Abstract
The biology of Sydney Brenner’s eponymous species of nematode, Caenorhabditis brenneri, is little known to science, despite its famous sibling Caenorhabditis elegans. Here we demonstrate that C. brenneri harbors the most molecular diversity of any eukaryote, with its 14.1% of polymorphic synonymous sites between individuals being 150-fold greater than humans and most comparable to hyperdiverse bacteria. This diversity is not an artifact of cryptic species divergence but reflects an enormous pan-tropical population, confirmed by fully viable genetic crosses between continents, extensive intralocus recombination, selection on codon use, and only weak geographic genetic structure. These findings in an animal galvanize tests of theory about the evolution of complexity in genomes and phenotypes and enable molecular population genetics methods to finely resolve uncharacterized functional noncoding elements.
Keywords: genetic variation, genome evolution, molecular evolution, biodiversity, nucleotide polymorphism
All of the diversity of life on earth originated from genetic variability within populations that transformed into differences between populations, species, and broad taxonomic groups. Yet, explaining the differences in molecular diversity among organisms is a continuing challenge in evolutionary genetics (1, 2). Although theory shows that, in general, large populations will retain more sequence variation than small populations (3), microbes appear to monopolize those species with extremely large and diverse populations (2, 4). This trend constrains the generality of tests of theory about the evolution of genome complexity, because disparities in genome architecture are most acute between prokaryotic microbes and metazoans (5). Can populations of sexually reproducing animals with complex development achieve within-species biodiversity comparable to hyperdiverse bacteria (6)? Such organisms would hold a keystone position for understanding genome evolution, by their twin virtues of multicellular eukaryotic biology and microbe-like population sizes.
The bactivorous nematode Caenorhabditis brenneri, a recently described relative of the animal model Caenorhabditis elegans, provides a candidate organism for high diversity (7–9). This pan-tropical species breeds by outcrossing obligatorily (unlike the highly selfing C. elegans) and is morphologically nearly indistinguishable from its sibling Caenorhabditis remanei, despite complete biogeographic and postmating reproductive isolation and high molecular divergence (7, 10). Its small size (∼1 mm long) and abundant food (bacteria in rotting fruit and vegetation) can support enormous population densities. Additionally, residual heterozygosity comprised an astonishing ∼40% of the C. brenneri reference genome sequence, with curated sets of allelic pairs showing high pairwise differences (8, 9).
In C. elegans, natural variation is exploited to dissect the molecular genetics causing intriguing trait differences, such as body size (11), copulatory plug deposition (12), pesticide resistance (13), and a paternal-effect killer toxin (14), in addition to deciphering how evolution creates genomic patterns of expression differences (15, 16). However, diversity in the wild for the highly self-fertilizing C. elegans is humble compared with relatives that outbreed obligatorily (17). High population diversity can serve practical purposes such as identifying and resolving to fine scale the location and selective pressures on noncoding regulatory elements (18). Given the experimental power of Caenorhabditis, this suggests that higher diversity in outbreeding species like C. brenneri (7) could also be leveraged to characterize complementary aspects of the evolutionary process and molecular mechanisms to those that are accessible to the C. elegans model system. Here we interrogate the C. brenneri genome to test for microbe-like within-species diversity for application to tests of evolutionary theory and the characterization of genome function.
