Abstract
Directional cell migration is a fundamental process in neural development. In Caenorhabditis elegans, Q neuroblasts on the left (QL) and right (QR) sides of the animal generate cells that migrate in opposite directions along the anteroposterior body axis. The homeobox (Hox) gene lin-39 promotes the anterior migration of QR descendants (QR.x), whereas the canonical Wnt signaling pathway activates another Hox gene, mab-5, to ensure the QL descendants’ (QL.x) posterior migration. However, the regulatory targets of LIN-39 and MAB-5 remain elusive. Here, we showed that MIG-13, an evolutionarily conserved transmembrane protein, cell-autonomously regulates the asymmetric distribution of the actin cytoskeleton in the leading migratory edge. We identified mig-13 as a cellular target of LIN-39 and MAB-5. LIN-39 establishes QR.x anterior polarity by binding to the mig-13 promoter and promoting mig-13 expression, whereas MAB-5 inhibits QL.x anterior polarity by associating with the lin-39 promoter and downregulating lin-39 and mig-13 expression. Thus, MIG-13 links the Wnt signaling and Hox genes that guide migrations, to the actin cytoskeleton, which executes the motility response in neuronal migration.
During neural development, migrating cells respond to extracellular cues and reach their final destinations by navigating along the dorsoventral or anteroposterior (A/P) body axes. Despite important advances that have been made in the identification of guidance cues, their receptors, and the intracellular protein machinery responsible for cell motility (1–4), comparatively little is known about the connection between the signaling pathways and the actin cytoskeleton. The Caenorhabditis elegans Q neuroblasts were born as bilaterally symmetric cells on the left (QL) and right (QR) sides in the posterior body region. These cells divide asymmetrically such that each generates three neurons and two apoptotic cells. QL and its descendants (QL.x) migrate to the posterior, whereas QR and its descendants (QR.x) move to the anterior (Fig. 1 A and B) (5). We developed live fluorescence imaging techniques to document C. elegans Q-cell migration (6, 7).
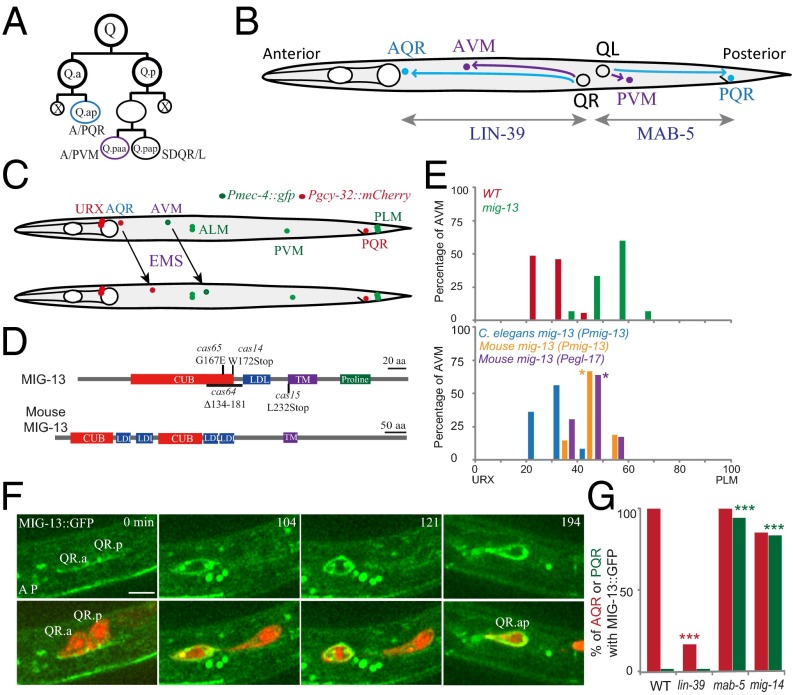
Fig. 1.
MIG-13 in C. elegans Q-neuroblast migration. (A) Q-neuroblast asymmetric divisions generate three neurons and two apoptotic cells (X in black). QL produces PQR, PVM, and SDQL, and QR produces AQR, AVM, and SDQR. (B) QR descendants (QR.x), AQR (cyan circle) and AVM (blue), migrate anteriorly, and the QL descendant (QL.x), PQR (cyan), migrates posteriorly. Lin-39 and mab-5 regulate Q-cell migration (two gray double arrow lines). (C) Genetic screens using zdIs5[Pmec-4::GFP] and casIs35[Pgcy-32::mCherry] for mutations defective in QR.x anterior migration. AVM and PVM (green) or AQR and PQR (red) from Q-cell lineages were marked by cell type-specific promoters. (D) C. elegans MIG-13 and mouse Lrp12 protein domains. The amino acid changes or deletion in mig-13 mutant alleles (cas14, cas15, cas64, cas65) are indicated. (E) Quantifications of AVM position of mig-13 (mu225) and the rescue experiments. Genetic backgrounds were indicated on the Upper Left. n = 60–100 from a single experiment. Statistical analysis is shown in Fig. S3B. (F) Still images of muIs62 [MIG-13::GFP] (green) during WT QR.x migration. Q-cell plasma membrane and chromosomes are imaged by mCherry. Merged images are shown at Bottom. The cell name is adjacent. The anterior of the cell is on the Left. Time is in minutes. (Scale bar, 5 µm.) (G) Percentage of Q.ap cells expressing mig-13 in lin-39 (n1760), mab-5 (e2088), or mig-14 (mu71) mutants. n = 12–30 from a single experiment. ***P < 0.001 by Student t test.
Distinct signaling pathways sequentially control Q-cell migration. Several transmembrane proteins, DPY-19, MIG-21, UNC-40/DCC (Deleted in Colorectal Cancer), and NF-κB-inducing kinase (NIK) MIG-15, regulate the initial Q-neuroblast polarization, and mutations of these genes reduced Q-neuroblast polarization (8–11). The C. elegans Wnt/β-catenin pathway activates the expression of the Antennapedia-like Homeobox(Hox) gene mab-5, which is necessary and sufficient to ensure the posterior migration of QL.a (the anterior descendant of QL) and QL.ap (the posterior descendant of QL.a) (12–17). Another Hox gene lin-39/Sex combs reduced, in combination with its two cofactors, CEH-20/Extradenticle and UNC-62/Homothorax, promotes QR.x anterior migration (18–20). MAB-5 and LIN-39 function cell-autonomously to guide cell migration, but their targets are largely unknown.
