Significance
The global problem of TB has worsened in recent years with the emergence of drug-resistant organisms, and new drugs are clearly needed. In a cell-based high-throughput screen, a small molecule, TCA1, was discovered that has activity against replicating and nonreplicating Mycobacterium tuberculosis. It is also efficacious in acute and chronic rodent models of TB alone or combined with frontline TB drugs. TCA1 functions by a unique mechanism, inhibiting enzymes involved in cell wall and molybdenum cofactor biosynthesis. This discovery represents a significant advance in the search for new agents to treat persistent and drug-resistant TB.
Keywords: drug resistance, dual mechanism
Abstract
A cell-based phenotypic screen for inhibitors of biofilm formation in mycobacteria identified the small molecule TCA1, which has bactericidal activity against both drug-susceptible and -resistant Mycobacterium tuberculosis (Mtb) and sterilizes Mtb in vitro combined with rifampicin or isoniazid. In addition, TCA1 has bactericidal activity against nonreplicating Mtb in vitro and is efficacious in acute and chronic Mtb infection mouse models both alone and combined with rifampicin or isoniazid. Transcriptional analysis revealed that TCA1 down-regulates genes known to be involved in Mtb persistence. Genetic and affinity-based methods identified decaprenyl-phosphoryl-β-D-ribofuranose oxidoreductase DprE1 and MoeW, enzymes involved in cell wall and molybdenum cofactor biosynthesis, respectively, as targets responsible for the activity of TCA1. These in vitro and in vivo results indicate that this compound functions by a unique mechanism and suggest that TCA1 may lead to the development of a class of antituberculosis agents.
One of the greatest needs in global health is the development of new drugs against tuberculosis (TB) that shorten the duration of TB chemotherapy and that are potent against drug-resistant strains of Mycobacterium tuberculosis (Mtb), for which current therapies are no longer effective (1). TB is exceptional among bacterial infections in that even drug-susceptible strains are difficult to treat rapidly and effectively. This challenge is, in part, because of the phenomenon of Mtb persistence, a state of phenotypic drug tolerance that is attributed to a quiescent or nonreplicating population of bacilli. Long treatment regimes make compliance problematic and lead to the emergence of drug-resistant mutants. Indeed, multidrug-resistant (MDR) and extensively drug-resistant (XDR) Mtb strains are becoming widespread, resulting in high failure rates, despite the use of second- and third-line antibiotics and longer treatment times (up to 2 y). A new drug in the drug regimen should shorten chemotherapy and overcome the emergence of resistance to have a real impact on TB.
Although numerous cell-based screens against Mtb have been performed, to date, most screens are designed to identify molecules that are active against rapidly growing mycobacteria under growth-optimal laboratory conditions and inherently biased to identifying bactericidal or bacteriostatic compounds against replicating Mtb (2). However, it is becoming apparent that the culture conditions used in a screen very much affect our ability to identify inhibitors that will be active in vivo (2, 3). This issue is a particular concern in the development of drugs targeting persistent Mtb. Both target- and cell-based screens have been carried out under conditions that are thought to simulate those Mtb encounters during a chronic infection (4, 5). For example, it has been shown that oxygen deprivation or nutrient starvation in Mtb cultures triggers metabolic changes, resulting in nonreplicating, phenotypically drug-resistant bacilli in vitro (6, 7). Indeed, anaerobic Mtb cultures are resistant to isoniazid (INH) and partly resistant to rifampicin (RIF) but highly sensitive to pyrazinamide (8), underscoring the differing drug sensitivities of Mtb in different metabolic states. Given the lack of clear consensus on cell culture conditions that best reflect the in vivo biology of Mtb, we carried out a screen based on in vitro biofilm formation in the hope of identifying compounds with new mechanisms of action that may be effective against drug-resistant and -persistent Mtb. Here, we report that the molecule TCA1 identified through this screen not only shows bactericidal activity against both replicating (WT and drug resistant) and nonreplicating Mtb but also, is efficacious in acute and chronic Mtb infection mouse models both alone and combined with INH or RIF. Moreover, genetic and biochemical studies show that TCA1 functions by inhibiting two distinct biosynthetic pathways with concomitant down-regulation of genes known to be involved in mycobacterial persistence.
Results and Discussion
High-Throughput Screen Under Biofilm Culture Conditions.
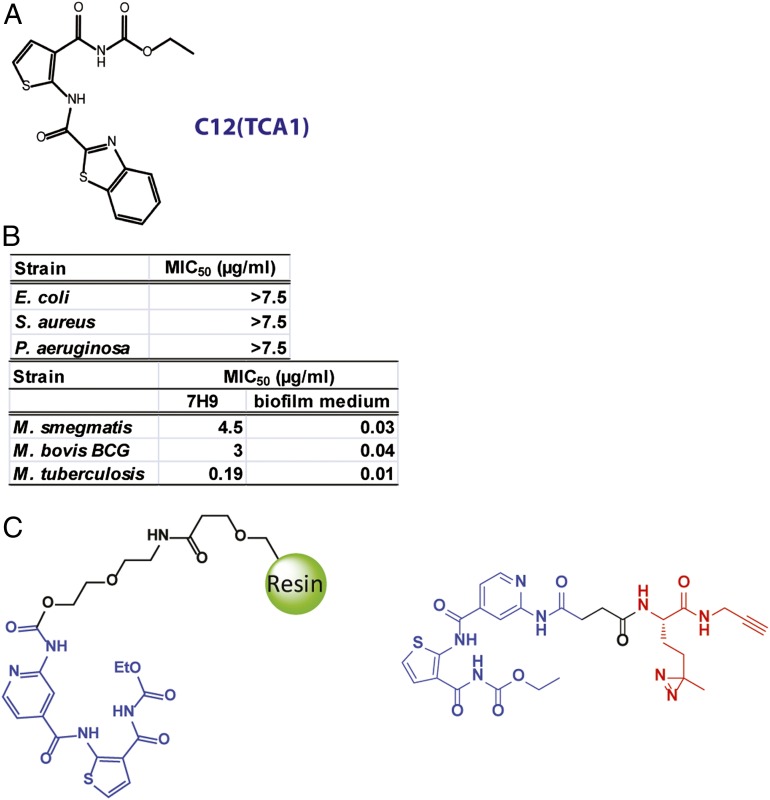
Pathogenic Mtb is not conducive to high-throughput screens involving automation, because these experiments would need to be carried out in a biosafety level 3 facility. However, M. smegmatis, a saprophytic, nonpathogenic mycobacteria that also forms in vitro biofilms (9) that induce drug tolerance (10), is amenable to high-throughput screening. Therefore, the primary cell-based screen was based on the inhibition of biofilm formation in M. smegmatis. We found that the in vitro biofilm, visualized as a pellicle that grows at the air–liquid interface, covers the whole surface of the well in a 384-well plate after formed, affording a high signal-to-noise ratio for positive hits. A diverse library of 70,000 heterocycles was screened (SI Materials and Methods), which afforded 17 compounds with minimum inhibitory concentrations (MIC50 values) of less than 10 µM in a biofilm inhibition assay. Two classes of compounds were identified: the first class inhibits the growth of mycobacteria under biofilm culture conditions, whereas the second class inhibits the formation of biofilms without significant growth inhibition. These hit compounds from the primary screen were then tested for their ability to inhibit in vitro biofilm growth in virulent Mtb H37Rv using a scaled-up 24-well assay as previously described (11). Two compounds, C7 and TCA1, were found to also inhibit biofilm formation by Mtb H37Rv (Fig. 1A). TCA1, which displayed potent inhibitory activity against Mtb under both biofilm and planktonic culture conditions, was selected for additional studies.
