Abstract
Bifidobacteria represent one of the dominant groups of microorganisms colonizing the human infant intestine. Commensal bacteria that interact with a eukaryotic host are believed to express adhesive molecules on their cell surface that bind to specific host cell receptors or soluble macromolecules. Whole-genome transcription profiling of Bifidobacterium bifidum PRL2010, a strain isolated from infant stool, revealed a small number of commonly expressed extracellular proteins, among which were genes that specify sortase-dependent pili. Expression of the coding sequences of these B. bifidum PRL2010 appendages in nonpiliated Lactococcus lactis enhanced adherence to human enterocytes through extracellular matrix protein and bacterial aggregation. Furthermore, such piliated L. lactis cells evoked a higher TNF-α response during murine colonization compared with their nonpiliated parent, suggesting that bifidobacterial sortase-dependent pili not only contribute to adherence but also display immunomodulatory activity.
Keywords: genomics, host–microbe interaction, probiotics, transcriptomics
The interaction between bacteria and their human host affects the latter in several ways. In some cases, this interaction may impact negatively on the health status of the host (pathogenesis), whereas in other cases may not influence host health at all (commensalism). In addition, certain bacterium–host interactions that represent symbiotic and probiotic relationships promote the health status of the host (1). Currently, the molecular mechanisms underlying the presumed health-promoting activities are largely unknown, despite the widely held view that microbial populations residing in the human gastrointestinal tract exert activities that positively affect host health (2). Bifidobacteria represent prominent commensals of the human infant gut (3) where they modulate metabolic and immune activities of their host (4–6). Through functional genomic approaches, significant progress has been made in unraveling bifidobacterial gut colonization strategies (1). In a recent study, we found that the genome of Bifidobacterium bifidum PRL2010 harbors an extensive gene set involved in the utilization of host-derived glycans, such as those found in the outermost layer of the intestinal mucosa (7). These findings are very suggestive of host-microbe coevolution, and signify B. bifidum PRL2010 as a bifidobacterial prototype for analysis of interactions between microbes and the intestinal mucosa. Many commensal and pathogenic microorganisms that interact with eukaryotic hosts express adhesive structures on their cell surface that mediate physical contacts between such bacteria and specific host cell receptors or soluble macromolecules (8). Bacterial surface appendages, such as pili or fimbrial adhesins in Gram-negative bacteria, have historically been considered to be the predominant bacterial structures involved in host–microbe interaction (8). Gene clusters responsible for the biosynthesis of pili have been identified in the genomes of many Gram-negative and Gram-positive bacteria, not only in pathogens (9–12) but also in gut inhabitants, such as Lactobacillus rhamnosus GG (13). Notably, sortase-dependent pili encoded by L. rhamnosus GG were demonstrated to be pivotal for efficient adherence and immunomodulatory interactions with human gut cells (14). In bifidobacteria, experimental evidence of the existence of sortase-dependent and type IV pili was reported only very recently (15, 16). Here, we describe in vivo analyses of the gut commensal B. bifidum PRL2010, with a focus on the role of sortase-dependent pili in host–microbe interaction.
Results and Discussion
Introduction of B. bifidum PRL2010 to the Murine Gut.
Conventional female BALB/c mice were administered a single daily dose of 109 CFU B. bifidum PRL2010 (SI Materials and Methods). Mice were a priori checked for the presence of bifidobacteria in fecal samples by PCR using Bifidobacterium-specific primers (17), which revealed that bifidobacteria were either absent or below the limit of detection. Animals were killed 12 d later, allowing sufficient time for several cycles of turnover of the intestinal epithelium and its overlying mucus layer (18). Microbial evaluation of the murine gut showed the presence of strain PRL2010 at stable numbers, reminiscent of at least transient colonization over time with the highest numbers of this strain recorded in the cecum and colon (Fig. S1). Given the robustness of its colonization of the distal gut, we focused on determining adaptations of PRL2010 to the caecal habitat.
B. bifidum PRL2010 Transcriptome Under in Vitro and in Vivo Conditions.
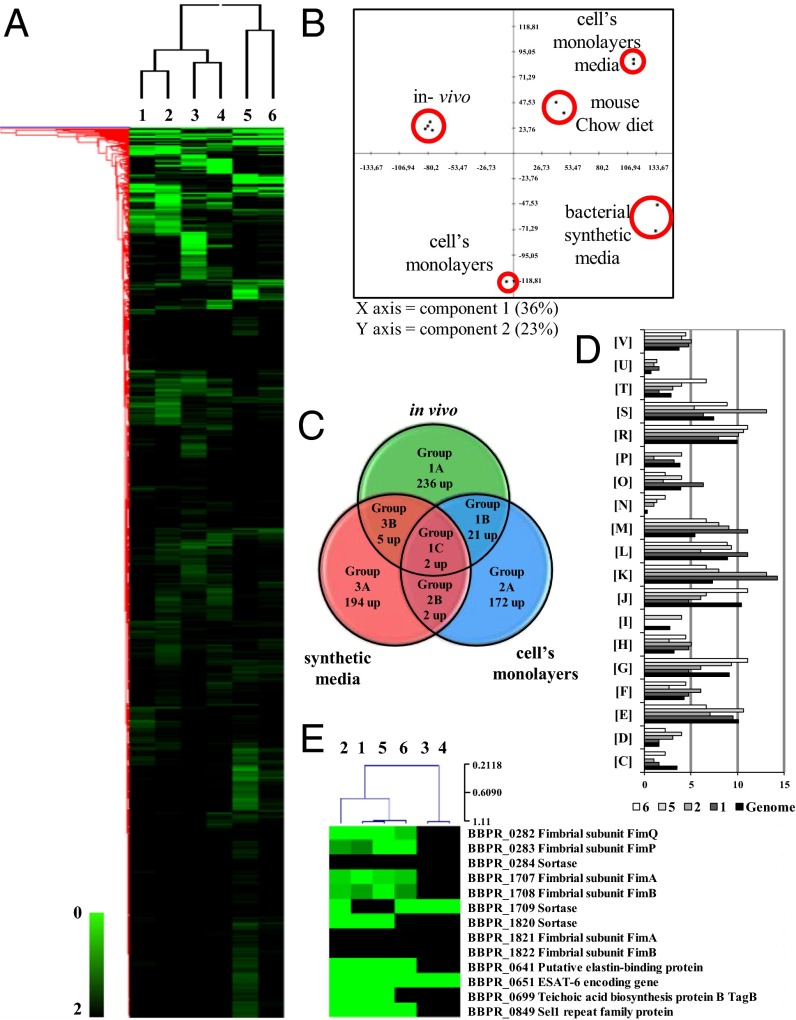
To investigate possible interactions of PRL2010 with its natural ecological niche, the gut, we performed global genome transcription profiling of this strain in an in vitro human gut model using HT29 cells, as well as upon colonization of PRL2010 of the murine gut using a custom-made B. bifidum PRL2010 array representing 90% of the identified genes of this organism (7). The global gene expression profile of B. bifidum PRL2010 was conserved in the caeca between different mice (Fig. 1A). According to a principal component analysis, these profiles were clearly different from the transcriptomes of PRL2010 obtained under in vitro conditions (HT29 monolayer or laboratory cultures) (Fig. 1B). This finding suggests, not unexpectedly, distinctly different transcriptional responses of PRL2010 to each of these environments. A total of 104 or 62 genes exhibited a ≥twofold change (P < 0.0005) in transcription upon bringing PRL2010 cells in contact with HT29 cells, using the transcriptome of PRL2010 grown in DMEM or Man-Rogosa-Sharp (MRS) synthetic medium as a