Abstract
The cysteine protease legumain plays important functions in immunity and cancer at different cellular locations, some of which appeared conflicting with its proteolytic activity and stability. Here, we report crystal structures of legumain in the zymogenic and fully activated form in complex with different substrate analogs. We show that the eponymous asparagine-specific endopeptidase activity is electrostatically generated by pH shift. Completely unexpectedly, the structure points toward a hidden carboxypeptidase activity that develops upon proteolytic activation with the release of an activation peptide. These activation routes reconcile the enigmatic pH stability of legumain, e.g., lysosomal, nuclear, and extracellular activities with relevance in immunology and cancer. Substrate access and turnover is controlled by selective protonation of the S1 pocket (KM) and the catalytic nucleophile (kcat), respectively. The multibranched and context-dependent activation process of legumain illustrates how proteases can act not only as signal transducers but as decision makers.
Keywords: allostery, context-dependent activities, death domain, kcat-substrate specificity, electrostatic stability switch
Biological signaling represents a highly complex decision matrix that must be reflected by the activity regulation of proteases, universal master switchers in health, and disease (1). The primarily endolysosomal cysteine protease legumain exemplifies how intricate multifactorial processes combine in zymogen activation and activity regulation (2–4). These principles are key to accomplish its demonstrated roles in (auto-)immunity and cancer, involving diverse processes such as antigen processing (5, 6); TLR processing and activation (7); or modulating tumor- and stroma-derived components of the cancer degradome (8). However, the activity regulation principles are poorly understood and seem to be partly conflicting with the localizations and pH requirements of legumain’s moonlighting activities (8–10). By a pH-induced activation route, legumain was shown to develop asparaginyl-specific endopeptidase (AEP) activity that is used synonymously for the legumain (3, 11). Importantly, we could show that active AEP becomes irreversibly denatured at pH > 6.0 (4), clearly indicating our fundamental lack of understanding of legumain’s activation process and stabilization mechanisms. This knowledge is critical to understand and refine experimental anticancer treatments that use macrophage-anchored legumain for prodrug activation (8, 12, 13). Therefore, we set out to determine the crystal structure of legumain and to decipher its intriguing activity regulation mechanisms.
Results
Overall Architecture.
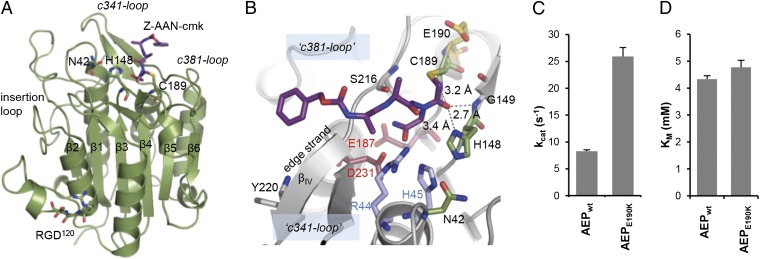
The crystal structure of active legumain revealed a caspase-like fold, with a central six-stranded β-sheet (β1–β6), flanked by five major α-helices (α1–α5). The 3D arrangement and connectivity of these secondary structure elements is topologically equivalent to caspases (Fig. 1, Fig. S1, and Table S1) (14). At the same time, we observed important differences such as a characteristic ∼30-aa insertion between strand β2 and helix α2 that controls zymogen activation, as to be described below; rigidity of the substrate specificity loops following strand β5 (“c341-loop” in caspase-1 numbering) and preceding strand β6 (“c381-loop”; Fig. S1 B and C); and, most strikingly, the monomeric state of active legumain, contrasting the situation in caspases where active forms are dimers (14). Although an analogous sheet extension would be sterically possible in legumain, there is no biological need for dimerization, which affects the substrate specificity loops c341 and c381 (15, 16); the equivalent loops are rigid in legumain, with the active site fully accessible as described below. Additionally, whereas the termini of the cleaved intersubunit linker strengthen the dimer interface in most caspases (14), the corresponding linker connecting strands β4 and β5 in legumain remains intact.
Fig. 1.
Substrate specificity and catalytic mechanism of legumain. (A) Legumain has a caspase-like overall structure. Active site residues (Cys189, His148, and Asn42) and the RGD120 motif are highlighted as green sticks, and the covalent Z-Ala-Ala-AzaAsn-chloromethylketone inhibitor in purple sticks. (B) Substrate specificity of legumain is determined by its zwitterionic S1 pocket. The Z-Ala-Ala-AzaAsn-cmk inhibitor (purple sticks) is covalently linked to the catalytic Cys189. The catalytic triad is represented by green sticks and residues forming the S1 pocket by red and blue sticks. Note the trivalent oxyanion pocket indicated by dashed lines. (C) Legumain activity critically depends on the local pKa of Cys189. The latter can be tuned by an E190K mutation, resulting in a fourfold increase in kcat at pH 5.5 as measured by the turnover of Bz-Asn-pNA. (D) KM values of active wild-type legumain (AEP) and E190K legumain toward the Bz-Asn-pNA substrate at pH 5.5 are virtually identical, indicating that the charge reversal affects exclusively the turnover number via pKa tuning of Cys189. Data in C and D are represented as mean ± SD of three experiments.
Catalytic Residues.
The catalytic Cys189 (Cysc285 in caspase-1) was confirmed by covalent labeling with peptidic chloromethylketone-based inhibitors. His148 (Hisc237) and Asn42 complete the catalytic triad (Oδ1Asn42–Nε2His148 ∼3.0 Å; Fig. 1B). Interestingly, caspases exhibit a carbonyl oxygen at position c177 that overlaps with the Asn42 side chain in legumain; this agreement supports the proposal that the carbonyl oxygen might participate in the catalysis of caspases (17), as well as paracaspases (18, 19) and metacaspases (20).
