Abstract
Human respiratory syncytial virus is a human pathogen that causes severe infection of the respiratory tract. Current information about the structure of the virus and its interaction with host cells is limited. We carried out an electron cryotomographic characterization of cell culture-grown human respiratory syncytial virus to determine the architecture of the virion. The particles ranged from 100 nm to 1,000 nm in diameter and were spherical, filamentous, or a combination of the two. The filamentous morphology correlated with the presence of a cylindrical matrix protein layer linked to the inner leaflet of the viral envelope and with local ordering of the glycoprotein spikes. Recombinant viruses with only the fusion protein in their envelope showed that these glycoproteins were predominantly in the postfusion conformation, but some were also in the prefusion form. The ribonucleocapsids were left-handed, randomly oriented, and curved inside the virions. In filamentous particles, they were often adjacent to an intermediate layer of protein assigned to M2-1 (an envelope-associated protein known to mediate association of ribonucleocapsids with the matrix protein). Our results indicate important differences in structure between the Paramyxovirinae and Pneumovirinae subfamilies within the Paramyxoviridae, and provide fresh insights into host cell exit of a serious pathogen.
Keywords: cryo-ET, paramyxovirus, virus structure
Human respiratory syncytial virus (HRSV) causes severe disease, especially in children and the elderly (1, 2). In a study covering data from 2005, it was found that HRSV was responsible for 22% of acute lower respiratory infections worldwide and 66,000–199,000 deaths in children under the age of 5 y (2). Treatment is currently limited to purine analogs (Ribavirin) and passive immune-prophylaxis with humanized monoclonal antibodies (Palivizumab) (3).
RSV belongs to the Paramyxoviridae, a large family of enveloped, negative-sense RNA viruses with many members from humans and animals. Together with human metapneumovirus, it constitutes the Pneumovirinae subfamily. Like other paramyxoviruses, it has a plasma-membrane-derived lipid envelope and a helical ribonucleocapsid (RNP) that contains a single viral RNA molecule associated with nucleoproteins (N), and an RNA-dependent RNA polymerase. The virion contains, in addition, a matrix protein (M) (4–7) and a transcription antiterminator, M2-1 (an envelope-associated protein known to mediate association of RNPs with the M protein) (8–11). The lipid envelope contains two transmembrane glycoproteins: the fusion protein, F, and a glycoprotein, G, used for attachment to host cells. In addition, HRSV contains a small hydrophobic protein, SH, of unclear function and proteins of cellular origin, such as actin and chaperones (12). The RNP and F have been analyzed using 3D-EM and X-ray crystallography (13, 14).
HRSV particles are pleomorphic with both spherical and filamentous particles of different sizes (15). Early electron microscopy studies showed the presence of a helical protein layer under the membrane of filamentous HRSV (15). This layer is likely to contain the M protein because M has been shown to play a critical role in formation of filamentous protrusions in infected cells and to form lattices and helices when incubated with lipids in vitro (16, 17). However, because the 3D structure of whole virions is not known, it remains unclear where M, other proteins, and RNP inside the virus are localized and how they are organized with respect to the membrane, and the glycoproteins.
Here, we examined the structure of cell culture-grown, purified HRSV using electron cryotomography (cryo-ET). The population of viruses was pleomorphic with the majority roughly spherical in shape. The M protein was found to form assemblies lining the virion membrane, but only in filamentous portions. The RNP was randomly arranged and formed a left-handed helix. On the surface, the F proteins were found in both pre- and postfusion conformations. Based on our observations and previous studies, we provide a model for HRSV budding.
Results
Particle Morphology Ranges from Spherical to Filamentous.
To get a general overview of viruses released from cells, we began our structural study by collecting low-magnification cryo-electron microscopy (cryo-EM) images and analyzing them by visual inspection. We studied A2 HRSV as well as viruses from two recombinant strains, rgRSVΔG and rgRSVΔSHΔG, which lacked the G protein and the G and SH proteins, respectively. The recombinant strains were derived from the A2 virus. The majority of particles observed in A2 and recombinant virus samples were close to spherical in shape, with a diameter ranging between 100 nm and 1 µm. A fraction (5.2%, n = 2,000) of the A2 particles examined had at least a partially tubular morphology with tubular areas of membrane interchanging with regions where the membrane was spherically curved (Figs. 1 A and B, 2 A and C, and Fig. S1). Although the shape of tubular viruses could be quite irregular, especially at the ends of the particle, we will refer to these viruses as filamentous. The length of the filamentous domain varied from 200 nm to 2 µm. The width was also variable ranging between 70 nm and 190 nm, with an average of 120 nm (n = 28). The recombinant viruses were similar to the A2 in overall morphology (Fig. S2). Although we did not systematically analyze the stability of the filamentous particles, it was apparent that they converted to spherical particles during storage and freezing, and some of the infectivity was lost (SI Materials and Methods and SI Results). Imaging of cells infected with A2 revealed that the periphery of the cell was decorated with many filaments, which are most likely the budding virus (Fig. S3).