Results
C. brenneri Is Hyperdiverse.
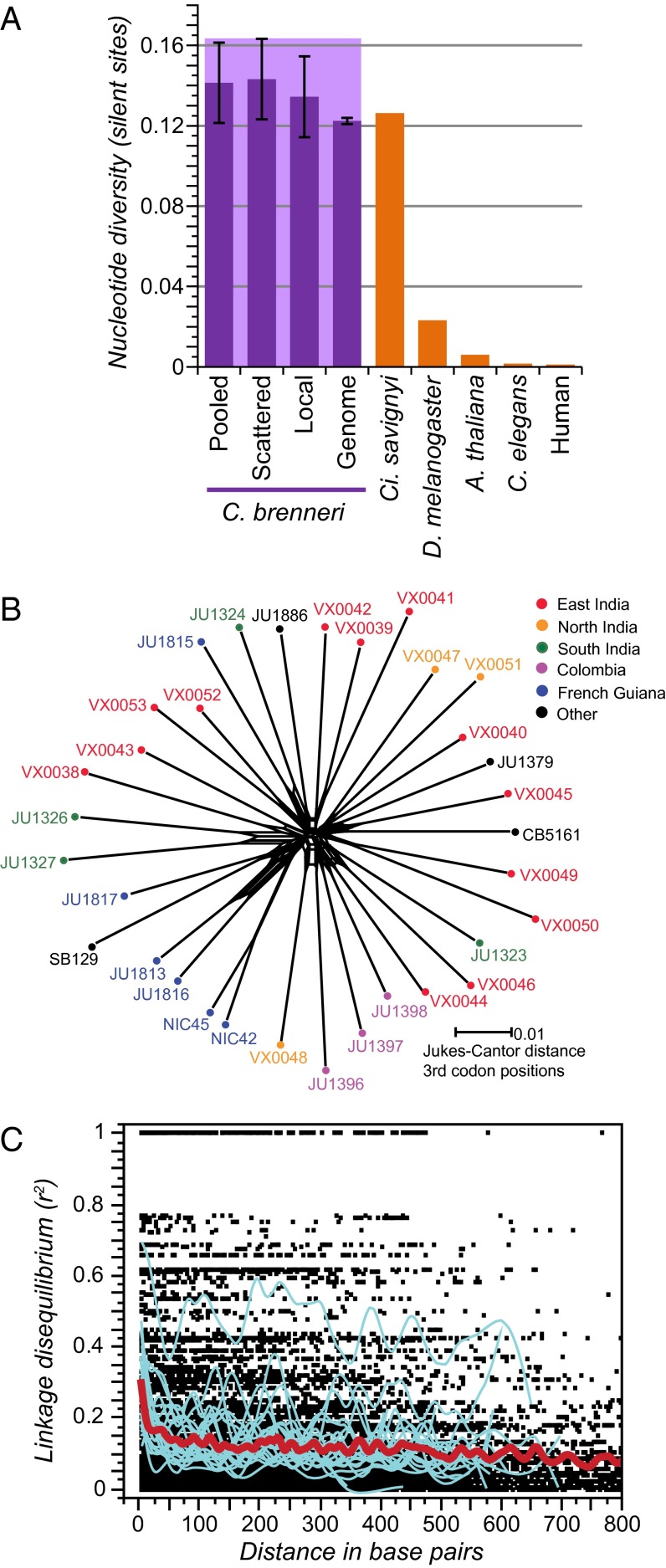
To assess the utility of natural polymorphisms in C. brenneri for understanding genome evolution and function, we investigated the scope of sequence polymorphism within and among collecting sites in South Asia and northern South America, among other tropical locations around the world (Table S1). We calculate an average diversity at replacement sites of πrep = 0.42% and at synonymous sites of πsyn-JC = 14.1%, with close correspondence of values from local population samples, globally pooled, and scattered samples of individuals (Fig. 1A; Table S2). More than half of all synonymous sites had an SNP, and an average of 8.0% of polymorphic sites segregate three or more variant nucleotides in our full sample, with 18 cases in which all four nucleotides are present as alleles. Moreover, we detect significant ongoing selection for preferred synonymous codons (4Nes = 0.44, P < 0.0001). Excluding loci most susceptible to this subtle source of natural selection that distinguishes fitness differences on the order of one in a million, we estimate that neutral polymorphism in C. brenneri is πneu = 16.4%. This finding implies that a pair of randomly chosen, selectively unconstrained sequences are expected to have experienced a mutation every 6 bp in their coalescent history. Such a level of nucleotide polymorphism is exceptionally high for a eukaryote and affirms the indications of high diversity implicated by residual heterozygosity in the highly inbred strain used for genome sequencing (8, 9) (Fig. 1A).
Fig. 1.
C. brenneri is hyperdiverse. (A) C. brenneri shows hyperdiverse nucleotide variability (πsyn-JC, the mean number of pairwise differences at synonymous sites with Jukes-Cantor correction for multiple hits; dark purple) for haplotypes pooled across the species range, for analysis restricted to a single haplotype per collection locality (scattered) or a single local population or when quantified for alleles seen in residual heterozygosity of the draft genome assembly. Pale bar behind C. brenneri values indicates estimated πneu = 0.164. Comparable measures of diversity are shown for other biological model organisms (orange). Error bars for C. brenneri indicate SEM (23 nuclear gene loci for pooled, scattered, and local samples; 3,265 gene loci for genome analysis). (B) The reticulated neighbor-network for globally sampled C. brenneri, based on concatenated sequence of 23 nuclear loci, indicates extensive recombination and little genetic structure among the strain collection locations (color coded). Nucleotide distances for third positions of codons (Jukes-Cantor corrected) exclude gaps. (C) Linkage disequilibrium (r2) decays rapidly with distance in the scattered sample of C. brenneri. Data for 23 nuclear loci are superimposed; solid red line indicates a loose spline fit for all loci; blue lines show loose spline fits for each locus separately.
To further confirm the generality of this result, we identified a set of 3,265 genes with residual heterozygosity comprising 4.2 Mb in the genome assembly of C. brenneri (8). We computed a mean πsyn-JC = 12.2% for this genomic sampling of two haplotypes per gene (πrep = 0.39%), from which we also estimate πneu = 16.4% after correcting for the influence of translational selection on synonymous sites. By comparison, the related species C. elegans has ∼100-fold less variation (17, 19), and C. brenneri diversity exceeds even that of many bacteria (6) and the previous eukaryotic exemplar of hyperdiversity, the ascidian sea squirt Ciona savignyi (2, 20). The human species has >150 times less variation than C. brenneri, and the population genetic model Drosophila melanogaster has approximately eightfold less (2).
