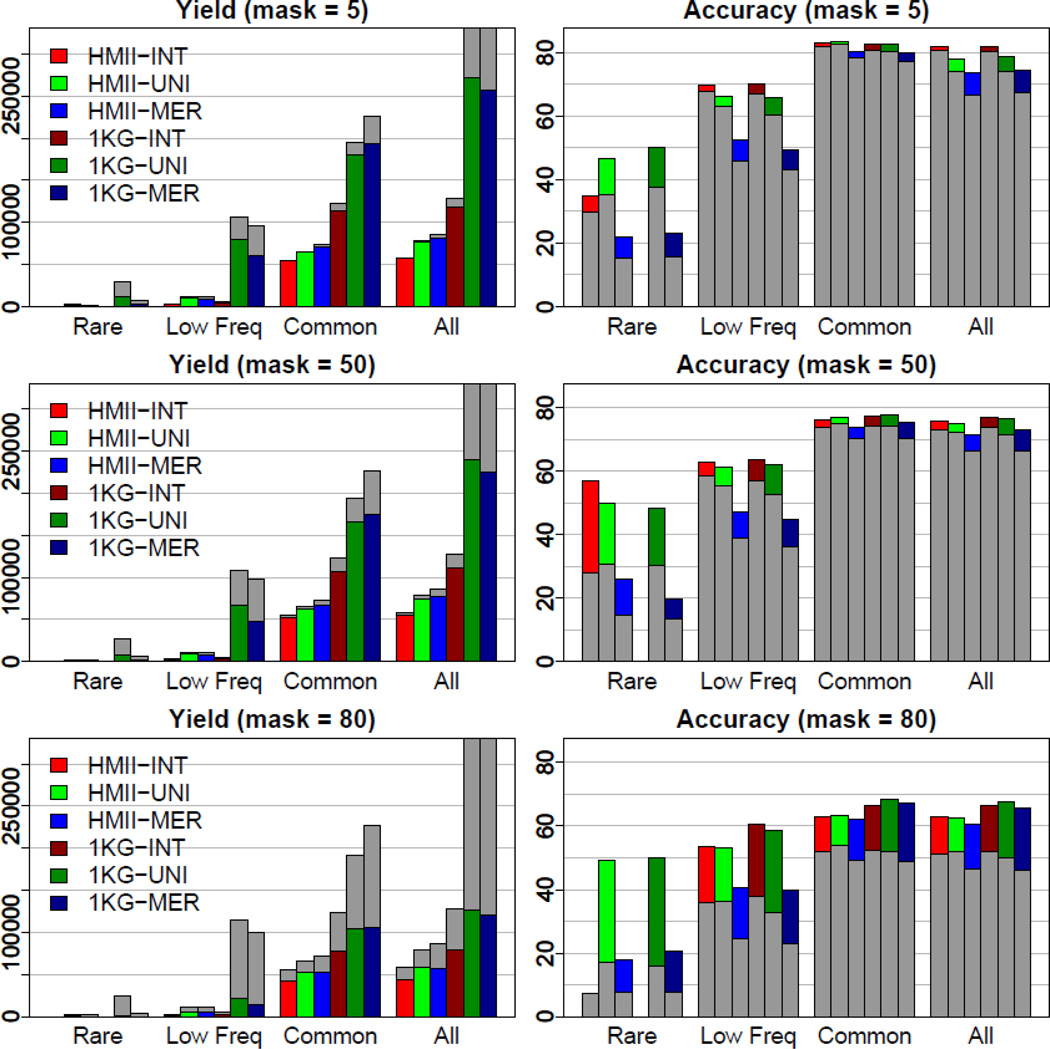
Imputations using 1KG data provided significantly higher yield (the number of filtered SNPs) than imputations using HapMap II (HMII) data. This higher yield is expected because the 1KG panel includes more SNPs. Table II and Figure 3 show overall yield and yields across the MAF spectrum for all three masked data. Figure 3 also shows a total number of imputed SNPs in gray color. For the 5% masked data, which corresponded to a typical imputation scenario, 1KG-UNI provided the highest yield (271K SNPs), and HMII-INT provided the lowest yield (57K SNPs). Among results using the 1KG data, UNI provided the highest yield, MER provided next highest and INT provided the lowest yield for all masking rates. In particular, 1KG-UNI provided twice as high yield as 1KG-INT (271K vs. 119K at 5% masking). HapMap-based imputations provided similar patterns, although difference between highest and lowest yields were less extreme (81K using HMII-MER and 57K using HMII-INT at 5% masking). As expected, imputation yield dropped with higher masking rates. However, decrease in imputation yield was surprisingly small for the 50% masked data and became significant for the 80% masked data.