Fig. 1.
Chemical structures of the affinity resin (TCAP1) and the photo-affinity probe (TCAP2) used in pull-down experiments. Hit compound from screen under biofilm culture condition. (A) Chemical structure of active compound. (B) TCA1 has selective activity against mycobacteria. (C) Chemical structures of the affinity resin and the photo-affinity probe used in pull-down experiments.
In Vitro Bactericidal Activity.
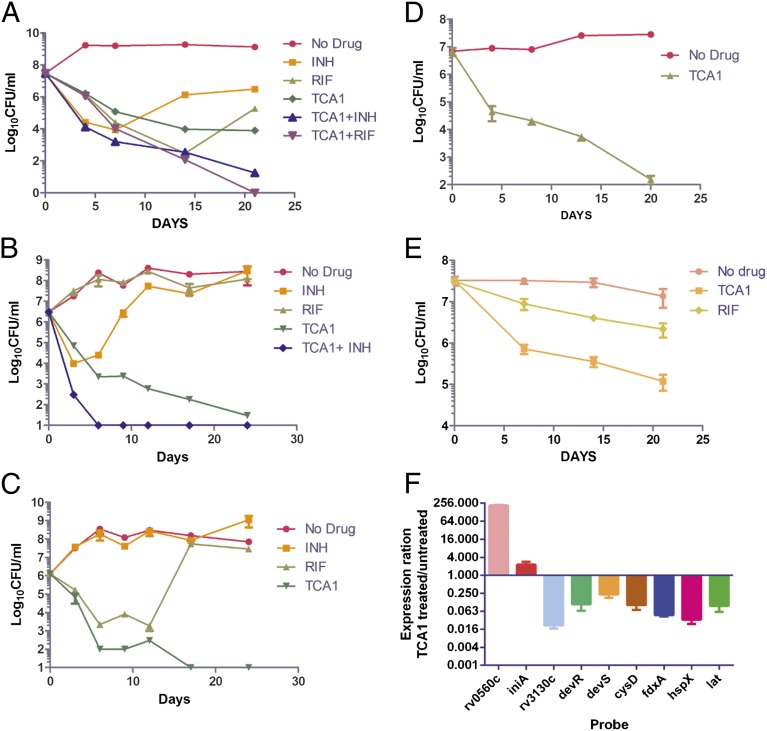
TCA1 shows selective inhibitory activity against bacterial growth—it is inactive against Escherichia coli, Staphylococcus aureus, and Pseudomonas aeruginosa, suggesting that the target for its bactericidal activity is specific to the genus Mycobacterium (Fig. 1B). Interestingly, the activities of TCA1 against M. smegmatis, M. bovis bacillus Calmette–Guérin, and Mtb are 20- to 150-fold higher in biofilm medium (MIC50 = 0.03, 0.04, and 0.01 µg/mL, respectively) than 7H9 medium (MIC50 = 4.5, 3, and 0.19 µg/mL, respectively). This observation underscores the variable efficacy of a drug in different growth media (3), which in part, may result from the expression of distinct target genes and metabolic pathways. TCA1 is bactericidal with an MIC99 values of 2.1 µg/mL in solid medium. To evaluate the bactericidal activity of TCA1 against Mtb compared with the two frontline TB drugs INH and RIF, we performed a 21-d kinetic killing assay using comparable levels of each of the three drugs (20× MIC50 of each of the three drugs). TCA1 is active by itself against exponentially growing virulent Mtb in 7H9 media, with a more than 3 log reduction in the number of bacilli over a treatment period of 21 d. Treatment with INH or RIF resulted in a comparable drop in cfu over the first 7 d of treatment, but the subsequent outgrowth of bacilli detected in INH- and RIF-treated cultures is absent in TCA1-treated cultures. Furthermore, TCA1 combined with either RIF or INH is able to sterilize an Mtb culture in ∼3 wk (Fig. 2A); removal of drug after 3 wk of combination drug treatment did not lead to Mtb outgrowth.
Fig. 2.
In vitro activity of TCA1. (A) Kill kinetics of Mtb by TCA1 (3.75 µg/mL) alone or combined with INH (1 µg/mL) or RIF (2 µg/mL) compared with RIF (2 µg/mL) and INH (1 µg/mL) alone in 7H9 medium. Activity of TCA1 (3.75 µg/mL) against (B) an RIF-resistant Mtb strain and (C) an INH-resistant Mtb strain in 7H9 medium. (D) Kill kinetics of an XDR-TB strain by TCA1 (7.5 µg/mL) in 7H9 medium. (E) Activity of TCA1 against nonreplicating Mtb under nutrient starvation conditions. (F) Quantitative PCR analysis of expression ratios of selected genes from TCA1-treated (3.75 µg/mL) and -untreated (0.1% DMSO) Mtb.