Analogous as in caspases, the reactive thiolate of Cys189 must form spontaneously and will depend on its local pKa. The structure suggested that the neighboring Glu190 stabilizes the protonated state of Cys189 (Fig. 1B). By contrast, an E190K mutant should lower the local pKa of Cys189. Indeed, this mutant showed a fourfold increase in kcat at pH 5.5 compared with the wild type, with KM unchanged (Fig. 1 C and D). This remarkable stimulation hints at a regulation potential that is implemented, yet untapped, in the isolated structure.
Substrate Recognition.
To delineate the principles of substrate recognition in fully activated legumain, we investigated peptidomimetic chloromethylketones with either aza-asparagine or aspartate in P1 position. Both inhibitors are covalently bound to the Cys189-Sγ via the methyl group of their electrophilic warhead (Fig. S2). Additionally, we found that aspartyl-chloromethylketone inhibitors could be displaced from the active site at pH 7.5 by soaking crystals with mercury, thus providing us with an uninhibited legumain structure. Notably, the conformation of the protease appears virtually unchanged upon ligand binding (Fig. S2 D and E).
The S1 specificity pocket has a zwitterionic character with its positively charged hemisphere built from Arg44 (Argc341) and His45 (Argc179), positioned at the entrance to helix α1 (Fig. 1B and Figs. S1B and S2); these residues interact with the oxygen Oδ1 of the P1 (Asn/Asp) side chain at 2.8–2.9 Å distance, similar as observed in caspases. Contrasting the situation in caspases, the S1 pocket is complemented by negatively charged residues Asp231 (Serc347; entrance α4) and Glu187 (Glnc283; exit β4) with ∼2.4 Å and ∼3.5 Å distance to the Asn Nδ2, respectively. Together with its steric restraints, the dual charge character of the S1 pocket makes it an ideal binding site for Asn in P1. Importantly, however, a virtually identical binding geometry was observed with Asp as a P1 residue, strongly suggesting that the Oδ2 of the P1 Asp is protonated within the S1 pocket. This conclusion is in line with our previous observation that aspartate in P1 is accepted at pH ≤ 4.5 only, consistent with its pKa of ∼4 (4).
The P1 amide nitrogen was hydrogen bonded to the carbonyl oxygen of Ser216 (Serc339). The βIV strand (Ser216–Tyr220) serves as the edge strand for nonprimed substrate recognition up to the P6 residue (Fig. 1B and Figs. S1B and S2). This antiparallel β-sheet formation dominates the nonprimed substrate recognition. However, some substrate preferences result from the side chains of Tyr217 (Trpc340) forming an open, partly hydrophobic S2 site; Tyr228 forming a polar S3 site; Tyr217 (Trpc340) and Trp232 forming a large hydrophobic (“aryl”) S4 binding site, and the glyco-asparagine Asn263 affecting the binding of residues upstream to P5 (Fig. S2). The structures further suggest that Gly149 (Glyc238), Ser150 (Ilec239), and Glu190 will form the S1′ and S2′ sites; this situation differs from caspases with their cleaved intersubunit linker (14).
Prolegumain Structure.
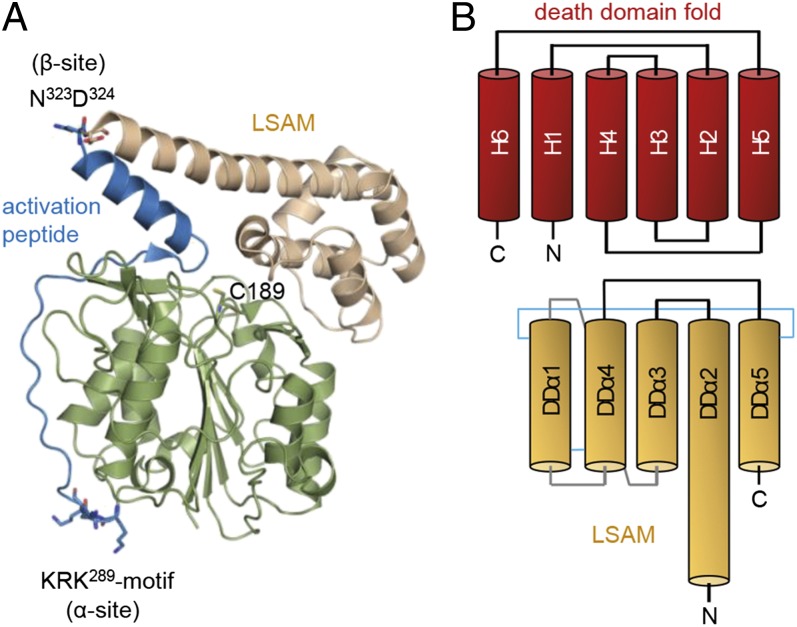
To understand how the C-terminal propeptide confers enzymatic latency and conformational stability to the protease, we determined the crystal structure of prolegumain. The structure reveals an overall helical “pro-domain” positioned on top of the protease domain and blocking access to the active site (Fig. 2A). The prodomain can be organized into an activation peptide (AP) spanning Lys287 to Asn323 and a C-terminal death domain-like fold (Asp324–Tyr433), denoted as legumain stabilization and activity modulation (LSAM) domain. The LSAM helices superpose onto death domain helices 1–5 with equivalent relative spatial arrangement and polarity, albeit partially different connectivity (Fig. 2B and Fig. S3). LSAM helices 1–5 correspond to death domain (DD) helices DDα2–DDα3–DDα1–DDα4–DDα5. As such, the LSAM domain corresponds closest to the open conformation of the Fas death domain and of the NLRC1 CARD domain [Protein Data Bank (PDB) ID codes 3EZQ and 4E9M], both of which have helix 6 integrated into a prolonged helix 5.
Fig. 2.