reference, respectively (Fig. S1B). Analysis of the transcriptome of PRL2010 obtained when this microorganism was present in the cecum of conventional BALB/c mice showed that transcription of 87 or 141 genes was increased more than twofold compared with their transcription level in PRL2010 cells when obtained from the caecal contents of mice fed on fresh Chow diet or when grown on MRS, respectively. The comparative analysis shown in Fig. 1A and Fig. S1B yielded three groups of regulated genes, including those specific to in vivo conditions (group 1), to in vitro experiments (i.e., exposure to a human cell line, group 2), and to growth in synthetic medium under laboratory conditions (group 3). We also categorized genes from these three groups into three subgroups based on whether the genes were contributed from a single environmental condition (i.e., human cell line model, in vivo, or synthetic media datasets) (subgroups A), or two conditions (subgroups B), or all three conditions (subgroup C) (Fig. 1C). Assignment of genes into these groups may reflect bacterial responses to differences in host structures. These differences could be species-specific (human vs. murine), tissue-specific (colon vs. cecal mucosa), or they could also be linked to differential carbohydrate availability or to the effect of the residential intestinal microorganisms. We used cluster of orthologous groups (COG) analysis to identify differentially transcribed genes that contribute to specific biological functions. As illustrated in Fig. 1D, carbohydrate metabolism, corresponding to COG category [G], is one of the COG functions of PRL2010 most significantly affected by the interaction with the murine host, which is probably because of a response to the presence of specific host glycans, in particular mucin (7). Various members of this COG function were significantly up-regulated (≥twofold; P < 0.0005) under in vivo conditions, encompassing genes involved in breakdown of glycoproteins (Fig. 1A).
Fig. 1.
Identification of B. bifidum PRL2010 differentially expressed genes by transcriptome analysis in response to contact with the host. (A) Heat-map displaying the change in PRL2010 gene expression upon colonization of murine caeca (lanes 1 and 2), when grown in DMEM synthetic medium (lanes 3) or in fresh chow diet (lane 4), and following incubation with human intestinal HT29 cells (lanes 5 and 6). Each row represents a separate transcript and each column represents a separate sample. Color legend is on the bottom of the microarray plot; green indicates increased transcription levels compared with the reference samples. The reference conditions used were as follows: lane 1, fresh chow diet; lanes 2–4 and 6, MRS medium; lane 5, DMEM. Dendrogram on the left margin of the heat-map represents the hierarchical clustering algorithm result based on average linkage and Euclidean distance of the gene dataset. (B) The clustering of PRL2010 transcriptomes under in vitro and in vivo conditions by principal component analysis. (C) Venn diagram showing the number of genes expressed during the different conditions: in vitro (exposure to a human cell line), in vivo, and DMEM synthetic medium. (D) Depiction of a functional annotation of the in vitro- and in vivo-expressed genes of B. bifidum PRL2010 according to their COG categories. Each COG family is identified by a one-letter abbreviation (National Center for Biotechnology Information database). For each category, the black bar represents the percentage of genes in that category as detected in the sequenced genome of PRL2010 (7). The other bar shows the percentages of genes transcribed during murine colonization by PRL2010 (conditions 1 and 2), and following exposure of PRL2010 cells to human intestinal cells (conditions 5 and 6). The percentage was calculated as the percentage of transcribed genes belonging to the indicated COG category with respect to all transcribed genes. (E) Selected genes that were up-regulated when PRL2010 were cultivated in the conditions indicated in A.
PRL2010 cell surface properties also appear to be modified in response to tissue contact, as indicated by the increased transcription of genes encoding several extracellular and membrane-spanning proteins, many of which are predicted to mediate interaction with eukaryotic cells (Fig. 1E). Adhesion of bacteria to human intestinal mucosa or extracellular matrix (ECM) proteins represents a key strategy for intestinal colonization, and bifidobacteria can indeed adhere to intestinal cells (19, 20). Genes that specify putative adhesion functions for PRL2010 include BBPR_0641, which specifies a putative elastin-binding protein and whose transcription was significantly induced under in vivo conditions as well as upon exposure to HT29 cells (18- to 24-fold). It is known that elastin-binding proteins promote recognition of mammalian ECM, thus allowing colonization of the host by gut bacteria (21). Furthermore, transcription of BBPR_0651, whose protein product displays similarity to early secretory antigen target 6 (ESAT-6), was shown to be highly induced following HT29 exposure and when PRL2010 was present in the murine gut. The small ESAT-6 protein appears to be of fundamental importance in virulence and protective immunity in Mycobacterium tuberculosis and Staphylococcus aureus (22), suggesting that the homologous protein of PRL2010 acts as a protective immunity determinant. PRL2010 contact with HT29 cells and its presence in mice also triggered the transcription of another gene (BBPR_0699), predicted to encode a protein involved in the biosynthesis of teichoic acids, which for Lactobacillus acidophilus NCFM have been shown to modulate host–microbe interaction (23). Two of the three pilus clusters identified on the PRL2010 genome (7, 