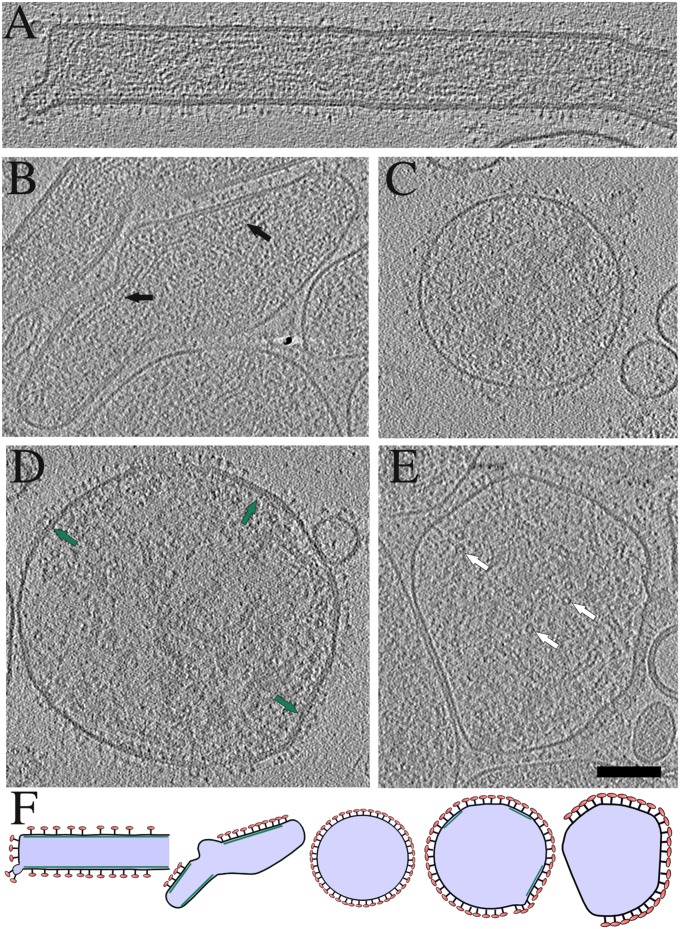
Fig. 1.
Tomography of A2 HRSV virions. Virion morphology ranges from completely filamentous (A) to completely spherical (C) with intermediate forms (B and D) that have some tubularly curved parts but are otherwise spherically curved. Spherical particles are highly deformable when in the proximity of other particles and the membrane proximal to the neighboring particle is free of glycoprotein spikes (E). Virions in A–E are illustrated schematically in F in alphabetical order. Black arrows: side views of the RNP; white arrows: top views of the RNP; green arrows: secondary density layer under the membrane in a spherical particle. (Scale bar, 100 nm.) Tomographic slices are 3.8-nm-thick.
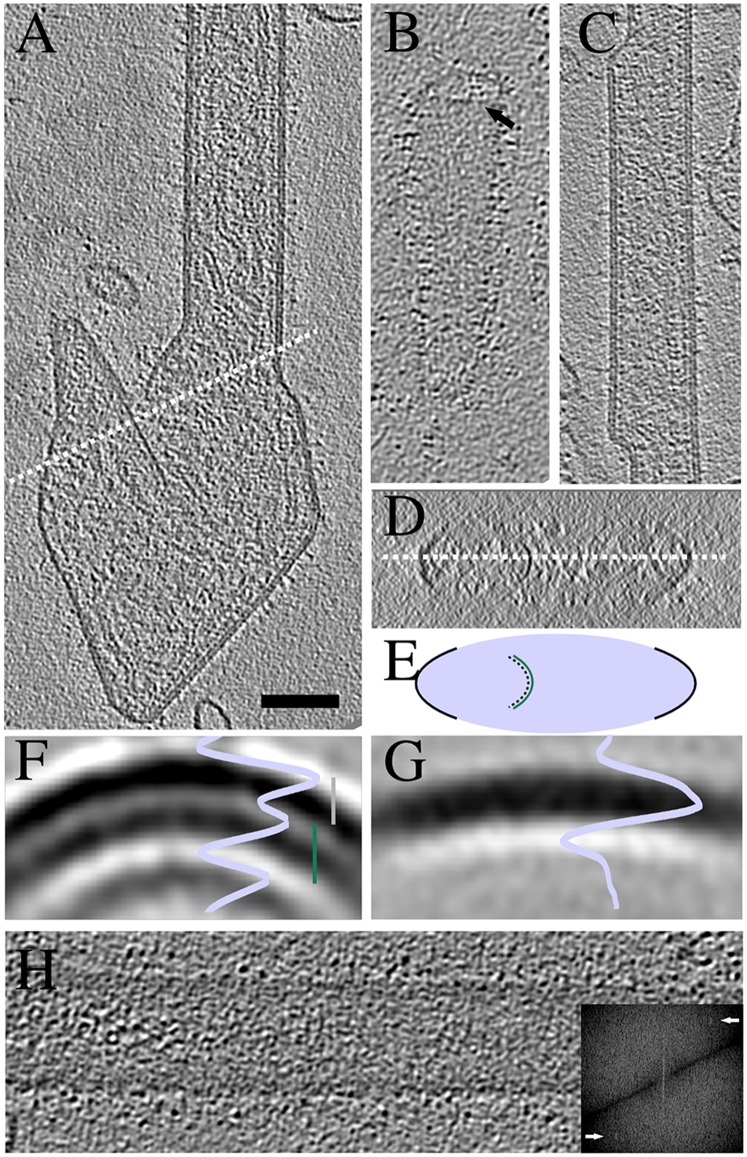
Fig. 2.
Assembly of M causes curvature of the virion membrane. (A) A tomographic slice of a filamentous A2 virion with a large irregular appendage at one end. White dotted line cuts through a curved M layer detached from the membrane. An orthogonal view around the dotted line in A is shown in D and schematically in E. (B) A tomographic slice of an A2 virion without M layer in the particle tip viewed from the top at the level of the M layer. Arrow indicates the end of the M layer. (C) A tomographic slice of an rgRSVΔG filamentous virion with varying diameter. (F) A radial profile and a slice of a subvolume average from A2 filamentous particles. Distance from membrane to M layer (gray bar) is 6.1 nm and from M to the third, innermost layer (green bar) 6.9 nm. (G) A radial profile and a slice of a subvolume average from A2 spherical particles. (H) A tomographic slice from the top of a filamentous virion. Fourier transform of the corresponding slice is shown in the Inset. Peaks corresponding to a layer line at ∼8 nm are shown with arrows. Slices are 0.77-nm-thick in A–C, F, and G; 11.5 nm in D, and 3.8 nm in H. (Scale bar, 100 nm for A–D and H.)
Filamentous Particles Have a Matrix Layer.