The C-terminal “propeptide” harbors a death domain like fold. (A) Prolegumain consists of a catalytic domain (green, Val18-Met286), an AP (blue, Lys287–Asn323), and a LSAM domain (wheat, Asp324–Tyr433). Cleavages at the α- (KRK289) and β-sites (N323D324) release the AP and, thus, renders the active site (C189) accessible. (B) Topology diagram of LSAM (colored in wheat) compared with classical death domains (colored in red).
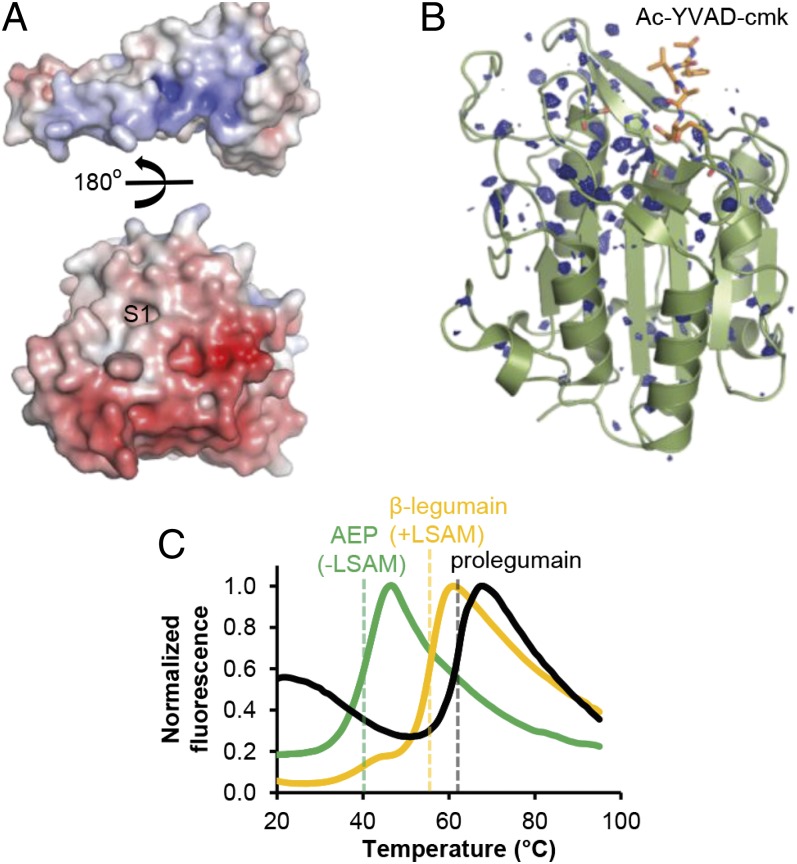
The interaction with the protease involves the complete activation peptide as well as the DDα3-DDα1 linker, DDα1 and DDα4 of the LSAM domain. Conversely, the protease domain contributes sheet βI–βIII as prodomain docking platform (Fig. 2A and Fig. S1B). The interaction is predominantly electrostatic in nature. The highly negatively charged protease surface is balanced by the positively charged prodomain interface (Fig. 3A). This charge balancing rationalizes (i) why prolegumain is stable at neutral pH; (ii) how protonation of the acidic protease surface will support the release of the prodomain; (iii) why pH-activated legumain (AEP) is stable at acidic pH with many of the protease surface-exposed Asp and Glu residues being protonated. And finally, (iv) it is comprehensible why deprotonation of the acidic residues at pH > 6.0 leads to electrostatic repulsion and irreversible conformational destabilization of AEP, an effect that can be partially delayed by active site ligands (4). The last conclusion (iv) was corroborated by a crystallographic difference analysis of legumain crystallized at pH 7.5 and pH 5.0. This comparison showed significant changes in the stability and dynamics localized at the “north” of legumain, near the active site (Fig. 3B), indicating that this surface patch serves as an electrostatically encoded stability switch (ESS) that can be triggered by ligands or pH.
Fig. 3.
The LSAM domain stabilizes legumain via electrostatic interactions. (A) Color-coded electrostatic surface potential of the catalytic domain (red; negative charge) and the LSAM domain including helix APαV (blue; positive charge) calculated at pH 7.0. The catalytic domain is rotated relative to the view in Fig. 2A by 90° around the horizontal x axis, and the LSAM domain has been separated and rotated by 180° relative to the catalytic domain. (B) Isomorphous σA-weighted difference density FpH7.5–FpH5.0 of legumain (AEP) crystallized at pH 7.5 and pH 5.0. In either case, legumain was covalently inhibited with Ac-YVAD-cmk (orange sticks). Strong changes in X-ray diffraction, as evident by the difference density, reflect changes in protein dynamics and stability. These changes cluster at the area surrounding the active site that corresponds to the LSAM domain binding interface; this confined mobility suggests that LSAM stabilizes the protease domain at neutral pH. Contour level: 2.9 σ over the mean; catalytic residues are indicated as green sticks. (C) Thermal denaturation curves show a stabilization of AEP by the LSAM domain. Melting curves of AEP (lacking the LSAM domain), β-legumain (cleaved at Asn323, LSAM domain remains noncovalently bound to the catalytic domain), and prolegumain were measured at pH 6.5 by the Thermofluor method. Melting points are indicated by dashed lines.
As a further, more direct approach to experimentally validate the ESS hypothesis with its impact on legumain activity, we performed thermal melting analyses with three different legumain species: single-chain zymogenic legumain, two-chain β-legumain (cleaved at Asn323–Asp324, with the LSAM domain noncovalently linked to the catalytic domain), and fully activated, single-chain AEP with the LSAM domain released (Fig. 3C). We found that the LSAM domain is instrumental in legumain stabilization at near neutral pH, as reflected by a ∼15 °C diminished melting temperature of AEP toward β-legumain (asparaginyl-specific carboxypeptidase; ACP) and more than 20 °C reduced toward zyomgenic prolegumain. The sharp melting curves document that each protein sample represented a distinct protein species, not a mixture of different forms.