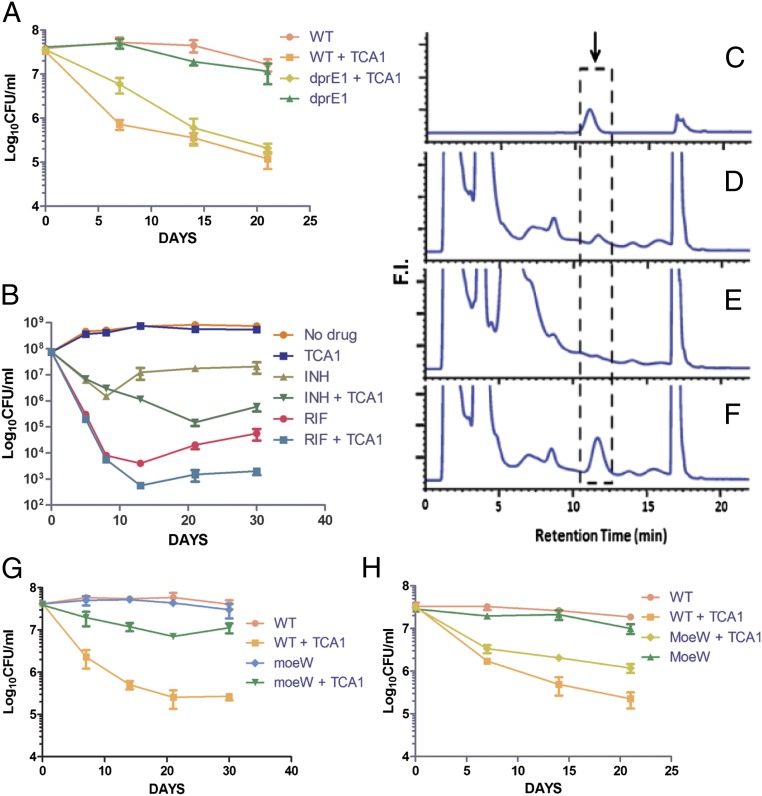
We also tested the activity of TCA1 on drug-resistant Mtb. RIF resistance is a marker for MDR-TB (90% of RIF-resistant strains are also MDR) and typically requires 18–24 mo of treatment. TCA1 by itself was active against a clinical strain that is resistant to RIF (because of a mutation in rpoB), and more importantly, combined with INH, it sterilized the cultures within 1 wk (Fig. 2B). Removal of both drugs after 3 wk of treatment did not result in outgrowth. TCA1 was also found to be bactericidal against a strain with a mutation in katG (resulting in resistance to INH) (Fig. 2C). Finally, we also tested TCA1 against an XDR-TB strain, mc28013, which is resistant to 10 TB drugs, including all frontline drugs (Table S1). TCA1 showed potent bactericidal activity against the XDR-TB strain (5 log cfu reduction in 3 wk) (Fig. 2D). The lack of cross-resistance to TCA1 in any of these drug-resistant strains suggests that TCA1 functions by a distinct mechanism.
We next tested the activity of TCA1 against nonreplicating Mtb in a nutrient starvation assay, a widely used in vitro model of the Mtb dormancy phenotype (12, 13). Under these conditions, Mtb enters a nonreplicating state and has been shown to become tolerant to drugs without acquiring heritable drug resistance-inducing mutations (13). TCA1 shows bactericidal activity against nonreplicating Mtb at a concentration of 7.5 µg/mL (40× MIC50 in 7H9 medium), reducing cfu by 3 logs in 3 wk (Fig. 2E). Under the same assay conditions, RIF (40× MIC50 in 7H9 medium) showed less bactericidal activity than TCA1. We also tested the activity of TCA1 in an intramacrophage cell culture system to determine whether it is active against intracellular mycobacteria, because in the mouse model of infection and in humans, Mtb is believed to reside mainly in macrophages. TCA1 was found to be quite potent in an intracellular cfu assay with an MIC50 value of 0.6 µg/mL [MIC50 (RIF) = 2.7 µg/mL, MIC50 (INH) = 0.2 µg/mL] (SI Materials and Methods). Finally, TCA1 shows no cytotoxicity against five mammalian cell lines (Huh7, 293T, K562, HepG2, and Vero cells) at the highest concentration tested (100 µM for Vero cells and 25 µM for others); an hERG assay indicated that TCA1 has no activity at 30 µM (Tables S2 and S3).
TCA1 Is Efficacious in Acute and Chronic Mtb Infection Mouse Models.
We next examined the activity of TCA1 in a mouse model of Mtb infection. We first determined the physical and pharmacokinetic characteristics of TCA1. It is stable to proteolytic activity in human or mouse plasma for up to 4 h. Moreover, a GSH trapping assay indicated that no GSH adduct was formed, and TCA1 has no inhibitory activity against four CYP enzymes. After i.v. administration, TCA1 exhibited a low clearance and steady-state volume of distribution, with an elimination half-life of 0.73 h. After oral administration of 20 and 50 mg/kg in solution formulation, TCA1 showed a high Cmax (2,122 and 5,653 nM, respectively), moderate exposure with oral bioavailability ranging from 19% to 46%, and a half-life of 1.8 h (Tables S2 and S3).
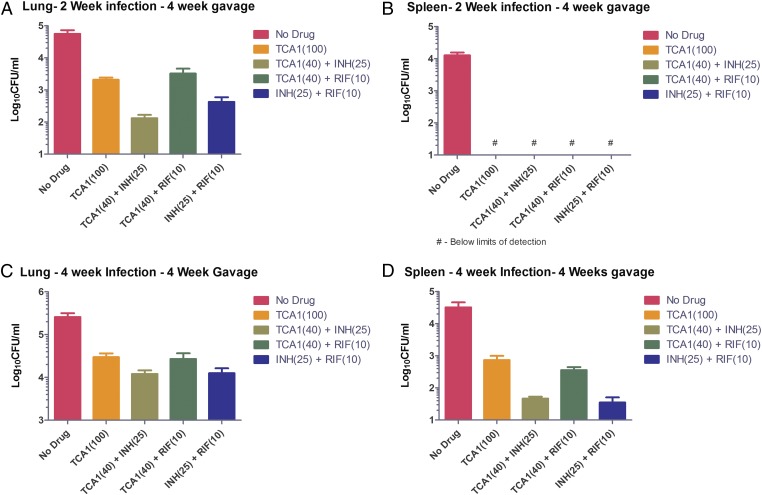
We first performed the in vivo efficacy experiments in an acute infection model with a low dose of TCA1. BALB/c mice were infected with a low dose of Mtb H37Rv (∼200 bacilli); 2 wk after infection, mice were treated with TCA1 (40 mg/kg), INH (25 mg/kg), or RIF (10 mg/kg) for 4 wk (dosed 1 time/d for 5 d/wk). The doses of INH and RIF are consistent with those doses published in the literature (14). After 4 wk of treatment with TCA1, the cfu dropped 0.5 log in lung and 1.5 logs in spleen, which is comparable with the potency of RIF but less than the potency of INH (Fig. S1). The gross pathology and histopathology also showed significant improvement in both tissues (Table S4). We also tested the in vivo efficacy of TCA1 (40 mg/kg) combined with INH (25 mg/kg) or RIF (10 mg/kg). In the acute infection model, TCA1+INH and TCA1+RIF showed nearly a 2 and 3 log cfu reduction in lung, respectively, and a more than 3 log cfu reduction in spleen (Fig. 3 A and B). There is a greater cfu drop in the lungs of mice treated by the combination of TCA1 and INH relative to the combination of INH and RIF. We next tested the drug in a mouse model of chronic TB infection. Mice were challenged with a low-dose aerosol infection, and treatment was initiated 4 wk after infection. Similar combination treatments were efficacious in the chronic infection model as well (Fig. 3 C and D).
Fig. 3.
In vivo efficacy of TCA1 in mouse models. In an acute Mtb infection mouse model (2 wk infection) followed by 4 wk of drug treatment, TCA1 showed significant bactericidal activity in (A) lungs and (B) spleen both alone (100 mg/kg) and combined with (40 mg/kg) INH (25 mg/kg) or RIF (10 mg/kg). The low activity of RIF as a monotherapy in this model is consistent with what has been observed in a previous study (14). In a chronic TB infection mouse model (4 wk low-dose infection) followed by 4 wk of drug treatment, TCA1 showed activity in (C) lungs and (D) spleen both alone (100 mg/kg) and combined with (40 mg/kg) INH (25 mg/kg) or RIF (10 mg/kg; P value < 0.05). Mice were gavaged with TCA1 1 time/d for 5 d/wk. RIF and INH were administered in drinking water.