MIG-13, a transmembrane protein, regulates QR.x anterior migration (21). Although the loss of function of UNC-71, a disintegrin and metalloprotease protein, partially suppresses the anterior migration defects of QR.x in mig-13 mutants (22), the mechanisms by which MIG-13 functions in cell migration remain elusive. A previous study showed that the C. elegans mig-13 is not expressed in the migrating cell and that mig-13 acts nonautonomously in cell migration (21). However, the mouse low density lipoprotein receptor-related protein 12 (Lrp12) gene, the worm mig-13 homolog, is expressed in the ventrally directed tangential migrating neurons in the preplate during corticogenesis (23), suggesting that Lrp12 may function autonomously in migrating neurons. Further experiments are thus required to distinguish whether MIG-13 and its homologs play autonomous or nonautonomous roles in neuronal migration.
This study provided multiple independent lines of evidence that C. elegans MIG-13 localizes on the plasma membrane of migrating QR.x and that mig-13 acts autonomously to direct the anterior neuronal migration. We showed that MIG-13 regulates the asymmetric distribution of actin cytoskeleton in the leading migratory edge. Using a combination of ChIP-sequencing (ChIP-seq) data analysis, genetic, and live-imaging approaches, we found that MIG-13 is a cellular target of the canonical Wnt signaling pathway and the Hox genes and that MIG-13 links extracellular signaling to the A/P polarity in cell migration.
Results and Discussion
MIG-13, a Conserved Transmembrane Protein, Regulates QR.x Anterior Migration.
We reasoned that loss-of-function mutations in downstream targets of LIN-39 and MAB-5 might phenocopy mutations in the transcription factors themselves. To identify the targets, we performed forward genetic screens for mutants that specifically reduce QR.x anterior migration but not QL.x posterior migration, and we isolated four alleles of mig-13 (Fig. 1 C and D). MIG-13 contains a C1r/C1s, Uegf, Bmp1 (CUB) domain, a low-density lipoprotein (LDL) receptor repeat, a transmembrane domain, and a proline-rich cytoplasmic tail (Fig. 1D). Among the four identified mig-13 alleles, cas65 changed a glycine 167 to glutamic acid and reduced the AVM neuron (differentiated from QR.paa) anterior migration, as did other alleles with premature STOP codons (cas14 and cas15) or a deletion (cas64) or the null allele (mu225) isolated in the previous screen (Fig. 1 D and E, and Fig. S1A) (21), indicating that G167 residue in the CUB domain is important for MIG-13 function.
The mouse homolog of mig-13, Lrp12, has two CUB domains and four LDL motifs (Fig. 1D) (23). We examined whether Lrp12 could rescue Q-cell migration defects in mig-13 (mu225) worms. We expressed the mouse Lrp12 cDNA using the mig-13 gene promoter (Pmig-13) or Pegl-17 promoter that activates gene expression in migrating Q cells. We found that both transgenes could partially move the position of Q-cell progeny AVM to the anterior in mig-13 mutants (Fig. 1E), suggesting that Lrp12 and mig-13 may have conserved function but they are not fully interchangeable.
MIG-13 Cell Autonomously Regulates Q-Cell Migration.
We first studied the cellular localization of MIG-13 during Q-cell migration. A previous study reported that the fluorescence of a functional MIG-13::GFP expressed by the mig-13 promoter could not be detected in Q cells (21). Using improved optics, we reexamined whether mig-13 is expressed in Q cells. We used genetic cross to introduce mCherry-labeled Q-cell markers into the MIG-13::GFP strain [muIs62 (Pmig-13::mig-13::gfp)], constructed in the original study. After identifying Q cells by the mCherry signal, we performed time-lapse imaging of MIG-13::GFP dynamics during migration. We did not observe any MIG-13::GFP fluorescence in QL.x (Fig. S1B, n = 11; Fig. S1 C and D, Right). However, we found that MIG-13::GFP is expressed in the migrating QR.x, and its GFP fluorescence is present on the cell surface (Fig. 1F, n = 23; Movie S1). Our time-lapse imaging revealed dynamic changes of mig-13 expression during migration: MIG-13::GFP was detectable at the initial stage of QR.x migration (Fig. 1F, 0 min); MIG-13::GFP gradually increased its fluorescence during migration and reached its maximum level at the end of migration (Fig. 1F, 194 min, Movie S1). We found that MIG-13::GFP fluorescence in QR.a/ap is stronger than that observed in QR.p/pa (Fig. 1F and Fig. S1 C and D, Left). Interestingly, QR.a/ap always migrates faster and further than QR.p/pa, indicating that Q cells with high migratory capacity produce more MIG-13. Consistent with this notion, we found that QR.ap migrated at 37.5 ± 8.1 µm/h (n = 11) in mig-13::gfp transgenic animals, which was faster than QR.ap migration (24.6 ± 6.3 µm/h; n = 27) in WT animals (P < 0.001 by Student t test). We confirmed the presence of MIG-13::GFP in QR.x using an independent transgenic integrant (muIs42) and our newly constructed transgenic line (Fig. S1 C and D, and Movie S2). These data argued against a cell-nonautonomous mechanism of mig-13.