Activation Peptide and the ACP.
Direct access to the active site is blocked by the AP that binds in a substrate-like manner to the active site. Specifically, Leu302–Pro306 mimic P6–P2 residues and bind to the nonprimed edge strand (Ser216–Tyr220) (Fig. 2A and see Fig. S7). Two proline residues, formally positioned at P2 and P1′, hinder the P1 residue (Ser307) from diving into the S1 pocket and explain why an in cis autolysis cannot occur.
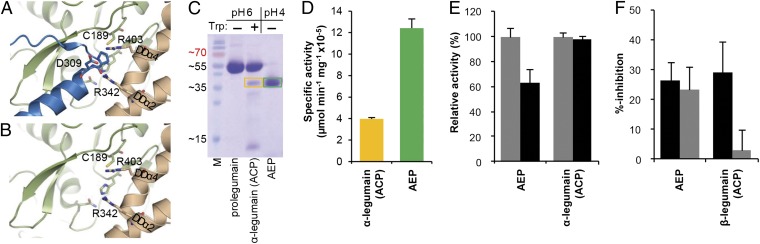
Of particular importance is the N-terminal anchoring of the AP helix (Asp309–Thr322) to a double arginine motif, Arg342 and Arg403, which are derived from the LSAM helices DDα2 and DDα4, respectively (Fig. 4A). Given the autoproteolytic cleavage site at Asn323–Asp324 (“β-cleavage”), it is conceivable that the activation peptide can be released from the zymogen while the C-terminal LSAM domain remains intact and bound to the protease (Fig. S4A). Complete dissociation of the activation peptide requires an additional cleavage at the N-terminal side of the activation peptide (“α-cleavage”). A putative cleavage site is the Lys-Arg-Lys289 motif because it is solvent exposed and flexible in the electron density (Fig. 2A). However, we should emphasize that the tissue-specific sugar moiety of glyco-Asn91 within the legumain-specific insertion loop is likely to restrict protease access and premature α-activation. Importantly, the proposed α-cleavage site is consistent in mass with the reported cellular processing of legumain (3). Upon release of the activation peptide, all nonprimed substrate recognition sites become accessible, as does the S1′ site, additionally flanked by the previously described double arginine-motif (Arg342, Arg403) that is characteristic for carboxypeptidases (Fig. 4B; ref. 21). In consequence, the structure suggests that a completely unexpected carboxypeptidase activity of legumain can be tapped upon selective release of the activation peptide. We refer to this asparaginyl-specific carboxypeptidase as ACP.
Fig. 4.
Prolegumain can develop carboxypeptidase activity upon cleavage of the AP. (A) The AP binds substrate-like to the nonprimed substrate binding cleft. The view to the active site is identical as in Fig. 1B. Catalytic residues are indicated as green sticks, and the double Arg-motif (Arg403 and Arg342) anchors the helix APαV via a salt bridge to Asp309. (B) View to the carboxypeptidase active site upon release of the AP. Arg403 and Arg342 are ideally positioned to anchor the C terminus of the substrate and, thereby, act as a molecular ruler for monopeptidyl peptidase activity, consistent with results from the peptide competition assay (Fig. S4 B and C). (C) SDS/PAGE after trypsin activation. After 2 h incubation of prolegumain with trypsin in a 1:50 molar ratio, approximately 12% of prolegumain were converted to an active enzyme (α-legumain, ∼36 kDa, orange box) as judged by using ImageJ (http://rsbweb.nih.gov/ij). In a control experiment, legumain was autoactivated via pH shift (AEP, green box). M, molecular mass marker in kilodaltons. (D) α-legumain carboxypeptidase activity can be generated via trypsin activation. Specific activity toward the fluorescent Z-AAN-AMC substrate is displayed. Orange, trypsin activated α-legumain; green, pH activated AEP. (E) Turnover of Z-AAN-AMC by pH 4.0 activated legumain (AEP) and trypsin activated α-legumain at pH 5.5 (gray bars) and pH 6.5 (black bars). Trypsin-activated legumain is not pH sensitive. Activity was normalized to maximum activity at pH 5.5. Data in D and E are represented as mean ± SD of three experiments. (F) Strict requirement for substrates with free carboxy terminus in ACP, but not AEP. Fluorogenic ACP activity can be competed off by peptides with a free P1' C terminus, H-AlaAlaAsn-Ala-OH; a single atom replacement (H-AlaAlaAsn-Ala-NH2; gray bars) completely abolishes the binding to ACP. By contrast, this exchange does not affect binding to the endopeptidase AEP.
To test this conclusion, we first used trypsin to cleave legumain at the KRK289 motif at mildly acidic pH (6.0), thus avoiding the release of the LSAM domain. The single-cleaved α-legumain developed enzymatic activity toward a fluorogenic legumain substrate, consistent with the proposed carboxypeptidase activity (ACP) (Fig. 4 C and D). α-Activation by trypsin was incomplete, consistent with glyco-Asn91 restricting the access to the KRK motif. To exclude that the observed activity arose from trace amounts of AEP, we tested the pH dependence of trypsin-activated α-legumain. α-Legumain retained full activity at pH 6.5, contrasting the behavior of the auto-activated endopeptidase (Fig. 4E). We therefore conclude that prolegumain can be differentially activated to a carboxypeptidase (ACP) or an endopeptidase (AEP).
Similarly, autoproteolytic activation at moderately acidic pH yielded ACP activity (β-legumain), with the LSAM domain noncovalently bound to the catalytic domain (Fig. S4A). Bona fide carboxypeptidases require substrates with a free carboxy terminus (21). We resorted to a peptide competition assay (11) to confirm that the free C terminus (H-AlaAlaAsn-Ala-OH) was strictly required to compete with substrate turnover; an analogous peptide with a C-terminal amide (H-AlaAlaAsn-Ala-NH2) showed no binding to ACP (Fig. 4F). By contrast, both peptides equally well bound to AEP, as is to be expected for an endopeptidase active site (Fig. 4F). We further varied the number of primed substrate residues, H-AlaAlaAsn-(Ala)n-OH, and found a preference for one primed amino acid, indicative for a preferential monocarboxypeptidase (Fig. S4 B and C).