For a more detailed structural analysis, we used cryo-ET, a method of choice for 3D reconstruction of pleomorphic, membrane-containing particles (5, 18–20). In a total of 41 tomograms from three separate preparations of A2 virus collected from the cell culture supernatant, we analyzed 103 virions. We also studied one preparation of virus released from infected cells by freezing and thawing, a common practice in the paramyxovirus field. As the cell-derived virions appeared similar, having both spherical and filamentous shapes, further analysis was limited to supernatant viruses.
The structural analysis of the HRSV indicated a clear difference in the fine-structure between filamentous and spherical particles. The filamentous domains where the membrane was tubular had a layer of density just under the membrane (Fig. 2 and Movie S1); it was missing in nonfilamentous regions. The flexibility of the filaments made them susceptible to flattening in the layer of vitreous water, preventing subvolume analysis of the structure of the layer. However, a radial density profile of aligned and averaged surface subvolumes could be calculated. The distance between the peak densities of the membrane and the underlying layer was 6.1 nm (Fig. 2F) in filamentous particles. No such layer was visible in a similar average from spherical particles (Fig. 2G).
Based on earlier results with HRSV and other paramyxoviruses, the layer was likely to be composed of M, a protein of 29 kDa (4, 15, 17). In spherical particles, an M layer was observed only in a few particles (5 of 80), where it covered limited areas. In a few of the filamentous particles, the density continued as a defined structure (a curved layer) into areas devoid of membrane contact indicating that the layer was stable without direct membrane contact (Fig. 2 A, D, and E, and Fig. S4).
The M layer normally did not extend to the tips of the filamentous particles. One example is shown in Fig. 2B and in Movie S1, where the edge of the M layer can be seen as a dotted line just before the tip of the particle. The shape of the tips in filamentous particles was varied; it ranged from hemispherical to nearly straight with almost 90° angles in the membrane where the M layer ended (Figs. 1 A and B and 2B).
The diameter of filamentous regions was variable. As shown in Fig. 2C, the thickness of the tubular domains varied even within a single particle. That the M protein can polymerize into different tubular structures implied flexibility in assembly. Some order was detected in patches of the M layer detectable as a peak in the Fourier transform between 7.7 and 8.2 nm (Fig. 2H). However, the tubes were not helical as they were all flattened.
In one rgRSVΔG mutant particle, the membrane formed an inside-out tube where the spikes were pointing toward the center (Fig. S5). The tube was suitable for subvolume analysis of the M layer. The average of 324 subvolumes (4.2-nm resolution) showed a 7-start helix with a ridge-to-ridge distance of 7.8 nm. Inside the helix a separate, smooth layer of density representing the membrane, was observed. Although this helical tube was a singular observation, the spacing was similar to that found in the commonly observed flattened tubes (Fig. 2). That the M can form tubular structures with inverted curvature suggested flexibility in M–M interactions in the lattice.
A further layer of density was observed in the filamentous particles. The peak of the density was 6.9-nm away from the peak density of the M layer (Fig. 2 C and F). This density seemed not to be as regularly arranged as the M layer, yet it was always detectable where the M layer was clearly visible. The average density was ∼75% of the M-layer density (Fig. 2F). The inner layer was also present in the recombinant rgRSVΔG and rgRSVΔGΔSH viruses.
Virion Surface Is Covered by F Glycoproteins in Both Pre- and Postfusion Conformations.
The number of glycoproteins present in the membranes of individual virions was variable. In a number of spherical particles, the envelope was covered by an almost uniform, dense layer of glycoproteins, whereas other particles had barely any (Fig. S6). In the spherical particles, the membrane regions close to other particles were free of spikes and conversely other regions of the membrane in the same virion were often very tightly packed with spikes (Fig. 1E). No regular arrangement of the spikes could be detected even in the most tightly packed areas of the spherical particle surfaces. The filamentous regions were often densely covered by spikes.
When comparing tomograms of A2 HRSV and recombinant rgRSVΔG and rgΔGΔSH viruses, we discerned two different types of spikes in the virion envelope. The most common had a long stalk and a small head (Fig. 3A), which was found in filamentous and spherical A2 virions, but any one particular virion appeared to contain mainly one type of spike. This long spike was similar to the spikes found in rgΔGΔSH virions, thus identifying it as F. A second type of spike found in some filamentous A2 particles, not clearly apparent by visual inspection in rgΔGΔSH virions, had a large bulky head and a short stalk (Fig. 3C). The short spikes were arranged roughly in rows nearly perpendicular to the filament’s longitudinal axis (Fig. 3D), whereas filaments with the long spikes did not have any regular long-range order (Fig. 3B). We designated the filamentous virions with mainly long spikes as A2(l) and those with mainly short spikes as A2(s). Based on this observation, we had to distinguish if the short spikes were F or G glycoproteins.
Fig. 3.
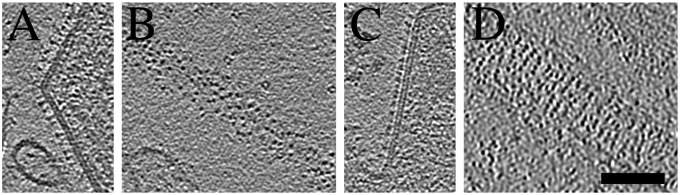
Virions have two types of spikes that are organized differently on the surface. Central tomographic slices from virions with long (A) or short (C) spikes. Top slices from virions with long (B) or short (D) spikes. Slices are 0.77-nm-thick. (Scale bar, 100 nm.)
