Abstract
We have recently developed a novel optical technology, partial wave spectroscopic (PWS) microscopy, which is exquisitely sensitive to the nanoarchitectural manifestation of the genetic/epigenetic alterations of field carcinogenesis. Our approach was to screen for lung cancer by assessing the cheek cells based on emerging genetic/epigenetic data which suggests that the buccal epithelium is altered in lung field carcinogenesis. We performed PWS analysis from microscopically normal buccal epithelial brushings from smokers with and without lung cancer (n = 135). The PWS parameter, disorder strength of cell nanoarchitecture (Ld), was markedly (>50%) elevated in patients harboring lung cancer compared with neoplasia-free smokers. The performance characteristic was excellent with an area under the receiver operator characteristic curve of >0.80 and was equivalent for both disease stage (early versus late) and histologies (small cell versus non–small cell lung cancers). An independent data set validated the findings with only a minimal degradation of performance characteristics. Our results offer proof of concept that buccal PWS may potentially herald a minimally intrusive prescreening test that could be integral to the success of lung cancer population screening programs.
Introduction
Lung malignancies are a major cause of morbidity and mortality among Americans with an estimated 219,440 new cases and 159,390 deaths in 2009 (1). Although early stage disease can be effectively managed by surgery, most patients come to medical attention because of symptoms (cough, hemoptysis, dyspnea, etc.) which are generally harbingers of advanced, incurable stage. This is the major reason for the dismal 5-year survival rate (~15%) and provides the impetus for screening the asymptomatic population (1).
Lung cancer would seem to be an ideal cancer for screening because the at-risk population is well-defined in that smokers (current or past) comprise ~90% of lung cancer patients (2). Indeed, the lifetime risk of smokers to develop lung cancer is quite significant (~10%; ref. 2). Attempts to screen for lung cancer have classically relied on the identification of tumor products (e.g., sputum cytology) or direct lesion visualization such as chest X-ray, low-dose computerized tomography or bronchoscopy (3). However, these approaches have been clinically unsatisfactory and none of these tests is currently sanctioned for population screening due to suboptimal efficacy, equivocal survival benefit, and numerous false-positives affecting the cost-effectiveness (3). This highlights the need for an accurate, minimally intrusive prescreening (risk-stratification) technique to identify the subset of patients who might benefit from more expensive or invasive tests.
One promising approach centers on field carcinogenesis, the notion that the environmental and genetic pathogenic factors that result in a focal neoplastic lesion in one area of the lung will affect the entire aerodigestive tract mucosa (4). This is substantiated by several lines of evidence including the demonstration of genetic/epigenetic concordance between the lung cancer and the distant histologically normal bronchial epithelium (4, 5). The buccal epithelium represents a particularly attractive target as a surrogate site for field carcinogenesis detection. In particular, given the emerging data that the buccal (cheek) epithelium serves as a “molecular mirror” for lung carcinogenesis, this represented an attractive surrogate site for field effect detection (6). However, clinical exploitation would necessitate the development of highly accurate biomarkers that can be assessed on readily accessible tissue.
Our group has recently developed a novel optical technique, partial wave spectroscopic (PWS) microscopy, that provides heretofore unattainable quantification of the nanoscale cellular organization (7). The main principle of PWS is the extraction of one-dimensional propagating waves from different parts of a cell. PWS measures the disorder strength (Ld) of the intracellular architecture that is proportional to the variance and the correlation length (e.g., the effective size of intracellular nanostructures) of the spatial fluctuations of local mass density. We have previously shown that Ld is exquisitely sensitive to subtle genetic/epigenetic alterations in microscopically normal cells thus supporting the utility for field effect detection. Indeed, we recently reported that PWS analysis of the microscopically normal rectal colonocytes was able to identify patients harboring adenomas elsewhere in the colon (8). In the current study, we investigated the ability of PWS interrogation of the buccal epithelium to identify lung field carcinogenesis and hence serve as a minimally intrusive screening test for pulmonary malignancies.
