Abstract
Sucrose nonfermenting 1 (SNF1)-related protein kinase 2s (SnRK2s) are central components of abscisic acid (ABA) signaling pathways. The snrk2.2/2.3/2.6 triple-mutant plants are nearly completely insensitive to ABA, suggesting that most of the molecular actions of ABA are triggered by the SnRK2s-mediated phosphorylation of substrate proteins. Only a few substrate proteins of the SnRK2s are known. To identify additional substrate proteins of the SnRK2s and provide insight into the molecular actions of ABA, we used quantitative phosphoproteomics to compare the global changes in phosphopeptides in WT and snrk2.2/2.3/2.6 triple mutant seedlings in response to ABA treatment. Among the 5,386 unique phosphorylated peptides identified in this study, we found that ABA can increase the phosphorylation of 166 peptides and decrease the phosphorylation of 117 peptides in WT seedlings. In the snrk2.2/2.3/2.6 triple mutant, 84 of the 166 peptides, representing 58 proteins, could not be phosphorylated, or phosphorylation was not increased under ABA treatment. In vitro kinase assays suggest that most of the 58 proteins can serve as substrates of the SnRK2s. The SnRK2 substrates include proteins involved in flowering time regulation, RNA and DNA binding, miRNA and epigenetic regulation, signal transduction, chloroplast function, and many other cellular processes. Consistent with the SnRK2 phosphorylation of flowering time regulators, the snrk2.2/2.3/2.6 triple mutant flowered significantly earlier than WT. These results shed new light on the role of the SnRK2 protein kinases and on the downstream effectors of ABA action, and improve our understanding of plant responses to adverse environments.
The phytohormone abscisic acid (ABA) plays important roles in plant development and responses to stressful environments (1, 2). Recently, the discovery of the PYR1 (Pyrabactin Resistance 1)/PYL (PYR1-Like)/RCAR (Regulatory Component of ABA Receptor) family of ABA receptors led to the elucidation of the core ABA signaling pathway. ABA binds to the PYLs, triggering the PYLs to interact with and inactivate clade A protein phosphatase 2Cs (PP2Cs). This releases Sucrose nonfermenting 1 (SNF1)-related protein kinase 2s (SnRK2s) from inhibition by the PP2Cs, allowing the kinases to phosphorylate downstream effectors of ABA responses (3–5).
SnRK2s are a plant-specific protein kinase family related to the yeast SNF1 and animal AMP-dependent protein kinase (AMPK) (6), and the family has 10 members (SnRK2.1–2.10) in Arabidopsis. ABA treatment can quickly activate SnRK2.2, 2.3 and 2.6 (7), and the snrk2.2/2.3/2.6 triple-knockout mutant has a very strong ABA-insensitive phenotype and shows little response to even very high concentrations of ABA in seed germination, root growth, and stomatal movement (8). In contrast, mutations in the other seven SnRK2 family members do not cause significant ABA insensitivity (9). Notwithstanding the key role of SnRK2.2/2.3/2.6 in ABA signaling, some ABA responses are possibly independent of the SnRK2s, because the PYL receptors and PP2Cs may have other protein targets besides the SnRK2s. Consistent with the crucial role of the PYL family of ABA receptors, the pyr1pyl1pyl2pyl4pyl5pyl8 sextuple mutant is also highly insensitive to ABA (10).
Thus far, only a few proteins have been identified as substrates of the SnRK2, including the bZIP transcription factors AREBs (ABA-Responsive Element Binding factors) that function in ABA-responsive gene regulation, the ion channels SLAC1 (Slow Anion Channel-Associated 1) and KAT1 (K+ channel in Arabidopsis thaliana 1) that are critical for ABA regulation of stomatal movement, and RBOHF (Respiratory Burst Oxidase Homolog F) that functions in reactive oxygen species (ROS) generation in response to ABA. These SnRK2 substrate proteins are important effectors of ABA action and are critical to our understanding of how ABA regulates oxidative burst, stomatal movement, and gene expression. Considering the very important roles of ABA in plant growth, development, and stress responses, many more effectors of ABA action are expected. The identification of these effector proteins will provide insight into how ABA functions in plants.
The transient property of phosphorylation and the possible functional redundancy of some substrate proteins limit the identification of SnRK2 substrates by biochemical and genetic approaches. In recent years, mass spectrometry-based global phosphorylation profiling has evolved to where it is able to dissect thousands of phosphorylation events simultaneously in vivo (14). To comprehensively examine the regulation by phosphorylation in response to ABA and the role of the SnRK2s, we quantitatively compared the phosphoproteomes of WT and snrk2.2/2.3/2.6 triple mutant under ABA treatment. We found 58 putative substrates (84 phosphopeptides) of ABA-activated SnRK2s. Most of the tested putative substrates can be phosphorylated by recombinant SnRK2.6 protein in vitro. An examination of the putative SnRK2 substrates revealed important roles of ABA in regulating not only gene transcription, but also RNA processing, epigenetic modifications, chloroplast processes, and control of flowering time. Consistent with the phosphorylation of several flowering time regulators by the SnRK2s, we found that the snrk2.2/2.3/2.6 triple mutant flowers early under both long- day and short-day conditions. Our study provides comprehensive information on how the SnRK2s mediate the many important functions of ABA in plants.
Results
Global Identification of Phosphopeptides in Arabidopsis.
To determine the changes in protein phosphorylation patterns on ABA treatment and to assess the role of ABA-activated SnRK2s, we applied a liquid chromatography-tandem mass spectrometry (LC-MS/MS) approach (Fig. S1) to compare phosphopeptides isolated from WT and snrk2.2/2.3/2.6 triple mutant with or without treatment with 50 µM ABA. Four independent biological replicates were performed for each genotype and treatment combination. In each replicate, proteins from four samples were extracted using methanol-chloroform precipitation, followed by in-solution trypsin digestion. Phosphopeptides were enriched by two polymer-based metal ion affinity chromatography (PolyMAC) techniques (12, 13), with titanium (PolyMAC-Ti) and zirconium (PolyMAC-Zr). The two enrichment methods are partially complementary, ensuring high coverage of the phosphopeptides. The enriched phosphopeptides were analyzed with an LTQ-Orbitrap Velos mass spectrometer (Thermo Scientific), which collects collision-induced dissociation (CID) spectra. The SEQUEST database search algorithm was used for peptide sequence identification and determination of modification sites. Ion intensity-based label-free quantitation (14) in the parent ion mode was performed to quantify the amount of phosphopeptides across four samples. Using this strategy, a total of 5,386 unique phosphopeptides (5,084 phosphosites) belonging to 2,243 unique phosphoproteins were identified with a 1% false discovery rate (Dataset S1).
