Abstract
G-protein-coupled receptors (GPCRs) mediate cellular responses to various hormones and neurotransmitters and are important targets for treating a wide spectrum of diseases. Although significant advances have been made in structural studies of GPCRs, details of their activation mechanism remain unclear. The X-ray crystal structure of the M2 muscarinic receptor, a key GPCR that regulates human heart rate and contractile forces of cardiomyocytes, was determined recently in an inactive antagonist-bound state. Here, activation of the M2 receptor is directly observed via accelerated molecular dynamics simulation, in contrast to previous microsecond-timescale conventional molecular dynamics simulations in which the receptor remained inactive. Receptor activation is characterized by formation of a Tyr2065.58–Tyr4407.53 hydrogen bond and ∼6-Å outward tilting of the cytoplasmic end of transmembrane α-helix 6, preceded by relocation of Trp4006.48 toward Phe1955.47 and Val1995.51 and flipping of Tyr4307.43 away from the ligand-binding cavity. Network analysis reveals that communication in the intracellular domains is greatly weakened during activation of the receptor. Together with the finding that residue motions in the ligand-binding and G-protein-coupling sites of the apo receptor are correlated, this result highlights a dynamic network for allosteric regulation of the M2 receptor activation.
Keywords: enhanced sampling, GPCR signaling, allosteric network, cross-correlations, drug design
Muscarinic acetylcholine receptors belong to the superfamily of G-protein-coupled receptors (GPCRs) that mediate cellular responses to hormones, neurotransmitters, and the senses of sight, olfaction, and taste. They play critical roles in both the central and parasympathetic nervous systems and are important targets for the treatment of a wide spectrum of diseases, including abnormal heart rate, asthma, chronic obstructive pulmonary disease, Alzheimer’s disease, Parkinson disease, and schizophrenia (1).
Muscarinic receptors are known to be constitutively active—i.e., they exhibit a certain level of basal activity even without binding any agonist (2). This characteristic suggests that there exists an ensemble of different conformations in muscarinic receptors. The conformational equilibrium is biased toward an active state when the receptors are bound by agonists. In contrast, the receptors are switched to an inactive state upon binding of inverse agonists. Additionally, they are able to bind neutral antagonists that have no signaling effects but block binding of other ligands, as well as partial agonists that induce only submaximal activity (2).
The M2 muscarinic receptor is widely distributed in mammalian tissues and is the only subtype found in the human heart. Its activation results in a decrease in heart rate and a reduction in heart contraction force (3). The receptor X-ray structure was determined recently in an inactive state with antagonist 3-quinuclidinyl-benzilate (QNB) bound (4). Starting from the N terminus on the extracellular side, the receptor traverses through the cell membrane with seven transmembrane (TM) α helices (referred as TM1 to TM7). Three extracellular loops (ECL1 to ECL3) and three intracellular loops (ICL1 to ICL3) are exposed on alternating sides of the membrane. The receptor ends with helix 8 at the C terminus on the intracellular side.
Previous X-ray studies have revealed active structures of only two GPCRs, opsin, activated rhodopsin (5, 6), and the β2-adrenergic receptor (β2AR) (7, 8). These structures are characterized by rearrangements of the TM5, TM6, and TM7 helices relative to the inactive configuration. The cytoplasmic end of TM6 in active β2AR is tilted outward by 14 and 11 Å when coupled with the G protein (7) and its mimetic nanobody (8), respectively. In comparison, a smaller TM6 movement (∼6–7 Å) is found in ligand-free opsin (5, 6). In both opsin and active β2AR, the salt bridge between Arg3.50 and Glu6.30 (“ionic lock” in many GPCRs) is broken, whereas Tyr5.58 and Tyr7.53 relocate their side chains toward each other in the intracellular pocket (5–8). Note that the residue superscripts denote Ballesteros–Weinstein numbering (9). The most conserved residue in helix N is assigned N.50, and the others are numbered decreasingly toward the N terminus and increasingly toward the C terminus.