Table II.
Imputation yield, number of filtered SNPs (with MACH Rsq over 0.3), using HapMap II (HMII) and 1KG data across MAF spectrum. Red boldfaced are highest imputation yield for each masked data.
| Masking Rate |
Public Data |
Strategy | MAF Spectrum | Polymorphic SNPs | Overall yield |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| Rare | Low Freq |
Common | CEU- specific |
YRI- specific |
in both panels |
|||||
| 5% | HMII | YRI | 625 | 7,504 | 66,118 | 0 | 17,552 | 56,695 | 74,247 | |
| CEU | 165 | 4,185 | 57,100 | 7,088 | 0 | 54,362 | 61,450 | |||
| INT | 50 | 3,054 | 53,994 | 0 | 0 | 57,098 | 57,098 | |||
| UNI | 1,645 | 10,435 | 64,358 | 5,582 | 13,525 | 57,331 | 76,438 | |||
| MER | 723 | 9,403 | 71,209 | 7,088 | 17,552 | 56,695 | 81,335 | |||
| 1KG | YRI | 2,308 | 53,004 | 172,743 | 0 | 112,454 | 115,601 | 228,05 | ||
| CEU | 391 | 9,943 | 128,995 | 28,628 | 0 | 110,701 | 139,329 | |||
| INT | 3 | 4,671 | 114,124 | 0 | 0 | 118,798 | 118,798 | |||
| UNI | 11,203 | 80,130 | 179,973 | 33,785 | 119,221 | 118,300 | 271,306 | |||
| MER | 2,655 | 59,948 | 194,080 | 28,628 | 112,454 | 115,601 | 256,683 | |||
| 50% | HMII | YRI | 492 | 7,064 | 62,712 | 0 | 16,251 | 54,017 | 70,268 | |
| CEU | 169 | 3,926 | 53,320 | 6,384 | 0 | 51,031 | 57,415 | |||
| INT | 47 | 2,673 | 52,175 | 0 | 0 | 54,895 | 54,895 | |||
| UNI | 1,298 | 9,561 | 62,871 | 4,971 | 12,937 | 55,822 | 73,730 | |||
| MER | 590 | 8,636 | 67,426 | 6,384 | 16,251 | 54,017 | 76,652 | |||
| 1KG | YRI | 1,479 | 42,017 | 156,866 | 0 | 94,185 | 106,177 | 200,362 | ||
| CEU | 317 | 8,603 | 117,250 | 24,325 | 0 | 101,845 | 126,170 | |||
| INT | 4 | 4,019 | 107,486 | 0 | 0 | 111,509 | 111,509 | |||
| UNI | 7,403 | 66,349 | 165,927 | 26,923 | 102,264 | 110,492 | 239,679 | |||
| MER | 1,740 | 47,688 | 175,259 | 24,325 | 94,185 | 106,177 | 224,687 | |||
| 80% | HMII | YRI | 227 | 4,236 | 48,649 | 0 | 11,421 | 41,691 | 53,112 | |
| CEU | 77 | 2,562 | 41,559 | 4,624 | 0 | 39,574 | 44,198 | |||
| INT | 15 | 1,558 | 42,124 | 0 | 0 | 43,697 | 43,697 | |||
| UNI | 536 | 5,955 | 51,959 | 3,204 | 9,485 | 45,761 | 58,450 | |||
| MER | 247 | 5,120 | 52,369 | 4,624 | 11,421 | 41,691 | 57,736 | |||
| 1KG | YRI | 99 | 11,860 | 94,253 | 0 | 37,679 | 68,533 | 106,212 | ||
| CEU | 70 | 4,438 | 81,416 | 14,152 | 0 | 71,772 | 85,924 | |||
| INT | 0 | 1,807 | 77,411 | 0 | 0 | 79,218 | 79,218 | |||
| UNI | 1,067 | 21,594 | 104,056 | 10,780 | 40,445 | 75,492 | 126,717 | |||
| MER | 150 | 14,343 | 105,871 | 14,152 | 37,679 | 68,533 | 120,364 | |||
Rare variants with MAF ≤ 0.01; Low frequency variants with 0.01 < MAF ≤ 0.05; Common variants with MAF > 0.05
INT refers the intersection strategy; UNI refers the union strategy; MER refers the merge strategy
CEU-specific SNPs are polymorphic only in CEU; YRI-specific SNPs are polymorphic only in YRI; SNPs in both panels are polymorphic in both CEU and YRI
Figure 3.