Materials and Methods
Human studies
The human studies were performed in accordance with the Institutional Review Board at NorthShore University HealthSystem. The cohort comprised 135 patients including 63 smokers (current or past) with histologically confirmed lung cancer including 10 small cell carcinomas and 53 non–small cell carcinomas (32 being adenocarcinomas). Twenty lung cancer patients had early stage disease (stage I or II). The controls included 37 smokers with chronic obstructive pulmonary disease (COPD), 13 smokers without COPD, and 22 nonsmokers. The primary comparator was COPD to approximate tobacco exposure of cancer patients. None of the controls (COPD, smokers without COPD, or nonsmokers) had any personal/family history of lung cancer.
Epithelial cells from the visually normal buccal mucosa were obtained using a cytologic brush, applied onto a cytologic slide, and immediately fixed in a 95% ethanol bath. The PWS analysis was then performed on ~25 to 30 cells randomly chosen by an observer blinded to the clinical diagnosis.
PWS system
The design of PWS instrument has been described in detail previously (7–9). Briefly, a broadband light with a low spatial coherence length of <l μm was focused onto the sample by a low numerical aperture objective (Edmund Optics, numerical aperture of objective = 0.4, numerical aperture of illumination = 0.2, numerical aperture of collection = 0.4). The illumination-beam diameter is ~120 μm and is much larger than the biological cells. The cells are located in the waist of the beam. The resulting backscattered image is projected in the far-field with a 60× magnification onto the slit of an imaging spectrograph (slit width = 10 μm) coupled with a CCD camera (Coolsnap HQ) and mounted on a motorized one-dimensional linear scanning stage (Zaber Technologies). The backscattering image was acquired by linearly scanning the slit of the spectrograph with 10-μm step. The corresponding size of a pixel in the object plane (cell pixel) is 100 × 165 nm. At each scanning step x, the CCD camera records a matrix with one axis corresponding to wavelength λ and the other to the spatial position of the image y resulting in a data cube (x, y, λ). For a given pixel (x, y), Ld was calculated as, where <R> and C(Δk) are the root mean square average and the autocorrelation function of the fluctuating part of backscattering spectrum (λ = 500–670 um), respectively, k is the wave number, and B is the normalization constant (more information regarding the data extraction procedure is provided in Supplementary Information). Thus, by using PWS, a two-dimensional map depicting the distribution of Ld(x, y) can be obtained for a particular cell. From these two-dimensional images, the mean intracellular disorder strength Ld(c) [the average Ld(x, y) over x and y] is obtained. The averages of Ld(c) over a group of cells, such as cells sampled from a particular patient, are termed the patient mean Ld(p). It is noted that the intrapatient variability of Ld(c) is ~26%, which probably reflects the “patchiness” of the field effect.
Statistical analysis
All statistical analyses were performed using Microsoft Excel and STATA data analysis and statistical software package (Statacorp LP). The mean Ld was compared between different patient groups using the two-sided Student’s t test. The effects of different demographic factors on PWS measurements were analyzed using analysis of covariance test. The training set contained 54 patients (COPD = 18 and cancer = 36), whereas the independent validation set contained 46 patients (COPD = 19 and cancer = 27).
Results
We first assessed whether PWS-measured Ld is altered in microscopically normal buccal cells in patients with lung cancer. Given the potential for smoking per se to alter the buccal mucosa ultrastructure, we wanted to assuage the concern that any relationship between buccal Ld and lung cancer would not be confounded by exposure to cigarette smoke. To this end, we chose as our primary comparator (control group) patients with chronic obstructive lung disease (COPD) because they have a similar tobacco exposure (measured in “pack-years”) as lung cancer patients. Additional control groups included smokers without COPD and nonsmokers. The characteristics of the cohorts are given in Table 1.
Table 1.