Computational simulations have previously been performed to investigate conformational ensembles and structural dynamics of GPCRs (10–12). In a landmark study by Dror et al. (10), deactivation of β2AR was modeled with microsecond-timescale conventional molecular dynamics (cMD) simulations by using a specialized supercomputer Anton. Starting from the active X-ray structure, β2AR transitioned into the inactive conformation upon removal of the G protein or its mimetic nanobody, and an intermediate was identified in the transition pathways. Moreover, Anton simulations of the M2 receptor that started from the QNB-removed X-ray structure captured binding of antagonist tiotropium (TTP) to an extracellular vestibule, but not to the orthosteric binding site (13). This finding suggests that the receptor stayed in a ligand-free (apo) form. Further investigation (described below) showed that the receptor remained inactive through the simulations. GPCR activation has not been observed, even in the longest cMD simulations (10), and has been shown experimentally to occur on millisecond timescales (14).
Active structures for most GPCRs are still lacking, and details of the GPCR activation mechanism remain unclear. For the M2 receptor, open questions include what different conformational states are involved during activation, how the allosteric regulation from the extracellular ligand-binding site to the intracellular G-protein-coupling site is achieved, and whether there are correlated motions between the two sites.
Accelerated molecular dynamics (aMD) is a biomolecular enhanced sampling technique that works by adding boost potential to the energy surface, effectively decreasing the energy barriers and thus accelerating transitions between the low-energy states (15–17). AMD has been successfully applied to a number of systems (18, 19), and hundreds-of-nanosecond aMD simulations have been shown to capture millisecond-timescale events (20).
In the present study, we apply aMD to simulate the M2 receptor and observe its activation from the inactive X-ray structure on removal of the QNB antagonist. Conformational changes of the receptor and the dynamic network behind its activation are analyzed through community network and generalized correlation analyses. Community network analysis identifies communities of highly connected residues and assesses the probability of information transfer between communities based on residue correlation and proximity (19, 21, 22), and generalized correlation analysis calculates cross-correlated residue motions in proteins (23).
Results
Starting from the X-ray structure of QNB-bound M2 receptor [Protein Data Bank ID: 3UON), we performed a 100-ns cMD simulation, followed by a 100-ns dihedral aMD simulation (see SI Appendix for simulation details). The receptor does not deviate substantially from the X-ray structure in the simulations, although the ECL3 region exhibits markedly higher fluctuations in dihedral aMD than in cMD, which agrees very well with a previous 16.4-μs Anton simulation in ref. 13. (SI Appendix, Fig. S2). This finding suggests that enhanced sampling is achieved by using dihedral aMD. Next, we removed the QNB antagonist to simulate the receptor in its apo form. In contrast to the QNB-bound form, the apo receptor shows increased dynamics in the ligand-binding regions of TM3, TM4, TM5, and TM6 in the dihedral aMD simulations (SI Appendix, Fig. S3A). Furthermore, fluctuations in ECL3 and part of ECL2 appear to be higher than those observed in a set of Anton simulations (one for 14.2 μs and two for 1 μs), in which antagonist TTP binds to an extracellular vestibule region formed by ECL2 and ECL3 (13) (SI Appendix, Fig. S3B).
Although increased dynamics are observed in the apo M2 receptor compared with the antagonist-bound form in dihedral aMD simulations, the receptor maintains a conformation similar to the inactive X-ray structure (Movie S1), similarly for the microsecond-timescale Anton simulations (Movie S2). Therefore, we applied dual-boost aMD, which provides greater enhanced sampling than dihedral aMD, to additional simulations of the apo M2 receptor. Restarting from the final structure of the 100-ns cMD simulation, five independent dual-boost aMD simulations (one for 400 ns and four for 200 ns) were performed with random atomic velocity initializations at 310 K (SI Appendix, Table S1). Significantly larger conformational space is sampled in the 400-ns dual-boost aMD simulation than in the cMD and dihedral aMD simulations (SI Appendix, Fig. S4). Such enhanced sampling enables direct observation of the receptor activation and identification of the intermediate and active states, which are distinct from the inactive X-ray conformation. In the four 200-ns dual-boost aMD simulations, the apo receptor visits only the intermediate state in simulation 8 (Sim8) and Sim9 and both the intermediate and active states in Sim10 and Sim11 (SI Appendix, Fig. S5). In comparison, in the QNB-bound form, the receptor maintains the inactive conformation through a 200-ns dual-boost aMD simulation (SI Appendix, Fig. S6).