C. brenneri Strains Do Not Comprise a Cryptic Species Complex.
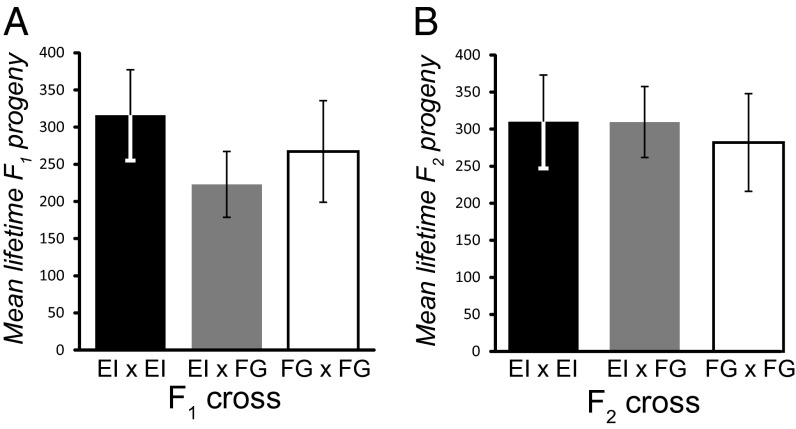
Given the exceptional diversity of C. brenneri, could it be that divergence between cryptic species or structured populations might be responsible? Population genetic patterns led to the discovery of the cryptic C. sp. 23 as a close relative to C. remanei (21), but we can rule out this possibility for our specimens of C. brenneri based on three lines of evidence. First, crosses between geographically separated strains yielded no detectable F1 or F2 hybrid dysfunction or Haldane’s rule (Fig. 2), unlike that observed for interspecies crosses of related species pairs (21–23). Second, we find that the diversity within a single sampling locality (πsyn-JC = 13.4%), or even heterozygosity in a single individual genome, is comparable to the global average (Fig. 1A) and that the distribution of variant frequencies is not consistent with a model of admixture of a few cryptic species (24) (mean scattered sample DTajima = −0.54 for synonymous sites). Third, we identify minimal structuring of genetic variation between sampling sites (mean FST = 0.092 with zero fixed differences between eastern India and French Guiana), and gene trees yield star-shaped topologies with extensive reticulation indicative of recombination (Fig. 1B). Within-locus recombination is pervasive, with four-gamete tests showing 95% of loci to have at least Rmin = 1 recombination event in their coalescent history (median Rmin = 20 per ∼700 bp locus), and the per-site population recombination parameter averages ρ = 0.085 within the east Indian local sample (ρ = 0.093 for the globally scattered sample). Linkage disequilibrium within loci is low and decays quickly over tens of base pairs (Fig. 1C), indicating rampant gene flow and recombination in nature. Consequently, we conclude that the specimens of C. brenneri analyzed here conform well to biological and genealogical notions of a single species.
Fig. 2.
No hybrid breakdown between strains from different continents. (A) F1 and (B) F2 progeny production does not differ significantly among crosses made within populations or between populations of C. brenneri (EI, East India; FG, French Guiana; F1 one-way ANOVA, F2,34 = 0.77, P = 0.47; F2 one-way ANOVA, F2,37 = 0.066, P = 0.94). Error bars indicate SEM.
Large Population Size, and Not High Mutation Rate, Best Explains High Diversity.
The hyperdiversity we find for C. brenneri could be due to it experiencing either an unusually high mutation rate or a particularly large effective population size. Branch lengths from phylogenetic analyses of the genus do not imply that it has exceptional rates of mutation (10, 25), and the evidence of low polymorphism in amino acid replacement sites and of potent selection for codon usage bias is consistent with a large effective population size (17). Assuming that C. brenneri has an equivalent mutation rate to C. elegans (26), we estimate that the long-term effective population size of C. brenneri is about 15.2 × 106 (±1 SE = 2.6 × 106) individuals, with census size several orders of magnitude larger (27).
Discussion
The nematode Caenorhabditis brenneri has the highest within-species biodiversity of any eukaryote, and we confirmed that this reflects the accumulation of genetic variation in a single species with an enormous population size (genetic effective population size Ne ∼ 15.2 × 106). Such a large effective population size implies that natural selection is unlikely to be limited by the influx of mutation and that adaptation might more likely operate via “soft” selective sweeps involving multiple alleles or alleles present at nonnegligible frequency in the population (28, 29). Beyond highlighting that eukaryotes, in addition to viruses and bacteria, can violate standard models of mutation that presume mutation to be irreversible (the “infinite sites” assumption) (30), the high density of polymorphisms in C. brenneri raises intriguing questions about how the cell’s recombination machinery can tolerate such high allelic variability to retain species integrity (31).