Impact of demographic factors on buccal Ld
| Demographic information |
Testing set | Validation set | Nonsmoking controls (n = 22) |
Smoking controls (n = 12) |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| COPD (n = 18) |
Cancer (n = 36) |
COPD vs. cancer (P) |
Effect of Ld (P) |
COPD (n = 19) |
Cancer (n = 27) |
COPD vs. cancer (P) |
Effect of Ld (P) |
|||
| Age (mean ± SO), pack years |
72 ± 10 | 69 ± 12 | 0.38 | 0.73 | 71 ± 8 | 71 ± 13 | 0.96 | 0.55 | 64 ± 12 | 50 ± 14 |
| Smoking (mean ± SO), pack years |
63 ± 32 | 59 ±27 | 0.73 | 0.95 | 65 ± 34 | 51 ± 36 | 0.17 | 0.62 | N/A | 23 ± 24 |
| Race (% white) | 95 | 92 | 0.98 | 0.41 | 95 | 85 | 0.27 | 0.10 | 100 | 75 |
| Gender (% male) | 61 | 44 | 0.26 | 0.90 | 50 | 48 | 0.65 | 0.88 | 36 | 50 |
NOTE: Demographic characteristics such as age, smoking history, race, and gender is shown for different patient groups and their effect on Ld. The P value is calculated using the analysis of covariance analysis in STATA. The data indicate that the diagnostic information calculated in the testing and validation set is not confounded by the patient demographic factors. The data also indicate that the demographic factors are not significant between COPD and cancer patients in both the training and validation set.
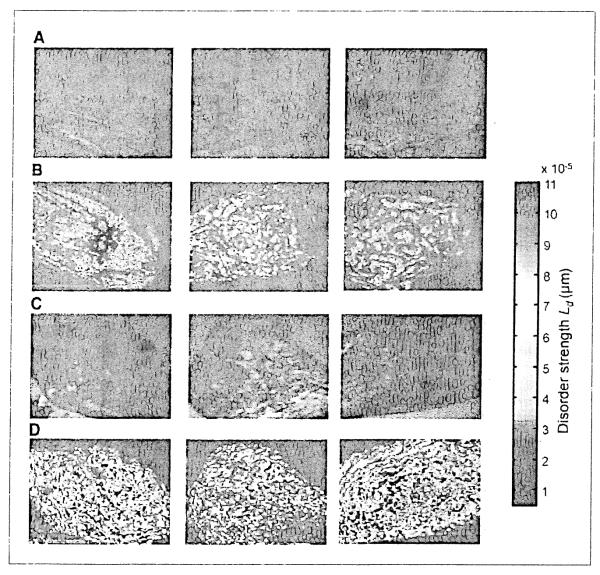
A detailed explanation of the patient cohorts and the PWS instrumentation is provided in Materials and Methods. For each cell, PWS generated a two-dimensional map of Ld as a function of location within the cells. Figure 1 A and C shows a representative bright-field image of six buccal cells obtained from patients with COPD and lung cancer, respectively. These images appear identical, suggesting no detectable alterations at the microscopic level (i.e., length scales <200 nm). However, when the pseudocolor map for Ld are displayed, there seems to be a marked difference indicative of nanoscale perturbations (Fig. lB versus D). The augmentation of Ld in the lung cancer patient occurred throughout the cell but seemed to be most pronounced in the nuclear and perinuclear regions.
Figure 1.
Representative buccal epithelial cells from COPD (neoplasia-free controls) and patients harboring lung cancer. A, bright-field from COPD control patient. B, PWS pseudocolor heatmap of Ld from COPD control patient. C, bright-field from lung cancer patient. D, PWS pseudocolor heatmap of Ld lung cancer patients. The bright-field images of buccal cells from COPD controls (A) and those with cancer (C) were identical. However, the disorder streng1h (Ld) of the same cells obtained from PWS analysis from the cancer patients (D) were markedly increased when compared with the COPD controls (B).