15) [i.e., pil2PRL2010 (BBPR_1707-BBPR_1709), and pil3PRL2010 (BBPR_282-BBPR_284)] were shown to be expressed under both in vitro and in vivo conditions. Notably, and in contrast to the adjacent pilin subunit-encoding genes (BBPR_1707 and BBPR_1708), BBPR_1709, which specifies a predicted sortase, was shown to be expressed when PRL2010 was grown in MRS medium, suggesting that this sortase also processes other cell wall-anchored proteins. This finding is supported by a previously reported finding that the BBPR_1709 gene is transcribed separately from the other components of this pilus gene cluster (15). The predicted pilus proteins are similar to subunits of so-called sortase-dependent pili, which are typically composed of a major pilin subunit (represented by FimAPRL2010 or FimPPRL2010 for the pil2PRL2010 and pil3PRL2010 clusters, respectively), and one or two ancillary minor pilin subunits (represented by FimBPRL2010 and FimQPRL2010 for the pil2PRL2010 and pil3PRL2010 clusters, respectively) (Fig. S2) (15). When we compared FimAPRL2010 to FimA homologs encoded by other B. bifidum strains, their amino acid sequences were shown to display much higher variability compared with the FimP homologs (Fig. S2). In addition, FimAPRL2010 contains a CnaB-type domain, which is described to act as a stalk in binding to components of the ECM of the host, such as fibronectin, collagen types I to XV, and laminin (24). Atomic force microscopy (AFM) assays of PRL2010 cells that had been exposed to human cell lines (Caco-2 or HT29) revealed a highly piliated cell morphology (Fig. S3B). Interestingly, we found by AFM that pili were also present in PRL2010 cultivated in liquid media or on agar plates, although pili appeared to be less abundant compared with the numbers seen upon contact with a human cell line (Fig. S3 A–D). Furthermore, Western blot analysis using antibodies that had been raised against the major subunit protein of the pil2 or pil3 loci, FimAPRL2010 and FimPPRL2010, respectively (Abpil2 or Abpil3) (SI Materials and Methods) was performed on PRL2010 cells that had been in contact with Caco-2 monolayers or cultivated on agar plates. Clear signals representing a protein of 55.87 kDa and 55.43 kDa were noticed when a crude extract of PRL2010 cells, previously exposed to Caco-2 monolayers, was probed in a Western blot using Abpil2 or Abpil3, respectively (Fig. S3E). In contrast, Western blot signals of lower intensity were observed using protein extracts from PRL2010 cultivated on agar plates or MRS plus lysine. The higher molecular weight signals above 100 kDa detected in each immunoblot image (Fig. S3E) likely represent the covalently linked polymers of FimA2010 and FimP2010, a typical feature of sortase-dependent pili (12, 25).
Differential Binding to Human Epithelial Cells Mediated by PRL2010 Pili.
To obtain further insight into the functional roles exerted by pili encoded by PRL2010, we expressed the pil2PRL2010 and pil3PRL2010 gene clusters in the Gram-positive host Lactococcus lactis NZ9000 (SI Materials and Methods), because genetic manipulation of PRL2010, such as creating knockout mutants, is currently not possible. L. lactis has previously been used successfully as a heterologous host for expression of bifidobacterial proteins (26, 27), as well as a host to display full-length forms of microbial surface structures (28). AFM analysis revealed that both L. lactis-pil2PRL2010 and L. lactis-pil3PRL2010 clones displayed evident piliated morphology when pilus expression had been induced with nisin, but no pili were observed in noninduced L. lactis controls (Fig. S4 A–C). Furthermore, the cell surface of (nisin-induced) L. lactis-pil2PRL2010 and L. lactis-pil3PRL2010 clones was less densely piliated than that of the wild strain B. bifidum PRL2010 (Figs. S3 A–D and S4 B and D).
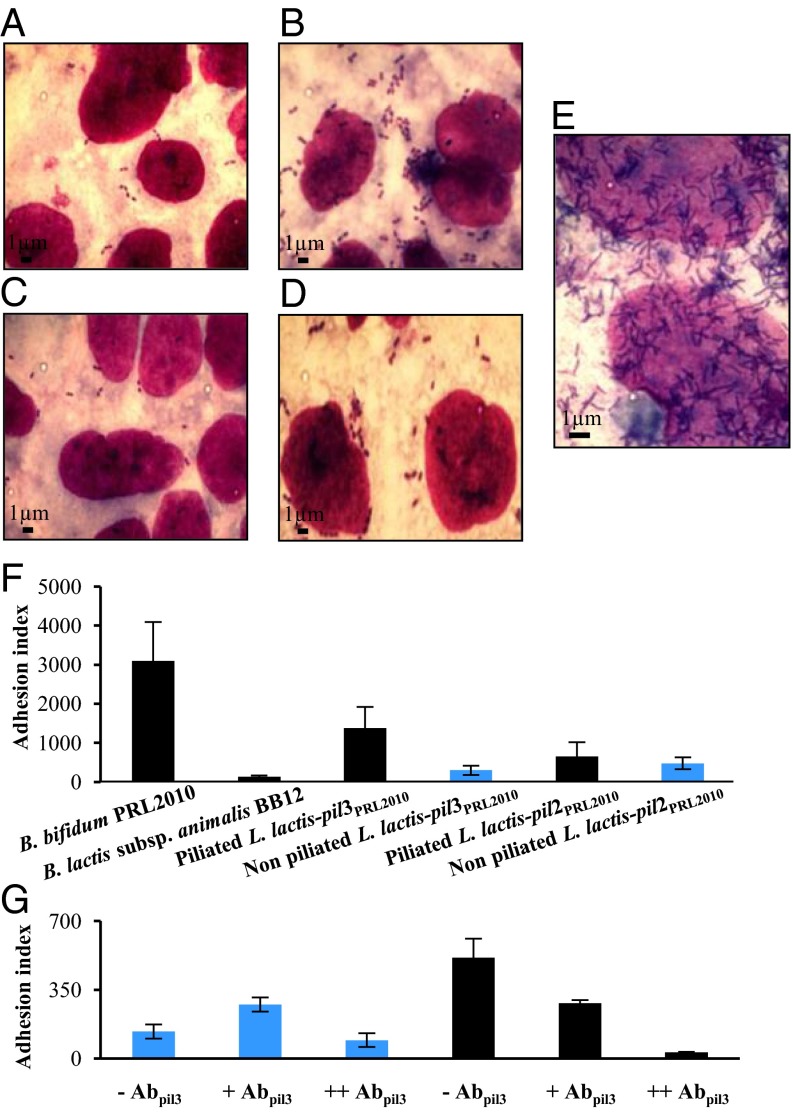
Based on previous findings for analogous structures in Actinomyces oris (29), we decided to evaluate possible interactions between B. bifidum PRL2010 pili structures and a Caco-2 human intestinal epithelial cell line, to establish if they are involved in bacterial adhesion to human enterocytes. To this aim, we used L. lactis-pil2PRL2010 and L. lactis-pil3PRL2010 cells in adhesion experiments using a Caco-2 differentiated cell layer. Remarkably, following nisin induction L. lactis-pil3PRL2010 displayed a significant enhancement in adhesion to eukaryotic cells compared with noninduced L. lactis-pil3PRL2010 (Fig. 2 A and B). In contrast, L. lactis-pil2PRL2010 cells did not display any significant change in adhesion properties under the conditions tested (Fig. 2 C and D). A clear adhesion phenotype was also noticed in the wild-type strain B. bifidum PRL2010 (Fig. 2 E and F); the observed Caco-2-adhesive behavior was displayed by PRL2010, which is clearly more pronounced than that observed for nisin-induced L. lactis-pil2PRL2010 cells, and which may be because of the elevated abundance of pili structures in PRL2010 cells compared with those produced in the heterologous host, or the result of the presence of additional adhesion promoting cell-surface molecules on PRL2010, such as BopA (19). These results therefore demonstrate direct involvement of bifidobacterial sortase-dependent pili structures in mediating adhesion to human intestinal cells, thus implicating these extracellular structures in host colonization by the infant intestinal commensal B. bifidum PRL2010. To substantiate these findings, competitive adhesion assays involving piliated L. lactis-pil3PRL2010 cells that had first been treated with Abpil3, were performed. This experiment showed that treatment with Abpil3 decreased adhesion of piliated L. lactis-pil3PRL2010 cells to Caco-2 cells by maximum 17-fold (P < 0.05) (Fig. 2G).