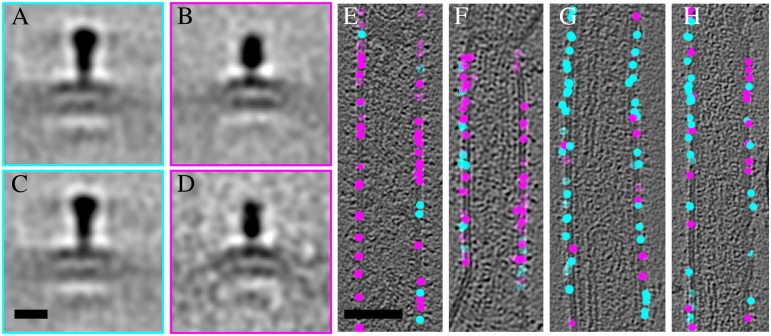
We carried out an objective analysis of the distribution of the two types of spikes in A2 and rgΔGΔSH virions by classification of the subvolumes using principal component analysis. To circumvent problems caused by the missing tomographic wedge, only spikes whose longitudinal axis was lying approximately on a plane perpendicular to the electron beam were included (Fig. 4 E–H). Two separate classification analyses were done to assess the distribution of the spikes: A2(s) against rgΔGΔSH, and A2(s) against A2(l). Classification of the A2(s) spikes against the rgΔGΔSH spikes was done into four classes. Class averages 1 and 2 both showed a spike on a membrane, differing primarily in the spike length and width (Fig. 4 A and B). The A2(s) contained mainly the short spike (Fig. 4 E and F) and the rgΔGΔSH virions contained mainly the long spike (Fig. 4 G and H). Class averages of rgΔGΔSH showed a long spike essentially identical to the class 1 average and a short spike resembling the class 2 average. Thus, it is likely that classes 1 and 2 represent two different states of F, because no other viral protein is present on the rgΔGΔSH surface. Classification of the A2(s) against A2(l) also resulted in separation of the two forms of F so that a particular virion contained almost exclusively one type of F (Fig. 4 and Fig. S7). This finding suggested that, once triggered, there is a systematic conversion of one form of F to the other within one virion. Given the arguments set out above, we did not assign a class average to the G protein.
Fig. 4.
F occurs in two different conformations on the virions. Averages of class 1 (253 subvolumes) representing the long (A) and class 2 (226 subvolumes) representing the short spike (B) from subvolumes extracted from both A2 and rgΔGΔSH virions. Class averages 1 (224 subvolumes) and 2 (65 subvolumes) including only subvolumes from rgΔGΔSH are shown (C and D). Positions of the classified spikes on the A2 virions (E and F) and on the rgΔGΔSH virions (G and H). Tomographic slices in E–H are transparent to show all spike positions. Cyan spheres correspond to class 1 spikes and magenta spheres to class 2 spikes. (A–D) Scale bar in (C) is 10 nm. (E–H) Scale bar in (E) is 100 nm.
To get a more detailed description of the F structures, alignment and averaging was carried out separately on subvolumes extracted from A2(s) and A2(l) tomograms. Spikes in all orientations in the tomograms were included for an even sampling of Fourier space. The spikes extracted from A2(s) were reconstructed from 828 subvolumes to 4.7-nm resolution (Fig. 5A). We also reconstructed spikes extracted from spherical A2 particles and found that the results were similar to those from the A2(l) virus. Hence, we pooled the data from A2(l) and spherical A2 particles, applied threefold averaging, and calculated an average of 2857 subvolumes to 3.6 nm resolution (Fourier shell correlation 0.5) (Fig. 5C). The dimensions of the long and short F reconstructions corresponded to those of the atomic model of HRSV F in the postfusion conformation (14) (PDB ID code 3RRT) and to those of the parainfluenza 5 (PDB ID code PIV5) F in prefusion conformation (21) (PDB ID code 4GIP), respectively. The atomic models fit well into the reconstructed densities within the limits of the resolution of our models (Fig. 5 B and D). The fits indicate that the cytoplasmic tail of F can reach the M layer. Thus, F on A2 virions was found in both pre- and postfusion conformations. Where F was in the prefusion form, it was found in rough rows (Fig. 3D).
Fig. 5.
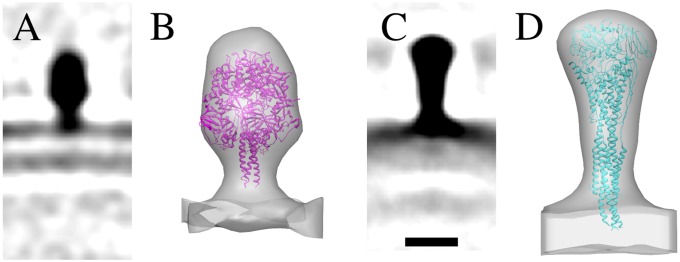
F spike structures in pre- and postfusion conformation. Central sections (0.77-nm-thick) of the subvolume averages of F from A2(s) (A) and A2(l) (C). Crystal structures of PIV5 F in the prefusion conformation (PDB ID code 4GIP) (21) and HRSV F in the postfusion conformation (PDB ID code 3RRT) (14) fitted into the subvolume average density of the short (B) and the long (D) spike. (Scale bar, 10 nm.) The isosurfaces in B and D were rendered at 2 σ and at 5 σ from the mean, respectively.
RNP Is Packed as Flexible Left-Handed Helix.
The RNPs were packed as multiple flexible helical filaments inside both spherical and filamentous virions (Fig. 6). The packing density varied between different spherical particles. In the filamentous particles, the RNPs were always packed at levels comparable to the most densely packed spherical particles (Fig. S8). Because of the tight packing and random arrangement, we could not follow individual RNPs to unambiguously say how many genome copies each particle contained. It was also apparent that some of the viruses contained filaments similar to the dimensions expected of F-actin, consistent with the demonstrated presence of actin in purified HRSV preparations (Fig. S6 and Movie S2) (12, 22).
Fig. 6.
The RNP of HRSV is left-handed. A central section (A) and an isosurface representation of the HRSV RNP (B). (Scale bar, 10 nm.) (C) A transparent tomographic slice showing the refined positions and orientations of the RNP subvolumes on the tomogram. Cyan triangles indicate the direction of the RNP helix. The isosurface was rendered at 1 σ from the mean.