Imputation yield (left) and accuracy (right) using filtered SNPs across the MAF spectrum for all three masked data using panels from both the HapMap II (HMII) and 1000 Genomes (1KG) Projects. Gray bars show total number of imputed SNPs (left) and accuracy rates (right) using all imputed SNPs.
Improvement of imputation yield using the 1KG data over the HMII data varied across the MAF spectrum. For the 5% masked data, imputation using 1KG-UNI provided three times as many common SNPs as HMII-UNI (180K vs. 64K), eight times as many low frequency SNPs (80K vs. 10K), and seven times as many rare SNPs (11K vs. 1.6K). We expect that, with non-masked data, an even larger number of rare and low frequency SNPs would be successfully imputed using the 1KG data. Note that Table II shows the MAF spectrum of imputed SNPs for the HERITAGE study, whereas Table I shows the MAF spectrum of SNPs present in the reference panels. We observed that the number of rare and low frequency variants in imputed results (shown in Table II) is usually larger than the number of these variants in reference panels (shown in Table I). With higher masking rates, imputation yield decreased more for rare and low frequency SNPs.
IMPUTATION ACCURACY
Table III shows these accuracy rates for filtered SNPs and Supplementary Table I for all imputed SNPs. Figure 3 shows accuracy using all imputed SNPs (in gray) and filtered SNPs (in color). Overall, INT provided the highest accuracy, UNI provided next highest and MER provided the lowest. Accuracy rates from HapMap-based imputations were slightly higher than those from 1KG-based imputation before filtering (74.2 vs. 74.0 at 5% masking; 72.4 vs. 71.4 at 50%; 51.8 vs. 50.1 at 80% using UNI) but slightly lower after filtering (78.0 vs. 78.9 at 5%; 75.1 vs. 76.4 at 50%; 62.7 vs. 67.7 at 80% using UNI). The patterns were similar in each MAF bin.
Table III.
Imputation accuracy rates (in percent) of filtered SNPs (with MACH Rsq over 0.3) using HapMap II (HMII) and 1000 Genomes (1KG) data across the MAF spectrum. Red boldfaced are highest accuracy rates for each masked data. Imputation accuracy rates of all imputed SNPs are shown in Supplementary Table I.
| Masking Rate |
Public Data |
Strategy | MAF spectrum | Polymorphic SNPs | Overall Accuracy | ||||
|---|---|---|---|---|---|---|---|---|---|
| Rare | Low Freq | Common | CEU- specific |
YRI- specific |
in both panels | ||||
| 5% | HMII | YRI | 17.8 | 64.0 | 80.9 | 75.6 | 80.1 | 79.3 | |
| CEU | 22.7 | 38.6 | 56.8 | 30.3 | 56.2 | 52.4 | |||
| INT | 34.7 | 69.8 | 83.0 | 82.0 | 82.0 | ||||
| UNI | 46.5 | 66.2 | 83.3 | 54.4 | 76.6 | 83.2 | 78.0 | ||
| MER | 22.1 | 52.6 | 80.1 | 30.3 | 75.6 | 80.1 | 73.5 | ||
| 1KG | YRI | 15.4 | 64.5 | 81.2 | 77.7 | 80.9 | 80.2 | ||
| CEU | 23.0 | 39.0 | 57.9 | 31.7 | 58.2 | 54.3 | |||
| INT | 70.1 | 82.5 | 82.0 | 82.0 | |||||
| UNI | 50.1 | 65.8 | 82.5 | 56.9 | 77.5 | 84.0 | 78.9 | ||
| MER | 23.0 | 49.5 | 80.0 | 31.7 | 77.7 | 80.9 | 74.5 | ||
| 50% | HMII | YRI | 24.9 | 56.9 | 74.5 | 70.5 | 74.1 | 73.5 | |
| CEU | 26.0 | 34.4 | 51.2 | 35.7 | 50.6 | 49.6 | |||
| INT | 56.9 | 62.7 | 76.2 | 75.6 | 75.6 | ||||
| UNI | 49.9 | 61.4 | 76.8 | 59.0 | 70.4 | 77.0 | 75.1 | ||
| MER | 26.0 | 47.3 | 73.9 | 35.7 | 70.5 | 74.1 | 71.4 | ||
| 1KG | YRI | 61.2 | 76.4 | 71.9 | 76.7 | 75.8 | |||
| CEU | 20.1 | 35.4 | 54.1 | 37.7 | 54.1 | 52.5 | |||
| INT | 63.7 | 77.4 | 77.1 | 77.1 | |||||
| UNI | 48.2 | 62.0 | 77.9 | 58.9 | 71.9 | 79.3 | 76.4 | ||
| MER | 19.7 | 45.0 | 75.4 | 37.7 | 71.9 | 76.7 | 72.9 | ||
| 80% | HMII | YRI | 50.0 | 62.5 | 58.7 | 62.6 | 62.0 | ||
| CEU | 18.9 | 30.3 | 42.0 | 32.2 | 41.6 | 41.0 | |||