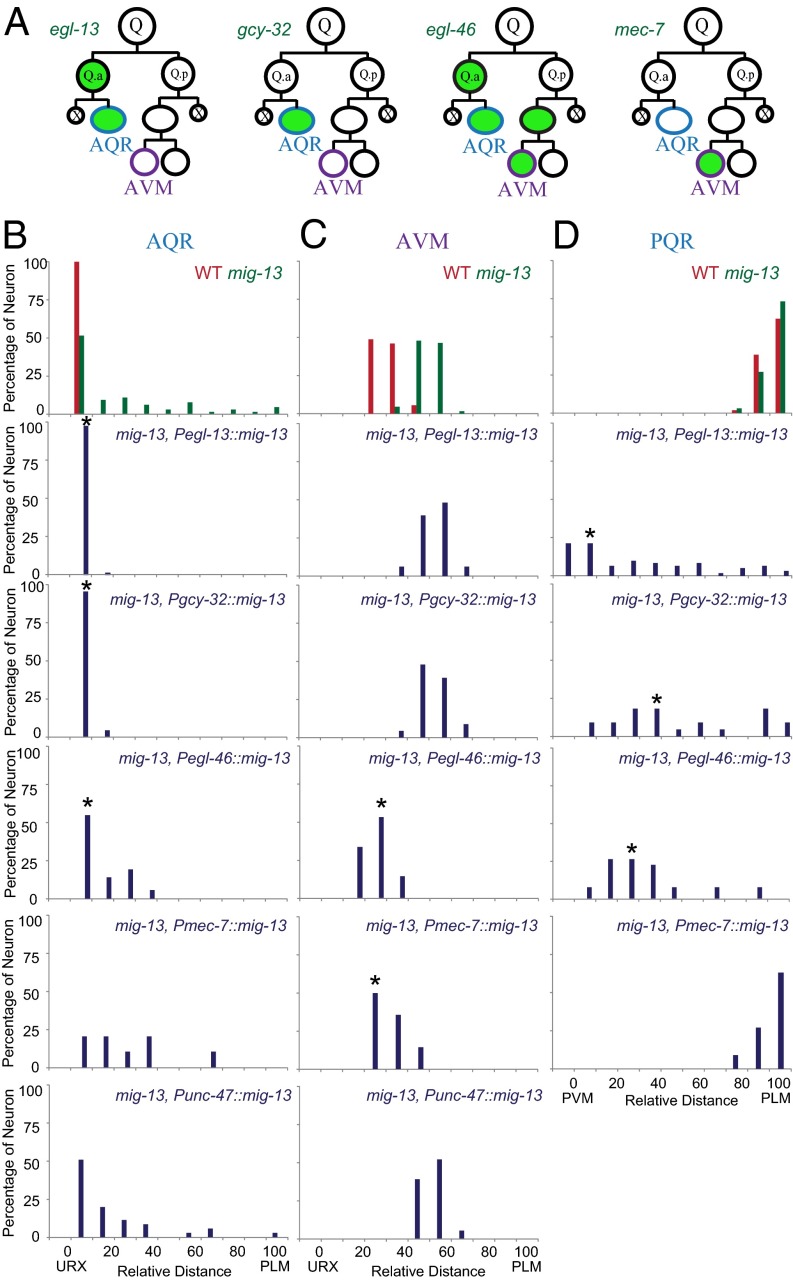
We next performed cell type-specific rescue experiments to examine the cell-autonomous function of mig-13 in cell migration. We used four distinct promoters to express the mig-13a isoform, the longest isoform of mig-13, in different Q-cell progenies: the egl-13 promoter (Pegl-13) specifically expresses in the Q.a lineage (Fig. S1F and Fig. S2E), Pgcy-32 expresses in Q.ap, Pegl-46 expresses in Q.a/Q.ap and Q.pa/Q.paa, and Pmec-7 expresses in Q.paa (24–26) (Fig. 2A). By quantifying the final positions of Q-cell progenies, we showed that AQR migration defects in mig-13 mutants can be completely or partially rescued by the expression of mig-13 under the control of QR.a/ap-specific promoters, but not QR.p/paa promoter (Fig. 2 B and C, and Fig. S3 C and D). The final positions of AVM in mig-13 mutants were consistently restored if mig-13 was expressed in AVM but not in AQR (Fig. 2 B and C, and Fig. S3 C and D). The previous study suggested that mig-13 acted in the motor neurons to promote QR.x migration (21). To examine this possibility, we expressed mig-13a using a motor neuron-specific promoter, Punc-47, in mig-13 mutants (27), and we found that Punc-47::mig-13 transgene failed to rescue the migration defects of AQR and AVM (Fig. 2 B and C). Our results indicated that mig-13 acts inside of migrating Q cells to regulate the anterior QR.x migration.
Fig. 2.
Q-cell position in mig-13 cell-specific rescue animals. (A) Schematics show four cell type-specific promoters. Green indicates expression from the promoter. Q.ap differentiates into A/PQR and Q.paa differentiates into A/PVM. (B–D) Quantifications of AQR (B), AVM (C), and PQR (D) position in mig-13-related genetic backgrounds (indicated on Top Right). n = 20–50 for each measurement. Statistical analysis is in Fig. S3 C and D. *P < 0.01 by Student t test.
To further examine the autonomy of MIG-13, we showed that the QL.ap/PQR posterior migration was reduced by ectopically expressing mig-13 in this cell (Fig. 2D). MIG-13 is not expressed in QL.ap, nor is it essential for QL.ap migration (Figs. 1G and 2D). When mig-13 was ectopically expressed in QL.ap, the final positions of PQR were spread between PVM and PLM, which are the landmarks of QL.ap birthplace and migration destination (Fig. 2D and Fig. S3D). Expression of mig-13 in PVM did not cause any migration defects in PQR (Fig. 2D and Fig. S3D), reinforcing a cell-autonomous function for mig-13 in cell migration. Thus, our live-imaging and genetic analyses showed that mig-13 acts cell autonomously in cell migration.
MIG-13 Controls Cell Polarity and the Actin Cytoskeleton During Cell Migration.
We studied the cellular mechanism by which MIG-13 regulates cell migration by visualizing the actin cytoskeleton in Q cells. We constructed a transgenic C. elegans line that expresses GFP-tagged COR-1, an ortholog of the mammalian actin-binding protein Coronin that localizes to the leading edge of migrating cells (28). C. elegans COR-1 and mouse Coronin 1B share 52% identities and 69% positives (BLAST, e−120), and they both have a conserved surface-exposed arginine residue on the N-terminal β propellers that is required for F-actin binding (28). We confirmed that COR-1::GFP is enriched at the leading edge of the Q cell, and our quantitative fluorescence analysis showed that the COR-1::GFP intensity was ∼1.7-fold as strong in the leading edge of migrating Q cells as in the lagging edge (Fig. 3 A and D, and Movie S3). A Rho family GTPase (MIG-2) and an integrin α subunit (INA-1) form two distinct cellular pathways to regulate Q-cell migration; MIG-2 controls the polarity but INA-1 contributes to the polarity-independent cellular process (6). We showed that COR-1::GFP polarity was disrupted in mig-2 mutants but was normal in ina-1 worms (Fig. S2 A–C and Movies S4, S5, and S6), indicating that COR-1::GFP properly illuminated the asymmetric distribution of actin cytoskeleton in migrating cells.