Although C. brenneri’s genetic variability is exceptional (πneu = 16.4%), other obligatorily outbreeding species of Caenorhabditis such as C. remanei and C. sp. 5 also appear hyperdiverse (32). C. brenneri has >150 times the density of polymorphisms as humans and approximately eightfold the density seen in Drosophila melanogaster (2). Literature surveys suggest that neutral nucleotide polymorphism >5% is rare among eukaryotes (2, 5), but is Caenorhabditis truly unusual? Sea squirts (e.g., Ciona savignyi) and some fruit flies (e.g., Drosophila recens) meet this threshold of high population variation (20, 33), but most phylogeographic studies use mitochondrial, microsatellite, or ribosomal subunit markers that do not easily permit direct population genetic comparisons across taxa. Accelerating application of genomic approaches to neglected organisms in molecular ecology will likely discover many more hyperdiverse species (32, 34).
Morphologically, C. brenneri is nearly indistinguishable from the related C. remanei—even in male reproductive structures (7)—despite the great molecular divergence between the species and the high molecular diversity within each species (25). The perseverance of form despite regulatory divergence (35) provides an intriguing contradistinction to popular views about the centrality of gene regulation in morphological disparity (36). This morphological conformity could be due to exceptionally effective stabilizing selection on nematode roundworm morphology as a consequence of large effective population size, providing a striking disconnect between morphological and molecular differentiation among species (37). Indeed, strong contemporary purifying selection is evident for coding sequences (average πrep/πneu = 0.026), implying that >97% of changes to amino acid replacement sites are detected by selection and purged from or fixed in the population. The modest genetic differentiation we calculated between continents does not preclude local adaptation involving particular loci, although we observed no fixed differences between strains isolated on opposite sides of the globe.
The extreme potency of selection to sift among mutations implicates a powerful means of exploiting population polymorphism in this species to characterize the identity and evolution of functional noncoding sequences: the high density of polymorphisms expected for unconstrained sequence permits characterization of constrained regulatory regions with fine resolution (18). The hyperdiversity of C. brenneri thus makes it an exceptional system for practical applications, such as interrogating gene promoter regions for cis-regulatory elements across the genome in addition to investigating evolutionary limits in very large populations (28).
What are the implications for the evolution of genome complexity, given that C. brenneri’s genome has diversity levels and population sizes most comparable to hyperdiverse bacteria (6)—keeping in mind that C. brenneri is a sexually reproducing animal with complex development? Theory predicts a pivotal role of population size differences in the evolution of many aspects of genome architecture, including the evolution of introns, transposable elements, gene regulation, mutation rates, and interactome complexity (5). However, the dearth of exemplar eukaryotes with large population sizes commensurate with prokaryotes limits some tests of theory about the role of population size in the evolution of complexity. C. brenneri and other hyperdiverse species help fill this gap and galvanize new and stronger tests of theory about genome evolution.
Methods
Nematode Isolation from Natural Populations.
We analyzed 33 isofemale strains of C. brenneri, mostly isolated from northern South America (French Guiana, Colombia) and India, among other tropical localities in the world (Table S1). Ten strains from Shyamnagar were used as a local sample and compared with six French Guiana samples in genetic differentiation analyses. We determined species identity from diagnostic morphology, mating tests with known species (fertile F1 progeny with strain CB5161), and sequences of 18S RNA (10).
Interpopulation Crosses.
To test for the presence of cryptic species, we conducted interpopulation F1 and F2 crosses of strains within and between eastern India and French Guiana. Intrapopulation crosses were performed between three distinct strains of each population to avoid any inbreeding depression confounds (38), including 8–11 replicates for intrapopulation crosses and 19 replicate interpopulation crosses to test for differential production of F1 and F2 offspring (JU1813, JU1816, NIC42, VX0038, VX0044, and VX0049). In each cross, one virgin female was allowed to mate with six males for 24 h on 35-mm-diameter dishes spotted with Escherichia coli strain OP50 on nematode growth medium (NGM)-lite agar, after which males were picked off and discarded, with progeny counted from daily transfers of the mother. All crosses were incubated at 25 °C.
Molecular Methods, Sequence Alignment, and Primary Analysis.