Integrin Binding.
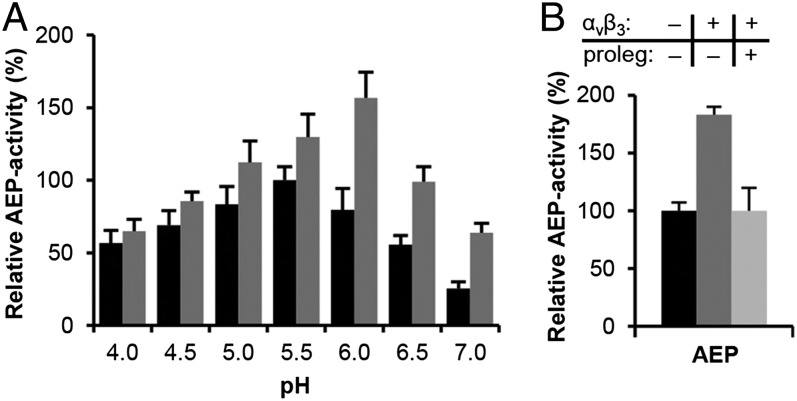
Legumain was reported to bind αVβ3 integrin (13). We determined pH-activity profiles of fully activated legumain (AEP) in the absence and the presence of αVβ3, revealing a shift of the pH optimum from pH 5.5 to 6.0 (Fig. 5A). The decrease of activity in isolated AEP with increasing pH (>5.5) correlates with its conformational destabilization (4), similar as has been reported for cathepsin B (22). Therefore, we concluded that the increase of AEP activity at pH 6.0 in the presence of αVβ3 was due to a conformational stabilization of AEP combined with a more frequently deprotonated Cys189 at increasing pH. Because legumain carries a characteristic integrin binding motif (RGD120) at the end of helix α2 (Fig. 1A and Fig. S1B), we tried to structurally interpret the legumain–αVβ3 complex by docking legumain to the RGD binding site of αVβ3, (PDB ID code 1L5G; Fig. S5). Following this docking model, αVβ3 should act as an allosteric ESS trigger (23), not interacting with the active site of legumain. This interpretation was corroborated by the binding of prolegumain to αVβ3 (Fig. 5B) and suggests mechanistic analogies with cathepsins anchored by tumor-associated macrophages (24, 25). These analogies with cathepsins could be further elaborated by referring to their pH-dependent activity profile (26).
Fig. 5.
Legumain and prolegumain interact with, and are stabilized by, integrin αVβ3. (A) Incubation of AEP with αVβ3 shifts its pH activity optimum to the neutral. Black bars, turnover of Bz-Asn-pNA by AEP at indicated pH values; gray bars, activity after incubation of legumain with αVβ3. Activity was normalized to maximum turnover of Bz-Asn-pNA at pH 5.5 in the absence of αVβ3. (B) Binding to αVβ3 integrin results in a twofold increase in enzymatic activity at pH 6.0. Black, active legumain only (AEP); dark gray, AEP activity boost in the presence of αVβ3, suggesting direct physical interaction between AEP and αVβ3. AEP binding to αVβ3 can be outcompeted by coincubation with excess (∼20-fold) prolegumain, abolishing the stimulating effect (light gray). Data in A and B are represented as mean ± SD of three experiments.
Discussion
Catalytic Mechanism.
The carbonyl oxygen of the P1 residue was bound to a trivalent oxyanion pocket formed by the amide nitrogens of Cys189 (Cysc285), Gly149 (Glyc238), and the Nδ1 of the catalytic His148 (Hisc237), all in hydrogen-bonding distance (Fig. 1B and Fig. S2B). All three interactions are also seen in related structures, e.g., caspases (14), gingipain (27), and the repeats in toxin (RTX) cysteine protease (28). The observed interactions have mechanistic implications: We propose that the triple-polarized carbonyl oxygen will be able to abstract the proton from the catalytic His148 (Hisc237) Nδ1, thereby transforming the C=O bond to a constitutively single C–O–H bonding; this proposition is consistent with conclusions derived for ketone-based caspase inhibitors (29). The resulting electrophilic carbonyl carbon is long lived to allow for the Cys189–Sγ to deprotonate and the subsequent nucleophilic attack. A classical keto-enol tautomerization induces the peptide bond rupture, allowing the primed product to dissociate. This proposed role of His148 (Hisc237) explains how legumain retains enzymatic activity at strongly acidic pH (4) and why caspase activity is drastically reduced in alkaline pH (30).
kcat Versus KM Substrate Specificity.
Proteases usually encode distinct substrate specificities by the shape and chemical nature of their substrate recognition sites. Only substrates that match and complement these sites will be able to efficiently and productively bind to a given active site. Therefore, substrate specificity primarily affects the KM of binding, although the binding affinity will usually couple to kcat effects, i.e., correlation of strength and productivity of binding.