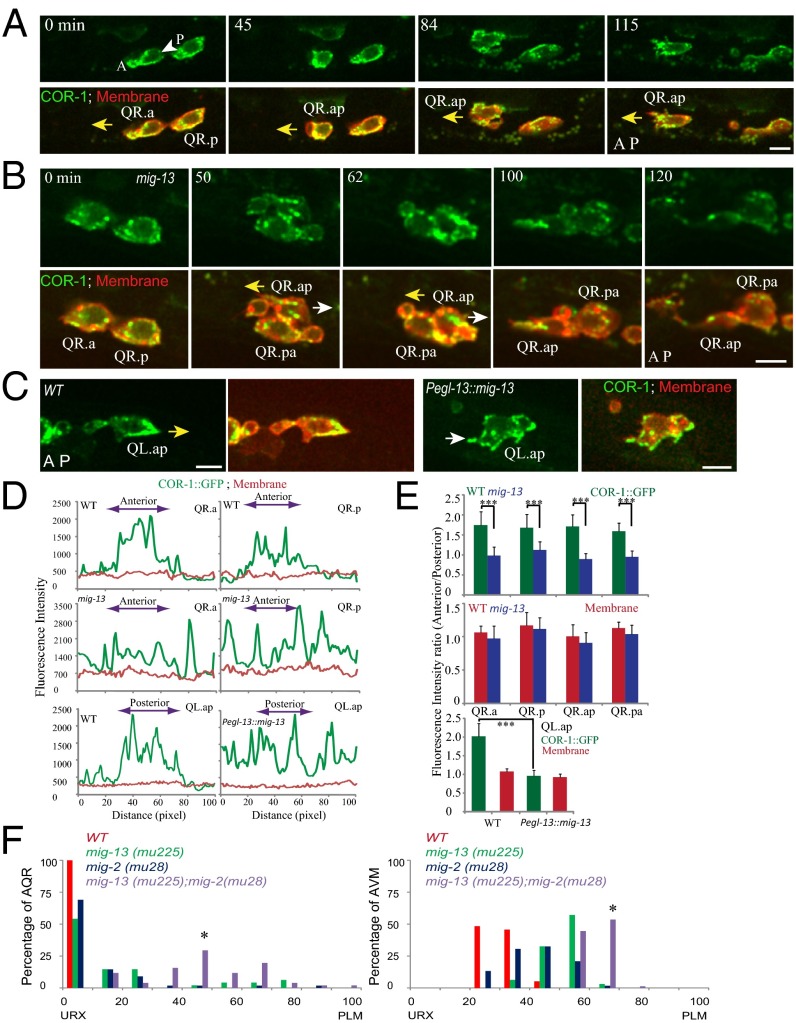
Fig. 3.
Dynamics of COR-1(Coronin)::GFP in Q cells. (A–C) Still images show the localization of casIs49 [COR-1::GFP] (green) in WT QR.x (A), mig-13 (cas15) QR.x (B), or Pegl-13::mig-13 QL.ap (C; more frames in Fig. S2E). The Q-cell plasma membrane is visualized with mCherry. The yellow arrows show the normal migration direction of QR.x or QL.ap. The white arrows show the defective migration direction, either posterior migration of QR.ap (B) or the anterior process of QL.ap (C, Right). Time is in minutes. (Scale bars, 5 µm.) (D) Traces are line-scan intensity plots (in arbitrary units) of the casIs49 [COR-1::GFP] (green) and Myri-mCherry (red) signal around the periphery of QR.a, QR.p, or QL.ap in WT or mig-13 (cas15) or mig-13 ectopic expression animals (Pegl-13::mig-13). Using QR.a in the Top Left of A as an example, the trace starts from the posterior of QR.a (P, white arrowhead) and moves counterclockwise along the cell periphery to the anterior (A) and back to the posterior (P). The genotype (Top Left) and cell type (Top Right) are noted. Double-headed arrows show QR.x anterior or QL.ap posterior. (E) Statistical analysis of COR-1::GFP asymmetric distribution in the anterior and posterior in mig-13 (cas15) QR.x (Top) or QL.ap with mig-13 ectopic expression worms (Pegl-13::mig-13) (Bottom). Error bars indicate SEM. ***P < 0.001 by Student t test. (F) Quantifications of AQR and AVM positions in mig-13 and mig-2-related genetic backgrounds (indicated on Top Right). Statistical analysis is in Fig. S4B; *P < 0.01 by Student t test.
We next quantified COR-1::GFP fluorescence in both WT cells and mig-13 mutants. Although COR-1::GFP showed significant enrichment in the leading edge in WT (Fig. 3 D, Top, and E, Top, for statistical analysis), COR-1::GFP did not show asymmetry in the migrating Q cells of mig-13 mutants (Fig. 3 B and D, Middle, and E, Top, and Movie S7), indicating that MIG-13 regulates the asymmetric distribution of the actin cytoskeleton. The membrane-bound mCherry control was distributed evenly in the QR.x of WT and mig-13 mutants (Fig. 3 A and B, and D and E). Strikingly, in certain mig-13 animals, QR.ap even switched its anterior migration (yellow arrow, correct direction) to the posterior (Fig. 3B, 50 and 62 min; white arrow, wrong direction, Movie S7). We found that COR-1::GFP was not concentrated to the posterior of the cell and that the QR.ap moved posteriorly for less than 5 µm and then turned back to the anterior (Fig. 3B, 100 min), suggesting that COR-1 asymmetry is important for the persistent directional migration. In 3 of 15 analyzed QR.ap cells in mig-13 worms, we observed the complete lack of COR-1 asymmetry and the randomized migration. As a result, these cells severely reduced their anterior migration and stayed close to QR.pa near their birthplaces (e.g., QR.ap in Fig. 3B, 120 min). The rest (12) of the QR.ap cells continued the anterior migration with the reduced COR-1 asymmetry, and their final positions were more anterior than those with the complete loss of COR-1 asymmetry, indicating that COR-1 asymmetry contributes to the QR.ap final position.
Given that mig-2 and mig-13 regulate the QR.x polarity, we performed double-mutant analysis to study their genetic interactions. Using null alleles of mig-2(mu28) and mig-13(mu225), we showed that the double mutations significantly enhanced the AQR and AVM migration defects compared with the single mutation (Fig. 3F and Fig. S4B). The additive defects were also observed in the double mutant constructed using the mig-2 constitutive active allele rh17 and mig-13(mu225) (Fig. S4 A and B). Consistently, we did not detect any change of MIG-2::GFP distribution in mig-13(mu225) or the change of MIG-13::GFP in mig-2(mu28) (Fig. S4C). Our results demonstrated that mig-2 and mig-13 function in two parallel pathways to regulate cell polarity during QR.x anterior migration.
We further examined whether an ectopically expression of MIG-13 could disrupt the organization of the actin cytoskeleton in PQR/QL.ap. COR-1::GFP normally accumulated to the posterior of QL.ap, and its intensity was approximately twice as strong in the leading edge as in the lagging edge (Fig. 3 C–E and Fig. S2D and Movie S8). When mig-13 was ectopically expressed, QL.ap still initiated its posterior migration (Fig. S2E, n = 10). However, QL.ap randomly extended cell processes to the anterior over time (Fig. 3 C–E and Fig. S2E and Movie S9). By examining COR-1::GFP, we found that the GFP fluorescence was equally distributed across the QL.ap membrane (Fig. 3 C–E and Fig. S2E), indicating that an ectopic gain of MIG-13 disrupted the asymmetric localization of the actin cytoskeleton toward the posterior.
LIN-39 Promotes mig-13 Expression During QR.x Anterior Migration.