We amplified genomic DNA from a single male worm of each isofemale line as described (21, 39), with subsequent locus-specific amplification by PCR and sequencing of both strands (Table S2). Sequence quality for the 14 kb across 23 loci was confirmed by eye and manually aligned and edited with the programs SEQUENCHER 4.10 and BioEdit 7.0.9. Five loci are predicted to be X-linked, with the rest being autosomal, having been selected arbitrarily from gene annotations that have long exons. Putative X-linked and autosomal loci did not differ significantly in polymorphism metrics and so were analyzed together without adjusting diversity values for X-linked loci; a 4/3 adjustment for the smaller Ne of sex chromosomes would elevate diversity estimates further, making our calculations conservative with respect to our conclusions. Given the high heterozygosity, repeated failures in PCR amplification and/or sequencing yielded missing data for 1–6 strains for 12 of the loci (mean sample size = 31.8 strains). Exclusion of these sequences may downwardly bias estimates of diversity, which is conservative with respect to our conclusions. After removing primer and short intron sequences, the program PHASE v 2.1.1 was used to generate sets of the most probable haplotypes for sequences with heterozygous base calls (40). Custom Perl scripts took files generated by SeqPHASE as input (41) to randomly pick one haplotype per strain, for each locus, according to local, pooled, and scattered sampling schemes (see below).
Sampling Schemes and Population Genetic Analysis.
We conducted population genetic analyses for C. brenneri strains using three sampling schemes to better interpret the demographic implications of the data (24, 42). First, in the pooled scheme, we pooled all 33 strains from all of the localities. Second, we used a local population scheme, in which we considered strains only within the Shyamnagar, India, locality that had the highest sample size. For the third scattered sampling scheme, we selected just a single strain from each sampling locality for analysis (43). We constructed 1,000 replicates of these scattered samples by randomly selecting one strain from each of the 15 localities in the dataset. The rationale behind choosing such a scattered sample approach is that it recapitulates a standard neutral coalescence process if each locality represents one of many demes (43).
PHASE v. 2.1.1 was unable to resolve unique pairs of haplotypes for all loci, so, for a given strain included in a sample, we randomly drew a haplotype for that strain in accordance with the PHASE-inferred probabilities for the corresponding locus. For all of the sampling schemes, one haplotype was selected randomly per locus per strain. This procedure was repeated 1,000 times for both pooled and scattered sampling schemes and 50 times for each local sample, to ensure that any particular selection of haplotypes or strains does not skew inference of population genetics summary statistics. We used the mean of these random selections of haplotypes as our estimate of population genetic metrics for a given sampling scheme. Because random selection of haplotypes generally gave very similar diversity and site frequency spectrum estimates, we restricted analysis of population differentiation and selection intensity on synonymous sites to a single arbitrarily chosen replicate.
We used Polymorphorama (44) to compute population genetics metrics for each random selection of haplotypes, including Jukes-Cantor adjusted measures of average nucleotide polymorphism at synonymous sites (πsyn-JC), replacement sites (πrep), and the site frequency spectrum for synonymous sites (Tajima’s DTaj). LDHat v. 2.2 was used to calculate the population recombination rate (ρ) and the minimum number of recombination events per locus (Rmin) (45, 46). Further, distributions of di-, tri-, and tetra-allelic polymorphic sites were identified for each replicate sample of haplotypes with LDHat. See Tables S3–S7 for summaries of these population genetic metrics.
For several analyses, we restricted our computations to a single replicate sample of haplotypes per locus. We quantified with DnaSP 5.10 the population genetic differentiation between the two local samples with the most strains (6 strains from French Guiana and 10 strains from Shyamnagar), computing FST and NST (Jukes-Cantor–corrected FST; Table S6). Neighbor-network trees were constructed using SPLITSTREE 4.10 (47) using all strains in the pooled sampling scheme, for each locus separately and also from a concatenation of all loci to explicitly display recombination in the ancestry of the strains by reticulating genealogies (Fig. S1). Nucleotide distances in genealogical networks (Jukes-Cantor corrected) exclude gaps and include only third base pair codon positions. Note that the star-shaped genealogical network topologies are generally not consistent with accidental sequencing of gene paralogs; two cases with atypical topologies (Cbn-nmy-1 and Cbn-Y25C1A.5) actually have lower than average diversity, suggesting that all haplotypes do correspond to the same gene. We used JMP v. 10 to fit a spline (λ = 10,000) to the decay of linkage disequilibrium (r2) with distance between polymorphic sites for a scattered sample replicate, fitting the linkage disequilibrium decay for each locus individually and for all loci combined. Distances between polymorphic sites were computed in DnaSP.
Estimates of nucleotide diversity for Fig. 1 were obtained from literature-reported values for species other than C. brenneri, including humans (48), Arabidopsis thaliana (49), and D. melanogaster (50). Joshua Shapiro (Bryn Mawr College, Bryn Mawr, PA) provided the value for C. elegans from ref. 19. Diversity for Ciona savignyi is that calculated in ref. 32 from Jukes-Cantor–corrected estimates of πsyn, also adjusted to account for codon use bias, derived from alignments in ref. 20.
Estimating Effective Population Size and Selection Intensity on Synonymous Sites.