By contrast, E190 in AEP represents a primarily kcat-driven mechanism of substrate specificity. By its negative charge, E190 stabilizes the catalytic Cys189 in the protonated state. Neutralization or charge reversal at the position 190 can be accomplished by site-directed mutagenesis (e.g., E190K; Fig. 1 C and D) or by virtue of charge shielding substrates: Substrates with positively charged residues at P1′ or P2′ position should be able to efficiently shield and neutralize the charge of E190. This charge neutralization should enhance AEP’s kcat with little effect on its binding, KM, similar as seen in the E190K mutant. On the other side, negative charges at P1′/P2′ will enhance the E190 effect on Cys189 and further decrease its pKa. Consequently, we expect peptide sequences with Asn(P1)-Asp/Glu(P1′)-Asp/Glu(P2′) to be slowly turned over at pH values >> pKa of Asp/Glu, i.e., >5.0. Importantly, the β-cleavage site (Asn323-Asp324) represents such a low-kcat recognition site. When probing AEP with the dead mutant prolegumain C189S as a substrate, we observed a drastically retarded turnover at pH 5.5 compared with pH < 4.5, despite the AEP pH optimum of 5.5 toward chromogenic/fluorogenic substrates with Asn at P1 (4). Substrates with Asp at P1 are preferred at pH ∼4.0, reflecting the requirement for the protonation of Asp to bind to the S1 pocket. In summary, legumain encodes two complementary mechanisms for pH-dependent recognition of substrates carrying Asp at P1 and Asp/Glu at P1′ or P2′. Substrates appear to exploit this potential to tune the local pKa by presenting charged residues at the proper position, i.e., the P1′ or P2′ site (31).
Conversely, E190 seems to contribute in anchoring the protein inhibitors cystatin C, E/M, and F (32) by electrostatic interactions, as suggested by docking studies (Fig. S6).
Electrostatic and Proteolytic Routes Lead to Complementary Legumain Activities.
We found two orthogonal ways to produce legumain activities with complementary specificity. Full endopeptidase activity can be obtained by pH reduction to pH 4.0 (2–4). The acidification is accompanied by autoproteolytic processing at the Asn323–Asp324 peptide bond. Although the autoproteolysis will accelerate the production of legumain endopeptidase activity, in light of the structures presented here, it becomes immediately evident that lowering of the pH alone is sufficient to generate AEP activity. At low pH, acidic residues on the AEP-LSAM interface become protonated, resulting in a release of the electrostatic clamp between the catalytic domain and prodomain. Indeed, an N323A mutation blocked the cleavage of the C-terminal propeptide (Asp324–Tyr433) but did not prevent the development of AEP activity, albeit at a strongly retarded time scale (3). A postulated involvement of the N-terminal propeptide (Val18–Asp25) in the activation process is neither supported by a Δ(Val18–Asp25) truncation mutant (4) nor by the structure (Fig. 1A).
Contrasting the pH-induced activation, proteolytically driven activation at near neutral pH will selectively release the activation peptide while keeping the LSAM–catalytic domain interaction intact, resulting in ACP formation. Complete release of the activation peptide requires an α-cleavage at the KRK289 motif by an as-yet-unknown protease and an autolytic β-cleavage of the Asn323–Asp324 peptide bond (Fig. 2A and Fig. S7). The amino acid sequence at the proposed α-site suggests several candidate activators, including trypsin or more specific prohormone convertases, e.g., PC2 (31). Although we showed that the α-cleavage suffices to generate fluorogenic activity, both α and β cleavages (αβ-legumain) may be necessary for C-terminal trimming of protein substrates. The β-cleavage at Asn323–Asp324 requires endopeptidase activity, which might appear to exclude ACP as a β-activator. However, negative charges at the primed side (here: Asp324 at P1′) can substitute for the substrate carboxy terminus and, thus, confer endopeptidase activity to carboxypeptidases, as demonstrated (21). Moreover, the tight kink following Asp324 enables docking of prolegumain into the active site of ACP with only minor side chain adjustments. The docking study together with the P1′-Asp imply that ACP can serve as a β-activator.
Legumain–Caspase Analogies.
Finally, we note that the recognition of the death domain-like fold within the C-terminal “pro-peptide” discloses an unforeseen analogy to caspases, carrying death domains N-terminal to the protease domain. Death domains are known for their homotypic interactions (33). Given the apparently convergent evolution of LSAM with death domain family proteins, it is conceivable that the legumain-LSAM exploits death domain interactions. Conversely, the interdomain interaction seen in prolegumain could serve as a template for the (in cis or in trans) interaction of a caspase protease domain with a CARD or DED domain, possibly to modulate caspase activity (Fig. S3).
The impact of the work presented here comes with many facets and on different levels. On an applied level, and given its pathophysiological relevance, the crystal structure of legumain will enable target-directed approaches for drug discovery. On a fundamental level, the analysis reveals the mechanisms of legumain’s zymogenicity, its electrostatically encoded stability switch, and its dual activation pathways with complementary activities. On a physiological level, this work rationalizes legumain’s context-dependent functions (Fig. 6). Although electrostatically activated AEP is stable and active in the acidic endolysosomal compartments, almost all or the remaining activities cannot be explained by AEP directly. We demonstrate here an allosteric and a direct mode of legumain stabilization by αVβ3 and the LSAM domain, respectively, both of which are key to explain its nonlysosomal functions, e.g., in bone formation, death-domain signaling, and tumor progression. Finally, on a conceptual level, our work changes the paradigm of proteases acting as binary switchers in signal transduction. Instead proteases can operate as decision makers. Put in engineering language, we find that proteases not only act as transistors but as logic gates and microprocessors.