We studied the pathway that activates MIG-13 in QR.x anterior migration. The Hox gene lin-39 promotes QR.x anterior migration (18). To examine whether LIN-39 could promote mig-13 expression, we compared the expression levels of Pmig-13::mig-13::gfp in the QR.ap of WT and lin-39 mutants. The MIG-13::GFP fluorescence was visible in the WT QR.ap (100%, n = 15) but not detectable in 75% of the QR.ap cells of lin-39 mutants (Fig. 1G and Fig. S1E; n = 12). We found that, in lin-39 worms, QR.ap without MIG-13::GFP expression stayed close to QR.pa near their birthplaces (Fig. S1I, Left) and that QR.ap expressing MIG-13::GFP migrated to the position more anterior than QR.pa (Fig. S1I, Right). Thus, LIN-39 positively regulates mig-13 expression, which is essential for QR.ap anterior migration.
We examined whether QR.ap polarity was affected in lin-39 (n1760) worms. To visualize the QR.ap cell polarity, we imaged COR-1::GFP dynamics in QR.ap of lin-39 mutants. The WT QR.ap cell polarized to the anterior by extending the lamellae toward the animal head (Fig. 3A). In eight of nine examined lin-39 worms, QR.ap was lack of COR-1 asymmetry and randomly extended processes to the dorsal/ventral side of the animal (Fig. 4A and Movie S10), indicating that LIN-39 regulates the anterior polarization of the actin cytoskeleton in QR.ap.
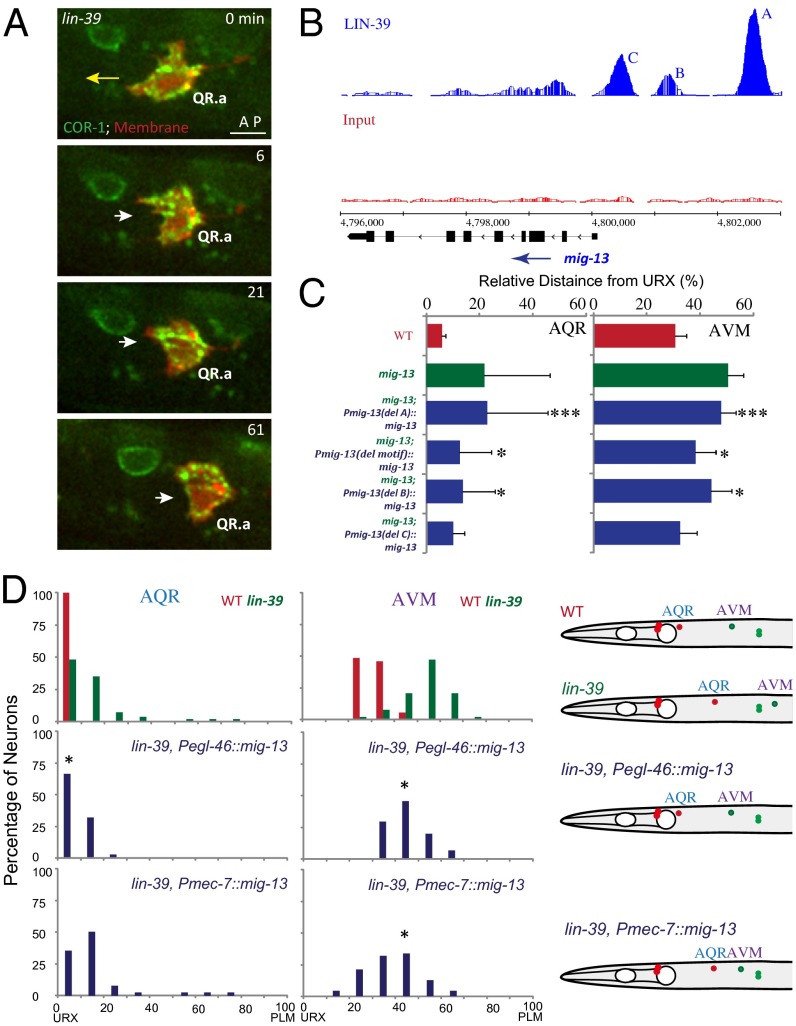
Fig. 4.
LIN-39 regulates the QR.x anterior polarity by promoting mig-13 expression. (A) Still images show QR.ap migration in lin-39(n1760) mutants. COR-1::GFP in green and Q-cell plasma membrane marked with mCherry in red. The long yellow arrow shows the anterior migration of WT QR.ap. The short white arrows show defects of QR.ap polarity in lin-39 (n1760) mutants. Time is in minutes. (Scale bar, 5 µm.) (B) ChIP-seq data show that LIN-39 binds to three sites (A–C, blue, Upper) in the mig-13 promoter. Input (the negative control in ChIP-Seq) is in red, middle. The mig-13 gene model is in black (Bottom). (C) AQR and AVM positions in WT, mig-13 (mu225), and mig-13 rescue experiments with LIN-39 binding site deletion. The position and range of binding site A, B, C, and motif (consensus DNA motif for LIN-39 target genes) are in the text. ***, nonrescue, P < 0.001; *, partial rescue, P < 0.01 by Student t test. (D) AQR (Left) and AVM (Center) positions in WT, lin-39 (n1760), and mig-13 rescue backgrounds. Schematics (Right) of AQR and AVM positions. *P < 0.01 by Student t test.
We addressed whether LIN-39 directly promotes mig-13 expression by binding to its promoter. The genome-wide binding sites of LIN-39 were determined by the modENCODE Consortium using ChIP coupled with high-throughput DNA sequencing (29). We uncovered three LIN-39 binding sites in the promoter of the mig-13 gene: site A (−3,197 to −2,685 upstream from ATG), site B (−1,729 to −1,420), and site C (−997 to −568) (Fig. 4B). We studied the functional significance of these sites by deleting them individually from a functional Pmig-13::mig-13::gfp DNA plasmid. We examined whether the deletion constructs still expressed mig-13::gfp in QR.x and rescued QR.x migration defects in mig-13 mutants. The deletion of binding site A completely abolished mig-13::gfp expression in QR.x (100%, three independent transgenic lines, 10–15 worms were examined from each line), whereas MIG-13::GFP fluorescence was visible in cells without binding sites B or C. Consistently, the construct containing the deletion of binding site A failed to rescue QR.x migration defects in mig-13 mutants, whereas those with the deletion of the binding site B or C partially or fully rescued QR.x migration defects (Fig. 4C). The bioinformatic analysis uncovered an enriched consensus DNA motif for LIN-39 target genes (29), and we found that site A contains such a motif [“CTCGAAAATGAGC” (−2,814 to −2,802 upstream of ATG)]. We showed that the deletion of this motif partially reduced the rescue of QR.x migration defects mig-13 mutants (Fig. 4C), indicating that the LIN-39 consensus binding motif modestly contributed to mig-13 expression.