Because the mice were able to tolerate 40 mg/kg TCA1, we increased the dose to 100 mg/kg using a similar protocol in the acute infection model. After 4 wk, the cfu dropped nearly 2 logs in lungs and more than 3 logs in spleen, showing that the in vivo bactericidal activity of TCA1 is dose-dependent (Fig. 3 A and B). The mice again showed no obvious adverse effects or weight loss after 4 wk of treatment. We also tested the drug in the chronic infection model at 100 mg/kg. Again, TCA1 showed efficacy in both lung (1 log cfu reduction) and spleen (1.4 log cfu reduction) (Fig. 3 C and D). These results show that the in vitro efficacy of TCA1 is recapitulated in vivo, suggesting that the in vitro mycobacterial biofilm is a useful phenotype to identify compounds effective against Mtb in vivo either alone or combined with existing TB drugs.
Mechanism of Action Studies of TCA1.
To gain insight into the mechanism of action of TCA1, we treated Mtb H37Rv with TCA1 (3.75 µg/mL) in 7H9 media and carried out genome-wide transcriptional analysis. Similar to INH and ethambutol (15), cell wall and fatty acid biosynthetic genes are affected by TCA1 treatment, suggesting that TCA1 likely interferes with these pathways. Unlike other known TB drugs, 10 of 86 genes differentially down-regulated compared with the DMSO control are genes previously implicated in TB dormancy, stress response, and RIF susceptibility. These genes include rv3130c–rv3134c, fdxA, and hspX (members of the dos regulon), cysD, and rv3288c–rv3290c (members of the sigF regulon). The microarray results were confirmed by quantitative PCR (Fig. 2F). Most of these genes are part of the dormancy regulon controlled by dosR (16) and up-regulated under hypoxic conditions or by nitric oxide exposure. For example, fdxA, a low-redox potential electron carrier, is highly up-regulated in Mtb under hypoxic conditions (17) but significantly down-regulated (>20-fold) in response to TCA1 treatment. Likewise, rv3130c is induced (>300-fold) under multiple stress conditions (18) but down-regulated (>30-fold) by TCA1. This down-regulation of genes involved in dormancy and drug tolerance seems to be unique to TCA1 and suggests that TCA1 may potentially sensitize Mtb to killing by antibiotics.
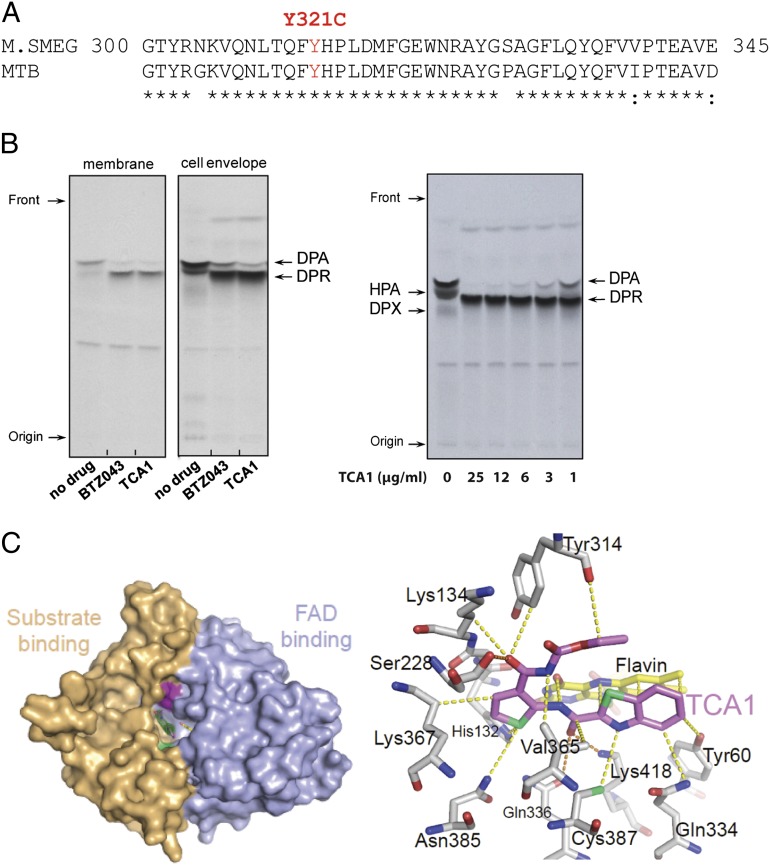
To further explore the mechanism of action of TCA1, a TCA1-resistant mutant that carries the cosmid (MSMEG_6379–MSMEG_6384) was isolated by selection of M. smegmatis transformed with a genomic cosmid library and grown in biofilm formation medium. Overexpression of each gene in this cosmid revealed that MSMEG_6382, which is homologous to rv3790 in the Mtb genome, confers high-level resistance to TCA1 (>20× MIC50) in both M. smegmatis and Mtb. We also managed to isolate spontaneous resistant mutants of M. smegmatis and Mtb, although the spontaneous mutation rate to TCA1 resistance is extremely low (10−8 to 10−9). Whole-genome sequencing of the genomic DNA of the resistant mutants revealed that they all have a single-point mutation, resulting in the amino acid replacement Tyr321Cys in MSMEG_6382 and Tyr314Cys in rv3790 (Fig. 4A). Rv3790 encodes DprE1, a component of the essential decaprenyl-phosphoryl-β-D-ribofuranose 2′-epimerase DprE1/DprE2 required for cell wall arabinan biosynthesis. Indeed, TCA1 suppressed the activity of M. smegmatis DprE1 in membrane and cell envelope enzymatic fractions in a dose-dependent manner (Fig. 4B). DprE1 was previously identified as the target of the benzothiazinones (BTZs) and a nitro-triazole molecule (3, 14). Both scaffolds contain active nitro-moieties and are believed to covalently modify Cys387 on activation. We performed a competitive binding assay using a fluorescently labeled BTZ analog. TCA1 potently competed with BTZ in binding to DprE1, suggesting that the binding site of TCA1 overlaps with BTZ (Fig. S2). However, TCA1 does not have an active nitro-moiety, and the Tyr314Cys mutant strain that is resistant to TCA1 is sensitive to BTZ, suggesting that the binding mechanism of TCA1 is different from these nitro-heterocycles.
Fig. 4.