Phosphopeptides Up-Regulated and Down-Regulated by ABA.
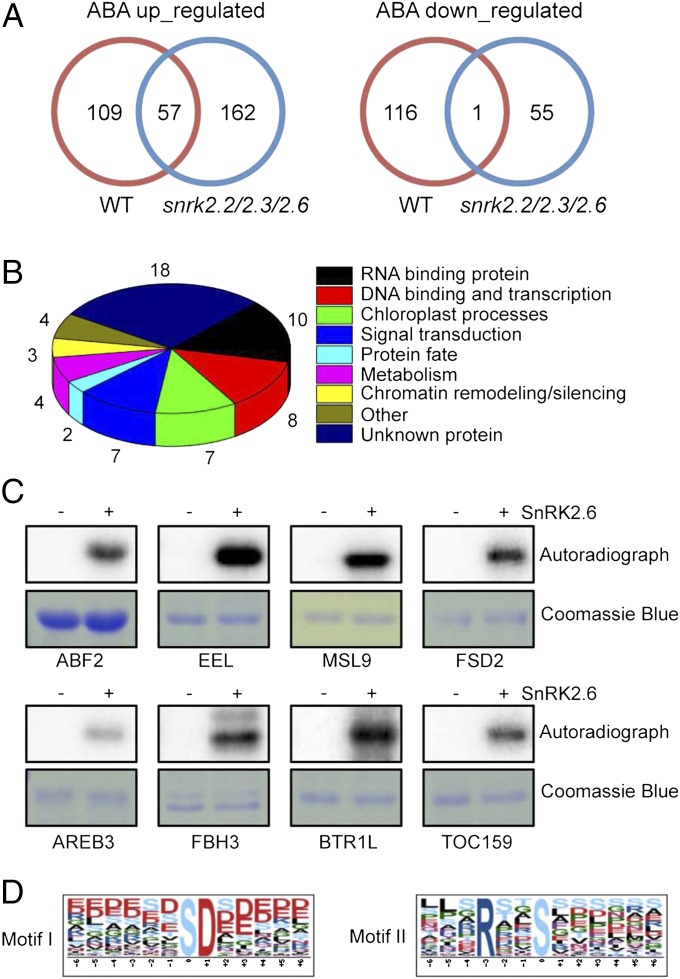
To study the ABA regulation of phosphorylation, we applied a pairwise comparison of phosphopeptide abundance between ABA treatment and control samples for both WT and the snrk2.2/2.3/2.6 triple mutant. An in-house label-free quantitation method using spike-in internal peptides (SI Materials and Methods) was used, with a twofold change set as the significance cutoff (Dataset S1). The peptides that were identified in at least two replicates in the ABA or control samples but not in any of the paring control or ABA samples were included in the lists of ABA up-regulated and down-regulated phosphopeptides, respectively. A total of 166 phosphopeptides (130 proteins) from the WT showed at least a twofold increase in ABA-treated sample relative to controls (Fig. 1A and Dataset S1), whereas 117 phosphopeptides showed at least a twofold decrease (Fig. 1A and Dataset S1). The ABA up-regulated phosphoproteins include proteins previously reported to be phosphorylated in response to ABA treatment, including SnRK2, AREBs, AT5G02240, nuclear transport factor 2, and phosphoribulokinase (11, 15). We also compared our result with data from a recent study on the phosphoproteome response to ABA treatment in Arabidopsis using isotope-labeling quantitative mass spectrometry. Our experiments identified 26 of the 50 reported phosphopeptides exhibiting significant up- or down-regulation in response to ABA (11). Moreover, 21 of these 26 phosphopeptides corresponded to the same phosphorylation changes (i.e., up or down), as reported previously (11). Our lists of ABA up-regulated and down-regulated phosphopeptides also include several proteins previously implicated in ABA responses but not known to be phosphorylated, including GUN5 (Genomes Uncoupled 5) (16), SNL2 (SIN3-like 2) (17), annexin 1 (18), and phospholipase D alpha 1 (PLDα1) (19).
Fig. 1.
(A) Venn diagram illustrating the overlap of up-regulated (Left) and down-regulated (Right) phosphopeptides identified in WT and snrk2.2/2.3/2.6 triple mutant. (B) GO catalogs of the 58 putative SnRK2 substrates. (C) In vitro phosphorylation of putative substrates by recombinant SnRK2.6. (D) Sequence logos (generated using Motif-X) of the phosphorylation sites seen only in ABA up-regulated phosphopeptides.
ABA treatment up-regulated 219 phosphopeptides and decreased the phosphorylation of 56 peptides in the snrk2.2/2.3/2.6 triple mutant (Fig. 1A and Dataset S1). No phosphopeptide from SnRK2.2/2.3/2.6 was found from the triple mutant, consistent with knockout of the three protein kinase genes in the mutant. Interestingly, a phosphopeptide representing the C-terminal domain II of SnRK2.1 was detected after ABA treatment in the snrk2.2/2.3/2.6 triple mutant. In SnRK2.2/2.3/2.6, this domain II (ABA box) is required for PP2C binding and for activation by ABA (20). The phosphorylation at this region of SnRK2.1 may represent an additional mechanism of regulating ABA-independent SnRK2s.
Identification of Putative SnRK2 Substrates.
The phosphopeptides that were up-regulated by ABA in WT but not detected or up-regulated in the snrk2.2/2.3/2.6 triple mutant were considered putative substrates of ABA-activated SnRK2.2/2.3/2.6. As a result, we recovered 84 phosphopeptides, representing 58 unique proteins that may be substrates of SnRK2.2/2.3/2.6 in ABA signaling. The SnRK2.2 itself, as well as several known SnRK2.6 substrates, including ABA responsive element binding factor 2 (ABF2) and AREB3, were included in the list (Dataset S1). We preformed Gene Ontology (GO) analysis to classify the possible functions of these putative SnRK2 substrate proteins. The largest GO categories are nucleotide binding, transcriptional regulation, chloroplast processes, and signal transduction (Fig. 1B). To validate that these proteins can be phosphorylated by the SnRK2s, we selected 15 of the 58 proteins for expression in Escherichia coli to produce recombinant proteins for in vitro phosphorylation assays. Nine of the 15 proteins could be expressed and purified. Seven of the nine proteins [Enhanced late Embryogenesis abundant Level (EEL), Mechanosensitive channel of Small conductance-Like 9 (MSL9), Fe Superoxide Dismutase 2 (FSD2), AREB3, Flowering Basic Helix-loop-helix-type transcription factor 3 (FBH3), Binding to Tomato mosaic virus RNA 1 Long from (BTR1L), and Translocon at the Outer envelope membrane of Chloroplasts 159 (TOC159)] could be phosphorylated by recombinant SnRK2.6, whereas two [arginine/serine-rich splicing factor 41 (RSP41) and Histone Deacetylase 2 B (HD2B)] could not (Fig. 1C). These results suggest that the majority of the 58 SnRK2.2/2.3/2.6-dependent phosphoproteins are direct substrates of the SnRK2s.
