Abstract
Macrophage migration inhibitory factor (MIF) is a proinflammatory cytokine. In addition to its known receptor-mediated biological activities, MIF possesses a catalytic site of unknown function between subunits of a homotrimer. Each subunit contributes three β-strands to adjacent subunits to form a core seven-stranded β-sheet for each monomer. MIF monomers, dimers, or trimers have been reported, but the active form that binds and activates the MIF receptor (CD74) is still a matter of debate. A cysteine mutant (N110C) that covalently locks MIF into a trimer by forming a disulfide with Cys-80 of an adjacent subunit is used to study this issue. Partial catalytic activity and receptor binding to CD74 are retained by N110C (locked trimer), but there is no cellular signaling. Wild-type MIF-induced cellular signaling, in vivo lung neutrophil accumulation, and alveolar permeability are inhibited with a fivefold excess of N110C. NMR and size-exclusion chromatography with light scattering reveal that N110C can form a higher-order oligomer in equilibrium with a single locked trimer. The X-ray structure confirms a local conformational change that disrupts the subunit interface and results in global changes responsible for the oligomeric form. The structure also confirms these changes are consistent for the partial catalytic and receptor binding activities. The absence of any potential monomer and the retention of partial catalytic and receptor binding activities despite changes in conformation (and dynamics) in the mutant support an endogenous MIF trimer that binds and activates CD74 at nanomolar concentrations. This conclusion has implications for therapeutic development.
Keywords: mechanism, thermostable variant, ERK-1/2 activation
Macrophage migration inhibitory factor (MIF) is a proinflammatory protein and an important regulator of innate and adaptive immunity (1). It localizes to the cytosol of all human nucleated cells and is released upon cellular stress to activate the receptor CD74 in complex with its signaling component CD44, or the chemokine receptors CXCR2 and CXCR4 to induce extracellular signal-regulated kinase 1/2 (ERK-1/2) activation (1–3). MIF promotes the cellular secretion of CXCL8, prostaglandin E2 (PGE2), and other proinflammatory mediators, and it inhibits the anti-inflammatory activities of glucocorticoids (4). In addition to inflammation, MIF plays important roles in other diseases and is a drug target in phase I clinical trials for solid tumors (5, 6). MIF is a trimer with extensive subunit–subunit interactions, including a seven-stranded β-sheet for each monomer with β-strands contributed by adjacent subunits (7–9). An enzymatic site with an unknown substrate and a presumed tautomerase/isomerase activity exists between subunits similar to other proteins in the MIF superfamily (10). A key feature of these proteins is an invariant N-terminal residue, Pro-1, that functions as a catalytic base (11).
Oligomerization plays an important role in the activity and regulation of a wide variety of proteins (12). Although the crystal structure of MIF indicates that it is a trimer, other methods have been used to identify MIF monomers or dimers, in addition to the trimer (13–22). Sedimentation velocity and equilibrium ultracentrifugation data found MIF to be 97–98% trimeric species with a small amount of a large aggregate and an “incompetent monomer” not in equilibrium with the trimer (23). One hypothesis is that MIF trimer functions as enzyme with three active sites between adjacent subunit in the cytosol where it is most concentrated, but dissociates to a monomer at nanomolar concentrations in the extracellular milieu to function as a cytokine (24). In support of this hypothesis, the compound ebselen, which disrupts the trimer, results in a hyperagonist chemotactic activity, presumably by the released monomer (18). The mechanism by which the site of action of ebselen was identified included the structure-based design of the mutant that substitutes asparagine (Asn-110) by a cysteine resulting in a mutant (N110C) that forms an intersubunit disulfide with Cys-80 from an adjacent subunit.
In the current study we use the N110C locked trimer (LT) mutant to resolve whether the trimer of MIF is the physiological form responsible for extracellular activity. Previous attempts to create a monomer mutant have been unsuccessful (20) and exclude comparison with the LT. Nevertheless, we report the enzymatic activity, receptor binding, thermostability, intracellular signaling, and in vivo recruitment of neutrophils to the lung of mice by the LT. Structural studies of the disulfide-based LT mutant explain why signaling is not induced by CD74 receptor binding. Our studies support an endogenous trimer activating the MIF receptor (CD74) at physiological concentrations.
Results
Enzymatic Activity, Receptor Binding, Signaling, and in Vivo Activity of the N110C MIF Mutant.