TCA1 is a DprE1 inhibitor. (A) Sequence alignment of DprE1 of M. smegmatis and Mtb. A Y321C (Y314C in Mtb) mutation was identified in both M. smegmatis and Mtb strains resistant to TCA1. (B) Inhibition of DprE1 by TCA1 in the cell-free assay for decaprenylphosphoryl arabinose (DPA) production analyzed by TLC and autoradiography. M. smegmatis membrane or cell envelope fractions were incubated with (Left) phospho-[14C]-ribose diphosphate and 25 µg/mL TCA1 or BTZ043 or (Right) TCA1 in a dose–response fashion. Both TCA1 and BTZ043 potently inhibit conversion of the substrate DPR to the product DPA by DprE1/DprE2 epimerase. (C) Molecular surface of Mtb DprE1 with the FAD domain in light blue and the substrate binding domain in beige. The surface areas in pale green and magenta indicate the positions of (Left) Cys387 and Tyr314. (Right) Noncovalent contacts between TCA1 and DprE1 are shown. Residues within a 4-Å radius of the inhibitor (violet) are shown as sticks, with FAD in yellow. Dashed lines indicate the shortest contacts (yellow, hydrophobic/van der Waals; orange, polar) between the residues and the inhibitor. Trp230, located within 4 Å of the carbamate moiety of TCA1, has been omitted for clarity.
To determine the molecular basis by which TCA1 inhibits DprE1, we determined the crystal structure of the enzyme bound to TCA1 (Table S5). The overall structure of the DprE1–TCA1 complex is largely unaltered compared with the structure of the ligand-free protein with the same crystal symmetry (19). The enzyme, which is structurally related to the vanillyl-alcohol oxidase family of flavoproteins (20), consists of an FAD binding and a substrate binding domain, with the flavin moiety of FAD positioned at the interface between the two domains (Fig. 4C, Left). The substrate binding domain includes two disordered loop regions (residues 269–283 and 316–330) that leave the active site open and accessible for inhibitors. TCA1 binds in the central cavity of the enzyme, adjacent to the isoalloxazine ring of FAD, in a boomerang-like conformation, with the thiophene moiety inserted deeply into the bottom of the active site (Fig. 4C, Right). The benzothiazole ring is oriented roughly parallel to the isoalloxazine of FAD. Noncovalent interactions between TCA1 and the enzyme are dominated by hydrophobic and van der Waals interactions (Fig. 4C), with the flavin contributing a large fraction of the total contact surface. Polar contacts are sparse but include the H bonds between the carboxamido group and thiazole nitrogen of TCA1 and Nζ of Lys418 (3.0 and 3.1 Å, respectively). The carbamate moiety makes van der Waals interactions with the phenyl ring of Tyr314, consistent with the observation that substituting this tyrosine with cysteine renders DprE1 insensitive to TCA1. Superimposition of the structures of DprE1 bound to the BTZ analog (CT325) (19) and TCA1 showed that the binding sites of these two inhibitors overlap significantly.
The above results suggest that DprE1 is a relevant target for the bactericidal activity of TCA1 against replicating bacteria, similar to BTZ. However, there are some clear distinctions between TCA1 and BTZ. First, BTZ is not active against nonreplicating Mtb (14), whereas TCA1 is active against replicating and nonreplicating Mtb. Second, the gene expression profiles of Mtb treated by two compounds are also very different—TCA1 down-regulates persistence genes that are usually up-regulated in Mtb-dormant models, whereas BTZ does not (14). The Mtb strain overexpressing DprE1 is resistant to TCA1 in 7H9 medium but still sensitive to TCA1 in the nutrient starvation model (Fig. 5A). Moreover, TCA1 still potentiates INH or RIF on this DprE1-overexpressing strain (Fig. 5B). These results suggest that TCA1 could potentially act on an additional mycobacterial target.
Fig. 5.
TCA1 inhibits MoCo biosynthesis. (A) Mtb overexpressing dprE1 is sensitive to TCA1 (7.5 µg/mL) under nutrient starvation conditions. (B) Mtb overexpressing dprE1 conferred resistance to TCA1 (3.75 µg/mL) in 7H9 medium. In the meantime, TCA1 acts synergistically with INH (1 µg/mL) or RIF (2 µg/mL) against the same strain. (C–F) MoCo inhibition assay. HPLC profiles of MoCo Form A dephospho standard and sample extracted from Mtb. Arrow indicates the position of MoCo Form A dephospho. (C) MoCo Form A dephospho standard from synthetic source. (D) Extracted sample from Mtb in the absence of TCA1. (E) Extracted sample from Mtb after treatment with TCA1 (7.5 µg/mL; 18 h). (F) Extracted sample from Mtb in the absence of TCA1 spiked with an MoCo Form A dephospho standard. (G) Kill kinetics of WT Mtb and Mtb overexpressing moeW by 7.5 µg/mL TCA1 in media using nitrate as the only nitrogen source. (H) Kill kinetics of WT Mtb and Mtb overexpressing moeW by 7.5 µg/mL TCA1 under nutrient starvation conditions.
Because TCA1 had diminished activity against the DprE1(Y314C) mutant in normal growth medium, it is possible that a second TCA1 target is not essential for Mtb growth under conditions of optimal growth. This complication makes the selection of relevant mutants more difficult, and therefore, affinity-based methods were used to identify additional potential targets. Among a group of analogs of TCA1, we found that a pyridyl analog, TCA17, has very similar in vitro activity to the activity of TCA1. TCA17 was immobilized on a resin through a linker moiety (TCAP1) (Fig. 1C) and used in a pull-down experiment with cell lysates from Mtb. A 35-kDa band was identified on an SDS/PAGE after silver staining, which disappeared in the presence of 50 μM TCA1 as a competitor. MS identified the band as MoeW, a protein involved in the biosynthesis of the molybdenum cofactor (MoCo) (Fig. S3) with homologs in only a few bacterial genomes. To confirm the binding of TCA1 to MoeW, we overexpressed moeW in E. coli (which lacks an moeW gene homolog in its genome) and treated this strain with a photoaffinity probe analogous to TCA1 (TCAP2) (Fig. 1C) followed by UV irradiation and cell lysis. As shown in Fig. S4, a band with the size of MoeW is present on an SDS/PAGE gel for the sample from the moeW-induced strain and absent in the uninduced control sample. These results show that TCA1 scaffold directly binds to MoeW.