| INT | 53.4 | 63.1 | 62.8 | 62.8 | |||||
| UNI | 49.3 | 53.3 | 63.4 | 55.8 | 57.5 | 63.8 | 62.7 | ||
| MER | 17.8 | 40.8 | 62.1 | 32.2 | 58.7 | 62.6 | 60.5 | ||
| 1KG | YRI | 58.3 | 68.3 | 65.0 | 68.6 | 68.0 | |||
| CEU | 22.6 | 33.4 | 47.2 | 34.7 | 47.3 | 46.3 | |||
| INT | 60.8 | 66.6 | 66.5 | 66.5 | |||||
| UNI | 50.1 | 58.7 | 68.2 | 56.6 | 66.0 | 68.8 | 67.7 | ||
| MER | 20.6 | 39.8 | 67.4 | 34.7 | 65.0 | 68.6 | 65.6 | ||
Rare variants with MAF ≤ 0.01; Low frequency variants with 0.01 < MAF ≤ 0.05; Common variants with MAF > 0.05
INT refers the intersection strategy; UNI refers the union strategy; MER refers the merge strategy
CEU-specific SNPs are polymorphic only in CEU; YRI-specific SNPs are polymorphic only in YRI; SNPs in both panels are polymorphic in both CEU and YRI
Within each strategy, accuracy rates were the highest for common variants and the lowest for rare variants (Figure 3 and Table III). In each MAF bin, these accuracy rates were reduced with higher masking rates. This pattern was more pronounced before filtering (as shown in Supplementary Table I). For example, accuracy rates for rare variants with HMII-UNI were 35.4%, 30.7%, and 17% at 5%, 50% and 80% masking, respectively, before filtering. The remarkably similar rates after filtering which were 46.5%, 49.9% and 49.3%, may seem surprising at first. However, these rates after filtering are confounded with the number of remaining SNPs (which are necessarily of higher quality). HMII-UNI provided imputations for 163 rare variants that were masked (as shown in Supplementary Table II). Out of 163 rare variants, 111, 83 and 36 variants remained with MACH Rsq over 0.3 at 5%, 50% and 80% masking rates. Because accuracy rates after filtering were based on these remaining SNPs, it is no surprise that accuracy rates based on much smaller numbers of SNPs turned out to be higher.
Imputation accuracy highly depended on whether each strategy included imputation for CEU-specific SNPs (polymorphic only in CEU) or not. When accuracy rates were restricted to SNPs that were polymorphic in both CEU and YRI, the UNI strategy provided the highest accuracy for all three masked data (Tables III). For YRI-specific SNPs (polymorphic only in YRI), accuracy rates were a little lower than those SNPs that were in both panels. For CEU-specific SNPs, however, accuracy rates were much lower; 57% using 1KG-UNI and 31% using 1KG-MER for the 5% masked data (Table III). Because INT did not provide imputations for either YRI-specific and CEU-specific SNPs, INT provided consistently higher accuracy than UNI. For the same reason, imputation using YRI alone provided higher accuracy than using the MER strategy.
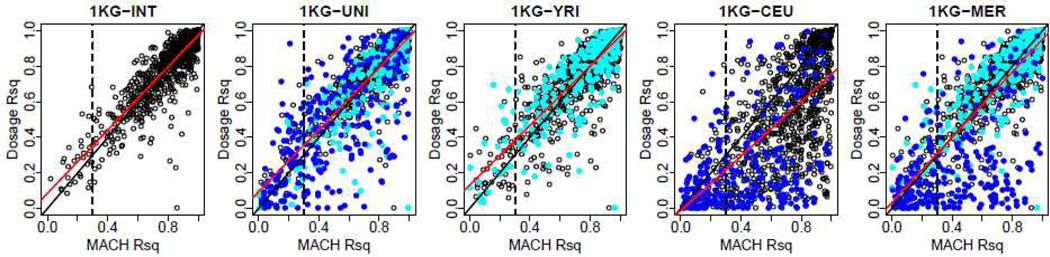
A possible explanation of this much lower imputation performance for CEU-specific SNPs is shown in Figure 4. The black line is where MACH Rsq values equal dosage Rsq values, and the red line is the actual regression line. For imputations using 1KG-INT, MACH Rsq values closely matched true dosage Rsq values. However, for imputations using 1KG-CEU, MACH Rsq values consistently underestimated true dosage Rsq values. This happened because of a mismatch between the reference panel and the study samples, hence it may not be a surprise. Imputations for African American subjects are never performed using the CEU panel only. However, this mismatch also affected imputations using 1KG-UNI, where MACH Rsq values often underestimated dosage Rsq values for CEU-specific SNPs. In consequence, CEU-specific SNPs had lower imputation accuracy than YRI-specific SNPs (57% vs. 77%, in Table III).