Motif Analysis of SnRK2 Substrate Proteins.
We used the foregoing datasets to investigate the potential phosphorylation site specificities of the SnRK2s. We found that two phosphor motifs, SD and RxxS, were enriched specifically in the WT ABA-treated samples at a significance level of 10−10 (Fig. 1D and Fig. S2). The RxxS is a known motif identified in SnRK2s substrate proteins (21, 22). The absence of this motif in the phosphopeptide datasets from the snrk2.2/2.3/2.6 triple mutant further supports its function as a phosphor motif specific for the SnRK2s. Moreover, the absence of RxxS in the WT control sample is consistent with the notion that ABA is required for activation of the SnRK2s.
SnRK2 Substrates Involved in Transcription, RNA Processing, and Chloroplast Processes.
It has been demonstrated that the AREBs ABF2, Abscisic Acid-Insensitive 5 (ABI5), and EEL can be phosphorylated by ABA-activated SnRK2s (23, 24). Our results demonstrate that one more AREB, AREB3, is also phosphorylated by SnRK2s in response to ABA treatment (Fig. 1C and Dataset S1). We found several other transcriptional regulators, including FBH3 and TATA-binding protein-associated factor TAF5, as substrates or putative substrates of the SnRK2s. Along with the DNA-binding proteins and transcriptional regulators, 10 RNA-binding proteins constitute a prominent group in our list of putative SnRK2 substrates (Dataset S1). Six of these proteins are related to RNA splicing. It was recently reported that the alternative splicing of ABI3 and ABI5, and thus some of the ABA responses, are regulated by splicing factors (25, 26). Therefore, regulation of RNA splicing is important for ABA responses, and our discovery of phosphorylation of several splicing factors by ABA-activated SnRK2s reveals how ABA regulates splicing. The RNA splicing factor BTR1L is a confirmed substrate of SnRK2.6 (Fig. 1C). BTR1L (27) and Modifier of SNC1, 3 (MOS3) (28), a putative substrate of ABA-activated SnRK2s, are known to be involved in plant immunity. These observations are consistent with the important functions of ABA in plant immunity (29) and suggest a mechanism of ABA regulation of plant immunity via phosphorylation by the SnRK2s. Another putative substrate in the category of RNA-binding proteins is the RNA-Recognition Motif (RRM) domain containing protein AT3G07810. AT3G07810 is an ortholog of Vicia faba Abscisic-acid-activated protein kinase (AAPK) interacting protein 1 (AKIP1), a substrate of the AAPK, an SnRK2.6 ortholog of Arabidopsis SnRK2.6 (30).
Another very prominent group in our list of putative SnRK2 substrates comprises proteins localized in chloroplasts or known to be involved in chloroplast processes. TOC159 is a protein translocation receptor required for importing nuclear-encoded chloroplast preproteins from the cytosol to chloroplasts. The acidic a-domain of TOC159 is hyperphosphorylated in Arabidopsis (31). In this domain, T692 is a putative SnRK2s-dependent phosphorylation site. The phosphorylation of the other two protein translocation receptors, TOC132 and TOC120, was also enhanced by ABA treatment only in the snrk2.2/2.3/2.6 triple mutant (Dataset S1). These results suggest a role of ABA, the SnRK2s, and possibly other ABA-activated protein kinases in regulating the translocation of nuclear-encoded chloroplast proteins. Another group of ABA up-regulated phosphoproteins related to chloroplast processes are the chloroplast motility-related proteins, including Plastid Movement Impaired 1 (PMI1), actin-bundling protein THRUMIN1, and the coiled-coil protein Weak chloroplast movement under Blue light 1 (WEB1) (32). Only PMI1 is in the list of putative SnRK2 substrates (Dataset S1). In addition, Constitutive Photomorphogenic 1 (COP1)-Interacting Protein 7 (CIP7), FSD2, and chloroplast SULFUR E, proteins involved in chloroplast development are also putative substrates of ABA-activated SnRK2s. Taken together, these results support important functions of ABA (33) and the SnRK2s in regulating chloroplast biogenesis and function.
SnRK2 Substrates Involved in Transcriptional Repression, Epigenetic Modification, and miRNA Regulation.
In higher plants, epigenetic modifications such as DNA methylation, histone acetylation, and chromatin remodeling are involved in the regulation of gene transcription. We found that the phosphorylation of some epigenetic-related proteins was regulated by ABA treatment. For example, phosphorylation of Chromatin Remodeling 2 (CHR2) at S1760 and S1762 were up-regulated by ABA in the WT, but not in the snrk2.2/2.3/2.6 triple mutant. CHR2 is known to directly bind to chromatin at the ABI5 locus and to repress ABI5 expression, and chr2 mutants show strong ABA hypersensitivity in postgermination seedling growth arrest (34). The epigenetics-related proteins in the list of putative SnRK2 substrates also include HD2B (35) and Nucleolin like 1, the latter of which regulates the DNA methylation and expression of rRNA genes (36) (Dataset S1). Interestingly, the phosphorylation of SNL2 (SIN3-Like 2), a component of the histone deacetylation complex, is up-regulated by ABA in both WT and the mutant seedlings (Dataset S1). SNL2 interacts with the histone deacetylase 19 (HDA19) and snl1/snl2 double mutant plants show increased acetylation levels of histone H3 lysine 9/18 (H3K9/18) and H3K14 (17). Because HDA19 is involved in ABA responses (37), our results suggest that the SnRK2s may regulate histone deacetylation and ABA-responsive transcriptional repression by phosphorylating SNL2. In addition, phosphorylation of the H3K4 trimethyltransferase SET [Su(var)3-9, Enhancer-of-zeste, Trithorax] domain protein 2 (SDG2) and H3K4 dimethylation and H3K9 acetylation-related protein Multicopy Suppressor of IRA1(aerial rosette 1) 4 (MSI4)/FVE was also up-regulated by ABA treatment in both WT and snrk2.2/2.3/2.6 triple-mutant plants.
miRNAs are small noncoding RNAs that repress gene expression by guiding the cleavage or translational inhibition of complementary mRNAs (38). The generation of miRNAs from their precursors and modulation of miRNA activity involve numerous proteins. The phosphorylation status of some of these proteins, including Exoribonuclease 2 (XRN2) and XRN3, SERRATE (SE), and importin β-like protein AT3G59020, was altered in response to ABA. Our findings of ABA and SnRK2 regulation of miRNA pathway-related proteins are consistent with previous reports that mutations in several miRNA pathway components, including hyponastic leaves 1 (hyl1), super sensitive to ABA and drought 2 (sad2), and argonaute 1 (ago1), cause ABA hypersensitivity during seed germination and root growth (39–41).