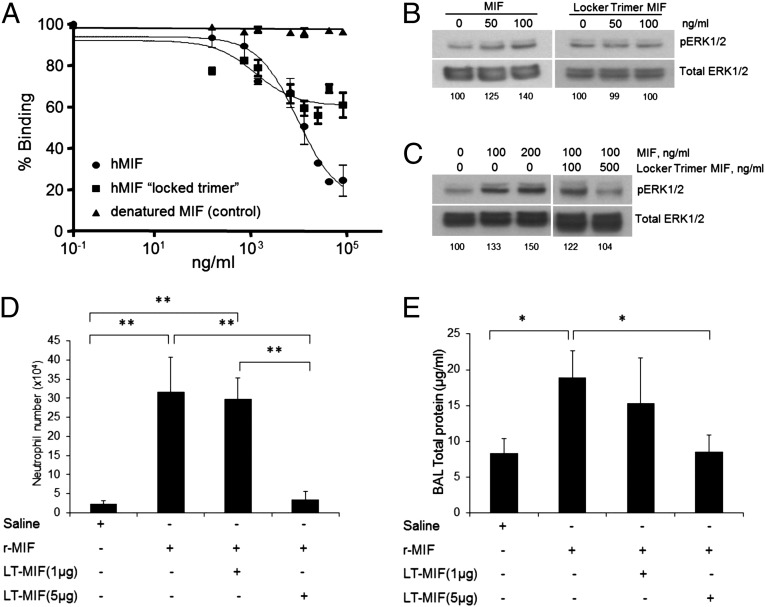
Size-exclusion chromatography (SEC) combined with multiangle laser light scattering (SEC/MALLS; Fig. S1) and SDS/PAGE (18) confirmed that the N110C mutant is a trimer with lower amounts of higher-order aggregates consisting of a dimer and trimer of the trimeric mutant. Michaelis–Menten kinetics using the substrate p-hydroxyphenylpyruvate on a keto/enol isomerization provided a kcat/Km of 1.27 mM−1s−1 for the mutant (Table 1), a 94% reduction compared with WT MIF. This large reduction is primarily due to a lower kcat. Receptor binding is also reduced, displacing human biotinylated WT MIF by 50% compared with 80% of WT MIF at the highest concentration (Fig. 1A). To eliminate the possibility that N110C interacts with WT MIF rather than competing for binding with CD74, elution of a mixture of N110C and WT MIF from SEC corresponded to the elution volume of each sample (MIF or N110C) alone (Fig. S2), indicating there is little, if any, direct interactions that could otherwise explain the competitive binding results.
Table 1.
Michaelis–Menten kinetic values for WT and LT MIF
| Kinetic parameters | WT MIF | LT MIF* |
| Vmax, μM s−1 | 3.6 ± 0.08 | 0.20 ± 0.03 |
| Km, mM | 1.73 ± 0.05 | 1.58 ± 0.04 |
| Kcat, s−1 | 36 | 2 |
| Kcat/Km, mM−1s−1 | 20.1 | 1.27 |
The nonenzymatic Vmax was 0.0201 ± 0.0002.
Fig. 1.
Binding of the N110C MIF mutant to CD74, signaling, and effects on lung neutrophil recruitment and alveolar permeability. (A) WT human MIF, the LT (N110C), and a denatured MIF control in a CD74 capture assay (40) that measures competitive receptor binding. (B) Western blot indicates WT human MIF induces a dose-dependent phosphorylation of ERK-1/2, but the LT (N110C) displays no signaling activity. The numbers at the bottom of the blot indicate the ratio of pERK-1/2 and total ERK normalized to 100 for no MIF. (C) The N110C LT mutant at fivefold concentration of WT MIF inhibits signaling activity. (D) MIF-induced neutrophil recruitment in the BAL fluid is inhibited by fivefold excess but not by an equimolar amount of N110C. (E) Alveolar permeability is also prevented by a fivefold excess but not equimolar amount of N110C, as measured by total protein concentration in the BAL. The BAL fluid samples for Fig. 1 D and E were collected from mice at 6 h postintratracheal instillation of 1 μg MIF alone or with the LT mutant (1 and 5 μg) in 50 μL saline, in addition to saline-only controls. *P < 0.01, **P < 0.001.
It was previously shown that WT MIF induces the phosphorylation of ERK-1/2. Although the LT binds to CD74, it has no signaling activity (Fig. 1B) and functions as a partial antagonist in cellular studies (Fig. 1C). Murine studies involving intratracheal administration of WT MIF and N110C to the lung similarly shows that the recruitment of neutrophils and total protein in the bronchoalveolar lavage (BAL) are inhibited by a fivefold excess of N110C (Fig. 1 D and E; Fig. S3) (25).
Temperature-Dependent Stabilization of the N110C MIF Mutant.
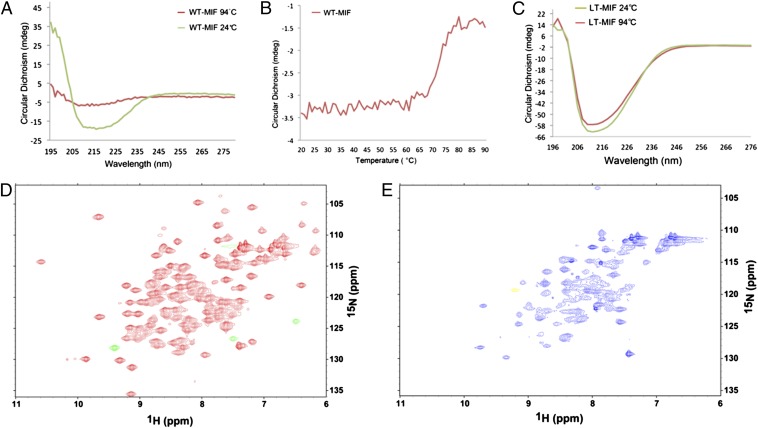
Despite reduced enzymatic activity and receptor binding, the disulfide mutant is unusually stable. In CD studies of WT MIF, a spectrum consistent with a mixed α-helix/β-sheet protein is observed that is totally unfolded at 94 °C (Fig. 2A). A temperature-dependent study indicates that WT MIF has a melting temperature (Tm) of 72 °C (Fig. 2B). Although there is a slight difference in the WT MIF and the N110C LT spectra, N110C retains its secondary structure at the instrument’s highest temperature, 94 °C (Fig. 2C). Because of the intersubunit disulfide bonds of the N110C mutant, the subunits cannot dissociate, and it is likely that the tertiary and quaternary structures are also retained given the contribution of the interdigitated β-strands by the two adjacent subunits to the core β-sheet that is present in each MIF monomer.