Activation of M2 Receptor Observed in aMD simulation.
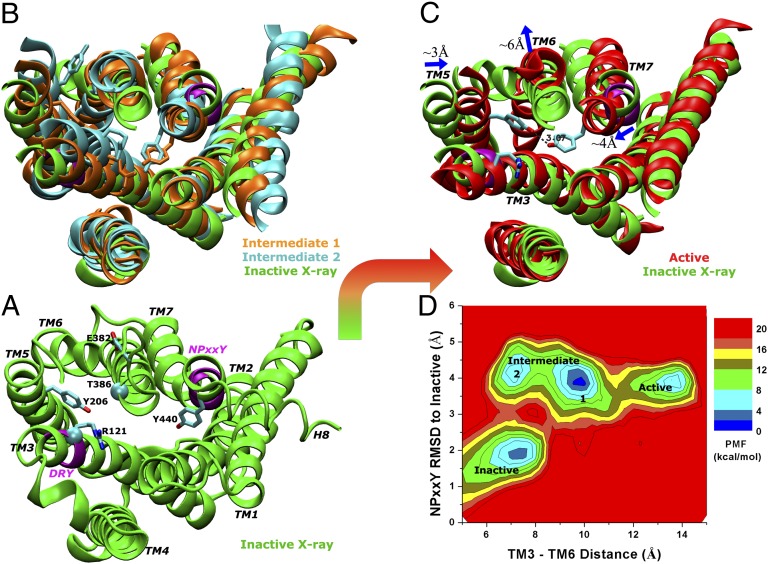
During the 400-ns dual-boost aMD simulation, activation of the apo M2 receptor from its inactive X-ray conformation is directly observed (Movie S3). The activation is characterized by formation of a Tyr2065.58–Tyr4407.53 hydrogen bond in the G-protein-coupling site and ∼6 Å outward tilting of the cytoplasmic end of TM6 as shown in Fig. 1. The Arg1213.50–Glu3826.30 salt bridge is broken during activation of the receptor (SI Appendix, Fig. S8A).
Fig. 1.
Activation of the apo M2 receptor is directly observed with dual-boost aMD simulation. (A) The starting X-ray structure (green), in which two structural motifs conserved among GPCRs (DRY in TM3 and NPxxY in TM7) are highlighted in purple; key residues including Arg1213.50, Glu3826.30, Tyr2065.58, and Tyr4407.53 are rendered as sticks; and the Cα atoms of Arg1213.50 and Thr3866.34 used for calculating the distance between cytoplasmic ends of TM3 and TM6 plotted in D are shown in spheres. (B) Two intermediate conformations, both of which exhibit inward displacement of the NPxxY motif and undistorted TM7, but differ in the orientation of the Tyr2065.58 side chain. (C) Activated receptor conformation showing ∼6-Å outward tilting of the TM6 cytoplasmic end and formation of a hydrogen bond between Tyr2065.58 and Tyr4407.53. The Arg1213.50–Glu3826.30 salt bridge (“ionic lock” identified in many GPCRs) is broken during activation of the receptor. (D) Reweighted potential of mean force calculated for the TM3–TM6 distance and RMSD of the NPxxY motif relative to the inactive X-ray structure.
The receptor initially changes from the inactive X-ray structure to an intermediate state, in which TM7 becomes undistorted with significant displacement in the intracellular NPxxY motif, ∼4 Å RMSD relative to the inactive conformation. The cytoplasmic end of TM5 exhibits high mobility, with Tyr2065.58 reorienting from the initial position between TM3 and TM6 to the lipid-exposed side of TM6. Two low-energy conformations are then observed in the intermediate state (Fig. 1B). Next, Tyr2065.58 and Tyr4407.53 relocate their side chains toward each other, forming a hydrogen bond in the intracellular pocket, and the cytoplasmic end of TM6 tilts outward by ∼6 Å (Fig. 1C). This change drives the apo receptor to an active state that resembles the X-ray structure of ligand-free opsin (5, 6), and the largely opened G-protein–coupling site can accommodate the GαCT peptide (SI Appendix, Fig. S7). Fig. 1D plots the potential of mean force (PMF) calculated for the TM3–TM6 distance vs. the RMSD of the NPxxY motif relative to the inactive X-ray structure. The PMF map was constructed by reweighting the aMD simulation according to the applied boost potential (SI Appendix, Eq. S3). It clearly depicts the inactive, intermediate, and active conformational states of the M2 receptor.