Involvement of the SnRK2s and Their Substrates in Flowering Time Regulation.
Several proteins important for flowering time regulation are putative substrates of ABA-activated SnRK2s. FBH3, a confirmed in vitro substrate of SnRK2.6 (Fig. 1C), is a basic helix-loop-helix type transcription factor that preferentially binds to the E-box cis-element in the CONSTANS (CO) promoter; overexpression of the FBH gene drastically elevates CO levels and causes early flowering regardless of photoperiod (42). MOS3 is a nucleoporin, a component of the nuclear pore complex, and is required for mRNA export from the nucleus. Mutations in MOS3 cause an early flowering phenotype (43). XRN3 is involved in the suppression of endogenous posttranscriptional gene silencing; Arabidopsis xrn2/xrn3 double-mutant plants show a late-flowering phenotype (44). MOS3 and XRN3 are two putative substrates of the ABA-activated SnRK2s (Dataset S1). The phosphorylation of several other proteins related to flowering time regulation is also altered by ABA, although this phosphorylation might not be dependent on the SnRK2s. These include FVE (45), Early Bolting in Short days (EBS) (45), Relative of Early Flowering 6 (REF6) (45), Nuclear Pore Anchor (NUA) (46), and Vernalization5/VIN3-like 1(VEL1) (47). These results suggest that the SnRK2s are important for flowering time regulation.
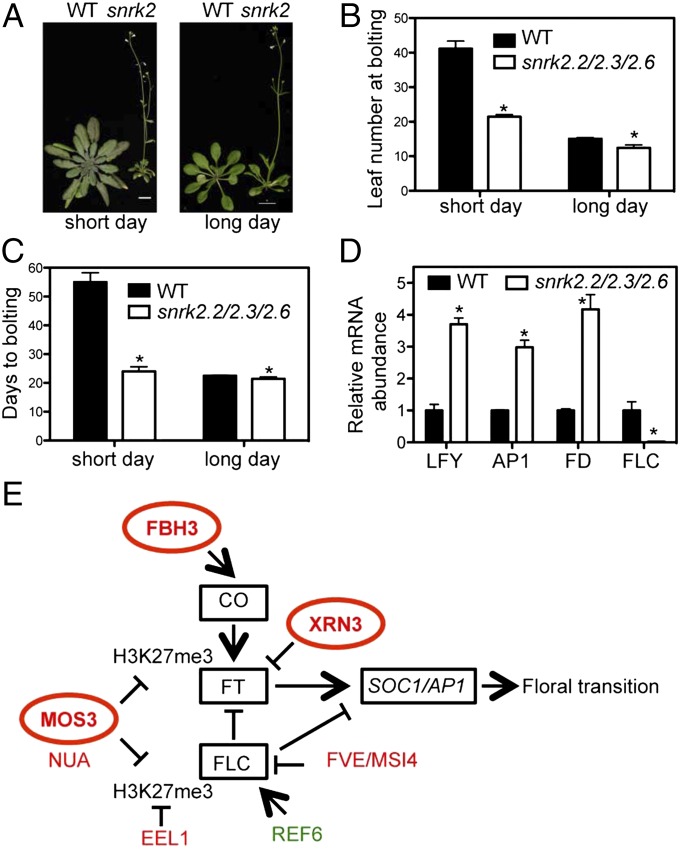
To test whether SnRK2.2/2.3/2.6 are indeed important for flowering time regulation, we compared the flowering times of WT and snrk2.2/2.3/2.6 mutant plants. As shown in Fig. 2 A–C, the snrk2.2/2.3/2.6 triple mutant began to bolt at 20.5 d, with approximately 12 rosette leaves, under long-day conditions, and at 24.0 d, with 15.1 leaves, under short-day conditions, whereas the WT bolted at 21.4 d, with 15.0 leaves, under long-day conditions, and at 55.5 d, with 41.0 leaves, under short-day conditions (P < 0.05; n > 25). Consistent with the early-flowering phenotype, the expression levels of several flowering time-related genes, including LEAFY (LFY), FD, and APETALA1 (AP1) were up-regulated in the snrk2.2/2.3/2.6 mutant, whereas the negative regulator Flowering Locus C (FLC) was repressed (Fig. 2D). Generally, ABA has been considered to repress flowering, and plants grown in ABA-treated media demonstrate delayed flowering (48). Early flowering is observed in the ABA biosynthesis mutant glucose insensitive 1 (gin1)/ABA deficient 2 (aba2) (49). Our findings of the early-flowering phenotype of the snrk2.2/2.3/2.6 triple mutant and the SnRK2s-mediated phosphorylation of several flowering time regulators are consistent with a repressive role of ABA in flowering time regulation. Although the role of phosphorylation in regulating the activity of these proteins has not yet been elucidated, our data suggest the existence of an ABA-dependent multitarget integrative pathway that regulates flowering time in Arabidopsis (Fig. 2E).
Fig. 2.
Role of SnRK2.2/2.3/2.6 and ABA up-regulated phosphoproteins in flowering time regulation. (A) Early flowering phenotype of the snrk2.2/2.3/2.6 triple mutant (snrk2) under short-day (Upper; 35-d-old plants) and long-day (Lower; 24-d-old plants) photoperiods. (Scale bars: 10 mm). (B and C) Number of days (B) and vegetative leaf number (C) at bolting of WT and the snrk2.2/2.3/2.6 triple mutant under long-day and short-day photoperiods. Error bars depict SE (n > 25). Asterisks denote significant differences (P < 0.05) between triple-mutant and WT plants. (D) Expression of the flowering time-related genes analyzed by qPCR in leaves of 21-d-old plants in WT and snrk2.2/2.3/2.6 triple mutant under long-day conditions. The experiment was repeated at least twice, with similar results obtained each time. (E) Model showing the function of flowering time-related phosphoproteins identified in this study. The phosphoproteins up-regulated and those down-regulated by ABA treatment are shown in red and green, respectively. Bold letters indicate putative SnRK2 substrates.