We tested the handedness of the RNP by tomography using subvolume alignment and averaging of segments of RNP extracted from a tomogram with a broken filamentous virion. A 3D model (4.0-nm resolution, 1,402 subvolumes) was generated from which the handedness could be determined (Fig. 6 A and B). The protocol used was similar to that which we have used previously for the measles virus RNP (5), which resulted in the correct handedness as determined earlier by other methods (23). We also used DNA origami gold nanoparticle helices of known handedness as a control sample (Movie S3) (24). From the subvolume average, it is clear that the HRSV RNP is left-handed like other paramyxoviral RNPs. The structure is otherwise similar (approximate diameter of 17 nm and ridge-to-ridge distance of 7 nm) to the one determined earlier from cryo-negative stained micrographs by helical processing (13). Placing the subvolume average back into a tomogram in the refined positions showed that most of the RNP helix segments analyzed pointed in the same direction (Fig. 6C).
Discussion
Like most other paramyxoviruses, the HRSV virions are quite heterogeneous in shape and size. In addition to this pleomorphicity, there are substantial challenges in obtaining high-titer HRSV preparations, making it particularly challenging to study this virus. Our results form a basis for developing a 3D structural understanding of HRSV. Whether isolated from the cells or the culture supernatant, the particles we observed had mostly spherical but also filamentous (tubular) and mixed morphologies. The size of the spherical particles was comparable to that observed earlier for Sendai virus (SeV) and measles virus (MV) using cryo-EM (5, 6). However, very large particles with diameters up to 1 µm were also present. Because these were too large for the RNP to be visualized, it was difficult to judge whether they were true virions or copurifying vesicles. Because the filamentous population of particles had a maximal length of ∼2 µm, they were significantly shorter than the 10-µm particles described earlier (15). The width of the filaments was between 70 nm and 190 nm. That the recombinant rgRSVΔG and rgRSVΔGΔSH viruses were similar to the A2 virions indicated that G and SH did not play a role in defining overall particle size and morphology. Although not essential under tissue-culture conditions, these proteins have been reported to elevate infectivity (25).
We found that the filamentous virions had an M layer juxtaposed to the envelope. Occasionally virions with other shapes also had such a layer, but mainly in places where the membrane was tubularly curved. We found the M layer in tubular domains of varying width, suggesting that the assembly of M protein to form these assemblies did not require any particular helical symmetry. In some particles, the M continued as a curved, sheet-like extension without contact with the membrane, showing that the structure possessed a degree of stability independent of membrane association.
The tips of filamentous viruses were generally devoid of an M layer, but they did contain spikes. Thus, it is likely that assembled M was not required to initiate a filamentous bud in the plasma membrane of an infected cell, but responsible for generating, extending, and maintaining the filamentous, tubular shape of the budding virus. This theory is supported by the observations that M is known to be required for elongation of filamentous buds in cells (16), and that purified M can assemble into filaments on lipid layers in vitro (17).
We observed that after freezing, virus preparations had a lower percentage of filamentous particles. Similarly, incubation at 37 °C and room temperature resulted in a drop in infectivity and decrease in the fraction of filamentous particles (SI Results). It seemed likely that the filamentous particles converted to spherical particles and lost infectivity. M in the spherical particles was disordered, probably dissociated from the membrane. However, M did not assemble in large aggregates as reported for low-pH treated influenza A (19), nor was it coating the RNPs as seen in MV (5).
We observed that filamentous particles had more densely packed helical RNPs than the spherical particles, suggesting a role for M in the longitudinal organization of the RNP. We also identified an internal layer 6.9 nm under the M-layer. Because this layer was also present in viruses devoid of G and SH, we propose that it is composed of the M2-1 protein. A location between M and the RNP is consistent with the earlier observation that M2-1 mediates the interaction of the RNP and M, and is required for inclusion of RNPs into virions (10).
We determined the absolute hand of the RNP by tilting and found that it consists of a left-handed helix, thus disproving a previous conjecture that the RNPs are arranged as right-handed helices based on modeling with a ring form of N bound to RNA (13). Thus, HRSV follows the general trend of left-handed nucleocapsids within the order Mononegavirales (18). As the RNA is wrapped around the outside of the complex, changing the hand of the helical model affects the residues of both RNA and N that will interact between subunits. Because most of the RNPs seem to be oriented in the same direction in filamentous particles (Fig. 6C), this implies that there is preferential directional interaction of the RNP with the budding tip of the virion and the M2-1 layer.
HRSV is unique so far among paramyxoviruses in requiring two cleavages by furin-like proteases to render the F protein fusion competent (26). The F spikes in the virus used in our studies had only one of these cleavages, which meant that the F protein was still inactive (22). We have found that the second cleavage occurs only after internalization by endocytosis. Therefore, it was surprising to us that the majority of the F spikes that could be resolved by reconstruction in the virus envelope showed a golf-tee shaped structure similar to the atomic model of isolated, postfusion F spikes (Fig. 5) (14). Similar spikes have also been reported on parainfluenza virus 5 (27).
The smaller spikes with a short, narrow stalk and a bulky head detected in some of the filamentous virions were most likely prefusion F (Fig. 5) because they were also found in small numbers on mutants devoid of G and SH (Fig. 4 and Fig S7). The regular spacing of these spikes, seen in Fig. 3, suggests an interaction with the closely packed M layer below perhaps by insertion into the lattice (4). That the regular spacing is observed only on virions with prefusion F, suggests that the interaction between F and M or lateral F–F interaction is lost upon triggering of F. This result in turn could lead to the disassembly of the M layer, and gradual transition to the spherical form (Fig. 7).
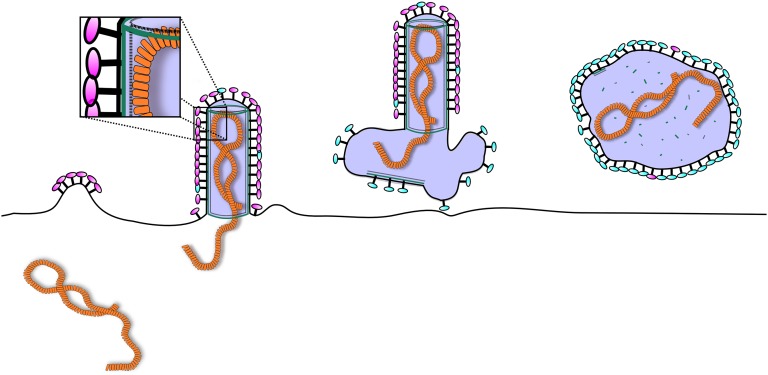
Fig. 7.