Fig. 2.
Thermostability and NMR of WT MIF and LT MIF. (A) CD spectra of WT MIF at 24 °C (green) and 94 °C (red). (B) Temperature-dependent CD scanning results in a Tm of 72 °C for WT MIF. (C) CD analysis of the N110C LT mutant at 24 °C (green) and 94 °C (red) shows a slight loss of secondary structure. 1H-15N HSQC of (D) WT and (E) N110C at 30 °C, each with identical buffer contoured to account for differences in scans and in concentration (2 mM for N110C and 1.78 mM for WT).
N110C Single-Site Mutation Alters the Oligomerization of MIF.
To further investigate the structure of this mutant, 15N-labeled MIF of the LT was examined for changes in the heteronuclear single-quantum coherence (HSQC) spectrum in a solution of 20 mM phosphate buffer (pH 7.0). It was expected that the spectrum would be well-resolved due to the thermostability of the mutant. Surprisingly, a spectrum of less than 50 resonances was observed. These observed peaks do not correlate well with the assigned WT MIF resonances, suggesting changes throughout the LT mutant (Fig. 2 D and E). In addition, the resonances of N110C are significantly line-broadened, indicative of a molecule greater than a trimer. WT MIF has a 15N R2 of ∼20 s–1, which corresponds well to a ∼30-kDa protein. The R2 for the N110C mutant cannot be measured because the signal decays too fast for accurate determination, but it can be estimated based on linewidths. The 15N linewidth averages ∼20 Hz for an R2 = 65 s–1. An R2 of this magnitude is more consistent with a protein with a rotational correlation time 20–30 ns compared with 13 ns for WT trimer, suggesting the N110C mutant is possibly a dimer or trimer of MIF trimers in solution. Alternatively, this type of line-broadening could be attributed to a protein-wide conformational exchange process in the millisecond timescale, but this scenario seems extremely unlikely. The NMR results are supported by the SEC/MALLS studies that show the formation of aggregates of two and three trimers for N110C (Fig. S1).
Crystal Structural Determination.
The N110C mutant was crystallized and its structure was determined by molecular replacement to 2.33 Å resolution at an Rfree of 0.27 (Table S1). Initial attempts to solve the structure in P212121 by molecular replacement using Phaser produced two trimers with high Z′ scores (>25) (21). Electron density from these phases indicated various loops, and the C-terminal region (100–114) of each subunit did not correspond to the electron density. Deletion of these regions did not improve the results during refinement with the Rfree remaining in the 0.40–0.50 range. Moreover, there was poor electron density for another trimer in the asymmetric unit; this was supported by the Matthews coefficient that is consistent with either two or three trimers in the asymmetric unit. A different strategy was used to solve the structure. The data were rescaled to P21, and a monomer of MIF with deleted loops and the C-terminal region was used with Phaser to produce a solution consisting of 18 monomers. A trimer was chosen from visual inspection and used as a search model in the original space group (P212121). This procedure produced three trimers that were identified by molecular replacement. Multiple cycles of refinement were used to model the deleted loops and the C-terminal region into density (Fig. S4). The final model consists of residues shown in Table S2 (column 2).
Structure of the N110C LT Mutant.
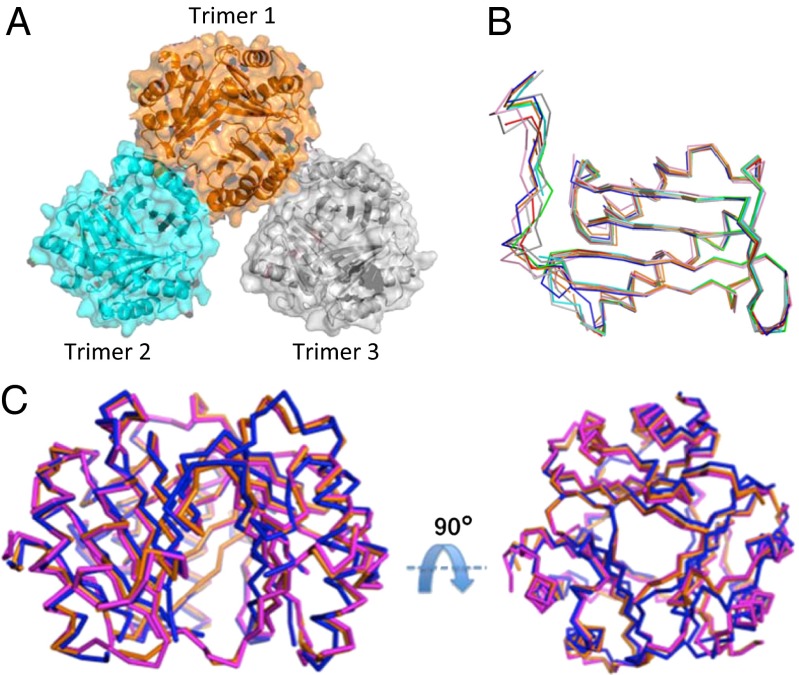
The intersubunit disulfide was verified for each of the nine monomers (Fig. S5). The three trimers in the asymmetric unit consist of one trimer (trimer 1) that makes significant interactions with trimer 2 with a buried surface area of ∼860 Å2 (Fig. 3A) but forms only two hydrogen bonds between the side chain of Arg-73 and the carbonyl of Gly-50 of trimer 3 (Fig. S6). The interactions between trimer 1 and 2 are from the C-terminal region of trimer 1 (monomer A) and both helices from a monomer (monomer F) of trimer 2. An additional interaction from the side chains of Arg-73 and Ser-20 occurs from an adjacent monomer of trimer 1 (monomer C) and monomer F of trimer 2. It is interesting that the majority of these trimer–trimer interactions involve regions from the unnatural intersubunit disulfide between Cys-80 in helix II in one monomer and the mutant N110C in the C-terminal region in another monomer (see Fig. S6 for specific interactions). Comparison of the three N110C trimers in the asymmetric unit reveals significant variability with rmsd values of 0.91–1.50 Å, and 0.48–1.50 Å for monomers with the C-terminal region playing a significant role in this variability (Fig. 3 B and C; Tables S2 and S3).