Discussion
Protein phosphorylation is a widely used mechanism of posttranslational modification that controls the protein activity, stability, turnover, subcellular localization, and interaction with partner proteins. In the core ABA signaling pathway, ABA binding to the PYL family of receptors causes the receptor to interact with and inhibit the PP2Cs, leading to release of suppression and activation of the SnRK2s. Our study suggests that the activated SnRK2s phosphorylate at least several dozens of downstream effector proteins of ABA action. These phosphorylated effector proteins function in regulating gene expression at various steps, including epigenetic and transcription factor regulation, RNA splicing, and mRNA cleavage or translational repression by miRNAs.
Our results suggest that posttranscriptional gene regulation is a very important part of ABA action. The putative substrates of the SnRK2s include 10 RNA-binding proteins (mainly splicing factors) and several factors related to miRNA regulation. In addition, a significant proportion of the putative SnRK2 substrates are proteins involved in chloroplast processes, including protein translocation and chloroplast development and function. This implies that an important aspect of ABA action is the regulation of chloroplast function, which likely includes photosynthesis, regulation of ROS, and retrograde signaling. The regulation of chloroplasts also may be important for stomatal control and for abiotic stress tolerance in general.
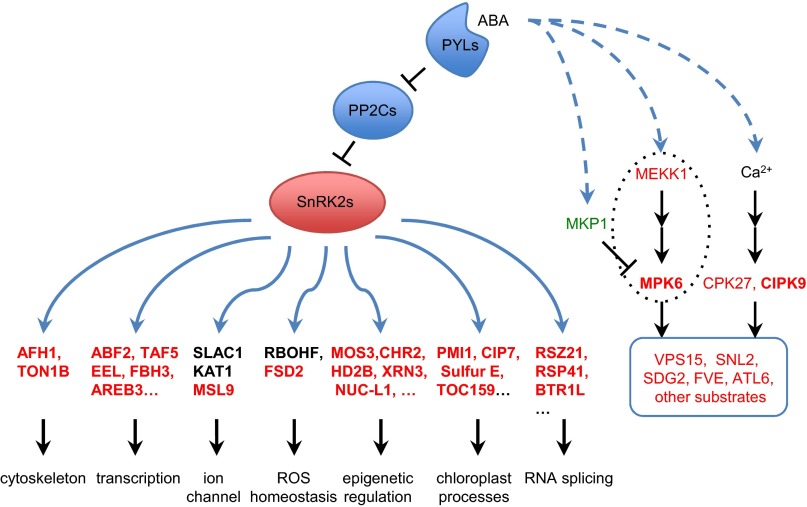
In this work, we identified 84 phosphopeptides (58 proteins) as putative substrates of ABA-activated SnRK2.2/2.3/2.6 (Dataset S1). Our in vitro kinase assays suggested that most of these proteins are direct substrates of the SnRK2s (Fig. 1C). Some previously identified substrates, including several AREB transcription factors, are included in our list. We have identified many substrates that appear to function in transcription, RNA splicing, chloroplast processes, cytoskeleton, flowering time control, epigenetic regulation, the miRNA pathway, and various other cellular processes (Fig. 3 and Dataset S1). Because our method does not enrich for membrane proteins (Fig. S3), several known membrane protein substrates of the SnRK2s (i.e., SLAC1, KAT1, and RBOHF) were not identified in our experiments. Nevertheless, we found that the membrane channel protein, MSL9, is a substrate of SnRK2.6 (Fig. 1C and Dataset S1). Considering the important roles of membrane proteins in ABA signaling and stomatal regulation, it would be important to identify additional membrane protein substrates of the SnRK2s using methods involving membrane protein enrichment.
Fig. 3.
Model showing the regulation and functional grouping of putative SnRK2 substrates in ABA response pathways. The phosphoproteins up-regulated or down-regulated by ABA treatment are shown in red and green, respectively. Bold letters indicate putative SnRK2 substrates.
Out of the 166 ABA up-regulated phosphopeptides in the WT, 57 were present in ABA-treated mutant sample as well (Fig. 1A). This suggests that along with SnRK2.2, 2.3, and 2.6, ABA also may activate other protein kinases to phosphorylate these proteins. Previous studies have shown that SnRK2.4, 2.7, and 2.8 could be weakly activated by ABA (7, 14). However, genetic evidence demonstrated that in planta ABA sensitivity depends almost entirely on SnRK2.2/2.3/2.6, and that SnRK2.4/2.7/2.8 have little influence (8, 9). It is possible that the 57 apparent SnRK2.2/2.3/2.6-independent phosphopeptides are normally phosphorylated by SnRK2.2/2.3/2.6 as well, but other protein kinases may assume this role when the SnRK2.2/2.3/2.6 are knocked out. This notion of compensation by other protein kinases in the snrk2.2/2.3/2.6 triple mutant also may help explain our finding of even more ABA up-regulated phosphopeptides in the triple mutant than in the WT.
The involvement of other protein kinases in ABA signaling has been implicated in previous studies. For instance, some members in the SnRK3 family [i.e., the SOS2(Salt Overly Sensitive 2)-like protein kinases (PKS)/CBL(calcineurin B-like calcium sensors)-interacting protein kinase (CIPKs)] can function as negative regulators of ABA signaling (50). Several calcium-dependent protein kinases (CDPKs/CPKs) are important for ABA responses in seed germination, stomatal movement, and early seedling growth (51, 52). Our phosphoproteomics data reported here show that ABA increased the phosphorylation of CIPK9 and CPK27 (Dataset S1). Moreover, a reported CDPK substrate, Arabidopsis Toxicos en Levadura 6 (ATL6), showed an ABA-induced phosphorylation in our dataset (Dataset S1).