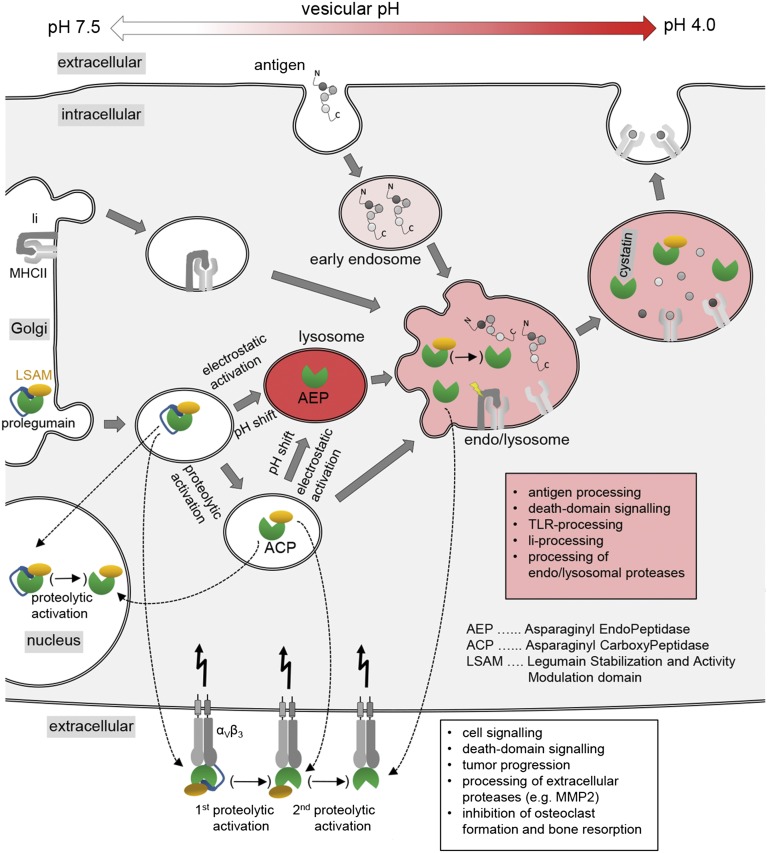
Fig. 6.
Multiple functions at multiple locations. Enzymatically active legumain was reported to exist in locations that are incompatible with AEP stability, i.e., pH ≥ 6. This contradiction can be reconciled by legumains three-modular architecture and its allosteric stabilization by the αVβ3 integrin receptor. After its posttranslational modification within the Golgi apparatus and vesiculation, prolegumain can be loaded onto αVβ3 receptors where a first proteolytic activation to ACP can take place, as necessary for its extracellular tasks (white text box). Integrin-stabilized AEP could be obtained by a second proteolytic activation event that would have to degrade the LSAM domain. Alternatively, transfer/fusion of prolegumain to lysosomal compartments with acidic pH (<5) will result in electrostatic release, and subsequent cleavage, of the prodomain resulting in AEP. By contrast, within near neutral compartments proteolytic activation generates ACP which will participate in endolysosomal roles (red text box).
Methods
Legumain preparation, crystallization, and assaying was done as described (4) and in SI Methods. Initial crystals of N272Q prolegumain were obtained with 35% (vol/vol) 2-ethoxyethanol and 100 mM cacodylate at pH 6.5 as precipitant. Equal volumes (0.2 µL) of protein solution (10 mg/mL) and precipitant were mixed and equilibrated against 60 µL of reservoir solution in 96-well INTELI-PLATEs (Art Robbins Instruments) at 277 K. Ethylmercuryphosphate (EMP)-soaked Ac-YVAD-cmk–inhibited legumain crystals were exploited for initial SIRAS phasing of active legumain. Molecular replacement followed by de novo building of the C-terminal prodomain was carried out to solve the structure of prolegumain. A detailed description of data collection, structure solution, and docking studies can be found in the SI Methods.
Supplementary Material
Acknowledgments
We thank Uli Demuth for providing the inhibitor Z-Ala-Ala-AzaAsn-cmk, Doriano Lamba for outstanding help and discussion on crystal handling, and the staff at the European Synchrotron Radiation Facility (ESRF, Grenoble) and the ELETTRA synchrotron (Trieste) for expert help in data collection. Funding was provided by Austrian Academy of Sciences ÖAW Project 22866 and Austrian Science Fund FWF Project P_23454-B11.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4aw9, 4awa, 4awb, and 4fgu).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1300686110/-/DCSupplemental.
References
- 1.Mason SD, Joyce JA. Proteolytic networks in cancer. Trends Cell Biol. 2011;21(4):228–237. doi: 10.1016/j.tcb.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen JM, Fortunato M, Barrett AJ. Activation of human prolegumain by cleavage at a C-terminal asparagine residue. Biochem J. 2000;352(Pt 2):327–334. [PMC free article] [PubMed] [Google Scholar]
- 3.Li DN, Matthews SP, Antoniou AN, Mazzeo D, Watts C. Multistep autoactivation of asparaginyl endopeptidase in vitro and in vivo. J Biol Chem. 2003;278(40):38980–38990. doi: 10.1074/jbc.M305930200. [DOI] [PubMed] [Google Scholar]
- 4.Dall E, Brandstetter H. Activation of legumain involves proteolytic and conformational events, resulting in a context- and substrate-dependent activity profile. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2012;68(Pt 1):24–31. doi: 10.1107/S1744309111048020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manoury B, et al. An asparaginyl endopeptidase processes a microbial antigen for class II MHC presentation. Nature. 1998;396(6712):695–699. doi: 10.1038/25379. [DOI] [PubMed] [Google Scholar]
- 6.Manoury B, et al. Destructive processing by asparagine endopeptidase limits presentation of a dominant T cell epitope in MBP. Nat Immunol. 2002;3(2):169–174. doi: 10.1038/ni754. [DOI] [PubMed] [Google Scholar]
- 7.Sepulveda FE, et al. Critical role for asparagine endopeptidase in endocytic Toll-like receptor signaling in dendritic cells. Immunity. 2009;31(5):737–748. doi: 10.1016/j.immuni.2009.09.013. [DOI] [PubMed] [Google Scholar]
- 8.Luo Y, et al. Targeting tumor-associated macrophages as a novel strategy against breast cancer. J Clin Invest. 2006;116(8):2132–2141. doi: 10.1172/JCI27648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith R, et al. Intra- and extracellular regulation of activity and processing of legumain by cystatin E/M. Biochimie. 2012;94(12):2590–2599. doi: 10.1016/j.biochi.2012.07.026. [DOI] [PubMed] [Google Scholar]
- 10.Lee J, Bogyo M. Development of near-infrared fluorophore (NIRF)-labeled activity-based probes for in vivo imaging of legumain. ACS Chem Biol. 2010;5(2):233–243. doi: 10.1021/cb900232a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwarz G, et al. Characterization of legumain. Biol Chem. 2002;383(11):1813–1816. doi: 10.1515/BC.2002.203. [DOI] [PubMed] [Google Scholar]