Figure 4.
Imputation accuracy (Dosage Rsq) values vs MACH Rsq values for masked SNPs at 5% masking rates using panels from the 1000 Genomes (1KG) Project. Plots for HapMap-based imputations were similar. Blue points are SNPs polymorphic only in CEU, and cyan points are SNPs polymorphic only in YRI. Black line is where MACH Rsq equals Dosage Rsq. Red line is the regression line. Vertical dashed line is the filtering rule that we used. Note MER equals YRI plus blue points from CEU.
DISCUSSION
In this paper, we evaluated imputation strategies for African Americans using data from the HapMap II and 1000 Genomes (1KG) Projects. We used 23,707 SNPs from chromosomes 21 and 22 on the Affymetrix SNP Array 6.0 genotyped for 1,075 HyperGEN African Americans. To impute untyped markers in African American subjects, we considered three strategies. The intersection strategy (INT) used a combined panel consisting of SNPs polymorphic in both CEU and YRI panels. The union strategy (UNI) used a panel consisting of SNPs polymorphic in either CEU or YRI. The merge strategy (MER) merged results from two separate imputations, one using CEU and the other using YRI. Our MER strategy used YRI-based imputations for SNPs polymorphic in YRI and otherwise used CEU-based imputations.
Genotype imputation is commonly used to increase power of individual association studies and to provide a uniform set of variants for meta-analysis of multiple studies. We observed that imputation accuracy for rare variants was low before filtering which improved after filtering. Therefore, so long as filtering is used to exclude poor quality imputations, the primary goals for imputing in the first place do not appear to be compromised. On average, rare variants had lower imputation quality measures. Therefore, a large proportion of rare variants were filtered out. Because genotype imputation uses LD across SNPs, it is no surprise that rare variants are usually not imputed as accurately as common variants. These are consistent with other studies [Huang et al 2009; Pei et al 2008]. In 1KG-based imputations, a large proportion of variants were filtered out. It is unclear whether this happened because 1KG data contained a large number of rare and low frequency variants or because haplotypes were derived from low-depth sequencing data. Because of much higher SNP density, however, even after filtering, 1KG-based imputations provided about three times as many common variants and eight times as many rare and low frequency variants as HapMap-based imputations. Our findings suggest that 1KG-based imputations can increase the opportunity to discover significant associations for SNPs across the whole MAF spectrum.
Our most important finding was that 1KG-based imputations provided a substantially larger number of variants than HapMap-based imputations with nearly identical accuracy. Accuracy rates using 1KG data were slightly lower than those using HapMap data before filtering, but slightly higher after filtering. Our results support those from the 1000 Genomes Project Consortium [2010] that 1KG-based imputations had lower accuracy than HapMap-based imputations. The 1000 Genomes Project Consortium used a much larger number of rare and low frequency variants discovered from high-depth sequencing data in their trio project. Because we used mostly common SNPs on Affymetrix Array 6.0, our accuracy for rare and low frequency variants may not be as precise. However, we used 1,075 African Americans for imputations, whereas they used one CEU subject and one YRI subject. Due to larger sample size, our accuracy could be more precise.
The UNI strategy provided the highest imputation yield with next highest accuracy. The INT strategy provided the lowest imputation yield but the highest accuracy. The MER strategy provided the lowest imputation accuracy. We observed that accuracy highly depends on whether a strategy provides imputation for CEU-specific SNPs (polymorphic only in CEU). CEU-specific SNPs had much lower accuracy. The UNI strategy had the highest accuracy at SNPs polymorphic in both CEU and YRI. However, UNI had slightly lower overall accuracy than INT, because UNI included CEU-specific SNPs. Furthermore, we observed that MACH Rsq values consistently underestimated true dosage Rsq values at CEU-specific SNPs, due to mismatch between reference panels and study samples at these SNPs.