MoeW is predicted to contain an FAD/NAD binding domain by protein sequence analysis, but its function has yet to be determined. The gene encoding MoeW is only conserved in Mtb and bacillus Calmette–Guérin and not M. smegmatis or other mycobacterial species, although it is homologous to moeB, another gene involved in MoCo biosynthesis pathway and conserved in all mycobacteria species and many other bacteria (21). All molybdenum-using enzymes identified to date contain MoCo. MoCo is essential for the nitrate respiratory and assimilatory function of Mtb nitro-reductase. Some of these nitro-reductases have been found to be involved in the response of Mtb to hypoxia and nitric oxide (21, 22). To determine if TCA1 can block the biosynthesis of MoCo, we analyzed cell extracts from Mtb treated with TCA1 by detection of dephosphorylated MoCo Form A using HPLC with fluorescence detection. Indeed, TCA1 (7.5 µg/mL) completely abolished the formation of MoCo in Mtb (Fig. 5 C–F). It is known that MoCo is indispensable for nitrate assimilation by Mtb and thus, essential for Mtb to survive in media that uses nitrate as the only nitrogen source (designated as nitrate media). We generated an Mtb strain overexpressing moeW and found it to confer resistance to TCA1 in nitrate media over 30 d of treatment (Fig. 5G). These results clearly show that TCA1 asserts its activity against Mtb by inhibition of MoCo biosynthesis through interaction with MoeW. The Mtb strain overexpressing moeW conferred resistance to TCA1 in a nutrient starvation model over 21 d of treatment as well (Fig. 5H), but the lower level of resistance to TCA1 in the nutrient starvation model than nitrate media suggests that the mechanism of action of TCA1 is more complicated under the former condition. Nevertheless, the biochemical and genetic results clearly show that MoeW is a relevant target of TCA1.
Conclusion
We have developed a cell-based screen involving the growth of mycobacteria as an in vitro biofilm (a pellicle). The natural mode of growth of Mtb in liquid culture in the absence of detergent is as a pellicle at the liquid–air interface. Indeed, bacillus Calmette–Guérin is grown as a pellicle for vaccine production. This assay allowed us to identify a potent inhibitor TCA1 against both replicating and nonreplicating Mtb as well as drug-resistant Mtb. TCA1 functions by a unique mechanism involving down-regulation of persistence genes and inhibition of both cell wall and MoCo biosynthesis. Moreover, TCA1 showed excellent in vivo efficacy in both acute and chronic TB infection mouse models, suggesting that this compound may serve as a lead for the development of a class of drugs against persistent and drug-resistant Mtb. Indeed, we have subsequently identified a compound with good serum half-life that has excellent activities under both aerobic and anaerobic conditions (MIC50 values are 0.3 and 1.5 µg/mL, respectively). Future work will focus on additional improvements in the in vivo activity of this molecule and detailed mechanistic studies, including attempts to isolate additional resistant mutants under varied growth conditions. This work underscores the power of cell-based phenotypic screens to uncover molecules with mechanisms of action that provide unique approaches to the treatment of human disease.
Materials and Methods
High-Throughput Screen for Inhibitors of Biofilm Formation Inhibition.
A diverse chemical library (∼70,000 compounds) was used for the primary screen. This in-house compound library was created based on a chemoinformatic analysis of scaffold chemical diversity, historical proprietary screen hit rates [>300 Mio data points from the high throughput (HTS) database], and commercial availability; 105/mL M. smegmatis cells were plated in 384-well plates in biofilm formation medium of M63 salts minimal medium supplemented with 2% glucose, 0.5% Casamino Acids, 1 mM MgSO4, and 0.7 mM CaCl2. RIF and TMC207 were used as positive controls, and DMSO (0.1%) was used as a negative control. Cells were treated with 10 µM compound and incubated for 3 d, and the OD of each well was determined with an EnVision Multilabel Reader. The average Z′ and coefficient values are 0.512 and 8.7%, respectively. We used a high-stringency cutoff (threefold inhibition) to pick hits that are most likely growth inhibitors (hit rate = 0.03%) and a low-stringency cutoff (twofold inhibition) to include hits that inhibit biofilm formation without significant growth inhibition (hit rate = 0.17%).
In Vitro Activity Assays.
For kinetic killing assays, exponentially growing cultures of mycobacteria were diluted in fresh media to an OD600 of 0.1–0.2. Various drugs were added to the culture at the indicated concentrations. The number of cfus at the start of the experiment was estimated by plating appropriate dilutions of the culture onto 7H10 agar plates. The effect of drug was monitored by plating for cfus at the indicated time points. All experiments were carried out in triplicate. MICs were determined by a turbidity assay. Threefold serial dilutions in DMSO were prepared for each compound. Mtb cultures (OD = 0.04) were incubated with compounds at 37 °C for 5 d, and OD600 was determined with an Envision plate reader. All experiments were carried out in duplicate. For assays under starvation conditions, a log-phase growing Mtb culture was centrifuged, and the cell pellet was washed two times with PBS, resuspended in PBS with Tyloxapol (0.05%; OD = 0.3), and incubated with DMSO, TCA1 (7.5 µg/mL), and RIF (2 µg/mL). All experiments were carried out in triplicate. For intracellular macrophage assays, J744.1 murine macrophage cells were infected with Mtb at a multiplicity of infection (MOI) of 1:3 and incubated for 2 h at 37 °C. After washing the cell monolayer three times, 20 μM amikacin was added, and the culture was incubated for an additional 2 h to kill the remaining extracellular bacteria. Infected cells were then incubated in the presence of serial dilutions of compounds for 5 d. Cells were washed three times and lysed in each well; the lysate was transferred to a 96-well plate for serial dilution and then plated on 7H11 agar medium for cfu assays. All experiments were carried out in triplicate.
In Vivo Efficacy Experiments.
Six- to eight-week-old female BALB/c mice (US National Cancer Institute) were infected by aerosol with a low dose (∼50 bacilli) of Mtb H37Rv. Infection dose was verified by plating the inoculum and the whole-lung homogenates onto 7H10 plates at 24 h postinfection. Treatment of BALB/c mice began at either 2 or 4 wk postinfection, with RIF (10 mg/kg) and INH (25 mg/kg) administered ad libitum in drinking water (changed one time every 2 d). TCA1 was administered by oral gavage one time daily for 5 d/wk at a dosage of either 40 or 100 mg/kg for the indicated durations. At predetermined time points or humane endpoints, animals were heavily sedated and euthanized, and tissues were collected for culture and pathology. Treatment efficacy was assessed on the basis of cfu in the lungs and spleen of treated mice compared with untreated controls and bacterial burden in these organs before treatment start. Organs were homogenized in PBS containing Tween-80 [0.05% (vol/vol)], and various dilutions were placed on 7H10 plates. Plates were incubated at 37 °C for 3 wk, and cfus on the various plates were recorded. All animal experimental protocols were approved by the Animal Care and Use Committee of AECOM.
Genome-Wide Transcriptional Analysis.
Triplicate 10-mL cultures of mycobacteria were grown to log phase for transcriptional profiling of planktonic cells or for 3 wk in pellicle media for transcriptional profiling of pellicle cells. For TCA1 treatment, log-phase cultures were treated with 3.75 μg/mL TCA1 or DMSO vehicle for 12 h. Cells were harvested, washed, and resuspended in 1 mL RNA Protect reagent (Qiagen) and incubated 4 h at room temperature (21 °C). All transcriptional profiling procedures, including RNA extraction, DNase treatment, cDNA synthesis, labeling, microarray hybridization, washing, scanning, and data analysis, were performed as previously described (23). Microarray data have been deposited in the US National Center for Biotechnology Information Gene Expression Omnibus (accession no. GSE42151). For quantitative PCR experiments, diluted cDNA was used as a template at 50 ng per reaction for real-time PCR reactions containing primer sets designed by Primer 3 and SYBR Green PCR Master Mix (Applied Biosystems) in accordance with the manufacturers’ instructions. These reactions were carried out on an ABI 9700HT real-time PCR cycler (Applied Biosystems).