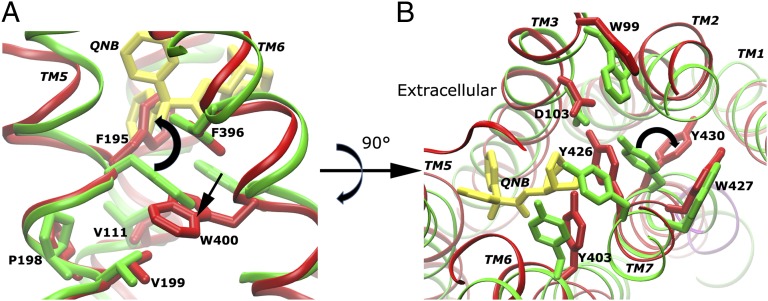
During activation of the M2 receptor, the most invariant region (“core”) is found to be TM3 by using the Bio3d program (24). Prominent structural changes occur in the ligand-binding site apart from rearrangements of the TM5, TM6, and TM7 helices in the G-protein-coupling site as described above. When receptor transitions to the intermediate state, Trp4006.48 relocates its side chain toward Phe1955.47 and Val1995.51, and Phe1955.47 flips the phenyl ring into the space that was originally occupied by the QNB antagonist in the X-ray structure (Fig. 2A). Meanwhile, Tyr4307.43 breaks the hydrogen bond with Asp1033.32, which is subsequently stabilized by Tyr4267.39, and flips the side chain from the ligand-binding cavity to the TM7–TM2 interface toward the cytoplasmic side (Fig. 2B). This change appears to correlate with displacement of the NPxxY motif in the intracellular domain of TM7 at ∼80 ns in the simulation (SI Appendix, Fig. S8 E and F).
Fig. 2.
Residue conformational changes are observed in the ligand-binding site during activation of the apo M2 receptor. (A) Trp4006.48 relocates toward Phe-1955.47 and Val-1995.51, and Phe1955.47 flips the phenyl ring into the space that was originally occupied by QNB in the X-ray structure as shown by superimposing the receptor TM bundle. (B) Tyr4307.43 flips the side chain from the ligand-binding cavity to the TM7–TM2 interface, and its hydrogen-bonding interaction with Asp1033.32 is replaced by Tyr4267.39.
Highly Dynamic Allosteric Network in the M2 Receptor.
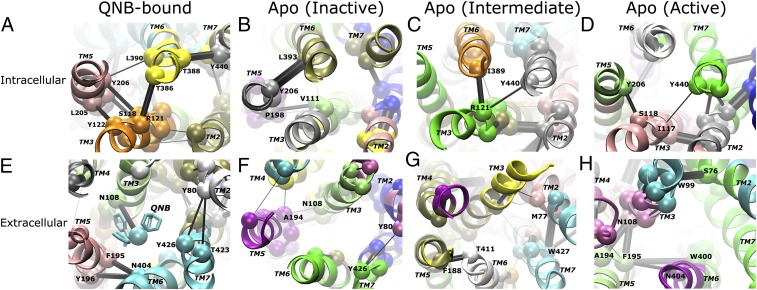
A highly dynamic network is revealed in the M2 receptor via community network analysis (SI Appendix, Methods). The receptor exhibits significant differences in the network between the QNB-bound form and the inactive, intermediate, and active states of the apo form. The distribution of residues into highly connected clusters (communities) evolves in these different receptor states. Additionally, the communication strength between communities in the ligand-binding and G-protein-coupling sites appears to be dynamically modulated by the conformational transitions.