Fig. 3.
Structure and comparison of the three N110C LTs. (A) Structures of the three trimers displayed as transparent surface areas (aqua, orange, and gray) with ribbon structures that are present in one asymmetric unit. (B) Superposition of the nine monomers illustrating that the greatest region of variation is at the C terminus (residues 100–114). (C) Superposition of the three LTs.
Comparison of the N110C Mutant to WT MIF.
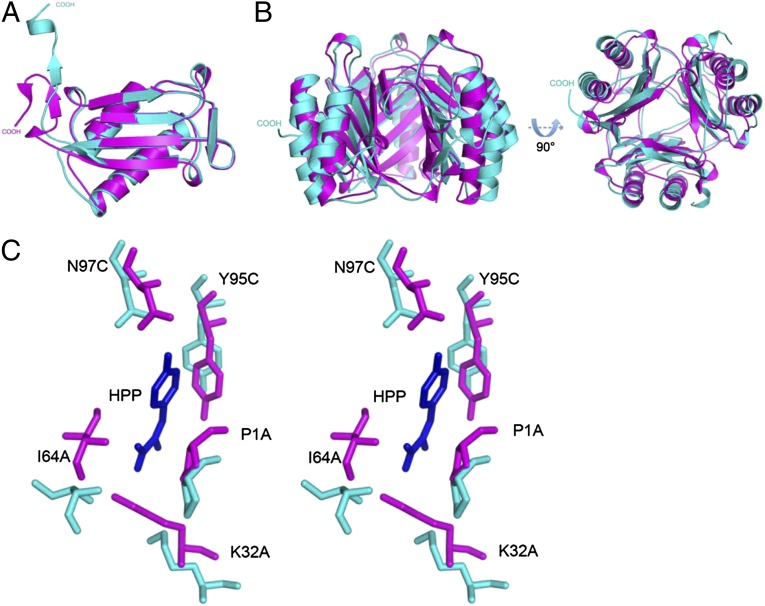
The N110C mutant has significant differences with the monomer and trimer of WT MIF (Fig. 4 A and B; Tables S2 and S3). In the monomer there are changes in the regions around Cys-80 and the mutated Cys-110. In addition to the formation of the disulfide, there are major changes in loop 5 (residues 65–68), helix II (residues 67–87), and the C-terminal region (residues 101–114) that forms a β-strand–loop–β-strand motif at the seven-stranded β-sheet core for the adjacent subunit. Helix II forms a slight kink at Lys-77 either to accommodate the disulfide formation with Cys-80 or as a result of disulfide formation. Loop 5 connecting helix II to a β-strand has a translational movement due to this kink. The most significant change is at the C-terminal region, particularly residues 108–114 that normally interact with helix II of the adjacent subunit and mediate part of the subunit–subunit contacts. This region is ejected from the WT position and protrudes into the solvent (Fig. 4 A and B). The absence of these subunit–subunit interactions has structural consequences, relaxing the tight interactions among subunits and resulting in a radial movement for the trimer perpendicular to the threefold axis. This movement is evident by an increase in the surface area of the N110C mutant trimer (15,390 Å2) compared with WT MIF (13,273 Å2). The expansion of the trimer alters the positions of atoms and explains the decrease in the enzyme activity (Table 1; Fig. 4C), receptor binding (Fig. 1A), and the absence of ERK-1/2 phosphorylation (Fig. 1B). The absence of signaling could also be due to restricted movements by the disulfide that may be required for activation upon MIF binding to CD74 and its signaling partner CD44 (3). The changes in atomic positions as well as the ejection of the C-terminal region normally intertwined with an adjacent subunit increase the propensity to form an oligomer of two trimers. The major interactions between the two trimers involve two regions of the cysteines that form the disulfide: the ejected C-terminal region from one trimer and helix II of the second trimer.
Fig. 4.
Comparison of the N110C LT mutant with WT MIF. (A) Superposition of a representative monomer from WT MIF (magenta) and the N110C LT MIF (aqua) indicates a large change at the β-strand−loop−β-strand of WT MIF relative to the mutant. (B) Superposition of the WT (magenta) and N110C (aqua) MIF trimers. There is an extension of the of the ribbon diagram of the N110C mutant compared with WT MIF. (C) Stereoview of superimposition of the N110C enzymatic active site onto that of WT MIF. The substrate p-hydroxyphenylpyruvate, the N110C atoms, and the WT MIF atoms are in blue, aqua, and magenta, respectively. The large movements away from the original positions for the catalytic residues explain the large decrease in enzymatic activity.