We estimated effective population size for C. brenneri from neutral polymorphism inferred from scattered samples for each of the 17 loci with the lowest codon bias, assuming population equilibrium (πneu = θ = 4Neμ) and mutation rate estimates from C. elegans (μ = 2.7 × 10−9 per site per generation) (26).
Using the C. elegans optimal codon table implemented in DnaSP 5.10 on one random sample from the scattered and Shyamnagar local population sampling schemes, we identified preferred-unpreferred (PU), unpreferred-unpreferred (UU), and preferred-preferred (PP) variants for synonymous site polymorphisms. We then estimated the selection intensity (4Nes) using the method from refs. 39 and 51. For the local sample from Shyamnagar, the maximum likelihood estimate was 4Nes = 0.4375 (2lnL interval, 0.24–0.65). Analysis of a scattered sample yielded a similar result (4Nes = 0.4125; 2lnL interval, 0.25–0.58; Table S7).
Analyses of Heterozygosity in the C. brenneri Draft Genome Assembly.
To identify allelic haplotypes present within the C. brenneri genome assembly (WS230 wormbase release), we aligned the predicted coding sequences in the assembly to each other using the cross_match program in the phred/phrap software suite with default parameters (52). We then used custom Perl scripts to discard self-hits and alignments involving genes belonging to the same contig, as these do not likely represent allelic variants of the same gene. Next, we used genic microsynteny to eliminate duplicated DNA segments from the dataset by requiring alignments to involve >90% of the length of one or both genes identified by cross_match, and then computed the fraction of genes on a given contig involved in a cross_match alignment. Because microsynteny is likely to be conserved between contigs representing allelic variants, and not for spurious pairs involving distinct members of multigene families, we only considered those contig pairs in which at least half of the genes aligned between the contig pair. Finally, we extracted the alignments for genes in the retained contig pairs and filtered them to reject those involving differing frames or reverse complements of each other.
These steps generated 9,179 in-frame alignments of coding genes, derived from 1,035 contigs in the C. brenneri WS230 assembly. We passed these sequence alignment pairs through the Polymorphorama program (44), which successfully computed Jukes-Cantor–corrected pairwise distances at synonymous sites (πsyn-JC) for 5,747 of them; the remaining sequences that failed this step likely represent spurious sequences for our purposes, such as sequences from multigene families. We further filtered this collection of sequence pairs to obtain a conservative set of 3,449 genes for which we conclude have two allelic haplotypes represented in the draft genome assembly. Specifically, we excluded sequences with <100 synonymous sites, with >2% pairwise differences at replacement sites, or with the ratio of replacement: synonymous site differences were >20%. Although some of these excluded sequence pairs might represent true alleles, some may represent duplicated sequences or multigene families; in general, sequences that were excluded based on these criteria tended to have higher numbers of pairwise differences and would therefore increase our estimates of nucleotide diversity even further.
We also computed the effective number of codons (ENC) metric of codon use bias for the sequences using CodonW (http://codonw.sourceforge.net). Among the full set of genes, 3,265 had relatively weak codon bias (ENC > 39) and were used to report πsyn-JC. We also performed a linear regression of πsyn-JC on ENC for the full set of genes. We then applied the resulting prediction equation to estimate πneu = 0.164 at the point where codon use is unbiased (ENC = 61) and therefore translational selection on synonymous sites is no longer expected to bias polymorphism levels downward (π = −0.064464 + 0.003748 × ENC; Fig. S2).
Supplementary Material
Acknowledgments
We thank M.-A. Felix, C. Braendle, and the Caenorhabditis Genetics Center for providing strains. We are grateful to Dr. S. P. Banik, S. Dey, and K. Chakra for assistance with field collections and to Dr. S. Khowala for providing laboratory facilities in India. This work was supported by a grant from the Natural Sciences and Engineering Research Council of Canada.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. KF004311–KF005040).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1303057110/-/DCSupplemental.