In the 200-ns dual-boost aMD simulation of the QNB-bound receptor, a strong network is identified between intracellular domains of the TM3, TM5, TM6, and TM7 helices through the Arg1213.50–Thr3866.34, Ser1183.47–Tyr2065.58, Tyr1223.51–Leu2055.57, Tyr2065.58–Leu3906.38, and Thr3886.36–Tyr4407.53 edge interactions (Fig. 3A). In the ligand-binding site, the QNB antagonist, which is clustered in the same community as the TM6 and TM7 extracellular domains, connects to TM3 strongly via Asn1083.37. Strong communication is also found between TM5 and TM6 via the Phe1955.47–Asn4046.52–Tyr1965.48 interactions, for which mutation of Asn4046.52 has been suggested to reduce antagonist binding affinity by >10-fold (4). Weaker communication appears between TM2 and TM7 through the Tyr4267.39–Tyr802.61–Thr4237.36 interactions (Fig. 3E). In the 100-ns cMD simulation of the QNB-bound receptor, similar strong intracellular and extracellular networks connecting the TM domains are also observed (SI Appendix, Fig. S9).
Fig. 3.
A highly dynamic network is identified in the M2 receptor through community network analysis. Intracellular views of the G-protein-coupling site for the QNB-bound form (A) and the apo form in inactive (B), intermediate (C), and active (D) states are shown, and the corresponding extracellular views of the ligand-binding site are shown in E–H. The receptor exhibits significant differences in its allosteric network between the QNB-bound form and the inactive, intermediate, and active states of the apo form. Notably, the network strength between communities in the intracellular domains is greatly weakened during activation of the apo receptor. Network communities are colored separately by their ID number; critical nodes located at the interface of neighboring communities are rendered as spheres and labeled by residue number; and the connecting edges are represented by black lines with their width weighted by betweenness, the probability of information transfer between communities. The TM helices are labeled in italics.
During the 400-ns dual-boost aMD simulation of the apo receptor, the hydrogen bond between the Tyr2065.58 and Tyr4407.53 side chains is formed twice at ∼120–150 and ∼360 ns, indicating activation of the receptor (SI Appendix, Fig. S6B). After clustering simulation snapshots into the different receptor states, the three longest time windows that approximately correspond to the inactive (0–60 ns), intermediate (180–300 ns), and active (120–150 ns) states, respectively, are extracted from the simulation to analyze the apo receptor community network as follows.
With removal of the QNB antagonist, the levels of network communication are altered, even when the apo receptor remains in the inactive state. In the G-protein-coupling site, the intracellular domains of TM3 and TM5 merge into a single community and TM6 and TM7 into another, and the two communities are strongly connected via the Tyr2065.58–Leu3936.41 interaction (Fig. 3B). In the “connector” region, which is located between the ligand-binding and G-protein-coupling sites (10), moderate network strength is found between TM3 and TM5 via the Val1113.40–Pro1985.50 interaction. However, in the ligand-binding site, the extracellular domains of the TM3, TM5, TM6, and TM7 helices become only loosely connected to each other (Fig. 3F).
In the intermediate state of the apo receptor, the intracellular domains of TM6 and TM7 break into two separate communities in the G-protein-coupling site. TM6 possesses close contact with TM3 via the Arg1213.50–Thr3866.34 interaction, and TM7 connects to TM3 via the Arg1213.50–Tyr4407.53 interaction with inward displacement of the NPxxY motif (Fig. 3C). The extracellular domains of TM5 and TM6 are connected via the Phe1885.40–Thr4116.59 interaction, and all extracellular domains of the TM5, TM6, and TM7 helices are loosely connected to that of TM3 (Fig. 3G).
As the receptor transitions to the active state, in the G-protein-coupling site connectivity is observed between TM3 and TM5 via the Ser1183.47–Tyr2065.58 interaction and between TM3 and TM7 via the Ile1173.46–Tyr4407.53interaction, largely due to the inward movement of the Tyr206 and Tyr440 residues (Fig. 3D). However, the overall network strength between intracellular domains of the TM helices in the active apo receptor is significantly weaker than in the inactive and intermediate states. Notably, TM6 becomes loosely connected to TM3, TM5, and TM7. In the connector region at the base of the ligand-binding site, TM6 connects to TM5 via the Try4006.48–Phe1955.47–Asn4046.52 interactions, and TM5 connects to TM3 via the Phe1955.47–Asn1083.37–Ala1945.46 interactions (Fig. 3H). These interactions tend to tighten the connector region of the TM helices, in contrast to the concomitant reduced interactions between intracellular domains in the G-protein-coupling site. Therefore, the network strength between communities in the M2 receptor appears to be dynamically modulated during activation of the receptor and network of the intracellular domains is greatly weakened in the active state.