To validate the role of LIN-39 in the transcriptional regulation of mig-13 further, we examined whether an expression of mig-13 by LIN-39 independent promoters (e.g., Pegl-46 or Pmec-7; Fig. S1 G and H) could rescue Q-cell migration defects in lin-39 mutants. We introduced transgenes that express mig-13 under the control of Pegl-46 (in AQR and AVM) or Pmec-7 (in AVM only) into lin-39 mutants. By examining the final position of AQR and AVM, we found that the Pegl-46::mig-13 transgene partially rescued AQR and AVM migration defects and that the Pmec-7::mig-13 transgene restored the AVM migration, indicating that bypassing LIN-39’s regulation of the mig-13 promoter could partially rescue QR.x migration defects in lin-39 mutants (Fig. 4D and Fig. S3E). Taken together, LIN-39 regulates QR.x anterior migration by directly promoting mig-13 expression.
Wnt Signaling and MAB-5 Inhibit lin-39 and mig-13 Expression in QL.ap.
The absence of MIG-13 in QL.ap is critical for its posterior migration (Figs. 1G and 2D), and we investigated the mechanism of mig-13 repression in QL.ap. The canonical Wnt signaling pathway and its downstream Hox gene mab-5 direct QL.ap posterior migration, and in mab-5 or mig-14 [Wntless ortholog essential for Wnt secretion (30)] mutants, QL.ap reverses its direction to the anterior. To test whether Wnt signaling and MAB-5 inhibit mig-13 expression, we examined Pmig-13::mig-13::gfp expression in QL.ap of mab-5 or mig-14 mutants. In WT animals, none of the QL.ap expresses MIG-13::GFP (Fig. 1G), but 95% of the QL.ap in mab-5 mutants or 88% of the QL.ap in mig-14 mutants showed MIG-13::GFP fluorescence (Fig. 1G and Fig. S1E).
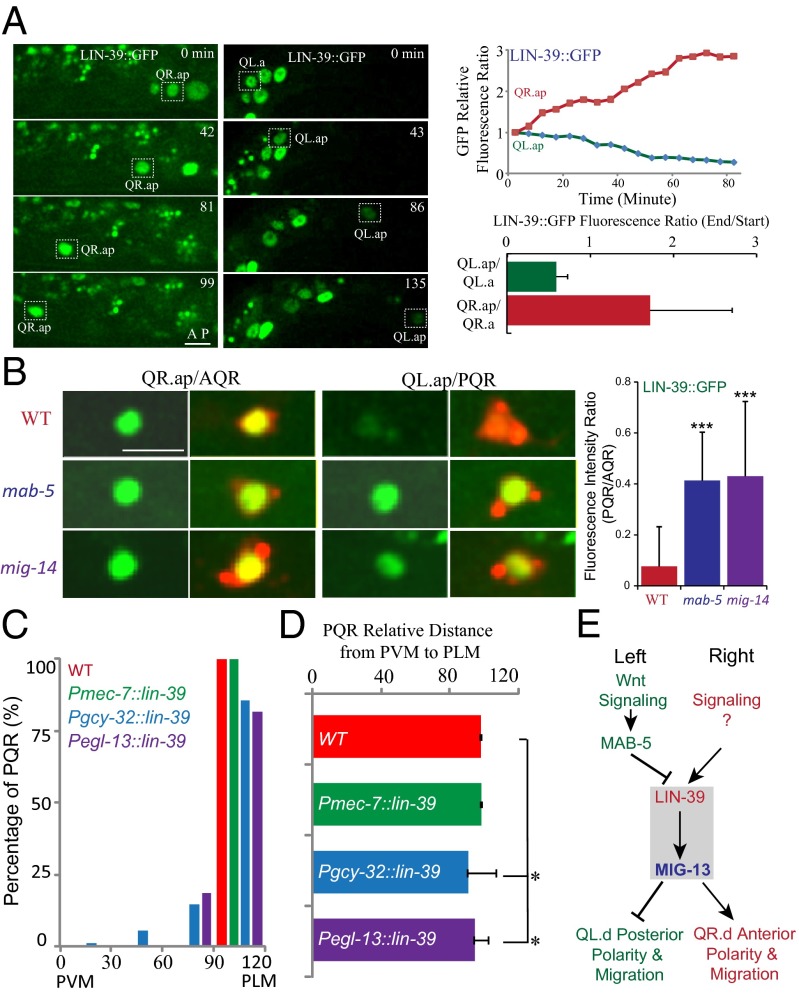
How do Wnt signaling and MAB-5 repress mig-13 expression? ChIP-seq data, provided by the modENCODE Consortium, showed that MAB-5 does not bind to the mig-13 promoter but binds to the lin-39 promoter (Fig. S3F) (29). We proposed that MAB-5 might indirectly repress mig-13 expression by negatively regulating lin-39 expression. We examined LIN-39 protein dynamics in Q-cell migration by imaging LIN-39::GFP expressed from the lin-39 promoter. We found that LIN-39::GFP fluorescence increased ∼1.8-fold in QR.ap but decreased ∼0.6-fold in QL.ap over the course of 1 h during their migrations (Fig. 5A and Movies S11 and S12), which was consistent with a specific role for LIN-39 in promoting QR.x anterior migration. We showed that LIN-39::GFP fluorescence ratio (PQR/AQR) is less than 0.1 (Fig. 5B). However, in mab-5 or mig-14 mutants, LIN-39::GFP was visible in PQR, and the fluorescence ratio (PQR/AQR) increased to ∼0.4 (Fig. 5B).
Fig. 5.
Wnt signaling and MAB-5 inhibit lin-39 and mig-13 expression and a proposed model. (A) Still images show wgIs18 [LIN-39::GFP] expression in the migrating QR.x (Left) or QL.x (Right). Upper Right shows the quantification of LIN-39::GFP fluorescence. Lower Right shows the fluorescence intensity ratio change of LIN-39::GFP during migration (n = 10–12 in each measurement). (B) wgIs18 [LIN-39::GFP] (green) expression in WT, mab-5 (e2088) and mig-14 (mu71) mutants. Q cells are visualized by mCherry. Quantifications are in the Right. n = 11–15 from a single experiment. ***P < 0.001 by Student t test. (C) PQR positions in WT or animals that ectopically express lin-39. (D) Statistical analysis of data in C. *P < 0.01 by Student t test. (E) A proposed model of the interaction among Wnt signaling, Hox genes, and MIG-13 in cell migration.