Schematic model of assembly of HRSV. Initially, the F glycoproteins (magenta) gather at the plasma membrane enriching in lipid rafts and initiate a bud. M protein (green) is recruited to the budding site via interactions with the glycoprotein tails and the membrane. Interactions between M and G promote incorporation of G into the bud. Assembly of M into tubular structure provides the force for the elongation of the virion and recruits the RNP into the nascent filament. This recruitment can be mediated by the M2-1 protein (dashed line). Once the RNP is inside the budding virion, release occurs via a currently unknown endosomal sorting complexes required for transport-independent mechanism. The virion draws a variable amount of membrane with it and this membrane remains as an appendage or a larger membrane sack in the end of the filament. Some of the virions then convert into roughly spherical forms as the M disassembles from the membrane with conversion of F to the postfusion-like conformation (cyan).
Within the paramyxovirus family, whole virion structures in 3D are available in addition to HRSV, for MV, Newcastle disease virus (NDV), and SeV (4–6). The results suggest differences between the Paramyxovirinae and Pneumovirinae subfamilies. The most striking are the location and mode of assembly of the M protein. In HRSV, NDV, and SeV, M is often found as a layer partially covering the inner surface of the membrane. In MV, it is mainly organized as left-handed helices around the RNP. Furthermore in MV and SeV, the envelope is densely covered by glycoproteins and it is difficult to distinguish the individual spikes. Where the M-layer is visible, the glycoproteins of NDV are also densely packed, suggesting an interaction between M and the glycoproteins (4). However, in contrast to previous paramyxovirus studies, we were able to distinguish the glycoproteins in the HRSV envelope and thus we could visualize their organization especially in the filamentous domains.
Together with published data from others, our structural analysis suggests an M-dependent mechanism for HRSV morphogenesis during budding in infected host cells illustrated in Fig. 7. The starting point can be considered as the accumulation of F protein in lipid rafts in the plasma membrane where it can generate (perhaps through F–F interactions) outward curvature to generate the tip of a budding virus (Fig. S3) (15, 16, 28, 29). Next, RNPs are likely to be transported to the site together with M in a process that depends on M2-1 (10). Next, interaction with the cytoplasmic tails of F (28) may increase the local concentration of M in the growing bud promoting M–M and M–lipid interactions (Figs. 1–5, and Figs. S4 and S7). The formation of an M-containing helical sheet tightly associated with the membrane can then begin to drive the extension of the bud by providing the force required for filament elongation (17, 30), clearly seen as filamentous cell-associated virus (Fig. S3). F and G may be incorporated into the growing filament via their cytoplasmic tails (Figs. 1–4, and Fig. S7) (31). In the meantime, interactions between M and RNP, whether mediated by an additional layer of M2-1 or by direct interaction, may promote the incorporation of the RNP into the growing particle (Fig. 2).
Materials and Methods
Cells and Viruses.
HEp-2 cells were obtained from the ATCC and cultured in complete medium (DMEM supplemented with 5–10% (vol/vol) FCS, 1 mM Hepes, 1% (vol/vol) Glutamax) (Invitrogen). HRSV-A2 was purchased from ATCC. Recombinant HRSV strains expressing GFP (rgRSVΔG, rgRSVΔSHΔG) were kindly provided by M. Peeples (Ohio State University, Columbus, OH) and P. Collins (National Institute of Allergy and Infectious Diseases, Bethesda, MD).
Virus Growth, Purification, and Vitrification.
HEp-2 cells (50–60% confluent) in T175 flasks were infected with HRSV (multiplicity of infection 0.1) in 8 mL serum free DMEM-Hepes medium (DMEM, 1 mM Hepes) for 1 h at room temperature while gently shaking. Virus inoculum was replaced with 25 mL of complete medium, and cultures were placed in a 37 °C humidified, 5% CO2 incubator. After 48 h, the cell supernatant was collected and clarified by centrifugation (3,000 rpm, 10 min, 4 °C, S4180 rotor (Beckman Coulter Inc., Indianapolis), Beckman Allegra 2IR centrifuge). The clarified supernatant was centrifuged (20,000 rpm, 90 min, 4 °C, SW32 Ti rotor, Beckman Optima 90-K ultracentrifuge) through an 8 mL 30% (wt/vol) sucrose cushion in HBSS-Hepes buffer (HBSS, 25 mM Hepes, pH 7.0–7.4). Pellets were gently washed and resuspended in 200 µL of HBSS-Hepes buffer for each T175 flask, virus stocks were snap-frozen in liquid nitrogen and stored at −80 °C or kept in 4 °C before analysis. To obtain a sample of cell-associated virus, the infected cells were washed with PBS, scraped, and resuspended in 5 mL of HBSS-Hepes buffer per T175 flask. The cells were then snap-frozen twice in liquid nitrogen, and the released viruses purified as described above.
All HRSV stocks were titered by infecting HEp-2 cells with serial dilutions of the virus in 96-well plates. Infection was allowed to proceed for 18–22 h at 37 °C. Fixed cells were assessed by microscopy for GFP expression, or stained with HRSV anti-N antibody (AF-488; Millipore) to detect infected cells of rgRSV or HRSV-A2, respectively. Titers for purified viruses after storage at -80 °C were 0.5–1.9 × 108 pfu/mL for A2 virus and 0.1–1.8 × 106 pfu/mL for recombinant viruses. Infectivity ratio measurements are described in SI Materials and Methods.