We also observed that the phosphorylation of several components of the Mitogen-Activated Protein Kinase (MAPK) cascade (53) was regulated by ABA. MPK6 is present in our list of putative SnRK2s substrates (Dataset S1), whereas ABA induced the phosphorylation of MAPK/ERK Kinase Kianse 1 (MEKK1) only in the snrk2.2/2.3/2/6 triple mutant. Furthermore, ABA repressed the phosphorylation of MAPK phosphatase 1 (MKP1), a negative regulator of MPK3/6 (53), only in the snrk2.2/2.3/2/6 triple mutant. Several studies have reported that MPK1/2, MPK3/6, and MPK9/12 can be activated by ABA (54, 55), although only the mpk9/mpk12 double mutant, and not other mpk mutants, show ABA-related phenotypes (54). ROS treatment also can activate the MAPKs, even more strongly than ABA. In the mpk9/mpk12 double mutant, ROS-induced stomatal movement is disrupted (54). Because ABA can trigger the rapid accumulation of ROS (56), it is possible that ABA activates the MAPK cascade at least partly by inducing ROS generation. In any case, the ABA induction of phosphorylation changes in the MAP kinase cascade in snrk2.2/2.3/2.6 supports the existence of another SnRK2-independent pathway, possibly caused by increases in the phosphorylation of MEKK1 as well as in the dephosphorylation of the negative regulator MKP1 (Fig. 3). Enhanced activation of the MAPK cascade in the snrk2.2/2.3/2.6 triple mutant may serve as one of the compensatory mechanisms in ABA signaling. Another well-known kinase, cyclin-dependent kinase F1 (CDKF1), also can be phosphorylated under ABA treatment in the WT, although its role in ABA signaling remains undefined. Both MAPKs and CDKs can phosphorylate the S/TP motif that we detected (Fig. S2).
In addition to inducing or up-regulating the phosphorylation of many proteins, we found that ABA also repressed the phosphorylation of 117 peptides (93 proteins) in the WT (Fig. 1A). The ABA-repressed phosphoproteins include many metabolic enzymes and various putative regulatory proteins (Dataset S1). For example, ABA down-regulated the phosphorylation of several carbohydrate metabolic enzymes, including phosphoglycerate mutase, UDP-glucose-6-dehydrogenase, cytosolic invertase, and fructose-6-phosphate 2-kinase (Dataset S1). Nearly all of the ABA-repressed phosphoproteins were dependent on the SnRK2s (Fig. 1A). Interestingly, some of the putative substrates of ABA-activated SnRK2s also appeared on the list of ABA-repressed phosphoproteins, but the ABA-activated and ABA-repressed phosphorylations occurred at distinct sites. This indicates that the regulation of some proteins requires a combination of new phosphorylation at certain sites and dephosphorylation at other sites. How ABA or the SnRK2s cause dephosphorylation is unknown. It is possible that the SnRK2s activate protein phosphatases and/or inhibit kinases directly or indirectly via phosphorylation. ABA also caused dephosphorylation in the snrk2.2/2.3/2.6 triple mutant, suggesting that some dephosphorylation also could occur independent of the SnRK2s.
While this manuscript was in preparation, Umezawa et al. (57) reported a phosphoproteomics study using the snrk2.2/2.3/2.6 triple mutant. The ABA-responsive phosphoproteome data of Umezawa et al. (57) show little overlap with our data here or with that reported by Kline et al. (14). Of the 53 ABA up-regulated phosphopeptides (56 proteins) detected in the study of Umezawa et al. (57), only 3 proteins (SnKR2.2, SnKR2.6, and ABF2) were also present in the dataset of Kline et al. (14), and only six overlapped with our data.
Through our phosphoproteomics analysis, we discovered many SnRK2 substrate proteins that mediate ABA responses by participating in various biological processes. Among these effector proteins of ABA action, the transcription factors AREBs/ABFs have well-characterized functions in ABA responses, but for the majority of the substrate proteins, future work is needed to understand the significance of their phosphorylation in ABA responses. As a case study, we began to investigate the potential significance of phosphorylation of the group of flowering time regulators. We found that the snrk2.2/2.3/2.6 triple mutant flowered significantly earlier than the WT, especially under short-day conditions. Consistent with this phenotype, the mutant showed altered expression levels of several flowering-related genes (Fig. 2). Plants evolved to time their reproduction according to environmental conditions. Considering that ABA is the most critical phytohormone for coping with abiotic stresses, our results suggest that the SnRK2-mediated phosphorylation of flowering time regulators is an important mechanism in the timing of plant reproduction to cope with stressful environments.
Materials and Methods
Plant Growth and Materials.
Seedlings of A. thaliana were grown in 40 mL of half-strength Murashige and Skoog medium at 22 °C in continuous light on a rotary shaker set at 100 rpm. Twelve-day-old seedlings were treated with 50 μM ABA in ethanol or with ethanol as the control for 30 min. For each sample, total protein was extracted from 2 g of seedlings by grinding in 2 mL of extraction buffer [100 mM Tris⋅HCl (pH 7.5), 250 mM NaCl, and 5 mM EDTA] with phosphatase inhibitor mixture 3 (Sigma-Aldrich). After centrifugation at 14,000 × g for 20 min, the supernatant was used for further analysis.
For analysis of the flowering time phenotype, plants were grown in soil at 22 °C under long-day (16 h light/8 h dark) or short-day (8 h light/16 h dark) conditions. Seeds were stratified at 4 °C for 2∼4 d and then transferred to 22 °C; this was defined as day 0. For expression analysis, plants were grown on medium with 1/2 strength Murashige and Skoog salts supplemented with 1% sucrose under long-day conditions.
Phosphopeptide Enrichment and Mass Spectrometry Analysis.
Details on protein extraction, digestion, phosphopeptide enrichment, mass spectrometry data acquisition, data analysis, and label-free quantitation are provided in SI Materials and Methods.
Recombinant Protein Expression and in Vitro Kinase Assay.
The in vitro kinase assay was performed as described previously (23); details are provided in SI Materials and Methods.
RNA Extraction and Quantitative PCR.
RNA from leaves of 3-wk-old plants grown under long-day conditions (16 h light/8 h dark) was extracted using TRIzol (Invitrogen). First-strand cDNA synthesis was performed using 3 µg of DNase-treated total RNA. Real-time RT reactions, using 1 µL of a 1:15 dilution of the RT reaction, were carried out with an iQ5 quantitative PCR (qPCR) detection system and SYBR Green (Bio-Rad). Relative expression levels were calculated using Actin2 as an internal standard and the ΔΔCt method for relative quantification. The primers used are listed in Dataset S1.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants R01 GM059138 (to J.-K.Z.) and R01 GM088317 (to W.A.T.). G.B. was supported by the International Exchange Program of the University of Naples Federico II.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1308974110/-/DCSupplemental.