Our initial focus was on comparing Ld in smokers with COPD (controls) versus cancer patients that we reasoned would be well-matched with regards to smoking intensity. As can be seen in Fig. 2A, the mean Ld(p) calculated from patients with lung cancer was ~50% greater than patients with COPD (P < 0.001). To assess the performance characteristics of Ld as a biomarker, we constructed a receiver operator characteristic (ROC) curve (Table 2). The area under ROC curve (AUROC) was 0.81, indicating a robust ability to discriminate between patients with COPD and cancer, one of the most clinically challenging scenarios (10).
Figure 2.
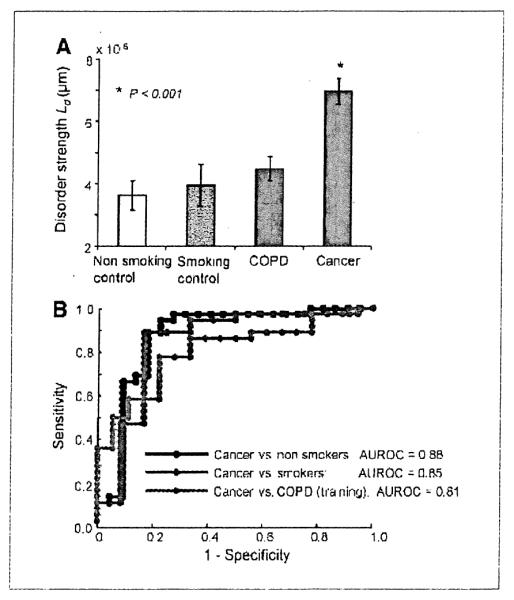
A, Ld quantification of buccal epithelium in patients with and without lung cancer. There was a stepwise increase in Ld with nonsmoking controls (n = 22), to smoking controls (n = 12), to neoplasia-free patients with COPD (n = 18), and then to patients harboring lung cancer (n = 36). B, ROC curve for the discrimination between nonneoplasia controls (either nonsmokers or those with COPD) and patients with lung cancer. The AUROC was 0.88 for nonsmokers and lung cancer, 0.85 for smokers vs. lung cancer, and 0.81 for COPD vs. lung cancer (training set).
Table 2.
Performance characteristics of buccal Ld for discriminating between control (no neoplasia) group and cases (those harboring lung cancer)
| Sensitivity (%) | Specificity (%) | AUROC | |
|---|---|---|---|
| Nonsmoker vs. cancer | 95 | 78 | 0.88 |
| Smoker vs. cancer | 89 | 83 | 0.85 |
| COPD vs. cancer (training) | 78 | 78 | 0.81 |
| COPD vs. cancer (validation) | 75 | 69 | N/A |
NOTE: Sensitivity, specificity, and the AUROC is shown for nonsmoking controls, smoking controls, and COPD controls (both training and validation set) compared with cancer patients. It is noted that the validation set does not contain a ROC curve as a single-cutoff value of Ld derived from the training set was applied.
We therefore evaluated buccal PWS with two other control groups: smokers without diagnosed COPD (probably the most relevant comparator) and age-matched nonsmokers. As seen in Fig. 2A, the microscopically normal buccal mucosa showed a threshold effect of mean Ld(p) from lung cancer dramatically greater than the control groups. Although the control groups varied slightly (e.g., smokers with COPD had Ld of 4.8 × 10−5 ± 2.1 × 10−5 whereas that for smokers without COPD was 4.0 × 10−5 ± 2.3 × 10−5), this failed to achieve statistical significance. On the other hand, the difference between cancer patients and either smoking or nonsmoking controls was dramatic and highly statistically significant (P < 0.001). Thus, the marginal effect of smoking and COPD on Ld was dwarfed by the presence of cancer. The AUROC for discriminating cancer patients from all control groups was excellent (0.81, 0.85, and 0.88 versus smokers with COPD, smokers without COPD, and nonsmoker controls, respectively) underscoring the potential of PWS in screening for patients with risk of lung cancer. The positive and negative predictive values were 88% and 64%, respectively.