- 12.Liu C, Sun C, Huang H, Janda K, Edgington T. Overexpression of legumain in tumors is significant for invasion/metastasis and a candidate enzymatic target for prodrug therapy. Cancer Res. 2003;63(11):2957–2964. [PubMed] [Google Scholar]
- 13.Liu Y, Bajjuri KM, Liu C, Sinha SC. Targeting cell surface alpha(v)beta(3) integrin increases therapeutic efficacies of a legumain protease-activated auristatin prodrug. Mol Pharm. 2012;9(1):168–175. doi: 10.1021/mp200434n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuentes-Prior P, Salvesen GS. The protein structures that shape caspase activity, specificity, activation and inhibition. Biochem J. 2004;384(Pt 2):201–232. doi: 10.1042/BJ20041142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Renatus M, Stennicke HR, Scott FL, Liddington RC, Salvesen GS. Dimer formation drives the activation of the cell death protease caspase 9. Proc Natl Acad Sci USA. 2001;98(25):14250–14255. doi: 10.1073/pnas.231465798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Witkowski WA, Hardy JA. L2′ loop is critical for caspase-7 active site formation. Protein Sci. 2009;18(7):1459–1468. doi: 10.1002/pro.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilson KP, et al. Structure and mechanism of interleukin-1 beta converting enzyme. Nature. 1994;370(6487):270–275. doi: 10.1038/370270a0. [DOI] [PubMed] [Google Scholar]
- 18.Wiesmann C, et al. Structural determinants of MALT1 protease activity. J Mol Biol. 2012;419(1-2):4–21. doi: 10.1016/j.jmb.2012.02.018. [DOI] [PubMed] [Google Scholar]
- 19.Yu JW, Jeffrey PD, Ha JY, Yang X, Shi Y. Crystal structure of the mucosa-associated lymphoid tissue lymphoma translocation 1 (MALT1) paracaspase region. Proc Natl Acad Sci USA. 2011;108(52):21004–21009. doi: 10.1073/pnas.1111708108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McLuskey K, et al. Crystal structure of a Trypanosoma brucei metacaspase. Proc Natl Acad Sci USA. 2012;109(19):7469–7474. doi: 10.1073/pnas.1200885109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brandstetter H, Kim JS, Groll M, Huber R. Crystal structure of the tricorn protease reveals a protein disassembly line. Nature. 2001;414(6862):466–470. doi: 10.1038/35106609. [DOI] [PubMed] [Google Scholar]
- 22.Buck MR, Karustis DG, Day NA, Honn KV, Sloane BF. Degradation of extracellular-matrix proteins by human cathepsin B from normal and tumour tissues. Biochem J. 1992;282(Pt 1):273–278. doi: 10.1042/bj2820273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hardy JA, Lam J, Nguyen JT, O’Brien T, Wells JA. Discovery of an allosteric site in the caspases. Proc Natl Acad Sci USA. 2004;101(34):12461–12466. doi: 10.1073/pnas.0404781101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gocheva V, et al. IL-4 induces cathepsin protease activity in tumor-associated macrophages to promote cancer growth and invasion. Genes Dev. 2010;24(3):241–255. doi: 10.1101/gad.1874010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lechner AM, et al. RGD-dependent binding of procathepsin X to integrin alphavbeta3 mediates cell-adhesive properties. J Biol Chem. 2006;281(51):39588–39597. doi: 10.1074/jbc.M513439200. [DOI] [PubMed] [Google Scholar]
- 26.Stachowiak K, Tokmina M, Karpińska A, Sosnowska R, Wiczk W. Fluorogenic peptide substrates for carboxydipeptidase activity of cathepsin B. Acta Biochim Pol. 2004;51(1):81–92. [PubMed] [Google Scholar]
- 27.Eichinger A, et al. Crystal structure of gingipain R: An Arg-specific bacterial cysteine proteinase with a caspase-like fold. EMBO J. 1999;18(20):5453–5462. doi: 10.1093/emboj/18.20.5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lupardus PJ, Shen A, Bogyo M, Garcia KC. Small molecule-induced allosteric activation of the Vibrio cholerae RTX cysteine protease domain. Science. 2008;322(5899):265–268. doi: 10.1126/science.1162403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brady KD, et al. A catalytic mechanism for caspase-1 and for bimodal inhibition of caspase-1 by activated aspartic ketones. Bioorg Med Chem. 1999;7(4):621–631. doi: 10.1016/s0968-0896(99)00009-7. [DOI] [PubMed] [Google Scholar]
- 30.Stennicke HR, Salvesen GS. Caspase assays. Methods Enzymol. 2000;322:91–100. doi: 10.1016/s0076-6879(00)22010-7. [DOI] [PubMed] [Google Scholar]
- 31.Rawlings ND, Barrett AJ, Bateman A. MEROPS: The database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2012;40(Database issue):D343–D350. doi: 10.1093/nar/gkr987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alvarez-Fernandez M, et al. Inhibition of mammalian legumain by some cystatins is due to a novel second reactive site. J Biol Chem. 1999;274(27):19195–19203. doi: 10.1074/jbc.274.27.19195. [DOI] [PubMed] [Google Scholar]
- 33.Kersse K, Verspurten J, Vanden Berghe T, Vandenabeele P. The death-fold superfamily of homotypic interaction motifs. Trends Biochem Sci. 2011;36(10):541–552. doi: 10.1016/j.tibs.2011.06.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.