References
- 1.Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. Abscisic acid: Emergence of a core signaling network. Annu Rev Plant Biol. 2010;61:651–679. doi: 10.1146/annurev-arplant-042809-112122. [DOI] [PubMed] [Google Scholar]
- 2.Klingler JP, Batelli G, Zhu JK. ABA receptors: The START of a new paradigm in phytohormone signalling. J Exp Bot. 2010;61(12):3199–3210. doi: 10.1093/jxb/erq151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fujii H, et al. In vitro reconstitution of an abscisic acid signalling pathway. Nature. 2009;462(7273):660–664. doi: 10.1038/nature08599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ma Y, et al. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. 2009;324(5930):1064–1068. doi: 10.1126/science.1172408. [DOI] [PubMed] [Google Scholar]
- 5.Park SY, et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324(5930):1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fujii H, Zhu JK. Osmotic stress signaling via protein kinases. Cell Mol Life Sci. 2012;69(19):3165–3173. doi: 10.1007/s00018-012-1087-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boudsocq M, Barbier-Brygoo H, Laurière C. Identification of nine sucrose nonfermenting 1-related protein kinases 2 activated by hyperosmotic and saline stresses in Arabidopsis thaliana. J Biol Chem. 2004;279(40):41758–41766. doi: 10.1074/jbc.M405259200. [DOI] [PubMed] [Google Scholar]
- 8.Fujii H, Zhu JK. Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc Natl Acad Sci USA. 2009;106(20):8380–8385. doi: 10.1073/pnas.0903144106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fujii H, Verslues PE, Zhu JK. Arabidopsis decuple mutant reveals the importance of SnRK2 kinases in osmotic stress responses in vivo. Proc Natl Acad Sci USA. 2011;108(4):1717–1722. doi: 10.1073/pnas.1018367108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gonzalez-Guzman M, et al. Arabidopsis PYR/PYL/RCAR receptors play a major role in quantitative regulation of stomatal aperture and transcriptional response to abscisic acid. Plant Cell. 2012;24(6):2483–2496. doi: 10.1105/tpc.112.098574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kline KG, Barrett-Wilt GA, Sussman MR. In planta changes in protein phosphorylation induced by the plant hormone abscisic acid. Proc Natl Acad Sci USA. 2010;107(36):15986–15991. doi: 10.1073/pnas.1007879107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Illiuk AB, Martin VA, Alicie BM, Geahlen RL, Tao WA. In-depth analyses of kinase-dependent tyrosine phosphoproteomes based on metal ion-functionalized soluble nanopolymers. Mol Cell Proteomics. 2010;9(10):2162–2172. doi: 10.1074/mcp.M110.000091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xue L, et al. Sensitive kinase assay linked with phosphoproteomics for identifying direct kinase substrates. Proc Natl Acad Sci USA. 2012;109(15):5615–5620. doi: 10.1073/pnas.1119418109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xue L, et al. Quantitative measurement of phosphoproteome response to osmotic stress in Arabidopsis based on Library-Assisted eXtracted Ion Chromatogram (LAXIC) Mol Cell Proteomics, 2013 doi: 10.1074/mcp.O113.027284. 10.1074/mcp.O113.027284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghelis T, et al. Protein tyrosine kinases and protein tyrosine phosphatases are involved in abscisic acid-dependent processes in Arabidopsis seeds and suspension cells. Plant Physiol. 2008;148(3):1668–1680. doi: 10.1104/pp.108.124594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen YY, et al. The Mg-chelatase H subunit is an abscisic acid receptor. Nature. 2006;443(7113):823–826. doi: 10.1038/nature05176. [DOI] [PubMed] [Google Scholar]
- 17.Wang Z, et al. Arabidopsis paired amphipathic helix proteins SNL1 and SNL2 redundantly regulate primary seed dormancy via abscisic acid-ethylene antagonism mediated by histone deacetylation. Plant Cell. 2013;25(1):149–166. doi: 10.1105/tpc.112.108191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee S, et al. Proteomic identification of annexins, calcium-dependent membrane binding proteins that mediate osmotic stress and abscisic acid signal transduction in Arabidopsis. Plant Cell. 2004;16(6):1378–1391. doi: 10.1105/tpc.021683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uraji M, et al. Cooperative function of PLDδ and PLDα1 in abscisic acid-induced stomatal closure in Arabidopsis. Plant Physiol. 2012;159(1):450–460. doi: 10.1104/pp.112.195578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soon FF, et al. Molecular mimicry regulates ABA signaling by SnRK2 kinases and PP2C phosphatases. Science. 2012;335(6064):85–88. doi: 10.1126/science.1215106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Furihata T, et al. Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc Natl Acad Sci USA. 2006;103(6):1988–1993. doi: 10.1073/pnas.0505667103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sirichandra C, et al. The Arabidopsis ABA-activated kinase OST1 phosphorylates the bZIP transcription factor ABF3 and creates a 14-3-3 binding site involved in its turnover. PLoS ONE. 2010;5(11):e13935. doi: 10.1371/journal.pone.0013935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fujii H, Verslues PE, Zhu JK. Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell. 2007;19(2):485–494. doi: 10.1105/tpc.106.048538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakashima K, et al. Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol. 2009;50(7):1345–1363. doi: 10.1093/pcp/pcp083. [DOI] [PubMed] [Google Scholar]
- 25.Sugliani M, Brambilla V, Clerkx EJM, Koornneef M, Soppe WJJ. The conserved splicing factor SUA controls alternative splicing of the developmental regulator ABI3 in Arabidopsis. Plant Cell. 2010;22(6):1936–1946. doi: 10.1105/tpc.110.074674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carvalho RF, Carvalho SD, Duque P. The plant-specific SR45 protein negatively regulates glucose and ABA signaling during early seedling development in Arabidopsis. Plant Physiol. 2010;154(2):772–783. doi: 10.1104/pp.110.155523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fujisaki K, Ishikawa M. Identification of an Arabidopsis thaliana protein that binds to tomato mosaic virus genomic RNA and inhibits its multiplication. Virology. 2008;380(2):402–411. doi: 10.1016/j.virol.2008.07.033. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Y, Li X. A putative nucleoporin 96 Is required for both basal defense and constitutive resistance responses mediated by suppressor of npr1-1,constitutive 1. Plant Cell. 2005;17(4):1306–1316. doi: 10.1105/tpc.104.029926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ton J, Flors V, Mauch-Mani B. The multifaceted role of ABA in disease resistance. Trends Plant Sci. 2009;14(6):310–317. doi: 10.1016/j.tplants.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 30.Li J, et al. Modulation of an RNA-binding protein by abscisic-acid–activated protein kinase. Nature. 2002;418(6899):793–797. doi: 10.1038/nature00936. [DOI] [PubMed] [Google Scholar]