Discussion
Previous studies indicate WT MIF is present as a monomer, dimer, or trimer, but most of these experiments were performed at concentrations much higher than the low nanomolar concentrations used physiologically for MIF activation of CD74 (13–20). Mutagenesis at the subunit interface has not been successful at producing a soluble monomer to test for receptor binding and biological activity (20). The question of the active oligomeric state of MIF is linked to the relationship between the site for enzymatic activity and receptor binding. The enzymatic site occurs between subunits with some enzyme–inhibitor complexes, preventing receptor activation (26–28). The question of whether inhibitors stabilize the trimer at low concentrations by limiting dissociation to monomers or the MIF–inhibitor complex functions directly as an antagonist has never been studied. This question has implications for the development of therapeutics, because it will more appropriately guide inhibitor design (27, 29, 30).
The N110C mutant was initially used to determine the mechanism of action of ebselen, an inhibitor of MIF identified by a high-throughput screen (18). In analytical ultracentrifugation experiments with MIF and ebselen (at 1.5 µM), molecular masses of 36 kDa and 12 kDa were calculated from the sedimentation coefficient s of two peaks, whereas only the s value for 36 kDa was present in the absence of ebselen. Further experiments revealed that ebselen formed a covalent bond with Cys-80, which led to dissociation of trimers to monomers and the formation of aggregates (18).
The LT was an elegant tool to study the mechanism of action for ebselen inhibition. In the current study the LT is used to study whether the trimer binds to the MIF receptor CD74 with the expectation that if the monomer is active, the LT mutant could not bind CD74. This inference is based on the 3D structure of WT MIF, which shows a very stable trimer with extensive contributions of β-strands by the two adjacent subunits to the core β-sheet of each monomer. If the WT trimer disassociated into monomers, it is likely there would be large conformational changes for each monomer to accommodate the free β-strands originally located in adjacent subunits. These conformational changes are not possible in the N110C mutant because the intersubunit disulfides confine the structure to a trimeric state even at high temperatures, as shown by the CD experiments. The competitive binding between WT MIF and the LT mutant for CD74, despite the unanticipated conformation changes (see below), supports a WT trimer as the active oligomer for CD74 at physiological concentrations.
The structural studies of N110C reveal an unexpected local conformational change that leads to changes at the monomeric and oligomeric levels in solution and in the crystal. These changes appear to be a result of a longer-than-optimal distance for formation of a disulfide bond. In the initial structure-based design, the Cγ of Asn-110, the equivalent position of the thiol in the N110C mutant, is 4–5 Å from the thiol group of Cys-80. To form a disulfide there must be significant movement in protein atoms from the helix containing Cys-80 and the loop containing Cys-110 to reduce this distance to less than 2.3 Å [the usual cutoff for disulfides from Protein Data Bank (PDB) structures] with an optimal distance of 2.05 Å (31). The disulfide bond likely occurs during normal MIF dynamics upon oxidation when Escherichia coli is lysed. However, the number of changes revealed by the crystal structure suggests disulfide formation produces a protein that exists in an energetically unstable state, resulting in further conformational changes. The helical residue Lys-77 forms a kink. Consequently, loop 5 connected to this helix also moves. The most significant and unexpected change is the ejection of residues 108–114 (containing the mutated N110C) into the solvent from their native position involved in subunit–subunit interactions. The absence of these residues from their natural positions disrupts the adjacent β-strand and loop (residues 102–108). The lack of the native interface between subunits leads to a slight radial extension of the entire trimer (Fig. 4B). The ejected C-terminal region makes interactions with helices of another trimer leading to a monomer–dimer equilibrium shown by SEC/MALLS, broadened NMR resonances, and the crystal structure.
The residues of the catalytic site that interact with the model substrate p-hydroxyphenylpyruvate have the same physicochemical properties in the N110C mutant as in WT MIF. However, the optimal positions of these residues have changed due to the absence of the C-terminal region originally adjacent to some of the active site residues and the extension of the global trimer (Fig. 4 B and C). Surprisingly, these structural changes at the active site are not sufficient to totally overcome the catalytic properties of MIF, which suggests that the LT mutant retains enough flexibility to place the catalytic residues in a chemically competent conformation to achieve 6% of the enzymatic activity of WT MIF. Similarly, N110C maintains ∼50% CD74 receptor binding at the highest concentration (Fig. 1A), which, in the context of the structure, must be due to some inherent flexibility in the LT that allows binding but not signaling. Consequently, the LT functions as an antagonist. The antagonism observed in the signaling experiment shown in Fig. 1C was repeated in an in vivo study on MIF-induced accumulation of lung neutrophils in mice (25). The number of neutrophils accumulated by MIF alone was reduced to levels similar to control (saline) when a fivefold excess of LT was administered by intratracheal instillation.