Fig. 2.
Adhesion of B. bifidum PRL2010 sortase-assembled pili to human intestinal cells. (A and B) Display of the adhesion phenotype of nonpiliated (A) and piliated (B) L. lactis-pil3PRL2010 cells to the Caco-2 cell monolayer. (C and D) The adhesion phenotype of nonpiliated (C) and piliated (D) L. lactis-pil2PRL2010 cells to the Caco-2 cell monolayer. (E) The adhesion of B. bifidum PRL2010 cells to Caco-2 cell monolayer. (F) The adhesion efficiency (adhesion index) of B. bifidum PRL2010, piliated (black pillar) and nonpiliated (blue pillar) L. lactis-pil2PRL2010 cells, as well as L. lactis-pil3PRL2010 cells determined in terms number of adhered bacterial cells per 100 Caco-2 cells. The negative control is represented by B. animalis subsp. lactis BB12 cells. (G) Depiction of inhibition pili-mediated adhesion to the Caco-2 cell monolayer by L. lactis-pil3PRL2010 cells, which had first been treated with anti-pil3 antibodies (Abpil3). Piliated L. lactis-pil3PRL2010 cells (black pillar) and nonpiliated (blue pillar) L. lactis-pil2PRL2010 cells were used. Two different concentrations of antibodies were used. +, Represents the use of diluted (1:50) Abpil3; ++, undiluted Abpil3. Bars represent mean values of three independent experiments, and the error bars indicate the SD (P < 0.05).
Bacterial Aggregation and PRL2010 Pili.
Sortase-assembled pili are also known to promote bacterial coaggregation (30). We therefore investigated if a similar scenario would apply to the B. bifidum PRL2010 pili. We performed aggregation experiments involving piliated L. lactis-pil3PRL2010, L. lactis-pil2PRL2010, and their nonpiliated L. lactis equivalent strains. Notably, in the case of nisin-induced L. lactis-pil2PRL2010 and L. lactis-pil3PRL2010 cells (from here on referred as piliated L. lactis-pil2PRL2010 and L. lactis-pil3PRL2010 cells), aggregation levels enhanced 13- and 21-fold with respect to their uninduced equivalents (P < 0.05) (from here on referred as nonpilated lactis-pil2PRL2010 and L. lactis-pil3PRL2010 cells), respectively (Fig. S5). Taken together, these data implicate Pil2PRL2010 and Pil3PRL2010 as important factors that promote bacterial autoaggregation, possibly with a contributing role in gut colonization. These findings corroborate the view that bacterial aggregation represents a mechanism by which gastrointestinal commensals adhere to each other and, as a result, may colonize persistently in biofilms on the host’s mucosa (31).
Human Receptors for PRL2010 Pili.
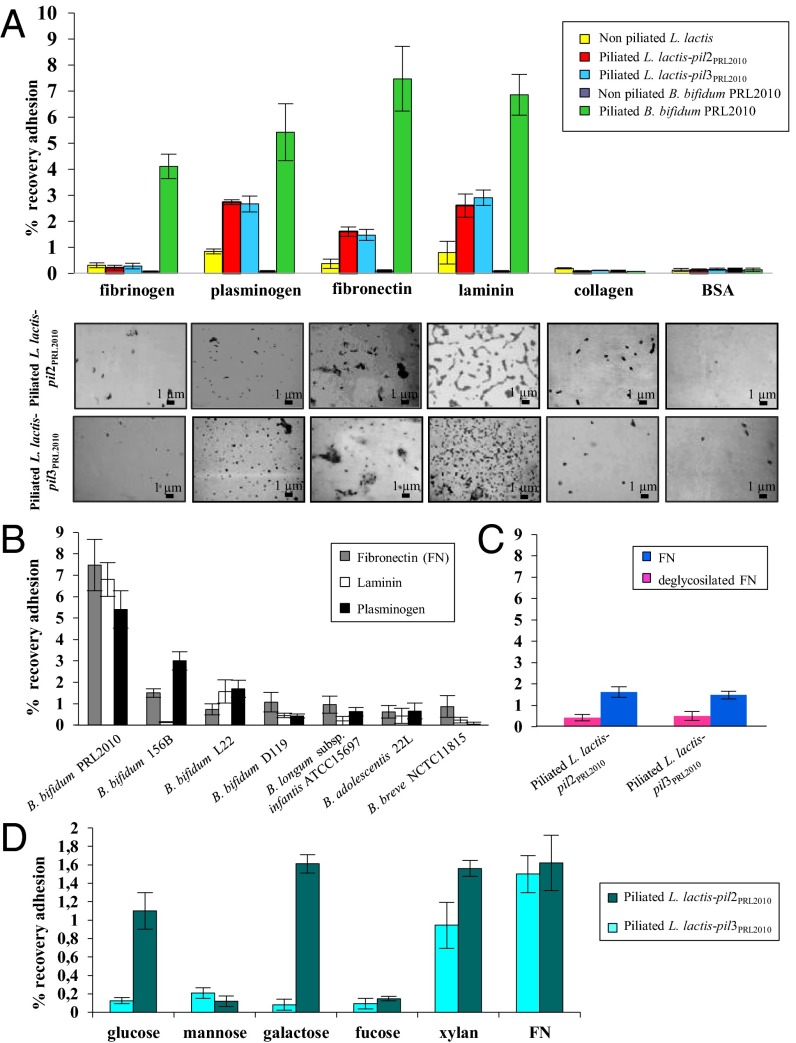
Although we identified sortase-dependent pili as a PRL2010 adhesion factor that mediates binding to epithelial cells, the receptors involved in their recognition are unknown. As enteropathogens are known to adhere to intestinal tissue by pili-mediated binding to ECM proteins (32–34), we examined the ability of PRL2010 to adhere to the ECM proteins fibrinogen, plasminogen, fibronectin, laminin, and collagen type IV. Notably, B. bifidum PRL2010 cells showed higher adhesion to fibronectin, plasminogen, and laminin, compared with the other ECM proteins (Fig. 3A). A similar scenario was noticed when piliated L. lactis-pil3PRL2010 as well as L. lactis-pil2PRL2010 clones were used. In contrast, nonpiliated L. lactis cells displayed very limited adhesion to these ECM substrates, as confirmed by microscopic examination of the samples (Fig. 3A). Binding of piliated L. lactis-pil3PRL2010 and L. lactis-pil2PRL2010 cells to serial dilutions of ECM substrates was evaluated, showing that saturation of binding already occurred at low concentrations of the ECM protein tested (Fig. S6). To evaluate whether the binding of B. bifidum PRL2010 to fibronectin, which appeared to be the most effective ECM substrate, occurs for strains besides our reference strain, other bifidobacterial strains were tested. Notably, within the B. bifidum species, strain PRL2010 displayed the highest adhesion level, whereas other bifidobacterial strains belonging to Bifidobacterium breve, Bifidobacterium adolescentis, and Bifidobacterium longum subsp. infantis, displayed much lower levels of binding to fibronectin, plasminogen, and laminin (Fig. 3B), which may reflect different strategies used by these bacteria to colonize the human gut. All these strains except B. longum subsp. infantis ATCC15697 encode putative sortase-dependent pili, and they were treated and cultivated under the same conditions as PRL2010. Considering that fibronectin is a glycoprotein and that carbohydrate residues have been shown to be involved in fimbrial binding to fibronectin (35), we addressed the possible involvement of fibronectin-associated glycans in PRL2010 pili binding. Fibronectin deglycosylation of O-linked and N-linked oligosaccharides caused a significant reduction in PRL2010 