We presented the performance of Affymetrix SNP Array 6.0 for imputing both HapMap and 1KG SNPs based on chromosomes 21 and 22. Because coverage of Affymetrix SNP Array 6.0 on chromosomes 21 and 22 was shown to be similar to coverage across the genome for both CEU and YRI [Li et al., 2008], we expect that our findings would hold for the entire genome. The performance of Affymetrix and Illumina arrays for imputing HapMap SNPs has been presented in several papers. SNPs on Affymetrix SNP Array 5.0 were selected based on sequence constraints of probes and, hence, were quasi-randomly distributed across the genome, ignoring LD. In contrast, SNPs on Illumina HumanMap300 were tag SNPs derived from over two million common variants in the HapMap data. Affymetrix Array 5.0 was shown to provide lower coverage than the Illumina array for CEU (65% vs. 75%) but higher coverage for YRI (41% vs. 28%) [Barrett and Cardon, 2006]. Affymetrix SNP Array 6.0 improved Affymetrix SNP Array 5.0 by adding tag SNPs [McCarroll et al., 2008]. Affymetrix SNP Array 6.0 was shown to provide lower coverage than the improved Illumina HumanMap650Y for CEU (83% vs. 87%) but higher coverage for YRI (62% vs. 60%) [Li et al., 2008]. Our findings about performance for imputing 1KG data should generalize to imputation using different genotype platforms.
In this paper, we investigated imputation strategies for African Americans using both HapMap and 1KG data. We found that the union strategy that contains SNPs that are polymorphic either in CEU and YRI performed the best. Furthermore, using the 1KG data had an additional advantage in imputing a large number of rare and low frequency variants. We found similar advantages when imputing subjects with European ancestry [Sung et al, 2012]. We used the 1KG Project Pilot 1 data, released in June 2010. To better handle low-depth sequencing data, these 1KG panels were constructed combining results from three independently developed calling methods that used sequencing data across samples and HapMap3 data: QCALL [Le and Durbin, 2010], Thunder [Li et al., 2011] and DePristo et al. [2011]. The most recently available 1KG data, released in June 2011, were constructed using the union strategy including 1,094 individuals of European, African and Asian ancestry. Furthermore, the 1KG Project is still underway, and genotype accuracy will be further improved due to increased sample sizes and a plan to directly genotype variants observed in the low-depth sequencing data. We expect that later versions will provide even better imputation performance. Hence we recommend using a newer version of the 1KG data for imputing other African Americans.
Supplementary Material
ACKNOWLEGEMENTS
We appreciate three anonymous reviewers for their constructive and insightful comments, which substantially improved the manuscript. The work was partly supported by NIH Grants HL55673, HL54473, HL72507 and GM28719.
REFERENCES
- Anderson CA, Pettersson FH, Barrett JC, Zhuang JJ, Ragoussis J, Cardon LR, Morris AP. Evaluating the effects of imputation on the power, coverage, and cost efficiency of genome-wide SNP platforms. Am J Hum Genet. 2008;83:112–119. doi: 10.1016/j.ajhg.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aulchenko YS, Struchalin MV, van Duijn CM. ProbABEL package for genome-wide association analysis of imputed data. BMC Bioinformatics. 2010;11:134. doi: 10.1186/1471-2105-11-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Cardon LR. Evaluating coverage of genome-wide association studies. Nat Genet. 2006;38:659–662. doi: 10.1038/ng1801. [DOI] [PubMed] [Google Scholar]
- Browning SR, Browning BL. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet. 2007;81:1084–1097. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bakker PIW, Ferreira MAR, Jia X, Neale BM, Raychaudhuri S, Voight BF. Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum Mol Genet. 2008;17:R122–R128. doi: 10.1093/hmg/ddn288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo M, Banks E, Poplin R, Garimella K, Maguire J, Hartl C, Philippakis A, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell T, Kernytsky A, Sivachenko A, Cibulskis K, Gabriel S, Altshuler D, Daly M. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellinghaus E, Ellinghaus D, Stuart PE, Nair RP, Debrus S, Raelson JV, Belouchi M, Fournier H, Reinhard C, Ding J, Li Y, Tejasvi T, Gudjonsson J, Stoll SW, Voorhees JJ, Lambert S, Weidinger S, Eberlein B, Kunz M, Rahman P, Gladman DD, Gieger C, Wichmann HE, Karlsen TH, Mayr G, Albrecht M, Kabelitz D, Mrowietz U, Abecasis GR, Elder JT, Schreiber S, Weichenthal M, Franke A. Genome-wide association study identifies a psoriasis susceptibility locus at TRAF3IP2. Nat Genet. 2010;42:991–995. doi: 10.1038/ng.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridley BL, Jenkins G, Devo-Svendsen ME, Hebbring S, Freimuth R. Utilizing genotype imputation for the augmentation of sequence data. PLoS One. 2010;5:e11018. doi: 10.1371/journal.pone.0011018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y, Stephens M. Practical issues in imputation-based association mapping. PLoS Genet. 2008;4:e1000279. doi: 10.1371/journal.pgen.1000279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao K, Chudin E, McElwee J, Schadt EE. Accuracy of genome-wide imputation of untyped markers and impacts on statistical power for association studies. BMC Genet. 2009;10:27. doi: 10.1186/1471-2156-10-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Li Y, Singleton AB, Hardy JA, Abecasis G, Rosenberg NA, Scheet P. Genotype-imputation accuracy across worldwide human populations. Am J Hum Genet. 2009;84:235–250. doi: 10.1016/j.ajhg.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le SQ, Durbin R. SNP detection and genotyping from low-coverage sequencing data on multiple diploid samples. Genomes Res. 2011;21:952–960. doi: 10.1101/gr.113084.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Li C, Guan W. Evaluation of coverage variation of SNP chips for genome-wide association studies. Eur J Hum Genet. 2008;16:635–643. doi: 10.1038/sj.ejhg.5202007. [DOI] [PubMed] [Google Scholar]
- Li Y, Abecasis GR. MACH 1.0: Rapid Haplotype Reconstruction and Missing Genotype Inference. Am J Hum Genet. 2006;S79:2290. [Google Scholar]
- Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Sidore C, Kang HM, Boehnke M, Abecasis GR. Low-coverage sequencing: Implications for design of complex trait association studies. Genome Res. 2011;21:940–951. doi: 10.1101/gr.117259.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JZ, Tozzi F, Waterworth DM, Pillai SG, Muglia P, Middleton L, Berrettini W, Knouff CW, Yuan X, Waeber G, Vollenweider P, Preisig M, Wareham NJ, Zhao JH, Loos RJ, Barroso I, Khaw KT, Grundy S, Barter P, Mahley R, Kesaniemi A, McPherson R, Vincent JB, Strauss J, Kennedy JL, Farmer A, McGuffin P, Day R, Matthews K, Bakke P, Gulsvik A, Lucae S, Ising M, Brueckl T, Horstmann S, Wichmann HE, Rawal R, Dahmen N, Lamina C, Polasek O, Zgaga L, Huffman J, Campbell S, Kooner J, Chambers JC, Burnett MS, Devaney JM, Pichard AD, Kent KM, Satler L, Lindsay JM, Waksman R, Epstein S, Wilson JF, Wild SH, Campbell H, Vitart V, Reilly MP, Li M, Qu L, Wilensky R, Matthai W, Hakonarson HH, Rader DJ, Franke A, Wittig M, Schäfer A, Uda M, Terracciano A, Xiao X, Busonero F, Scheet P, Schlessinger D, St Clair D, Rujescu D, Abecasis GR, Grabe HJ, Teumer A, Völzke H, Petersmann A, John U, Rudan I, Hayward C, Wright AF, Kolcic I, Wright BJ, Thompson JR, Balmforth AJ, Hall AS, Samani NJ, Anderson CA, Ahmad T, Mathew CG, Parkes M, Satsangi J, Caulfield M, Munroe PB, Farrall M, Dominiczak A, Worthington J, Thomson W, Eyre S, Barton A, Mooser V, Francks C, Marchini J Wellcome Trust Case Control Consortium. Meta-analysis and imputation refines the association of 15q25 with smoking quantity. Nat Genet. 2010;42:436–440. doi: 10.1038/ng.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat Rev Genet. 2010;11:499–511. doi: 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- McCarroll SA, Kuruvilla FG, Korn JM, Cawley S, Nemesh J, Wysoker A, Shapero MH, de Bakker PI, Maller JB, Kirby A, Elliott AL, Parkin M, Hubbell E, Webster T, Mei R, Veitch J, Collins PJ, Handsaker R, Lincoln S, Nizzari M, Blume J, Jones KW, Rava R, Daly MJ, Gabriel SB, Altshuler D. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat Genet. 2008;40:1166–1174. doi: 10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- Nothnagel M, Ellinghaus D, Schreiber S, Krawczak M, Franke A. A comprehensive evaluation of SNP genotype imputation. Hum Genet. 2009;125:163–171. doi: 10.1007/s00439-008-0606-5. [DOI] [PubMed] [Google Scholar]