The purified virus was examined fresh (storage at 4 °C), after incubation at 37 °C for 6.5 h followed by 18-h incubation in room temperature and after snap-freezing and storage at −80 °C. Samples for cryo-EM were prepared as previously described (5).
Collection of 2D Data.
Two-dimensional images for analysis of the number of filamentous particles were collected from vitrified specimens at liquid nitrogen temperature under low-dose conditions with an FEI F20 transmission electron microscope using a Gatan 914 cryo-holder and a Gatan Ultrascan 4000 CCD camera. Images were taken at 4,200× magnification and 200-µm underfocus.
Cryo-ET and Subvolume Processing,
Tilt series were collected and tomograms calculated essentially as reported previously (5) at a magnification of 39,400× with a 15-µm pixel size on the CCD resulting in a pixel size of 0.38 nm/pixel. The underfocus used for tilt series ranged from 4 µm to 6 µm. The data were binned to 0.77 nm/pixel to improve the signal for processing. Visual inspection of the M and RNP tomograms were carried out to investigate if there was any order in the data to justify subvolume averaging. All subvolume processing was carried out using 3dmod and PEET from the IMOD package (32–34). Bsoft was used for generating masks for subvolume processing and estimation of the subvolume average resolutions (35). Detailed methods for subvolume processing are described in the SI Materials and Methods.
Handedness Determination Controls.
The handedness of the helical averages was verified by carrying out similar tomographic data collection and reconstruction using negatively-stained, left-handed, silver-enhanced, DNA origami gold nanoparticle helices (24) as a control (kind gift of Anton Kuzyk, Aalto University, Espoo, Finland). The high contrast of the metal clusters allowed direct handedness determination of the structure without further subvolume processing (Movie S3).
The subtomographic volumes have been deposited in the Electron Microscopy Data Bank with the following accession numbers EMD-2391, EMD-2392, and EMD-2393.
Note Added in Proof.
Bakker et al. (36) corrected their initial assumption of the handedness of the HRSV nucleoprotein-RNA complex whilst this paper was in press.
Supplementary Material
Acknowledgments
We thank Drs. P. Collins and M. Peeples for providing us with recombinant human respiratory syncytial virus strains; Dr. Anton Kuzyk for the kind gift of DNA origami gold nanoparticle helices; Eevakaisa Vesanen and Roberta Mancini for technical assistance; Dr. John Heumann for advice in using PEET; Dr. Felix Rey and Dr. David Bhella for helpful discussions; and the Biocenter Finland National Cryo Electron Microscopy Unit, Institute of Biotechnology, Helsinki University for kindly providing facilities. This work was supported by the Academy of Finland Grant 1139178 (to S.J.B.); the Sigrid Juselius Foundation (S.J.B.); the Viikki Doctoral Programme in Molecular Biosciences (L.L.); European Research Council Grant VIRNA 2-73905-09 (to A.H.); and European Molecular Biology Organization Fellowship ALTF 349-2010 (to M.A.K.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The subvolume averages have been submitted to the Electron Microscopy Data Bank (www.emdatabank.org) (accession nos. EMD-2391, EMD-2392, and EMD-2393).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1309070110/-/DCSupplemental.
References
- 1.Falsey AR, Hennessey PA, Formica MA, Cox C, Walsh EE. Respiratory syncytial virus infection in elderly and high-risk adults. N Engl J Med. 2005;352(17):1749–1759. doi: 10.1056/NEJMoa043951. [DOI] [PubMed] [Google Scholar]
- 2.Nair H, et al. Global burden of acute lower respiratory infections due to respiratory syncytial virus in young children: A systematic review and meta-analysis. Lancet. 2010;375(9725):1545–1555. doi: 10.1016/S0140-6736(10)60206-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Group TI-RS Palivizumab, a humanized respiratory syncytial virus monoclonal antibody, reduces hospitalization from respiratory syncytial virus infection in high-risk infants. The IMpact-RSV Study Group. Pediatrics. 1998;102(3 Pt 1):531–537. [PubMed] [Google Scholar]
- 4.Battisti AJ, et al. Structure and assembly of a paramyxovirus matrix protein. Proc Natl Acad Sci USA. 2012;109(35):13996–14000. doi: 10.1073/pnas.1210275109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liljeroos L, Huiskonen JT, Ora A, Susi P, Butcher SJ. Electron cryotomography of measles virus reveals how matrix protein coats the ribonucleocapsid within intact virions. Proc Natl Acad Sci USA. 2011;108(44):18085–18090. doi: 10.1073/pnas.1105770108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Loney C, Mottet-Osman G, Roux L, Bhella D. Paramyxovirus ultrastructure and genome packaging: Cryo-electron tomography of Sendai virus. J Virol. 2009;83(16):8191–8197. doi: 10.1128/JVI.00693-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liljeroos L, Butcher SJ. Matrix proteins as centralized organizers of negative-sense RNA virions. Front Biosci. 2013;18:696–715. doi: 10.2741/4132. [DOI] [PubMed] [Google Scholar]
- 8.Collins PL, Wertz GW. The envelope-associated 22K protein of human respiratory syncytial virus: Nucleotide sequence of the mRNA and a related polytranscript. J Virol. 1985;54(1):65–71. doi: 10.1128/jvi.54.1.65-71.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang YT, Collins PL, Wertz GW. Characterization of the 10 proteins of human respiratory syncytial virus: Identification of a fourth envelope-associated protein. Virus Res. 1985;2(2):157–173. doi: 10.1016/0168-1702(85)90246-1. [DOI] [PubMed] [Google Scholar]