DprE1 Competition Assay.
DprE1 was incubated with serial dilutions of TCA1 for 15 min. BTZ-BODIPY was added, and the sample was incubated for 1 h at 37 °C. BTZ-BODIPY is a fluorescent BTZ derivative that reacts in the presence of farnesylphosphoribose, with DprE1 forming a covalent bond. Samples are then analyzed by SDS/PAGE (fluorescence and Coomassie staining).
Crystallization and Structure Determination.
Mtb DprE1 (Rv3790) was prepared for crystallization as described in the work by Batt et al. (19). Before setting up crystallization experiments, the TCA1 inhibitor (in DMSO) was incubated with concentrated protein (∼35 mg/mL) for 30 min at a molar ratio of 3 TCA1:1 DprE1. Crystals were grown by sitting drop vapor diffusion and appeared over a reservoir consisting of 40–43% (wt/vol) polypropylene glycol 400 and 0.1 M imidazole, pH 7.0. Crystals were mounted into nylon loops directly from the drop and frozen in liquid nitrogen. X-ray diffraction data to 2.6 Å resolution were recorded on beamline I02 of the Diamond Light Source (Table S5). The crystals were in space group P21, with two molecules of the complex in the crystallographic asymmetric unit. Initial phases were obtained by molecular replacement (PHASER) (24) using the apo structure of DprE1 (Protein Data Bank ID code 4FDP) (19) as a search model. After refinement of the molecular replacement solution, density for TCA1 was clearly visible in the active sites of the two crystallographically distinct copies of DprE1. Density shape and stereochemical constraints allowed us to unequivocally place the inhibitor in the active site of DprE1. Model rebuilding and structure refinement (COOT) (25), REFMAC5 (26), and PHENIX.REFINE (27) led to final R factors of 23.7% and 17.6% for the test and working sets, respectively.
Affinity-Based Proteomics and Photo-Affinity Labeling.
H37Ra cells were lysed with homogenization buffer (60 mM β-glycerophosphate, 15 mM p-nitrophenyl phosphate, 25 mM Mops, pH 7.2, 15 mM MgCl2, 1 mM DTT, protease inhibitors, 0.5% Nonidet P-40). Cell lysates were centrifuged at 16,000 × g for 20 min at 4 °C, and the supernatant was collected. Total protein concentration in the supernatant was determined by a BCA protein assay kit (Pierce). The lysates (1 mg) were then added to the affinity resin (30 μL), and the loading buffer (50 mM Tris⋅HCl, pH 7.4, 5 mM NaF, 250 mM NaCl, 5 mM EDTA, 5 mM EGTA, protease inhibitors, 0.1% Nonidet P-40) was added to a final volume of 1 mL (for the competition experiment, TCA1 was added to a final concentration of 50 μM). After rotating at 4 °C for 1 h, the mixture was centrifuged at 16,000 × g for 1 min at 4 °C, and the supernatant was removed. The affinity resin was then washed five times with cold loading buffer and eluted by boiling with Laemmli sample buffer (Invitrogen) at 95 °C for 3 min. Samples were loaded and separated on a 4–20% Tris⋅glycine gel (Invitrogen). The gel band was extracted and analyzed by proteomics. For the photo-affinity experiments, E. coli cells overexpressing MoeW and native cells were lysed, and the photo-affinity probe was added to cell lysates (1 mg) in 50 µL PBS and incubated for 2 h at room temperature followed by UV irradiation with a UV lamp for 20 min. The reaction mixtures were then subjected to click chemistry with rhodamine-azide (100 µM) and incubated for 2 h at room temperature with gentle mixing. The reactions were terminated by the addition of prechilled acetone (0.5 mL), placed at −20 °C for 30 min, and centrifuged at 16,000 × g for 10 min at 4 °C to precipitate proteins. The pellet was washed two times with 200 µL prechilled methanol, resuspended in 25 µL 1× standard reducing SDS loading buffer, and heated for 10 min at 95 °C; samples were loaded for separation by SDS/PAGE and then visualized by in-gel fluorescent scanning.
In Vitro Assays in Nitrate-Only Media.
An Mtb culture was resuspended under nitrogen-limiting conditions (22) (a basal medium [1 L basal medium contains 1 g KH2PO4, 2.5 g Na2HPO4, 2 g K2SO4, 2 mL trace elements; 1 L trace elements contained 40 mg ZnCl2, 200 mg FeCl30.6H2O, 10 mg CuCl20.4H2O, 10 mg MnCl20.4H2O, 10 mg Na2B4O70.10H2O, 10 mg (NH4)6Mo7O240.4H2O] supplemented with NaNO3 as the sole source of nitrogen, 0.5 mM MgCl2, 0.5 mM CaCl2, 10% ADS, 0.2% glycerol and 0.05% Tween 80) and incubated for 24 h; 7.5 µg/mL TCA1 was then added to the culture and incubated for 30 d. Cfu assay was used to determine the bacterial viability at each time point.
All experiments were carried out in triplicate.
MoCo Inhibition Assay.
The synthesis of MoCo Form A dephospho was carried out according to the procedures described. The 1H-NMR spectrum matches what has been reported in the literature (28, 29). Conversion of all sources of molybdopterin to Form A dephospho was performed by the methods previously reported with slight modifications (21, 30); 100 mL Mtb culture were harvested, and the pellet was resuspended in extraction solution (2 mL; 10 mM sodium ascorbate). The cells were lysed and centrifuged at 16,000 × g; the supernatant was collected and treated with acidic iodine solution at 95 °C for 25 min, and the excess iodine was removed by adding sodium ascorbate. After centrifugation, the solution was neutralized with ammonium hydroxide and then concentrated and dephosphorylated using calf intestinal phosphatase (NEB) at 37 °C for 3 h. HPLC analysis was performed using Agilent C18 column (150 × 4.6 mm, 10-µm particle size) with gradient elution by buffer A (50 mM ammonium acetate) and buffer B (MeOH; 97% A to 93% A in 14 min and 97% B wash from 15 to 22 min). Fluorescence detection was at 370/450 nm.
Supplementary Material
Acknowledgments
We thank Drs. Kelli Kuhen, Scott Robins, and Valerie Mizrahi for helpful discussions, Chun Li and Johnathan Chang for pharmacokinetic experiments, and Zhong Chen, Ivana Centarova, Emoke Kilacskova, and Dr. Baojie Wan for technical support. We also thank Dr. Stewart Cole and Dr. Vadim Makarov for providing BTZ043. This work was supported, in part, by European Community's Seventh Framework Programme Grant 260872 (to K.M.), the Global Alliance for TB Drug Development (P.G.S.), the National Institutes of Health Grants AI26170 and A10-97548, and the Albert Einstein College of Medicine Center for AIDS Research Grant AI0–51519.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors reported in this paper have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4KW5).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1309171110/-/DCSupplemental.