- Padmanabhan S, Melander O, Johnson T, Di Blasio AM, Lee WK, Gentilini D, Hastie E, Menni C, Monti MC, Delles C, Laing S, Corso B, Navis G, Kwakernaak AJ, van der Harst P, Bochud M, Maillard M, Burnier M, Hedner T, Kjeldsen S, Wahlstrand B, Sjögren M, Fava C, Montagnana M, Danese E, Torffvit O, Hedblad B, Snieder H, Connell JMC, Brown M, Samani NJ, Farrall M, Cesana G, Mancia G, Signorini S, Grassi G, Eyheramendy S, Wichmann HE, Laan M, Strachan DP, Sever P, Shields DC, Stanton A, Vollenweider P, Teumer A, Völzke H, Rettig R, Newton-Cheh C, Arora P, Zhang F, Soranzo N, Spector TD, Lucas G, Kathiresan S, Siscovick DS, Luan J, Loos RJF, Wareham NJ, Penninx BW, Nolte IM, McBride M, Miller WH, Nicklin SA, Baker AH, Graham D, McDonald RA, Pell JP, Sattar N, Welsh P, Consortium GB, Munroe P, Caulfield MJ, Zanchetti A, Dominiczak AF. Genome-Wide Association Study of Blood Pressure Extremes Identifies Variant near UMOD Associated with Hypertension. PLoS Genet. 2010;6:e1001177. doi: 10.1371/journal.pgen.1001177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra EJ, Marcini A, Akey J, Martinson J, Batzer MA, Cooper R, Forrester T, Allison DB, Deka R, Ferrel RE, Shriver MD. Estimating African American admixture proportions by use of population-specific alleles. Am J Hum Genet. 1998;63:1839–1851. doi: 10.1086/302148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei YF, Li J, Zhang L, Papasian CJ, Deng HW. Analyses and comparison of accuracy of different genotype imputation methods. PLoS One. 2008;3:e3551. doi: 10.1371/journal.pone.0003551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanna S, Pitzalis M, Zoledziewska M, Zara I, Sidore C, Murru R, Whalen MB, Busonero F, Maschio A, Costa G, Melis MC, Deidda F, Poddie F, Morelli L, Farina G, Li Y, Dei M, Lai S, Mulas A, Cuccuru G, Porcu E, Liang L, Zavattari P, Moi L, Deriu E, Urru MF, Bajorek M, Satta MA, Cocco E, Ferrigno P, Sotgiu S, Pugliatti M, Traccis S, Angius A, Melis M, Rosati G, Abecasis GR, Uda M, Marrosu MG, Schlessinger D, Cucca F. Variants within the immunoregulatory CBLB gene are associated with multiple sclerosis. Nat Genet. 2010;42:495–497. doi: 10.1038/ng.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheet P, Stephens M. A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase. Am J Hum Genet. 2006;78:629–644. doi: 10.1086/502802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servin B, Stephens M. Imputation-based analysis of association studies: candidate regions and quantitative traits. PLoS Genet. 2007;3:e114. doi: 10.1371/journal.pgen.0030114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL, Erdos MR, Stringham HM, Chines PS, Jackson AU, Prokunina-Olsson L, Ding CJ, Swift AJ, Narisu N, Hu T, Pruim R, Xiao R, Li XY, Conneely KN, Riebow NL, Sprau AG, Tong M, White PP, Hetrick KN, Barnhart MW, Bark CW, Goldstein JL, Watkins L, Xiang F, Saramies J, Buchanan TA, Watanabe RM, Valle TT, Kinnunen L, Abecasis GR, Pugh EW, Doheny KF, Bergman RN, Tuomilehto J, Collins FS, Boehnke M. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shriner D, Adeyemo A, Chen G, Rotimi CN. Practical considerations for imputation of untyped markers in admixed populations. Gen Epidemiol. 2010;34:258–265. doi: 10.1002/gepi.20457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung YJ, Wang L, Rankinen T, Bouchard C, Rao DC. Performance of Genotype Imputations Using Data from the 1000 Genomes Project. Hum Hered. 2012;73:18–25. doi: 10.1159/000334084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The 1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International HapMap Consortium. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RR, Rao DC, Ellison RC, Arnett DK, Heiss G, Oberman A, Eckfeldt JH, Leppert MF, Province MA, Mockrin SC, Hunt SC. NHLBI family blood pressure program: methodology and recruitment in the HyperGEN network. Hypertension genetic epidemiology network. Ann Epidemiol. 2000;10:389–400. doi: 10.1016/s1047-2797(00)00063-6. [DOI] [PubMed] [Google Scholar]
- Zhu X, Luke A, Cooper RS, Quertermous T, Hanis C, Mosley T, Gu CC, Tang H, Rao DC, Risch N, Weder A. Admixture mapping for hypertension loci with genome-scan markers. Nat Genet. 2005;37:177–181. doi: 10.1038/ng1510. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.