References
- 1.Lewontin RC. The Genetic Basis to Evolutionary Change. New York: Columbia Univ Press; 1974. [Google Scholar]
- 2.Leffler EM, et al. Revisiting an old riddle: What determines genetic diversity levels within species? PLoS Biol. 2012;10(9):e1001388. doi: 10.1371/journal.pbio.1001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kimura M. The Neutral Theory of Molecular Evolution. New York: Cambridge Univ Press; 1983. [Google Scholar]
- 4.Lynch M. The origins of eukaryotic gene structure. Mol Biol Evol. 2006;23(2):450–468. doi: 10.1093/molbev/msj050. [DOI] [PubMed] [Google Scholar]
- 5.Lynch M. The Origins of Genome Architecture. Sunderland, MA: Sinauer; 2007. [Google Scholar]
- 6.Schloissnig S, et al. Genomic variation landscape of the human gut microbiome. Nature. 2013;493(7430):45–50. doi: 10.1038/nature11711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sudhaus W, Kiontke K. Comparison of the cryptic nematode species Caenorhabditis brenneri sp. n. and C. remanei (Nematoda: Rhabditidae) with the stem species pattern of the Caenorhabditis Elegans group. Zootaxa. 2007;1456:45–62. [Google Scholar]
- 8.Barrière A, et al. Detecting heterozygosity in shotgun genome assemblies: Lessons from obligately outcrossing nematodes. Genome Res. 2009;19(3):470–480. doi: 10.1101/gr.081851.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jovelin R. Rapid sequence evolution of transcription factors controlling neuron differentiation in Caenorhabditis. Mol Biol Evol. 2009;26(10):2373–2386. doi: 10.1093/molbev/msp142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kiontke KC, et al. A phylogeny and molecular barcodes for Caenorhabditis, with numerous new species from rotting fruits. BMC Evol Biol. 2011;11(1):339. doi: 10.1186/1471-2148-11-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kammenga JE, et al. A Caenorhabditis elegans wild type defies the temperature-size rule owing to a single nucleotide polymorphism in tra-3. PLoS Genet. 2007;3(3):e34. doi: 10.1371/journal.pgen.0030034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palopoli MF, et al. Molecular basis of the copulatory plug polymorphism in Caenorhabditis elegans. Nature. 2008;454(7207):1019–1022. doi: 10.1038/nature07171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghosh R, Andersen EC, Shapiro JA, Gerke JP, Kruglyak L. Natural variation in a chloride channel subunit confers avermectin resistance in C. elegans. Science. 2012;335(6068):574–578. doi: 10.1126/science.1214318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seidel HS, Rockman MV, Kruglyak L. Widespread genetic incompatibility in C. elegans maintained by balancing selection. Science. 2008;319(5863):589–594. doi: 10.1126/science.1151107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rockman MV, Skrovanek SS, Kruglyak L. Selection at linked sites shapes heritable phenotypic variation in C. elegans. Science. 2010;330(6002):372–376. doi: 10.1126/science.1194208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Denver DR, et al. The transcriptional consequences of mutation and natural selection in Caenorhabditis elegans. Nat Genet. 2005;37(5):544–548. doi: 10.1038/ng1554. [DOI] [PubMed] [Google Scholar]
- 17.Cutter AD, Dey A, Murray RL. Evolution of the Caenorhabditis elegans genome. Mol Biol Evol. 2009;26(6):1199–1234. doi: 10.1093/molbev/msp048. [DOI] [PubMed] [Google Scholar]
- 18.Boffelli D, et al. Intraspecies sequence comparisons for annotating genomes. Genome Res. 2004;14(12):2406–2411. doi: 10.1101/gr.3199704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andersen EC, et al. Chromosome-scale selective sweeps shape Caenorhabditis elegans genomic diversity. Nat Genet. 2012;44(3):285–290. doi: 10.1038/ng.1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Small KS, Brudno M, Hill MM, Sidow A. Extreme genomic variation in a natural population. Proc Natl Acad Sci USA. 2007;104(13):5698–5703. doi: 10.1073/pnas.0700890104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dey A, Jeon Y, Wang G-X, Cutter AD. Global population genetic structure of Caenorhabditis remanei reveals incipient speciation. Genetics. 2012;191(4):1257–1269. doi: 10.1534/genetics.112.140418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woodruff GC, Eke O, Baird SE, Félix MA, Haag ES. Insights into species divergence and the evolution of hermaphroditism from fertile interspecies hybrids of Caenorhabditis nematodes. Genetics. 2010;186(3):997–1012. doi: 10.1534/genetics.110.120550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baird SE. Haldane’s rule by sexual transformation in Caenorhabditis. Genetics. 2002;161(3):1349–1353. doi: 10.1093/genetics/161.3.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Städler T, Haubold B, Merino C, Stephan W, Pfaffelhuber P. The impact of sampling schemes on the site frequency spectrum in nonequilibrium subdivided populations. Genetics. 2009;182(1):205–216. doi: 10.1534/genetics.108.094904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cutter AD. Divergence times in Caenorhabditis and Drosophila inferred from direct estimates of the neutral mutation rate. Mol Biol Evol. 2008;25(4):778–786. doi: 10.1093/molbev/msn024. [DOI] [PubMed] [Google Scholar]