We further validated buccal PWS in an independent data set of 46 patients (lung cancer versus COPD). For this validation data set, we applied a cutpoint (cancer present/absent) from analysis of the first 54 patients. As summarized in Table 2, the sensitivity and specificity in the validation set (sensitivity = 75% and specificity = 69%) well approximated the training set (sensitivity = 78% and specificity = 78%). It should be emphasized that this performance is based on a single biomarker (Ld) and it is probable that utilization of additional nanoarchitectural (PWS measured) markers would improve the performance characteristics.
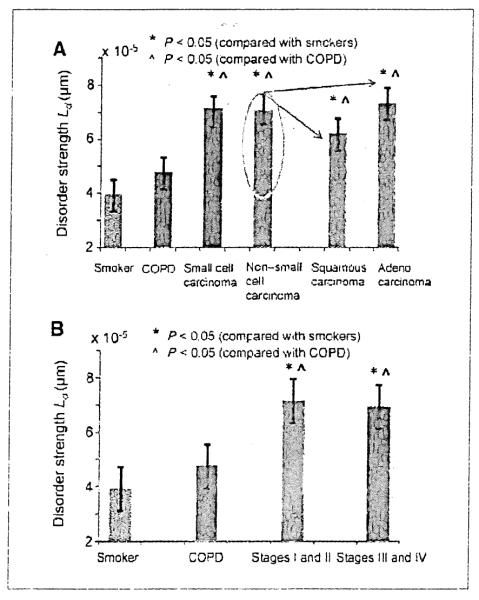
For buccal PWS as a risk-stratification tool to be clinically useful, it would need to be able to detect screen-relevant neoplasia (cancers diagnosed at a stage that could affect patient longevity and treatment). Therefore, we evaluated performance of Ld for both localized (stages I and II) and advanced disease (stages III and IV; Fig. 3A). The buccal Ld was similar for all stages of lung cancer (7.36 × 10−5 ± 3.38 × 10−5, 6.90 × 10−5 ± 2.4 × 10−5, 7.46 × 10−5 ± 2.91 × 10−5, and 6.32 × 10−5 ± 2.22 × 10−5 for stages I, II, III, and IV, respectively). The performance characteristics of Ld for stages I and II, and stages III and IV versus smokers were also equivalent (AUROC of 0.84 versus 0.83, respectively).
Figure 3.
A, buccal Ld was comparably elevated regardless of stage of lung cancer. Patients are divided into eary stage (stages I and II) vs. advanced (stages III and IV). Both were highly statistically significant (P < 0.01 vs. smokers). B, buccal Ld was equivalently increased irrespective of lung cancer histology. Buccal Ld was elevated in both small cell and non–small cell lung cancer (NSCLC) equivalently. Analysis of the major subtypes on NSCLC into squamous and adenocarcinoma revealed no important differences (P = 0.24) and both were still highly significantly different from the smoking comparator (P < 0.05).
Furthermore, because smoking could induce a variety of histologic Subtypes of lung cancer, it would be important to be sensitive to the entire spectrum of malignancies. For most studies, lung cancer is dichotomized into non–small cell lung cancer (NSCLC) and small cell lung cancer (the former comprising ~80% of cases). As shown in Fig. 3A, the Ld values were comparable in NSCLC and small cell lung cancer patients (7.1 × 10−5 ± 1.8 × 10−5 versus 7.0 × 10−5 ± 2.9 × 10−5). The performance characteristics were also excellent for both (0.83 versus 0.87 versus smokers, respectively). Further subdividing NSCLC into adenocarcinoma and squamous cancers showed that buccal Ld was sensitive to both major components with AUROC versus smokers of 0.84 and 0.83, respectively. An important insight derived from this comparison is that the location of cancer is unlikely to be important because small cell and squamous cancers are more typically central whereas adenocarcinomas are peripheral and yet buccal Ld was similarly elevated. Thus, the buccal screening approach seems to be robust over most clinical scenarios.