To further explore the down-regulation of lin-39 expression by MAB-5, we used MAB-5 independent promoters, Pgcy-32 and Pegl-13, to express lin-39 in QL.ap cells. We found that an ectopic addition of LIN-39 in QL.ap reduced its posterior migration such that QL.ap cells failed to arrive at their final destination (15% in Pgcy-32::lin-39, n = 89; 18% in Pegl-13::lin-39, n = 65; Fig. 5 C and D). The penetrance of the reduced QL.ap posterior migration was not high, likely due to the expression of LIN-39 by Pgcy-32 or Pegl-13 in the late stage of Q-cell development. Other transcription factors might also regulate the expression of mig-13 in QL.ap. However, we did not detect any defects in QL.ap migration in WT or in animals whose QL.paa/PVM ectopically expressed lin-39 by Pmec-7 (Fig. 5 C and D). We examined the MIG-13::GFP level in QL.ap with an ectopic gain of LIN-39, and we found that MIG-13::GFP was abnormally expressed in all of the QL.ap with the reduced posterior migration (n = 12). These results demonstrated that the down-regulation of lin-39 and mig-13 by MAB-5 is essential for the QL.ap posterior migration.
Conclusion
In summary, our study showed that C. elegans MIG-13 functions autonomously to regulate the anterior accumulation of the actin cytoskeleton in QR.x migration. This work identified the regulatory hierarchy in Q-cell migration; LIN-39 promotes mig-13 expression in QR.x to direct the anterior migration, whereas Wnt signaling and MAB-5 inhibit lin-39 and downstream mig-13 expression in QL.x to ensure the posterior migration (Fig. 5E).
Our work uncovered the autonomous function of mig-13 in cell migration. The previous study did not detect the fluorescence of MIG-13::GFP in Q-cell lineage (21), but our time-lapse imaging analysis showed that MIG-13 protein localizes on the membrane in QR.x but not in QL.x, which is consistent with MIG-13’s specific function in promoting QR.x anterior migration (Fig. 1F). The mosaic analysis using extrachromosomal arrays conducted in the published work can be subject to multiple losses in a cell lineage, which may complicate the analysis (21). Using cell type-specific promoters, we showed that the restricted expression of mig-13 in a subset of the Q-cell lineage only rescued the migration defects in Q cells expressing mig-13 (Fig. 2 B and C), indicating an autonomous function of mig-13 in cell migration. We showed that an ectopic gain of MIG-13 in QL.ap reduced its posterior migration, further strengthening the autonomy of mig-13. Interestingly, the previous study using the heat shock promoter to express mig-13 in the QL lineage moved the final position of QL.x anteriorly (21), which was consistent with our result that the ectopic mig-13 in the QL.ap reduced its posterior migration.
Although the expression of mig-13 under the control of Pgcy-32 and Pmec-7 promoters rescued mig-13 migration defects, these promoters are not active until the late migration stage. The addition of MIG-13 under a neuronal Punc-119 promoter could also consistently rescue Q-cell migration defects in mig-13 mutants (18), suggesting that the addition of MIG-13, even after migration, could rescue mig-13 migration defects. Alternatively, because MIG-13::GFP fluorescence is not strong when QR.x begins to migrate, Pgcy-32 and Pmec-7 may produce low but sufficient amounts of MIG-13 for the early migration. The nonautonomy of mig-13 may not be ruled out because the low level background expression of mig-13 in other tissues might be partially responsible for the rescue. However, the expression of mig-13 from the motor neurons failed to rescue QR.x migration defects in mig-13 mutants, and an ectopic expression of mig-13 in QL.ap reduced its posterior migration, further supporting the mig-13 autonomy (Fig. 2 B–D). Nevertheless, this study provided multiple independent lines of evidence to support that mig-13, at least, plays autonomous roles in cell migration.
MIG-13 may specifically regulate QR.x cell migration but not the cell fate. A/PQR is ciliated oxygen sensory neuron, and gcy-32 is a soluble guanylate cyclase specifically expressed in A/PQR and URX. The formation of cilium in A/PQR requires intraflagellar transport proteins (e.g., OSM-6), and Pgcy-32::mCherry and Posm-6::osm-6::gfp were used as the cell fate reporter for A/PQR. For the fate of A/PVM, mec-4 and mec-7 encode the touch neuron-specific channel protein and tubulin, respectively. We did not detect any abnormality of the expression of these markers in mig-13 mutants or animals with mig-13 ectopic expression (Fig. S5; n > 50 for each marker).
Although MIG-13 is required to establish polarity of migrating cells, MIG-13::GFP evenly distributes on the membrane of migrating QR.x cells (Fig. 1F and Movie S1), suggesting that the local activation of MIG-13 in the anterior might be required for its confined function. The ectopic addition of MIG-13 during QL.ap posterior migration also generated abnormal anterior processes (Fig. 3C), indicating that MIG-13 may be anteriorly activated inside the migrating cell regardless of its direction.
This work first identified mig-13 as the target of lin-39 and mab-5 in Q-cell migration. In mab-5 loss-of-function mutant (e2088), mig-13::gfp was reported to be ectopically expressed in the ventral cord neurons located in the posterior (21), which was consistent with the negative role of mab-5 in the expression of mig-13 in the QL lineage (Fig. 1G). The prior epistasis analysis of mab-5; lin-39 double mutants indicated that the lin-39 mutation could partially suppress the migration defects of QL.x in mab-5 mutants, suggesting that lin-39 may function downstream of mab-5 (19). Consistently, our data indicated that mab-5 negatively regulated the expression of lin-39 in QL.ap cells (Fig. 5B).
Materials and Methods
Maintenance of C. elegans strains, live cell imaging, and generation of transgenic animals were carried out as described previously (6, 7). Strains, PCR templates, primers, and plasmids are listed in Table S1–S3. Detailed methods and protocols can be found in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank the Caenorhabditis Genetics Center and Drs. C. Kenyon, M. Ding, X. Liu, and Z. Lu for strains, reagents, and bioinformatics. This work was supported by National Basic Research Program of China (973 Program, 2013CB945600, 2012CB945002, and 2012CB966800) (to G.O. and W.L.), the National Natural Science Foundation of China (31201048, 31222035, 31101002, 31100972, 31171295 and 31190063) (to X.W., W.L., Y.Y., and G.O.), the Natural Science Foundation of Beijing (5123045) (to X.W.), and the Junior Thousand Talents Program of China (G.O.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1301849110/-/DCSupplemental.