Correlated Motions Between the Ligand-Binding and G-Protein-Coupling Sites.
Correlated residue motions were identified between the ligand-binding and G-protein-coupling sites in the apo M2 receptor by using the generalized cross-correlation analysis (SI Appendix, Methods). The dynamic map of residue cross-correlations is compared to the QNB-bound and apo forms of the receptor in dual-boost aMD simulations as shown in Fig. 4.
Fig. 4.
Correlated motions are identified between residues in the extracellular ligand-binding and intracellular G-protein-coupling sites of the apo M2 receptor. Shown is a dynamic map of color-coded residue cross-correlations for the QNB-bound (lower triangle) and apo (upper triangle) forms of the M2 receptor calculated from the dual-boost aMD simulations. Residues in the TM1 to TM7 helices are indicated by bars on the top and right. Axis breaks correspond to residues 218–376 that are missing in the ICL3 region.
In the QNB-bound form, where the receptor stays in the inactive X-ray conformation, residue motions in different protein regions are poorly correlated with nearly all cross-correlation values <0.6 (lower triangle of Fig. 4). In comparison, in the apo form, where the receptor transitions between the inactive, intermediate, and active conformational states, residue motions exhibit significantly higher correlations across the entire protein (upper triangle of Fig. 4). Table 1 lists the protein regions that are involved in highly correlated residue motions with cross-correlations >0.6. Specifically, the ECL2 region is correlated with the extracellular domain of TM3 due to the Cys963.25–Cys176ECL2 disulfide bond and the neighboring extracellular domain of TM4. Other highly correlated protein regions are mostly located in the TM5, TM6, and TM7 helices.
Table 1.
List of protein regions that are involved in highly correlated residue motions with cross-correlations >0.6 in the dual-boost aMD simulation of the apo M2 receptor
| Region | Residues |
|
| Extracellular | Intracellular | |
| TM3 | 94–107 | 115–126 |
| TM4 | 156–164 | 138–152 |
| TM5 | 184–195 | 197–210 |
| TM6 | 400–410 | 385–399 |
| TM7 | 422–432 | 436–440 |
| ECL2 | 163–184 | — |
In the apo receptor, the intracellular domains of TM5, TM6, and TM7 are highly correlated with their corresponding extracellular domains. The intracellular domain of TM5 is also correlated to that of TM3, consistent with their close network interactions shown in Fig. 3 B–D. Moreover, the intracellular domain of TM6 that tilts outward upon receptor activation is correlated with the intracellular and extracellular domains of both TM5 and TM7. In the ligand-binding site, the extracellular domain of TM5 is correlated with those of TM6 and TM7, as well as the extracellular domains of TM6 and TM7. Similarly, increased cross-correlations of residue motions in the TM5, TM6, and TM7 helices of the apo form are also observed in cMD simulations (SI Appendix, Fig. S10).
Discussion
In this study, activation of the M2 receptor in the ligand-free (apo) form is directly observed through hundreds-of-nanosecond aMD simulation. This finding enables a detailed understanding of the GPCR activation mechanism at an atomistic level. The receptor activation is characterized by formation of a hydrogen bond between the intracellular domains of TM5 and TM7 (Tyr2065.58–Tyr4407.53) and also by ∼6-Å outward tilting of the cytoplasmic end of TM6. This result is in agreement with previous GPCR studies, e.g., TM6 has been suggested to be a switch for conformational transition between the inactive and active states of the M5 receptor (25), and similar structural rearrangements of TM5 and TM7 and the outward tilting of TM6 have been characterized in the active structures of rhodopsin (5, 6) and β2AR (7, 8).