Because screening would be performed predominantly on smokers, we investigated whether smoking per se could be a potential confounding factor. The cigarette consumption was not greater in cancer patients than COPD controls (59 ± 27 versus 63 ± 32 pack-years, respectively). Indeed, when stratified by clinical status, the effect of smoking history on Ld was in Significant (P = 0.95). Similarly, as noted in Table 1, demographic factors such as age, gender, and race did not seem to be a significant confounder (P = 0.73, P = 0.90, and P = 0.41, respectively). Lastly, in analysis of the control patients in the validation data set, we showed that the demographic factors did not significantly affect buccal Ld readings (Table 1).
Discussion
We show herein that buccal PWS analysis of cell nanoarchitecture has promise as a novel, minimally intrusive lung cancer screening tool. Indeed, the performance characteristics of the single marker, Ld, seemed to be very good and the addition of other nanoarchitectural, PWS-detectable markers would undoubtedly improve performance characteristics. From a proof-of-principle perspective, use of a single marker shows the power of the approach without raising concerns about overfitting. The robustness was further shown by both assessing an independent data set for validation and employing several control groups including current/past smokers (the most relevant group for targeted population screening), smokers with COPD (which matches for tobacco exposure but is confounded by the higher risk of future lung cancer) and nonsmokers. Importantly, buccal PWS was able to detect early curable lesions (stages I and II) equivalently to advanced stage lesions (stages III and IV).
Several lines of evidence support the biological plausibility of using buccal mucosa to detect lung cancer. It has been long recognized that buccal mucosa is part of the extended field carcinogenesis associated with lung cancer (11). Support of this “field of injury” concept comes from demonstrating that the smoking-induced transcriptosome alterations from bronchial and oral/nasal cavities are quite similar (12, 13). Furthermore, at a cytogenetic level, buccal epithelial losses at 9p21 (p16 locus), 17p13 (p53 locus), and 5q21 (adenomatous polyposis coli locus) occurred, by and large, only in smokers with concomitant lung cancer. Moreover, these changes were also noted in the premalignant bronchial epithelium, supporting its role in early lung carcinogenesis (14). Finally, data regarding DNA-based buccal cell image analysis showed a correlation with the presence of lung cancer (15). This has solidified the concept that the buccal epithelium is the “molecular mirror” of bronchial epithelium (6).
Therefore, although there is a clear biological precedence for using the buccal mucosa to gauge risk of lung cancer; heretofore, there have been no biomarkers of sufficient accuracy to consider translation into clinical practice. For instance, the genetic/epigenetic changes discussed above clearly lack adequate diagnostic abilities. The DNA-based buccal image analysis had a sensitivity and specificity of 61 %, which is clearly inadequate for population screening (16). However, the potential of field carcinogenesis as a diagnostic tool for lung cancer screening was highlighted by a seminal report that with an 80-gene microarray signature from the “normal” right main-stem bronchial epithelium, one could achieve an 80% sensitivity, 84% specificity for discriminating smokers with and without lung cancer (17). Similarly, another group developed a 14-antioxidant gene panel that, when applied to the microscopically normal bronchial epithelium, was able to achieve an AUROC of 0.82 for identifying lung cancer from controls (18). However, obtaining bronchial mucosa is somewhat invasive/uncomfortable precluding utilization of this approach for population screening.
PWS analysis provides the first definitive demonstration of nanoscale alterations in extended field carcinogenesis (8). These cells appear microscopically normal thus indicating that these fundamental architectural alterations sensed by Ld are determined by perturbations at length scales below the diffraction limited resolution of light microscopy (~200 nm). Previous work from our group has shown that Ld alterations during carcinogenesis might result in part from cytoskeletal dysregulation which, in turn, may alter the local concentration of particles such as ribosomes. mitochondria, and macromolecular complexes (all implicated in early carcinogenesis; refs. 19, 20). Whether these nanoarchitectural changes simply reflect the genetic/epigenetic changes or have functional consequences in neoplastic transformation is a key question that needs further investigation. However, the importance of Ld alteration is suggested by the observation that it is a common phenomenon in field carcinogenesis in a variety of organs (8).