pili-mediated binding ability compared with untreated fibronectin (Fig. 3c), suggesting that N- and O-linked glycoproteins are involved in adhesion of PRL2010 pili to fibronectin. Because it was previously shown that certain carbohydrates bind to sortase-dependent pili, thereby competing with the actual pilus receptor (32, 33, 35), we evaluated the effect of various carbohydrates on binding of PRL2010 Pil2 and Pil3 to fibronectin. Interestingly, we found that the binding of this ECM protein to piliated L. lactis-pil2PRL2010 was significantly reduced when mannose or fucose was present during the binding assay. Binding ability of piliated L. lactis-pil3PRL2010 was also affected by glucose and galactose, but not by the polysaccharide xylan (Fig. 3D). This finding suggests that mannose and fucose act as potential receptors for Pil2 of B. bifidum PRL2010 reminiscent of the pili behavior of enteric bacteria (32, 33, 35), whereas the putative receptors of Pil3 seem to include a wider spectrum of carbohydrates.
Fig. 3.
Involvement of pili produced by B. bifidum PRL2010 in binding to ECM proteins. (A) Adhesion of B. bifidum PRL2010 cells, L. lactis-pil2PRL2010 cells, and L. lactis-pil3PRL2010 cells to various ECM proteins. Below each pillar a picture is placed to indicate how piliated and nonpiliated L. lactis-pil2PRL2010 cells and L. lactis-pil3PRL2010 cells appeared under the microscope following exposure to ECM substrates. For each of these experiments, adhesion of microbial cells to BSA was used as negative control. Piliated PRL2010 represent PRL2010 cells grown under conditions that promote the production of sortase-dependent pili (cells grown on MRS agar or grown to stationary phase in MRS broth plus lysine), and nonpiliated PRL2010 represent PRL2010 cells grown under conditions that reduce production of sortase-dependent pili (cells grown in MRS broth to exponential phase). (B) Adhesion of different bifidobacterial strains to fibronectin, plasminogen and laminin. (C) Adhesion of piliated L. lactis-pil2PRL2010- and L. lactis-pil3PRL2010 cells to deglycosylated fibronectin. (D) Adhesion ability of piliated L. lactis-pil2PRL2010 and L. lactis-pil3PRL2010 to fibronectin in the presence of different carbohydrates. Bars represent mean values for three independent experiments, and the error bars indicate the SD (P < 0.05).
Immunomodulatory Activity Exerted by PRL2010 Pili.
Similar to other extracellular structures encoded by several enteropathogens colonizing the human gut (36), we wanted to explore the possible roles played by B. bifidum PRL2010 pili in triggering (aspects of) the immune system of its human host. When we assayed the impact of the L. lactis clones producing Pil2PRL2010 and Pil3PRL2010 on cytokine expression by human macrophage-like cell line U937, we noticed a different cytokine modulation exerted by Pil2 and Pil3. Notably, piliated L. lactis-pil3PRL2010 clones displayed a significant induction (10-fold; P < 0.05) of the TNF-α mRNA levels compared with nonpiliated L. lactis-pil3PRL2010 (Fig. S7A). In contrast, piliated L. lactis-pil2PRL2010 clones did not appear to have any effect on the expression of the four cytokines that we assessed (Fig. S7A). Presence or absence of PRL2010 pili might therefore explain the difference in TNF-α response. To test this possibility directly, the TNF-α response was measured in challenging mice treated with piliated L. lactis-pil3PRL2010 cells.
Pili of PRL2010 Affect the Cytokine Profiles in a Mouse Model.
To investigate the role of sortase-dependent pili in PRL2010 colonization in mammals, piliated L. lactis-pil3PRL2010 and non piliated L. lactis-pil3PRL2010 were used in murine models. To mimic the natural route of gut microbial colonization, a 109 CFU dose of microencapsulated lactococci (to prevent pili removal from L. lactis during gastric transit) was orally administered daily to 12-wk-old BALB/c mice. Production of pili was induced before microencapsulation of lactococci by the addition of nisin (SI Materials and Methods). Furthermore, to ensure proper delivery of piliated/nonpiliated lactococci, we used alginate microencapsulation, which is known to release encapsulated bacteria following gastric transit (37). Mice were killed 4 h following the last lactococcal administration, and cytokine expression profiles were determined (Fig. S7B). Notably, under these in vivo conditions the piliated L. lactis-pil3PRL2010 evoked TNF-α expression and a significantly lower IL-10 response compared with the nonpiliated L. lactis-pil3PRL2010 (Fig. S7B) in murine cecum mucosa samples. These results reinforce the notion that pili of PRL2010 can influence the host innate immunity in a similar manner as previously outlined for other human gut commensals, such as Bacteroides (38).