- 26.Denver DR, et al. A genome-wide view of Caenorhabditis elegans base-substitution mutation processes. Proc Natl Acad Sci USA. 2009;106(38):16310–16314. doi: 10.1073/pnas.0904895106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palstra FP, Ruzzante DE. Genetic estimates of contemporary effective population size: What can they tell us about the importance of genetic stochasticity for wild population persistence? Mol Ecol. 2008;17(15):3428–3447. doi: 10.1111/j.1365-294x.2008.03842.x. [DOI] [PubMed] [Google Scholar]
- 28.Barton N. Understanding adaptation in large populations. PLoS Genet. 2010;6(6):e1000987. doi: 10.1371/journal.pgen.1000987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hermisson J, Pennings PS. Soft sweeps: Molecular population genetics of adaptation from standing genetic variation. Genetics. 2005;169(4):2335–2352. doi: 10.1534/genetics.104.036947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Desai MM, Plotkin JB. The polymorphism frequency spectrum of finitely many sites under selection. Genetics. 2008;180(4):2175–2191. doi: 10.1534/genetics.108.087361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harfe BD, Jinks-Robertson S. DNA mismatch repair and genetic instability. Annu Rev Genet. 2000;34(1):359–399. doi: 10.1146/annurev.genet.34.1.359. [DOI] [PubMed] [Google Scholar]
- 32.Cutter AD, Jovelin R, Dey A. Molecular hyperdiversity and evolution in very large populations. Mol Ecol. 2013;22(8):2074–2095. doi: 10.1111/mec.12281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dyer KA, Charlesworth B, Jaenike J. Chromosome-wide linkage disequilibrium as a consequence of meiotic drive. Proc Natl Acad Sci USA. 2007;104(5):1587–1592. doi: 10.1073/pnas.0605578104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCormack JE, Hird SM, Zellmer AJ, Carstens BC, Brumfield RT. Applications of next-generation sequencing to phylogeography and phylogenetics. Mol Phylogenet Evol. 2013;66(2):526–538. doi: 10.1016/j.ympev.2011.12.007. [DOI] [PubMed] [Google Scholar]
- 35.Yanai I, Hunter CP. Comparison of diverse developmental transcriptomes reveals that coexpression of gene neighbors is not evolutionarily conserved. Genome Res. 2009;19(12):2214–2220. doi: 10.1101/gr.093815.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carroll SB. Evo-devo and an expanding evolutionary synthesis: A genetic theory of morphological evolution. Cell. 2008;134(1):25–36. doi: 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- 37.Barrière A, Gordon KL, Ruvinsky I. Coevolution within and between regulatory loci can preserve promoter function despite evolutionary rate acceleration. PLoS Genet. 2012;8(9):e1002961. doi: 10.1371/journal.pgen.1002961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dolgin ES, Charlesworth B, Baird SE, Cutter AD. Inbreeding and outbreeding depression in Caenorhabditis nematodes. Evolution. 2007;61(6):1339–1352. doi: 10.1111/j.1558-5646.2007.00118.x. [DOI] [PubMed] [Google Scholar]
- 39.Cutter AD. Multilocus patterns of polymorphism and selection across the X chromosome of Caenorhabditis remanei. Genetics. 2008;178(3):1661–1672. doi: 10.1534/genetics.107.085803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68(4):978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Flot JF. seqphase: A web tool for interconverting phase input/output files and fasta sequence alignments. Mol Ecol Resour. 2010;10(1):162–166. doi: 10.1111/j.1755-0998.2009.02732.x. [DOI] [PubMed] [Google Scholar]
- 42.Cutter AD, Wang G-X, Ai H, Peng Y. Influence of finite-sites mutation, population subdivision and sampling schemes on patterns of nucleotide polymorphism for species with molecular hyperdiversity. Mol Ecol. 2012;21(6):1345–1359. doi: 10.1111/j.1365-294X.2012.05475.x. [DOI] [PubMed] [Google Scholar]
- 43.Wakeley J. Nonequilibrium migration in human history. Genetics. 1999;153(4):1863–1871. doi: 10.1093/genetics/153.4.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bachtrog D, Andolfatto P. Selection, recombination and demographic history in Drosophila miranda. Genetics. 2006;174(4):2045–2059. doi: 10.1534/genetics.106.062760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hudson RR, Kaplan NL. Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics. 1985;111(1):147–164. doi: 10.1093/genetics/111.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McVean G, Awadalla P, Fearnhead P. A coalescent-based method for detecting and estimating recombination from gene sequences. Genetics. 2002;160(3):1231–1241. doi: 10.1093/genetics/160.3.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23(2):254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- 48.Perry GH, et al. Comparative RNA sequencing reveals substantial genetic variation in endangered primates. Genome Res. 2012;22(4):602–610. doi: 10.1101/gr.130468.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nordborg M, et al. The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol. 2005;3(7):e196. doi: 10.1371/journal.pbio.0030196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Langley CH, et al. Genomic variation in natural populations of Drosophila melanogaster. Genetics. 2012;192(2):533–598. doi: 10.1534/genetics.112.142018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cutter AD, Charlesworth B. Selection intensity on preferred codons correlates with overall codon usage bias in Caenorhabditis remanei. Curr Biol. 2006;16(20):2053–2057. doi: 10.1016/j.cub.2006.08.067. [DOI] [PubMed] [Google Scholar]
- 52.Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8(3):186–194. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.