We observed that buccal Ld mirrored lung carcinogenesis with a threshold that allowed discrimination between cancer and all controls (cancer > COPD/smokers/nonsmokers), which is critical for a potential population screening test. Moreover, the performance characteristic was excellent with an AUROC curve of 0.88 for nonsmokers, 0.85 for smokers, and 0.81 for COPD patients. In the final iteration of the screening test, we anticipate these performance characteristics to be even better by using other PWS markers. Even at present, the diagnostic ability rivals or exceeds other screening techniques.
There are several limitations/unresolved issues that need to be acknowledged. First, the control patients generally had not received lung cancer screening (because it is currently not the standard of care). Therefore, some controls (especially those with COPD) may later develop lung cancer thus having potential ramifications for the performance characteristics of buccal PWS (10). Second, lung cancer is a heterogeneous disease with several distinct histologies (squamous, adenocarcinoma, large cell, and small cell, etc.; ref. 21). Although our modest sample size precluded definitive conclusions for each subtype, preliminary analysis suggested no important differences. Third, we need to consider other aerodigestive malignancies in the tobacco field of injury that might affect the buccal markers. Head and neck cancer would not represent an important confounder because it should be readily visualized during buccal swabbing whereas esophageal cancer would be expected to be symptomatic. Future studies will address the buccal signatures in nonpulmonary malignancies. Fourth, there is a subset of patients (~10%) who develop lung cancer without a smoking history. These are biologically different (e.g., higher prevalence of epidermal growth factor receptor mutations) and may not be relevant to current screening strategies and field carcinogenesis and hence were not addressed in the current studies (21, 22). Fifth, a problem that has plagued screening approaches such as low-dose computerized tomography is the detection of insignificant lesions (nodules or early stage cancers that would have never progressed) that obligate aggressive follow-up and even surgery. Although we cannot comment on the performance of buccal PWS, it is reassuring to note that the effect size was similar for early and late disease. Finally, these preliminary studies were conducted on defined patients (those with and without cancer) and the sample size was modest. For validation, studies will be conducted on larger data sets in prospective studies of screening at-risk patients (e.g., smokers undergoing low-dose computerized tomography screening).
In conclusion, we provide the proof-of-principle that interrogation of the buccal epithelium with PWS can detect nanoscale architectural alterations of extended lung field carcinogenesis. If confirmed in large-scale validation trials, we would envision that this would be used as a “prescreen,” identifying patients at highest risk who are likely to benefit from more invasive or expensive testing (bronchoscopy or low-dose computerized tomography). This may herald a paradigm-shifting approach of employing PWS detection of extended field carcinogenesis for accurate risk-stratification and thereby bringing the era of “personalized medicine” to cancer screening.
Supplementary Material
Acknowledgments
Grant Support NSF grants CBET-0939778 and CBET-0937987 and NIH grants U01CA111257 and T32ESES007267.
Footnotes
Disclosure of Potential Conflicts of Interest Drs. H.K. Roy and V. Backman are cofounders and shareholders of American BioOptics LLC. All aspects of this study including data analysis and manuscript preparation were done under the supervision of the conflict of interest committee at Northwestern University.