The ejection of the C-terminal region from its normal position provided the opportunity for comparison with the results from C-terminal deletion mutants previously reported by El-Turk et al. (20) and Swope et al. (32). These deletion mutants lose all enzymatic activity. In El-Turk et al. (20), the deletion mutants retain all CD74 binding properties compared with ∼50% binding of N110C. NMR experiments from both deletion mutant studies indicate increased solvent accessibility based on 1H NMR (32), and peak broadening and destabilization based on 15N NMR studies (20). Unfortunately, there are no 3D structures for these deletion mutants to explain the lack of enzymatic activity and full retention of CD74 receptor binding, and the issue of the oligomeric structure at low nanomolar concentrations is not addressed. Nonetheless, there is qualitative similarity between the truncation mutants and the N110C LT mutant in the enzymatic activities (0% and 6%, respectively) and CD74 receptor binding (100% and 50%, respectively) based on the absence of subunit–subunit interactions, either through deletion or ejection of the C-terminal region. There is also a similarity at high concentrations with the deletion mutants (20) and N110C displaying a trimeric structure. The only remarkable difference between the deletion mutants and N110C is in the thermostability. The deletion mutants display a lower thermostability as assessed by CD, decreasing by 8–10 °C relative to WT MIF (20). In contrast, the N110C mutant has a significant gain in Tm of at least 20 °C in CD measurements, with only a slight difference in the CD spectrum at the maximum temperature available for experimentation (94 °C). This thermostability can be attributed to the intersubunit disulfides.
In summary, the N110C LT used for this study revealed unexpected results upon its characterization, which provided further insights about the mechanism of MIF. The decreased enzymatic activity and receptor binding properties upon disulfide bond formation is due to the conformational change at the C-terminal region, leading to an increased radial extension. The retention of these reduced activities is attributed to some (but not all) inherent conformational flexibility in WT MIF that remains at the enzymatic and receptor sites despite formation of an intersubunit disulfide. In this respect, extraordinary NMR HSQC chemical shifts in response to binding and mutations have been previously observed for WT MIF (18, 33, 34), suggesting an increased propensity for conformational flexibility of MIF compared with other soluble proteins. The absence of full flexibility in the LT explains the inability to activate the receptor. Most importantly, this study resolves the issue of the oligomeric form of MIF that activates CD74 at physiological concentrations and supports an active MIF trimer. We can also conclude that complexes of MIF with small molecule inhibitors are antagonists because they interact directly with CD74 and not because inhibitor binding to the trimer alters the equilibrium between trimer and monomer (26–28). This issue has important implications for therapeutics targeted to MIF.
Materials and Methods
Site-Directed Mutagenesis, Expression, and Purification of the N110C Mutant.
The QuikChange Site-Directed Mutagenesis Kit (Stratagene) was used to mutate WT MIF to the N110C in the pET11b(+)-MIF plasmid. The mutant and 15N-MIF was expressed and purified as previously described (8, 33).
Enzymatic Activity, Receptor Binding, and ERK Signaling Studies.
Enzymatic studies used p-hydroxyphenylpyruvate as the substrate, and the keto/enol tautomerase activity was monitored as previously described (33). To measure the binding of the N110C LT mutant to CD74, a capture assay was used (35). For signaling studies, the effect of MIF and/or N110C on ERK-1/2 activation was examined by Western blotting with Abs specific for phosphorylated (activated) ERK-1/2 (sc-7383; Santa Cruz Biotechnology) in human fibroblast-like synoviocytes (408RA-05a; Cell Applications Inc.).
Murine in Vivo Recruitment of Neutrophils to the Lung.
C57BL/6J mice (8–12 wk old) had recombinant murine MIF (1 μg in 50 μL normal saline) or LT (1 μg and 5 μg in 50 μL normal saline) directly administered via the intratracheal route (25). The differential cell counts were performed on cytocentrifuge preparations stained with HEMA 3 stain set (Fisher Scientific). A minimum of 200 cells were counted for the differential cell count (36). Total protein was measured using a DC Protein Assay (Bio-Rad) per the manufacturer's recommendations, as previously described (37).
CD Spectroscopy.
CD data were recorded using Applied Photophysics spectrometers with thermoelectric temperature control. CD and thermostability studies were recorded at 212 nm from 50 μg/mL LT MIF and WT MIF in 20 mM Tris⋅HCl, 20 mM NaCl (pH 7.5), with a temperature ramp of 1 °C/min.
Size Exclusion and Light Scattering.
The light-scattering data were collected using a Superdex 75 HR 10/30 SEC column (GE Healthcare) connected to the autosampler-equipped HPLC Agilent 1200. The elution from SEC was monitored with a photodiode array UV-visible light detector (Agilent Technologies), differential refractometer (Optilab rEx; Wyatt Corp.), and static and dynamic multiangle laser light scattering detector (HELEOS II with quasi-elastic light scattering capability; Wyatt Corp.). For comparison of elution peaks from WT MIF, N110C, and a mixture of the two proteins, Sepharose 75 16/60 was used.
NMR Spectra.
All NMR spectra were acquired at 14.1 T on a Varian Inova equipped with a triple-resonance gradient probe. Spectra were obtained at pH 7 in phosphate buffer.
Crystallization, Data Collection, Structure Determination, and Refinement.
Purified protein was concentrated to 22 mg/mL and crystallized by vapor diffusion from 0.2 M LiSO4, 0.1 M Tris (pH 8.5), and 33% (vol/vol) polyethylene glycol 3350. X-ray diffraction data sets from cryocooled crystals were collected at beamline X25 at the Brookhaven National Synchrotron Light Source. The diffraction data were integrated, scaled, and averaged using the HKL-2000 program suite (Table S1) (38). The atomic coordinates of WT MIF (1MIF) with some modifications was used as a search model for molecular replacement. Crystallographic refinement was performed using the program Phenix (39). After completion of refinement, the final Rwork and Rfree were calculated, respectively. The detailed statistics of data collection and refinement with the final model described below are provided in Table S1. The final model consisting residues for each monomer is listed in Table S2. All figures were prepared using PyMOL (DeLano Scientific). Atomic coordinates and structural factors have been deposited in the PDB (PDB ID code 4GUM).