Reportedly, and in accordance with our results, bifidobacteria can be strong inducers of TNF-α but weak inducers of other proinflammatory cytokines (39, 40), which are more specifically involved in mounting responses at systemic level, such as IL-12 (41, 42). Therefore, the immunomodulatory effects elicited by PRL2010 Pil3 may be delimited at local level, as previously suggested for other bifidobacterial strains, potentially because of insufficient induction of antigen-presenting cell maturation (41, 42). More specifically, a local induction of TNF-α could be important for the initiation of cross-talk among immune cells without causing any inflammation or detrimental effects (43).
Conclusions
Various ecological studies have demonstrated that bifidobacteria are a dominant bacterial group of the (human) infant gut microbiota, as well as part of the intestinal microbiota of an adult human being (3, 17). However, relatively little is known about the molecular basis sustaining their ability to colonize the human gut and to interact with the intestinal mucosa. Bifidobacteria arguably use a variety of mechanisms that may facilitate interactions with the intestinal mucosa at different life stages of the host, but perhaps also pertaining to different compartments of the gastrointestinal tract of the host (5, 16). The intimate attachment to the intestinal mucosa is presumed to be pivotal to allow colonization by gut commensals. Here, we describe the presence of a number of extracellular protein-encoding genes whose transcription is specifically up-regulated when our model bacterium B. bifidum PRL2010 was placed in contact with human cell lines or when present in the murine gut. Remarkably, among these specific host-induced genes we identified two loci that encode sortase-dependent pili. These appendages that decorate the bacterial surface are considered key molecules in mediating bacterial adherence to the host epithelium and may thus influence mucosal immune responses (12, 44). It was recently shown that type IV or Tad pili encoded by B. breve UCC2003 are required for colonization and persistence of this bacterium in the murine gut (16). However, in the case of B. bifidum PRL2010, the role of Tad pili in gut colonization and gut adhesion is currently not known. The genes that are responsible for the biosynthesis of the Tad pili are strictly conserved within bifidobacterial genomes, but the gene clusters that specify sortase-dependent pili production vary in number and sequence, and may thus represent strain-specific pili combinations. Thus, in contrast to Tad pili, sortase-dependent pili may impart unique features to a bifidobacterial strain among the complex microbiota present in the human gut, such as modulating specific host-microbe responses. In this context we noticed that piliated L. lactis-pil3PRL2010 clones evoked a higher TNF-α response during mouse colonization, compared with nonpiliated L. lactis-pil3PRL2010 clones, suggesting that PRL2010 pili not only contribute to adherence but also act as immunogenic effectors. Triggering of increased TNF-α production by pili encoded by B. bifidum PRL2010 may be an interesting feature of this species as one of the first colonizers of the human gut (3). In fact, cytokines belonging to the TNF-α superfamily are not only linked to the occurrence of inflammatory diseases (45), but also play a role in the rejection of tumors and the response to infections (46, 47). In addition, the induction of TNF-α may be important for the initiation of cross-talk among immune cells without causing any inflammation or detrimental effects (43). In fact, the infant’s immune system is immature and the presence of proinflammatory stimuli, such as those exerted by pili encoded by B. bifidum, may be crucial in developmental immunological programming. In this context, it is well known that decreased antigenic exposure has adverse effects on the budding immune system and increases the likelihood of developing atopic disorders (48). In addition, a transient inflammatory state could aid host defense. Thus, as recently suggested, the difference between pathogenic and gut commensal bacteria is in the magnitude of the immune response evoked, which can be defined as strong, intermediate or homeostatic (49).
The genome of B. bifidum PRL2010 encompasses three different loci encoding predicted sortase-dependent pili, of which only pil2 and pil3 appear to be functional as one of the pilus subunit-encoding genes of pil1 contains a frameshift (15). Here, we have shown that expression of pil2 and pil3 enhances adherence to enterocytes and modulates the host inflammatory response (for pil3), but it also promotes bacterial aggregation (for pil2). A possible model explaining the role of pili in host-PRL2010 interactions envisages an initial attachment of planctonic PRL2010 cells to the enterocytes by extending their pili, either sortase-dependent or Tad pili, toward the apical surface of the host cells. This initial adhesion to enterocytes is followed by a more intimate attachment driven by the establishment of the linkage between pili and specific host receptors, such as host-glycoproteins. In addition, pilus-mediated PRL2010 aggregation further assists the formation of a microbial community in the proximity of the colonized enterocytes. Pil2 and Pil3 were shown to exhibit different binding abilities with respect to carbohydrates, where Pil2 is able to adhere to typical mammalian gut carbohydrates, which appear to include fucose and mannose, and Pil3 showed an apparent ability to adhere to a wider set of carbohydrates, many of which would be expected to be present in the diet, thus suggesting that such diet-derived carbohydrates modulate PRL2010 gut colonization. Colonization as a result of a wide variety of host and bacterial factors, together with increased bacterial cell density, can lead to an enhanced innate immune response. This finding is in line with the notion that mammals depend on critical gene products from their gut microbiota to fully develop their immune system (50). Hence, the presence or absence of distinct B. bifidum sortase-dependent pili may represent pivotal molecules in colonization and persistence within the human gut, and may have a profound effect in terms of the developmental programming of the host immune system, which is in line with observations previously noted for other gut symbionts (51).
Materials and Methods
B. bifidum PRL2010 and L. lactis were manipulated and used as described in SI Materials and Methods. Detailed descriptions of bacterial strains, plasmids, and oligonucleotides (Table S1) used in this study, as well as methods for gene expression analyses, AFM investigations, bacterial adhesion assays, activation of human macrophage cell line, murine trials, and Western blot experiments are provided in SI Materials and Methods. The transcriptional array data have been deposited in the GEO database under accession no. GSE36442.
Supplementary Material
Acknowledgments
We thank GenProbio srl for financial support of the Laboratory of Probiogenomics. This work was supported in part by Fondazione Cariplo Grant 2010-0678 (to S.G. and V.T.); Irish Research Council for Science, Engineering, and Technology Embark postdoctoral Fellowship (to F.T.); and by PhD Fellowship Spinner 2013, Regione Emilia Romagna (to S.D.) and a Heath Research Board grant (to M.O.M.). D.v.S., M.O.M., and F.T. are members of The Alimentary Pharmabiotic Centre; D.v.S. is also a member of the Alimentary Glycoscience Research Cluster, both funded by Science Foundation Ireland through the Irish Government’s National Development Plan (Grant numbers 07/CE/B1368 and 08/SRC/B1393, respectively). M.O.M. is the recipient of a Health Research Board postdoctoral Fellowship Grant PDTM/20011/9.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE36442).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1303897110/-/DCSupplemental.