- 31.Agne B, et al. The acidic A-domain of Arabidopsis TOC159 occurs as a hyperphosphorylated protein. Plant Physiol. 2010;153(3):1016–1030. doi: 10.1104/pp.110.158048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morita MT, Nakamura M. Dynamic behavior of plastids related to environmental response. Curr Opin Plant Biol. 2012;15(6):722–728. doi: 10.1016/j.pbi.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 33.Maai E, et al. The avoidance and aggregative movements of mesophyll chloroplasts in C(4) monocots in response to blue light and abscisic acid. J Exp Bot. 2011;62(9):3213–3221. doi: 10.1093/jxb/err008. [DOI] [PubMed] [Google Scholar]
- 34.Han SK, et al. The SWI2/SNF2 chromatin remodeling ATPase BRAHMA represses abscisic acid responses in the absence of the stress stimulus in Arabidopsis. Plant Cell. 2012;24(12):4892–4906. doi: 10.1105/tpc.112.105114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luo M, et al. HD2C interacts with HDA6 and is involved in ABA and salt stress response in Arabidopsis. J Exp Bot. 2012;63(8):3297–3306. doi: 10.1093/jxb/ers059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pontvianne F, et al. Nucleolin is required for DNA methylation state and the expression of rRNA gene variants in Arabidopsis thaliana. PLoS Genet. 2010;6(11):e1001225. doi: 10.1371/journal.pgen.1001225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Song CP, et al. Role of an Arabidopsis AP2/EREBP-type transcriptional repressor in abscisic acid and drought stress responses. Plant Cell. 2005;17(8):2384–2396. doi: 10.1105/tpc.105.033043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu JK. Reconstituting plant miRNA biogenesis. Proc Natl Acad Sci USA. 2008;105(29):9851–9852. doi: 10.1073/pnas.0805207105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li W, et al. Transcriptional regulation of Arabidopsis MIR168a and argonaute1 homeostasis in abscisic acid and abiotic stress responses. Plant Physiol. 2012;158(3):1279–1292. doi: 10.1104/pp.111.188789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Verslues PE, Guo Y, Dong CH, Ma W, Zhu JK. Mutation of SAD2, an importin β-domain protein in Arabidopsis, alters abscisic acid sensitivity. Plant J. 2006;47(5):776–787. doi: 10.1111/j.1365-313X.2006.02833.x. [DOI] [PubMed] [Google Scholar]
- 41.Lu C, Fedoroff N. A mutation in the Arabidopsis HYL1 gene encoding a dsRNA-binding protein affects responses to abscisic acid, auxin, and cytokinin. Plant Cell. 2000;12(12):2351–2366. doi: 10.1105/tpc.12.12.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ito S, et al. FLOWERING BHLH transcriptional activators control expression of the photoperiodic flowering regulator CONSTANS in Arabidopsis. Proc Natl Acad Sci USA. 2012;109(9):3582–3587. doi: 10.1073/pnas.1118876109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parry G, Ward S, Cernac A, Dharmasiri S, Estelle M. The Arabidopsis SUPPRESSOR OF AUXIN RESISTANCE proteins are nucleoporins with an important role in hormone signaling and development. Plant Cell. 2006;18(7):1590–1603. doi: 10.1105/tpc.106.041566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim BH, von Arnim AG. FIERY1 regulates light-mediated repression of cell elongation and flowering time via its 3′(2′),5′-bisphosphate nucleotidase activity. Plant J. 2009;58(2):208–219. doi: 10.1111/j.1365-313X.2008.03770.x. [DOI] [PubMed] [Google Scholar]
- 45.Reyes JC. Chromatin modifiers that control plant development. Curr Opin Plant Biol. 2006;9(1):21–27. doi: 10.1016/j.pbi.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 46.Xu XM, et al. NUCLEAR PORE ANCHOR, the Arabidopsis homolog of Tpr/Mlp1/Mlp2/megator, is involved in mRNA export and SUMO homeostasis and affects diverse aspects of plant development. Plant Cell. 2007;19(5):1537–1548. doi: 10.1105/tpc.106.049239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim DH, Sung S. The Plant Homeo Domain finger protein, VIN3-LIKE 2, is necessary for photoperiod-mediated epigenetic regulation of the floral repressor, MAF5. Proc Natl Acad Sci USA. 2010;107(39):17029–17034. doi: 10.1073/pnas.1010834107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Achard P, et al. Integration of plant responses to environmentally activated phytohormonal signals. Science. 2006;311(5757):91–94. doi: 10.1126/science.1118642. [DOI] [PubMed] [Google Scholar]
- 49.Domagalska MA, Sarnowska E, Nagy F, Davis SJ. Genetic analyses of interactions among gibberellin, abscisic acid, and brassinosteroids in the control of flowering time in Arabidopsis thaliana. PLoS ONE. 2010;5(11):e14012. doi: 10.1371/journal.pone.0014012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guo Y, et al. A calcium sensor and its interacting protein kinase are global regulators of abscisic acid signaling in Arabidopsis. Dev Cell. 2002;3(2):233–244. doi: 10.1016/s1534-5807(02)00229-0. [DOI] [PubMed] [Google Scholar]
- 51.Mori IC, et al. CDPKs CPK6 and CPK3 function in ABA regulation of guard cell S-type anion- and Ca(2+)-permeable channels and stomatal closure. PLoS Biol. 2006;4(10):e327. doi: 10.1371/journal.pbio.0040327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu SY, et al. Two calcium-dependent protein kinases, CPK4 and CPK11, regulate abscisic acid signal transduction in Arabidopsis. Plant Cell. 2007;19(10):3019–3036. doi: 10.1105/tpc.107.050666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meng X, Zhang S. MAPK cascades in plant disease resistance signaling. Annu Rev Phytopathol. 2013 doi: 10.1146/annurev-phyto-082712-102314. in press. [DOI] [PubMed] [Google Scholar]
- 54.Jammes F, et al. MAP kinases MPK9 and MPK12 are preferentially expressed in guard cells and positively regulate ROS-mediated ABA signaling. Proc Natl Acad Sci USA. 2009;106(48):20520–20525. doi: 10.1073/pnas.0907205106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lu C, Han MH, Guevara-Garcia A, Fedoroff NV. Mitogen-activated protein kinase signaling in postgermination arrest of development by abscisic acid. Proc Natl Acad Sci USA. 2002;99(24):15812–15817. doi: 10.1073/pnas.242607499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pei ZM, et al. Calcium channels activated by hydrogen peroxide mediate abscisic acid signalling in guard cells. Nature. 2000;406(6797):731–734. doi: 10.1038/35021067. [DOI] [PubMed] [Google Scholar]
- 57.Umezawa T, et al. Genetics and phosphoproteomics reveal a protein phosphorylation network in the abscisic acid signaling pathway in Arabidopsis thaliana. Sci Signal. 2013;6(270):rs8. doi: 10.1126/scisignal.2003509. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.