References
- 1.Montell DJ, Yoon WH, Starz-Gaiano M. Group choreography: Mechanisms orchestrating the collective movement of border cells. Nat Rev Mol Cell Biol. 2012;13(10):631–645. doi: 10.1038/nrm3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112(4):453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 3.Ridley AJ, et al. Cell migration: Integrating signals from front to back. Science. 2003;302(5651):1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 4.Petrie RJ, Doyle AD, Yamada KM. Random versus directionally persistent cell migration. Nat Rev Mol Cell Biol. 2009;10(8):538–549. doi: 10.1038/nrm2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol. 1977;56(1):110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- 6.Ou G, Vale RD. Molecular signatures of cell migration in C. elegans Q neuroblasts. J Cell Biol. 2009;185(1):77–85. doi: 10.1083/jcb.200812077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chai Y, et al. Live imaging of cellular dynamics during Caenorhabditis elegans postembryonic development. Nat Protoc. 2012;7(12):2090–2102. doi: 10.1038/nprot.2012.128. [DOI] [PubMed] [Google Scholar]
- 8.Chapman JO, Li H, Lundquist EA. The MIG-15 NIK kinase acts cell-autonomously in neuroblast polarization and migration in C. elegans. Dev Biol. 2008;324(2):245–257. doi: 10.1016/j.ydbio.2008.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Honigberg L, Kenyon C. Establishment of left/right asymmetry in neuroblast migration by UNC-40/DCC, UNC-73/Trio and DPY-19 proteins in C. elegans. Development. 2000;127(21):4655–4668. doi: 10.1242/dev.127.21.4655. [DOI] [PubMed] [Google Scholar]
- 10.Middelkoop TC, et al. The thrombospondin repeat containing protein MIG-21 controls a left-right asymmetric Wnt signaling response in migrating C. elegans neuroblasts. Dev Biol. 2012;361(2):338–348. doi: 10.1016/j.ydbio.2011.10.029. [DOI] [PubMed] [Google Scholar]
- 11. Sundararajan L, Lundquist EA (2012) The transmembrane proteins UNC-40/DCC, PTP-3/LAR, and MIG-21 control anterior-posterior neuroblast migration with left-right functional asymmetry in Caenorhabditis elegans. Genetics 192(4):1373–1388. [DOI] [PMC free article] [PubMed]
- 12.Silhankova M, Korswagen HC. Migration of neuronal cells along the anterior-posterior body axis of C. elegans: Wnts are in control. Curr Opin Genet Dev. 2007;17(4):320–325. doi: 10.1016/j.gde.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 13.Whangbo J, Kenyon C. A Wnt signaling system that specifies two patterns of cell migration in C. elegans. Mol Cell. 1999;4(5):851–858. doi: 10.1016/s1097-2765(00)80394-9. [DOI] [PubMed] [Google Scholar]
- 14.Kenyon C. A gene involved in the development of the posterior body region of C. elegans. Cell. 1986;46(3):477–487. doi: 10.1016/0092-8674(86)90668-9. [DOI] [PubMed] [Google Scholar]
- 15.Maloof JN, Whangbo J, Harris JM, Jongeward GD, Kenyon C. A Wnt signaling pathway controls hox gene expression and neuroblast migration in C. elegans. Development. 1999;126(1):37–49. doi: 10.1242/dev.126.1.37. [DOI] [PubMed] [Google Scholar]
- 16.Harris J, Honigberg L, Robinson N, Kenyon C. Neuronal cell migration in C. elegans: Regulation of Hox gene expression and cell position. Development. 1996;122(10):3117–3131. doi: 10.1242/dev.122.10.3117. [DOI] [PubMed] [Google Scholar]
- 17.Salser SJ, Kenyon C. Activation of a C. elegans Antennapedia homologue in migrating cells controls their direction of migration. Nature. 1992;355(6357):255–258. doi: 10.1038/355255a0. [DOI] [PubMed] [Google Scholar]
- 18.Yang L, Sym M, Kenyon C. The roles of two C. elegans HOX co-factor orthologs in cell migration and vulva development. Development. 2005;132(6):1413–1428. doi: 10.1242/dev.01569. [DOI] [PubMed] [Google Scholar]
- 19.Clark SG, Chisholm AD, Horvitz HR. Control of cell fates in the central body region of C. elegans by the homeobox gene lin-39. Cell. 1993;74(1):43–55. doi: 10.1016/0092-8674(93)90293-y. [DOI] [PubMed] [Google Scholar]
- 20.Wang BB, et al. A homeotic gene cluster patterns the anteroposterior body axis of C. elegans. Cell. 1993;74(1):29–42. doi: 10.1016/0092-8674(93)90292-x. [DOI] [PubMed] [Google Scholar]
- 21.Sym M, Robinson N, Kenyon C. MIG-13 positions migrating cells along the anteroposterior body axis of C. elegans. Cell. 1999;98(1):25–36. doi: 10.1016/S0092-8674(00)80603-0. [DOI] [PubMed] [Google Scholar]
- 22.Masuda H, et al. MIG-13 controls anteroposterior cell migration by interacting with UNC-71/ADM-1 and SRC-1 in Caenorhabditis elegans. FEBS Lett. 2012;586(6):740–746. doi: 10.1016/j.febslet.2012.01.031. [DOI] [PubMed] [Google Scholar]
- 23.Schneider S, Gulacsi A, Hatten ME. Lrp12/Mig13a reveals changing patterns of preplate neuronal polarity during corticogenesis that are absent in reeler mutant mice. Cereb Cortex. 2011;21(1):134–144. doi: 10.1093/cercor/bhq070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qin H, Powell-Coffman JA. The Caenorhabditis elegans aryl hydrocarbon receptor, AHR-1, regulates neuronal development. Dev Biol. 2004;270(1):64–75. doi: 10.1016/j.ydbio.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 25.Wu J, Duggan A, Chalfie M. Inhibition of touch cell fate by egl-44 and egl-46 in C. elegans. Genes Dev. 2001;15(6):789–802. doi: 10.1101/gad.857401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Savage C, et al. mec-7 is a beta-tubulin gene required for the production of 15-protofilament microtubules in Caenorhabditis elegans. Genes Dev. 1989;3(6):870–881. doi: 10.1101/gad.3.6.870. [DOI] [PubMed] [Google Scholar]
- 27.McIntire SL, Reimer RJ, Schuske K, Edwards RH, Jorgensen EM. Identification and characterization of the vesicular GABA transporter. Nature. 1997;389(6653):870–876. doi: 10.1038/39908. [DOI] [PubMed] [Google Scholar]
- 28.Cai L, Makhov AM, Bear JE. F-actin binding is essential for coronin 1B function in vivo. J Cell Sci. 2007;120(Pt 10):1779–1790. doi: 10.1242/jcs.007641. [DOI] [PubMed] [Google Scholar]
- 29.Niu W, et al. Diverse transcription factor binding features revealed by genome-wide ChIP-seq in C. elegans. Genome Res. 2011;21(2):245–254. doi: 10.1101/gr.114587.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang PT, et al. Wnt signaling requires retromer-dependent recycling of MIG-14/Wntless in Wnt-producing cells. Dev Cell. 2008;14(1):140–147. doi: 10.1016/j.devcel.2007.12.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.