For all mice experiments, the use and care of animals was approved by the Institutional Animal Care and Use Committee at Yale University. For additional details, see SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Ewa Folta-Stogniew of the Keck Biophysics Resource at Yale University for assistance with SEC light scattering, UV absorbance, and refractive index detector instrumentation, as well as data analysis. We also thank the staff at the X25 beamline of National Synchrotron Light Source at Brookhaven National Laboratory for assistance in data collection, and Drs. Michael Hodsdon and Camille Keeler for scientific discussions. Financial support for this project was provided by National Institutes of Health Grants AI065029, AR050498, and 1S10RR023748.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4GUM).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1221817110/-/DCSupplemental.
References
- 1.Leng L, et al. MIF signal transduction initiated by binding to CD74. J Exp Med. 2003;197(11):1467–1476. doi: 10.1084/jem.20030286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernhagen J, et al. MIF is a noncognate ligand of CXC chemokine receptors in inflammatory and atherogenic cell recruitment. Nat Med. 2007;13(5):587–596. doi: 10.1038/nm1567. [DOI] [PubMed] [Google Scholar]
- 3.Shi X, et al. CD44 is the signaling component of the macrophage migration inhibitory factor-CD74 receptor complex. Immunity. 2006;25(4):595–606. doi: 10.1016/j.immuni.2006.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calandra T, et al. MIF as a glucocorticoid-induced modulator of cytokine production. Nature. 1995;377(6544):68–71. doi: 10.1038/377068a0. [DOI] [PubMed] [Google Scholar]
- 5.Lolis E. Glucocorticoid counter regulation: Macrophage migration inhibitory factor as a target for drug discovery. Curr Opin Pharmacol. 2001;1(6):662–668. doi: 10.1016/s1471-4892(01)00112-6. [DOI] [PubMed] [Google Scholar]
- 6.Bucala R, Lolis E. Macrophage migration inhibitory factor: A critical component of autoimmune inflammatory diseases. Drug News Perspect. 2005;18(7):417–426. doi: 10.1358/dnp.2005.18.7.939345. [DOI] [PubMed] [Google Scholar]
- 7.Kato Y, et al. The crystal structure of human glycosylation-inhibiting factor is a trimeric barrel with three 6-stranded beta-sheets. Proc Natl Acad Sci USA. 1996;93(7):3007–3010. doi: 10.1073/pnas.93.7.3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun HW, Bernhagen J, Bucala R, Lolis E. Crystal structure at 2.6-A resolution of human macrophage migration inhibitory factor. Proc Natl Acad Sci USA. 1996;93(11):5191–5196. doi: 10.1073/pnas.93.11.5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suzuki M, et al. Crystal structure of the macrophage migration inhibitory factor from rat liver. Nat Struct Biol. 1996;3(3):259–266. doi: 10.1038/nsb0396-259. [DOI] [PubMed] [Google Scholar]
- 10.Poelarends GJ, Veetil VP, Whitman CP. The chemical versatility of the β-α-β fold: Catalytic promiscuity and divergent evolution in the tautomerase superfamily. Cell Mol Life Sci. 2008;65(22):3606–3618. doi: 10.1007/s00018-008-8285-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Subramanya HS, et al. Enzymatic ketonization of 2-hydroxymuconate: Specificity and mechanism investigated by the crystal structures of two isomerases. Biochemistry. 1996;35(3):792–802. doi: 10.1021/bi951732k. [DOI] [PubMed] [Google Scholar]
- 12.Hashimoto K, Nishi H, Bryant S, Panchenko AR. Caught in self-interaction: Evolutionary and functional mechanisms of protein homooligomerization. Phys Biol. 2011;8(3):035007. doi: 10.1088/1478-3975/8/3/035007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bendrat K, et al. Biochemical and mutational investigations of the enzymatic activity of macrophage migration inhibitory factor. Biochemistry. 1997;36(49):15356–15362. doi: 10.1021/bi971153a. [DOI] [PubMed] [Google Scholar]
- 14.Blocki FA, Schlievert PM, Wackett LP. Rat liver protein linking chemical and immunological detoxification systems. Nature. 1992;360(6401):269–270. doi: 10.1038/360269a0. [DOI] [PubMed] [Google Scholar]
- 15.Galat A, Rivière S, Bouet F, Ménez A. A diversified family of 12-kDa proteins with a high amino acid sequence similarity to macrophage migration-inhibitory factor (MIF) Eur J Biochem. 1994;224(2):417–421. doi: 10.1111/j.1432-1033.1994.00417.x. [DOI] [PubMed] [Google Scholar]
- 16.Mischke R, Kleemann R, Brunner H, Bernhagen J. Cross-linking and mutational analysis of the oligomerization state of the cytokine macrophage migration inhibitory factor (MIF) FEBS Lett. 1998;427(1):85–90. doi: 10.1016/s0014-5793(98)00400-1. [DOI] [PubMed] [Google Scholar]
- 17.Nishihira J, et al. Purification and characterization of human macrophage migration inhibitory factor: Evidence for specific binding to glutathione and formation of subunit structure. Biochem Mol Biol Int. 1993;31(5):841–850. [PubMed] [Google Scholar]
- 18.Ouertatani-Sakouhi H, et al. Identification and characterization of novel classes of macrophage migration inhibitory factor (MIF) inhibitors with distinct mechanisms of action. J Biol Chem. 2010;285(34):26581–26598. doi: 10.1074/jbc.M110.113951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun HW, et al. The subunit structure of human macrophage migration inhibitory factor: Evidence for a trimer. Protein Eng. 1996;9(8):631–635. doi: 10.1093/protein/9.8.631. [DOI] [PubMed] [Google Scholar]