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–49. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Alberg AJ, Ford JG, Samet JM. Epidemiology of lung cancer: ACCP evidence-based clinical practice guidelines (2nd edition) Chest. 2007;132:29–55S. doi: 10.1378/chest.07-1347. [DOI] [PubMed] [Google Scholar]
- 3.Bach PB, Silvestri GA, Hanger M, Jett JR. Screening for lung cancer: ACCP evidence-based clinical practice guidelines (2nd edition) Chest. 2007;132:69–77S. doi: 10.1378/chest.07-1349. [DOI] [PubMed] [Google Scholar]
- 4.Stelling K, Ryan J, Brody JS, Spira A. The field of tissue injury in the lung and airway. Cancer Prev Res. 2008;1:396–403. doi: 10.1158/1940-6207.CAPR-08-0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guo M, House MG, Hooker C, et al. Promoter hypermethylation of resected bronchial margins: a field defect of changes? Clin Cancer Res. 2004;10:5131–6. doi: 10.1158/1078-0432.CCR-03-0763. [DOI] [PubMed] [Google Scholar]
- 6.Sidransky D. The oral cavity as a molecular mirror of lung carcinogenesis. Cancer Prev Res. 2008;1:12–4. doi: 10.1158/1940-6207.CAPR-08-0093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Subramanian H, Pradhan P, Liu Y, et al. Optical methodology for detecting histologically unapparent nanoscale consequences of genetic alterations in biological cells. Proc Natl Acad Sci USA. 2008;105:20118–23. doi: 10.1073/pnas.0804723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Subramanian H, Roy HK, Pradhan P, et al. Nanoscale cellular changes in field carcinogenesis detected by partial wave spectroscopy. Cancer Res. 2009;69:5357–63. doi: 10.1158/0008-5472.CAN-08-3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Subramanian H, Pradhan P, Liu Y, et al. Partial-wave microscopic spectroscopy detects subwavelength refractive index fluctuations: an application to cancer diagnosis. Opt Lett. 2009;34:518–20. doi: 10.1364/ol.34.000518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spitz MR, Hong WK, Amos CI, et al. A risk model for prediction of lung cancer. J Natl Cancer Inst. 2007;99:715–26. doi: 10.1093/jnci/djk153. [DOI] [PubMed] [Google Scholar]
- 11.Kopelovich L, Henson DE, Gazdar AF, et al. Surrogate anatomic/functional sites for evaluating cancer risk: an extension of the field effect. Clin Cancer Res. 1999;5:3899–905. [PubMed] [Google Scholar]
- 12.Sridhar S, Schembri F, Zeskind J, et al. Smoking-induced gene expression changes in the bronchial airway are reflected in nasal and buccal epithelium. BMC Genomics. 2008;9:259. doi: 10.1186/1471-2164-9-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boyle JO, Gumus ZH, Kacker A, et al. Effects of cigarette smoke on the human oral mucosal transcriptome. Cancer Prev Res (Phila Pa. 2010;3:266–78. doi: 10.1158/1940-6207.CAPR-09-0192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanz-Ortega J, Roig F, AI-Mousa MM, et al. 17p13 (p53 locus), 5q21 (APC locus) and 9p21 (p16 locus) allelic deletions are frequently found in oral exfoliative cytology cells from smoker patients with non-small-cell lung cancer. Histol Histopathol. 2007;22:541–5. doi: 10.14670/HH-22.541. [DOI] [PubMed] [Google Scholar]
- 15.Us-Krasovec M, Erzen J, Zganec M, et al. Malignancy associated changes in epithelial cells of buccal mucosa: a potential cancer detection test. Anal Quant Cytol Histol. 2005;27:254–62. [PubMed] [Google Scholar]
- 16.Tercelj M, Ales A, Rott T, et al. DNA-based sputum cell image analysis for lung cancer in a clinical setting. Acta Cytol. 2008;52:584–90. doi: 10.1159/000325602. [DOI] [PubMed] [Google Scholar]
- 17.Spira A, Beane JE, Shah V, et al. Airway epithelial gene expression in the diagnostic evaluation of smokers with suspect lung cancer. Nat Med. 2007;13:361–6. doi: 10.1038/nm1556. [DOI] [PubMed] [Google Scholar]
- 18.Blomquist T, Crawford EL, Mullins D, et al. Pattern of antioxidant and DNA repair gene expression in normal airway epithelium associated with lung cancer diagnosis. Cancer Res. 2009;69:8629–35. doi: 10.1158/0008-5472.CAN-09-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–33. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dai MS, Lu H. Crosstalk between c-Myc and ribosome in ribosomal biogenesis and cancer. J Cell Biochem. 2008;105:670–7. doi: 10.1002/jcb.21895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med. 2008;359:1367–80. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gazdar AF. Personalized medicine and inhibition of EGFR signaling in lung cancer. N Engl J Med. 2009;361:1018–20. doi: 10.1056/NEJMe0905763. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.