References
- 1.Gandhi NR, et al. HIV coinfection in multidrug- and extensively drug-resistant tuberculosis results in high early mortality. Am J Respir Crit Care Med. 2010;181(1):80–86. doi: 10.1164/rccm.200907-0989OC. [DOI] [PubMed] [Google Scholar]
- 2.Pethe K, et al. A chemical genetic screen in Mycobacterium tuberculosis identifies carbon-source-dependent growth inhibitors devoid of in vivo efficacy. Nat Commun. 2010;1:57. doi: 10.1038/ncomms1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stanley SA, et al. Identification of novel inhibitors of M. tuberculosis growth using whole cell based high-throughput screening. ACS Chem Biol. 2012;7(8):1377–1384. doi: 10.1021/cb300151m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mak PA, et al. A high-throughput screen to identify inhibitors of ATP homeostasis in non-replicating Mycobacterium tuberculosis. ACS Chem Biol. 2012;7(7):1190–1197. doi: 10.1021/cb2004884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gold B, et al. Nonsteroidal anti-inflammatory drug sensitizes Mycobacterium tuberculosis to endogenous and exogenous antimicrobials. Proc Natl Acad Sci USA. 2012;109(40):16004–16011. doi: 10.1073/pnas.1214188109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wayne LG, Hayes LG. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect Immun. 1996;64(6):2062–2069. doi: 10.1128/iai.64.6.2062-2069.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wayne LG, Sohaskey CD. Nonreplicating persistence of mycobacterium tuberculosis. Annu Rev Microbiol. 2001;55:139–163. doi: 10.1146/annurev.micro.55.1.139. [DOI] [PubMed] [Google Scholar]
- 8.Mitchison DA, Coates AR. Predictive in vitro models of the sterilizing activity of anti-tuberculosis drugs. Curr Pharm Des. 2004;10(26):3285–3295. doi: 10.2174/1381612043383269. [DOI] [PubMed] [Google Scholar]
- 9.Ojha A, et al. GroEL1: A dedicated chaperone involved in mycolic acid biosynthesis during biofilm formation in mycobacteria. Cell. 2005;123(5):861–873. doi: 10.1016/j.cell.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 10.Teng R, Dick T. Isoniazid resistance of exponentially growing Mycobacterium smegmatis biofilm culture. FEMS Microbiol Lett. 2003;227(2):171–174. doi: 10.1016/S0378-1097(03)00584-6. [DOI] [PubMed] [Google Scholar]
- 11.Ojha AK, et al. Growth of Mycobacterium tuberculosis biofilms containing free mycolic acids and harbouring drug-tolerant bacteria. Mol Microbiol. 2008;69(1):164–174. doi: 10.1111/j.1365-2958.2008.06274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gengenbacher M, Rao SP, Pethe K, Dick T. Nutrient-starved, non-replicating Mycobacterium tuberculosis requires respiration, ATP synthase and isocitrate lyase for maintenance of ATP homeostasis and viability. Microbiology. 2010;156(Pt 1):81–87. doi: 10.1099/mic.0.033084-0. [DOI] [PubMed] [Google Scholar]
- 13.Betts JC, Lukey PT, Robb LC, McAdam RA, Duncan K. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol Microbiol. 2002;43(3):717–731. doi: 10.1046/j.1365-2958.2002.02779.x. [DOI] [PubMed] [Google Scholar]
- 14.Makarov V, et al. Benzothiazinones kill Mycobacterium tuberculosis by blocking arabinan synthesis. Science. 2009;324(5928):801–804. doi: 10.1126/science.1171583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boshoff HI, et al. The transcriptional responses of Mycobacterium tuberculosis to inhibitors of metabolism: Novel insights into drug mechanisms of action. J Biol Chem. 2004;279(38):40174–40184. doi: 10.1074/jbc.M406796200. [DOI] [PubMed] [Google Scholar]
- 16.Voskuil MI, et al. Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J Exp Med. 2003;198(5):705–713. doi: 10.1084/jem.20030205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muttucumaru DG, Roberts G, Hinds J, Stabler RA, Parish T. Gene expression profile of Mycobacterium tuberculosis in a non-replicating state. Tuberculosis (Edinb) 2004;84(3–4):239–246. doi: 10.1016/j.tube.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 18.Deb C, et al. A novel in vitro multiple-stress dormancy model for Mycobacterium tuberculosis generates a lipid-loaded, drug-tolerant, dormant pathogen. PLoS One. 2009;4(6):e6077. doi: 10.1371/journal.pone.0006077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Batt SM, et al. Structural basis of inhibition of Mycobacterium tuberculosis DprE1 by benzothiazinone inhibitors. Proc Natl Acad Sci USA. 2012;109(28):11354–11359. doi: 10.1073/pnas.1205735109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mattevi A, Fraaije MW, Coda A, van Berkel WJ. Crystallization and preliminary X-ray analysis of the flavoenzyme vanillyl-alcohol oxidase from Penicillium simplicissimum. Proteins. 1997;27(4):601–603. doi: 10.1002/(sici)1097-0134(199704)27:4<601::aid-prot12>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 21.Williams MJ, Kana BD, Mizrahi V. Functional analysis of molybdopterin biosynthesis in mycobacteria identifies a fused molybdopterin synthase in Mycobacterium tuberculosis. J Bacteriol. 2011;193(1):98–106. doi: 10.1128/JB.00774-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malm S, et al. The roles of the nitrate reductase NarGHJI, the nitrite reductase NirBD and the response regulator GlnR in nitrate assimilation of Mycobacterium tuberculosis. Microbiology. 2009;155(Pt 4):1332–1339. doi: 10.1099/mic.0.023275-0. [DOI] [PubMed] [Google Scholar]
- 23.Vilchèze C, Weinrick B, Wong KW, Chen B, Jacobs WR., Jr NAD+ auxotrophy is bacteriocidal for the tubercle bacilli. Mol Microbiol. 2010;76(2):365–377. doi: 10.1111/j.1365-2958.2010.07099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taylor EC, Ray PS, Darwish IS. Studies on the molybdenum cofactor. Determination of the structure and absolute configuration of form A. J Am Chem Soc. 1989;111(19):7664–7665. [Google Scholar]
- 29.Mohr D, Kazimierczuk Z, Pfleiderer W. Pteridines. Part XCVII. Synthesis and properties of 6-thioxanthopterin and 7-thioisoxanthopterin. Helv Chim Acta. 1992;75(7):2317–2326. [Google Scholar]
- 30.Johnson ME, Rajagopalan KV. Involvement of chlA, E, M, and N loci in Escherichia coli molybdopterin biosynthesis. J Bacteriol. 1987;169(1):117–125. doi: 10.1128/jb.169.1.117-125.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.