- 20.El-Turk F, et al. The conformational flexibility of the carboxy terminal residues 105–114 is a key modulator of the catalytic activity and stability of macrophage migration inhibitory factor. Biochemistry. 2008;47(40):10740–10756. doi: 10.1021/bi800603x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cherepkova OA, Lutova EM, Gurvits BY. Charge heterogeneity of bovine brain macrophage migration inhibitory factor. Neurochem Res. 2005;30(1):151–158. doi: 10.1007/s11064-004-9696-1. [DOI] [PubMed] [Google Scholar]
- 22.Reidy T, et al. Homotrimeric macrophage migration inhibitory factor (MIF) drives inflammatory responses in the corneal epithelium by promoting caveolin-rich platform assembly in response to infection. J Biol Chem. 2013;288(12):8269–8278. doi: 10.1074/jbc.M112.351064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Philo JS, Yang TH, LaBarre M. Re-examining the oligomerization state of macrophage migration inhibitory factor (MIF) in solution. Biophys Chem. 2004;108(1-3):77–87. doi: 10.1016/j.bpc.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 24.Swope MD, Lolis E. Macrophage migration inhibitory factor: Cytokine, hormone, or enzyme? Rev Physiol Biochem Pharmacol. 1999;139:1–32. doi: 10.1007/BFb0033647. [DOI] [PubMed] [Google Scholar]
- 25.Takahashi K, et al. Macrophage CD74 contributes to MIF-induced pulmonary inflammation. Respir Res. 2009;10:33. doi: 10.1186/1465-9921-10-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amin MA, et al. Migration inhibitory factor up-regulates vascular cell adhesion molecule-1 and intercellular adhesion molecule-1 via Src, PI3 kinase, and NFkappaB. Blood. 2006;107(6):2252–2261. doi: 10.1182/blood-2005-05-2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cournia Z, et al. Discovery of human macrophage migration inhibitory factor (MIF)-CD74 antagonists via virtual screening. J Med Chem. 2009;52(2):416–424. doi: 10.1021/jm801100v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cho Y, et al. Drug repositioning and pharmacophore identification in the discovery of hookworm MIF inhibitors. Chem Biol. 2011;18(9):1089–1101. doi: 10.1016/j.chembiol.2011.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crichlow GV, et al. Alternative chemical modifications reverse the binding orientation of a pharmacophore scaffold in the active site of macrophage migration inhibitory factor. J Biol Chem. 2007;282(32):23089–23095. doi: 10.1074/jbc.M701825200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lubetsky JB, et al. The tautomerase active site of macrophage migration inhibitory factor is a potential target for discovery of novel anti-inflammatory agents. J Biol Chem. 2002;277(28):24976–24982. doi: 10.1074/jbc.M203220200. [DOI] [PubMed] [Google Scholar]
- 31.Bhattacharyya R, Pal D, Chakrabarti P. Disulfide bonds, their stereospecific environment and conservation in protein structures. Protein Eng Des Sel. 2004;17(11):795–808. doi: 10.1093/protein/gzh093. [DOI] [PubMed] [Google Scholar]
- 32.Swope M. 1998. Cytokine, hormone, or enzyme? An investigation of the biochemical and biophysical properties of macrophage migration inhibitory factor. PhD dissertation (Yale University, New Haven, CT)
- 33.Cho Y, et al. Allosteric inhibition of macrophage migration inhibitory factor revealed by ibudilast. Proc Natl Acad Sci USA. 2010;107(25):11313–11318. doi: 10.1073/pnas.1002716107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Crichlow GV, Fan C, Keeler C, Hodsdon M, Lolis EJ. Structural interactions dictate the kinetics of macrophage migration inhibitory factor inhibition by different cancer-preventive isothiocyanates. Biochemistry. 2012;51(38):7506–7514. doi: 10.1021/bi3005494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fingerle-Rowson G, et al. A tautomerase-null macrophage migration-inhibitory factor (MIF) gene knock-in mouse reveals that protein interactions and not enzymatic activity mediate MIF-dependent growth regulation. Mol Cell Biol. 2009;29(7):1922–1932. doi: 10.1128/MCB.01907-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rajasekaran D, et al. A model of GAG/MIP-2/CXCR2 interfaces and its functional effects. Biochemistry. 2012;51(28):5642–5654. doi: 10.1021/bi3001566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bhandari V, et al. Increased hyperoxia-induced lung injury in nitric oxide synthase 2 null mice is mediated via angiopoietin 2. Am J Respir Cell Mol Biol. 2012;46(5):668–676. doi: 10.1165/rcmb.2011-0074OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–325. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 39.Terwilliger TC, et al. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr D Biol Crystallogr. 2008;64(Pt 1):61–69. doi: 10.1107/S090744490705024X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cho Y, et al. Structural and functional characterization of a secreted hookworm macrophage migration inhibitory factor (MIF) that interacts with the human MIF receptor CD74. J Biol Chem. 2007;282(32):23447–23456. doi: 10.1074/jbc.M702950200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.