References
- 1.Ventura M, et al. Genome-scale analyses of health-promoting bacteria: Probiogenomics. Nat Rev Microbiol. 2009;7(1):61–71. doi: 10.1038/nrmicro2047. [DOI] [PubMed] [Google Scholar]
- 2.Guarner F, Malagelada JR. Gut flora in health and disease. Lancet. 2003;361(9356):512–519. doi: 10.1016/S0140-6736(03)12489-0. [DOI] [PubMed] [Google Scholar]
- 3.Turroni F, et al. Diversity of bifidobacteria within the infant gut microbiota. PLoS ONE. 2012;7(5):e36957. doi: 10.1371/journal.pone.0036957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ventura M, Turroni F, Motherway MO, MacSharry J, van Sinderen D. Host-microbe interactions that facilitate gut colonization by commensal bifidobacteria. Trends Microbiol. 2012;20(10):467–476. doi: 10.1016/j.tim.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 5.Fanning S, et al. Bifidobacterial surface-exopolysaccharide facilitates commensal-host interaction through immune modulation and pathogen protection. Proc Natl Acad Sci USA. 2012;109(6):2108–2113. doi: 10.1073/pnas.1115621109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Macfarlane S, Bahrami B, Macfarlane GT. Mucosal biofilm communities in the human intestinal tract. Adv Appl Microbiol. 2011;75:111–143. doi: 10.1016/B978-0-12-387046-9.00005-0. [DOI] [PubMed] [Google Scholar]
- 7.Turroni F, et al. Genome analysis of Bifidobacterium bifidum PRL2010 reveals metabolic pathways for host-derived glycan foraging. Proc Natl Acad Sci USA. 2010;107(45):19514–19519. doi: 10.1073/pnas.1011100107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kline KA, Fälker S, Dahlberg S, Normark S, Henriques-Normark B. Bacterial adhesins in host-microbe interactions. Cell Host Microbe. 2009;5(6):580–592. doi: 10.1016/j.chom.2009.05.011. [DOI] [PubMed] [Google Scholar]
- 9.Ton-That H, Schneewind O. Assembly of pili in Gram-positive bacteria. Trends Microbiol. 2004;12(5):228–234. doi: 10.1016/j.tim.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 10.Nallapareddy SR, et al. Endocarditis and biofilm-associated pili of Enterococcus faecalis. J Clin Invest. 2006;116(10):2799–2807. doi: 10.1172/JCI29021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scott JR, Zähner D. Pili with strong attachments: Gram-positive bacteria do it differently. Mol Microbiol. 2006;62(2):320–330. doi: 10.1111/j.1365-2958.2006.05279.x. [DOI] [PubMed] [Google Scholar]
- 12.Telford JL, Barocchi MA, Margarit I, Rappuoli R, Grandi G. Pili in gram-positive pathogens. Nat Rev Microbiol. 2006;4(7):509–519. doi: 10.1038/nrmicro1443. [DOI] [PubMed] [Google Scholar]
- 13.Kankainen M, et al. Comparative genomic analysis of Lactobacillus rhamnosus GG reveals pili containing a human-mucus binding protein. Proc Natl Acad Sci USA. 2009;106(40):17193–17198. doi: 10.1073/pnas.0908876106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lebeer S, et al. Functional analysis of Lactobacillus rhamnosus GG pili in relation to adhesion and immunomodulatory interactions with intestinal epithelial cells. Appl Environ Microbiol. 2012;78(1):185–193. doi: 10.1128/AEM.06192-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foroni E, et al. Genetic analysis and morphological identification of pilus-like structures in members of the genus Bifidobacterium. Microb Cell Fact. 2011;10(Suppl 1):S16. doi: 10.1186/1475-2859-10-S1-S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Connell Motherway M, et al. Functional genome analysis of Bifidobacterium breve UCC2003 reveals type IVb tight adherence (Tad) pili as an essential and conserved host-colonization factor. Proc Natl Acad Sci USA. 2011;108(27):11217–11222. doi: 10.1073/pnas.1105380108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Turroni F, et al. Exploring the diversity of the bifidobacterial population in the human intestinal tract. Appl Environ Microbiol. 2009;75(6):1534–1545. doi: 10.1128/AEM.02216-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wolfensohn S, Lloyd M. Handbook of Laboratory Animal Managment and Welfare. Blackwell, Oxford: 2003. [Google Scholar]
- 19.Guglielmetti S, et al. Implication of an outer surface lipoprotein in adhesion of Bifidobacterium bifidum to Caco-2 cells. Appl Environ Microbiol. 2008;74(15):4695–4702. doi: 10.1128/AEM.00124-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guglielmetti S, et al. Study of the adhesion of Bifidobacterium bifidum MIMBb75 to human intestinal cell lines. Curr Microbiol. 2009;59(2):167–172. doi: 10.1007/s00284-009-9415-x. [DOI] [PubMed] [Google Scholar]
- 21.Walsh EJ, O’Brien LM, Liang X, Hook M, Foster TJ. Clumping factor B, a fibrinogen-binding MSCRAMM (microbial surface components recognizing adhesive matrix molecules) adhesin of Staphylococcus aureus, also binds to the tail region of type I cytokeratin 10. J Biol Chem. 2004;279(49):50691–50699. doi: 10.1074/jbc.M408713200. [DOI] [PubMed] [Google Scholar]
- 22.Pallen MJ. The ESAT-6/WXG100 superfamily—And a new Gram-positive secretion system? Trends Microbiol. 2002;10(5):209–212. doi: 10.1016/s0966-842x(02)02345-4. [DOI] [PubMed] [Google Scholar]
- 23.Mohamadzadeh M, et al. Regulation of induced colonic inflammation by Lactobacillus acidophilus deficient in lipoteichoic acid. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4623–4630. doi: 10.1073/pnas.1005066107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deivanayagam CC, et al. Novel fold and assembly of the repetitive B region of the Staphylococcus aureus collagen-binding surface protein. Structure. 2000;8(1):67–78. doi: 10.1016/s0969-2126(00)00081-2. [DOI] [PubMed] [Google Scholar]
- 25.Ton-That H, Marraffini LA, Schneewind O. Sortases and pilin elements involved in pilus assembly of Corynebacterium diphtheriae. Mol Microbiol. 2004;53(1):251–261. doi: 10.1111/j.1365-2958.2004.04117.x. [DOI] [PubMed] [Google Scholar]