- 10.Li D, et al. Association of respiratory syncytial virus M protein with viral nucleocapsids is mediated by the M2-1 protein. J Virol. 2008;82(17):8863–8870. doi: 10.1128/JVI.00343-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fearns R, Collins PL. Role of the M2-1 transcription antitermination protein of respiratory syncytial virus in sequential transcription. J Virol. 1999;73(7):5852–5864. doi: 10.1128/jvi.73.7.5852-5864.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Radhakrishnan A, et al. Protein analysis of purified respiratory syncytial virus particles reveals an important role for heat shock protein 90 in virus particle assembly. Mol Cell Proteomics. 2010;9(9):1829–1848. doi: 10.1074/mcp.M110.001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tawar RG, et al. Crystal structure of a nucleocapsid-like nucleoprotein-RNA complex of respiratory syncytial virus. Science. 2009;326(5957):1279–1283. doi: 10.1126/science.1177634. [DOI] [PubMed] [Google Scholar]
- 14.McLellan JS, Yang Y, Graham BS, Kwong PD. Structure of respiratory syncytial virus fusion glycoprotein in the postfusion conformation reveals preservation of neutralizing epitopes. J Virol. 2011;85(15):7788–7796. doi: 10.1128/JVI.00555-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bächi T, Howe C. Morphogenesis and ultrastructure of respiratory syncytial virus. J Virol. 1973;12(5):1173–1180. doi: 10.1128/jvi.12.5.1173-1180.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mitra R, Baviskar P, Duncan-Decocq RR, Patel D, Oomens AG. The human respiratory syncytial virus matrix protein is required for maturation of viral filaments. J Virol. 2012;86(8):4432–4443. doi: 10.1128/JVI.06744-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McPhee HK, et al. Influence of lipids on the interfacial disposition of respiratory syncytical virus matrix protein. Langmuir. 2011;27(1):304–311. doi: 10.1021/la104041n. [DOI] [PubMed] [Google Scholar]
- 18.Bharat TA, et al. Cryo-electron tomography of Marburg virus particles and their morphogenesis within infected cells. PLoS Biol. 2011;9(11):e1001196. doi: 10.1371/journal.pbio.1001196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Calder LJ, Wasilewski S, Berriman JA, Rosenthal PB. Structural organization of a filamentous influenza A virus. Proc Natl Acad Sci USA. 2010;107(23):10685–10690. doi: 10.1073/pnas.1002123107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pietilä MK, et al. Virion architecture unifies globally distributed pleolipoviruses infecting halophilic archaea. J Virol. 2012;86(9):5067–5079. doi: 10.1128/JVI.06915-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Welch BD, et al. Structure of the cleavage-activated prefusion form of the parainfluenza virus 5 fusion protein. Proc Natl Acad Sci USA. 2012;109(41):16672–16677. doi: 10.1073/pnas.1213802109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krzyzaniak MA, Zumstein MT, Gerez JA, Picotti P, Helenius A. Host cell entry of respiratory syncytial virus involves macropinocytosis followed by proteolytic activation of the f protein. PLoS Pathog. 2013;9(4):e1003309. doi: 10.1371/journal.ppat.1003309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schoehn G, et al. The 12 A structure of trypsin-treated measles virus N-RNA. J Mol Biol. 2004;339(2):301–312. doi: 10.1016/j.jmb.2004.03.073. [DOI] [PubMed] [Google Scholar]
- 24.Kuzyk A, et al. DNA-based self-assembly of chiral plasmonic nanostructures with tailored optical response. Nature. 2012;483(7389):311–314. doi: 10.1038/nature10889. [DOI] [PubMed] [Google Scholar]
- 25.Techaarpornkul S, Barretto N, Peeples ME. Functional analysis of recombinant respiratory syncytial virus deletion mutants lacking the small hydrophobic and/or attachment glycoprotein gene. J Virol. 2001;75(15):6825–6834. doi: 10.1128/JVI.75.15.6825-6834.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zimmer G, Budz L, Herrler G. Proteolytic activation of respiratory syncytial virus fusion protein. Cleavage at two furin consensus sequences. J Biol Chem. 2001;276(34):31642–31650. doi: 10.1074/jbc.M102633200. [DOI] [PubMed] [Google Scholar]
- 27.Ludwig K, et al. Electron cryomicroscopy reveals different F1+F2 protein States in intact parainfluenza virions. J Virol. 2008;82(7):3775–3781. doi: 10.1128/JVI.02154-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shaikh FY, et al. A critical phenylalanine residue in the respiratory syncytial virus fusion protein cytoplasmic tail mediates assembly of internal viral proteins into viral filaments and particles. MBio. 2012;3(1):e00270–11. doi: 10.1128/mBio.00270-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fleming EH, Kolokoltsov AA, Davey RA, Nichols JE, Roberts NJ., Jr Respiratory syncytial virus F envelope protein associates with lipid rafts without a requirement for other virus proteins. J Virol. 2006;80(24):12160–12170. doi: 10.1128/JVI.00643-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saarikangas J, et al. Molecular mechanisms of membrane deformation by I-BAR domain proteins. Curr Biol. 2009;19(2):95–107. doi: 10.1016/j.cub.2008.12.029. [DOI] [PubMed] [Google Scholar]
- 31.Ghildyal R, et al. Interaction between the respiratory syncytial virus G glycoprotein cytoplasmic domain and the matrix protein. J Gen Virol. 2005;86(Pt 7):1879–1884. doi: 10.1099/vir.0.80829-0. [DOI] [PubMed] [Google Scholar]
- 32.Heumann JM, Hoenger A, Mastronarde DN. Clustering and variance maps for cryo-electron tomography using wedge-masked differences. J Struct Biol. 2011;175(3):288–299. doi: 10.1016/j.jsb.2011.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116(1):71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 34.Nicastro D, et al. The molecular architecture of axonemes revealed by cryoelectron tomography. Science. 2006;313(5789):944–948. doi: 10.1126/science.1128618. [DOI] [PubMed] [Google Scholar]
- 35.Heymann JB, Belnap DM. Bsoft: Image processing and molecular modeling for electron microscopy. J Struct Biol. 2007;157(1):3–18. doi: 10.1016/j.jsb.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 36.Bakker SE, et al. The Respiratory Syncytial Virus nucleoprotein-RNA complex forms a left-handed helical nucleocapsid. J Gen Virol. 2013 doi: 10.1099/vir.0.053025-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.