- 26.Alvarez-Martin P, et al. A two-component regulatory system controls autoregulated serpin expression in Bifidobacterium breve UCC2003. Appl Environ Microbiol. 2012;78(19):7032–7041. doi: 10.1128/AEM.01776-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruiz L, Zomer A, O’Connell-Motherway M, van Sinderen D, Margolles A. Discovering novel bile protection systems in Bifidobacterium breve UCC2003 through functional genomics. Appl Environ Microbiol. 2012;78(4):1123–1131. doi: 10.1128/AEM.06060-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Corrigan RM, Miajlovic H, Foster TJ. Surface proteins that promote adherence of Staphylococcus aureus to human desquamated nasal epithelial cells. BMC Microbiol. 2009;9:22. doi: 10.1186/1471-2180-9-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mishra A, et al. The Actinomyces oris type 2 fimbrial shaft FimA mediates co-aggregation with oral streptococci, adherence to red blood cells and biofilm development. Mol Microbiol. 2010;7:841–854. doi: 10.1111/j.1365-2958.2010.07252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yeung MK. Actinomyces: Surface macromolecules and bacteria-host interactions. In: Fischetti VA, Novick RP, Ferretti JJ, Portnoy DA, Rood JI, editors. Gram-Positive Pathogens. Washington, DC: American Society for Microbiology; 2000. pp. 583–593. [Google Scholar]
- 31.Kolenbrander PE. Coaggregations among oral bacteria. Methods Enzymol. 1995;253:385–397. doi: 10.1016/s0076-6879(95)53033-9. [DOI] [PubMed] [Google Scholar]
- 32.Farfan MJ, Cantero L, Vidal R, Botkin DJ, Torres AG. Long polar fimbriae of enterohemorrhagic Escherichia coli O157:H7 bind to extracellular matrix proteins. Infect Immun. 2011;79(9):3744–3750. doi: 10.1128/IAI.05317-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Torres AG, Slater TM, Patel SD, Popov VL, Arenas-Hernández MM. Contribution of the Ler- and H-NS-regulated long polar fimbriae of Escherichia coli O157:H7 during binding to tissue-cultured cells. Infect Immun. 2008;76(11):5062–5071. doi: 10.1128/IAI.00654-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Farfan MJ, Inman KG, Nataro JP. The major pilin subunit of the AAF/II fimbriae from enteroaggregative Escherichia coli mediates binding to extracellular matrix proteins. Infect Immun. 2008;76(10):4378–4384. doi: 10.1128/IAI.00439-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saarela S, Westerlund-Wikström B, Rhen M, Korhonen TK. The GafD protein of the G (F17) fimbrial complex confers adhesiveness of Escherichia coli to laminin. Infect Immun. 1996;64(7):2857–2860. doi: 10.1128/iai.64.7.2857-2860.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sampaio SC, et al. The flagella of an atypical enteropathogenic Escherichia coli strain are required for efficient interaction with and stimulation of interleukin-8 production by enterocytes in vitro. Infect Immun. 2009;77(10):4406–4413. doi: 10.1128/IAI.00177-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Islam MA, Yun CH, Choi YJ, Cho CS. Microencapsulation of live probiotic bacteria. J Microbiol Biotechnol. 2010;20(10):1367–1377. doi: 10.4014/jmb.1003.03020. [DOI] [PubMed] [Google Scholar]
- 38.Sonnenburg JL, Chen CT, Gordon JI. Genomic and metabolic studies of the impact of probiotics on a model gut symbiont and host. PLoS Biol. 2006;4(12):e413. doi: 10.1371/journal.pbio.0040413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Okada Y, et al. Anti-inflammatory effects of the genus Bifidobacterium on macrophages by modification of phospho-I kappaB and SOCS gene expression. Int J Exp Pathol. 2009;90(2):131–140. doi: 10.1111/j.1365-2613.2008.00632.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weiss G, et al. Bifidobacterium bifidum actively changes the gene expression profile induced by Lactobacillus acidophilus in murine dendritic cells. PLoS ONE. 2010;5(6):e11065. doi: 10.1371/journal.pone.0011065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fink LN, et al. Distinct gut-derived lactic acid bacteria elicit divergent dendritic cell-mediated NK cell responses. Int Immunol. 2007;19(12):1319–1327. doi: 10.1093/intimm/dxm103. [DOI] [PubMed] [Google Scholar]
- 42.Fink LN, Zeuthen LH, Ferlazzo G, Frøkiaer H. Human antigen-presenting cells respond differently to gut-derived probiotic bacteria but mediate similar strain-dependent NK and T cell activation. FEMS Immunol Med Microbiol. 2007;51(3):535–546. doi: 10.1111/j.1574-695X.2007.00333.x. [DOI] [PubMed] [Google Scholar]
- 43.Galdeano CM, de Moreno de LeBlanc A, Vinderola G, Bonet ME, Perdigón G. Proposed model: Mechanisms of immunomodulation induced by probiotic bacteria. Clin Vaccine Immunol. 2007;14(5):485–492. doi: 10.1128/CVI.00406-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mandlik A, Swierczynski A, Das A, Ton-That H. Pili in Gram-positive bacteria: Assembly, involvement in colonization and biofilm development. Trends Microbiol. 2008;16(1):33–40. doi: 10.1016/j.tim.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yasutake N, et al. The role of tumor necrosis factor (TNF)-alpha in the antitumor effect of intrapleural injection of Lactobacillus casei strain Shirota in mice. Med Microbiol Immunol (Berl) 1999;188(1):9–14. doi: 10.1007/s004300050099. [DOI] [PubMed] [Google Scholar]
- 46.Wajant H, Pfizenmaier K, Scheurich P. Tumor necrosis factor signaling. Cell Death Differ. 2003;10(1):45–65. doi: 10.1038/sj.cdd.4401189. [DOI] [PubMed] [Google Scholar]
- 47.Lebeer S, Vanderleyden J, De Keersmaecker SC. Host interactions of probiotic bacterial surface molecules: Comparison with commensals and pathogens. Nat Rev Microbiol. 2010;8(3):171–184. doi: 10.1038/nrmicro2297. [DOI] [PubMed] [Google Scholar]
- 48.Bach JF. The effect of infections on susceptibility to autoimmune and allergic diseases. N Engl J Med. 2002;347(12):911–920. doi: 10.1056/NEJMra020100. [DOI] [PubMed] [Google Scholar]
- 49.Otte JM, et al. Probiotics regulate the expression of COX-2 in intestinal epithelial cells. Nutr Cancer. 2009;61(1):103–113. doi: 10.1080/01635580802372625. [DOI] [PubMed] [Google Scholar]
- 50.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453(7195):620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 51.Kaplan JL, Shi HN, Walker WA. The role of microbes in developmental immunologic programming. Pediatr Res. 2011;69(6):465–472. doi: 10.1203/PDR.0b013e318217638a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.