The observed active M2 receptor resembles the ligand-free opsin with its G-protein–coupling site open to accommodate the GαCT peptide (5) (SI Appendix, Fig. S7). The M2 intermediate conformations (Fig. 1B) appear to be different from those of β2AR identified in earlier Anton simulations (10), largely due to the two different processes simulated—i.e., the deactivation of β2AR from the G-protein/nanobody-coupled conformation and the activation of the M2 receptor in a ligand-free form. TM3 is identified to be the most invariant “core” domain during activation of the M2 receptor, which is consistent with earlier findings that it serves as a conserved structural and functional hub across diverse GPCRs (26).
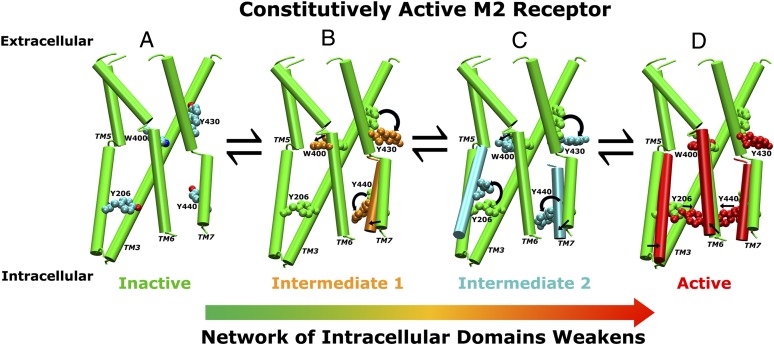
By examining conformational changes of key residues and TM domains in the aMD simulation, we are able to identify an allosteric activation pathway in the M2 receptor as shown in Fig. 5. As a constitutively active GPCR, the receptor exists in a conformational equilibrium of the inactive, intermediate, and active states. When the receptor transitions from the inactive to the intermediate state, Trp4006.48 relocates toward Phe1955.47 and Val1995.51, and the phenyl ring of Phe1955.47 flips into the space that was originally occupied by QNB in the X-ray structure (Fig. 2A); Tyr4307.43 flips the side chain from the ligand-binding cavity to the TM7–TM2 interface (Fig. 2B); and Tyr4307.43 reorients the side chain with concomitant inward displacement of the NPxxY motif in the intracellular domain of TM7. The side chain of Tyr2065.58 can reorient from the initial position between TM3 and TM6 to the lipid-exposed side of TM6, resulting in an alternative intermediate conformation (Fig. 5C). During final transition to the active sate, Tyr2065.58 and Tyr4407.53 relocate the side chains toward each other, forming hydrogen-bonding interaction, and the cytoplasmic end of TM6 tilts outward by ∼6 Å (Fig. 5D).
Fig. 5.
An allosteric activation pathway of the M2 receptor derived from aMD simulations. As a constitutively active GPCR, the apo M2 receptor exists in a conformational equilibrium of inactive, intermediate, and active states. (A) The inactive state with the TM3, TM5, TM6, and TM7 helices shown in cartoons and key residues Trp4006.48, Tyr4307.43, Tyr2065.58, and Tyr4407.53 in CPK representation. (B) Trp4006.48, Tyr4307.43, and Tyr4407.53 relocate their side chains during the receptor transition to the intermediate state. (C) Tyr2065.58 reorients the side chain from the initial position between TM3 and TM6 to the lipid-exposed side of TM6, resulting in an alternative intermediate conformation. (D) During final transition to the active sate, Tyr2065.58 and Tyr4407.53 relocate their side chains toward each other, forming a hydrogen bond, and the cytoplasmic end of TM6 tilts ∼6 Å outward away from the TM bundle. Activation of the receptor significantly reduces the network strength of the intracellular domains in the G-protein-coupling site, which apparently facilitates association of the G protein and further stabilization of the receptor active conformation.
With antagonist QNB bound in the extracellular ligand-binding site, the M2 receptor stays in the inactive state. Intracellular domains of the TM helices are strongly connected to each other via noncovalent residue interactions, precluding association of the G protein. In contrast, the apo receptor network of the intracellular domains is significantly weakened during the receptor activation (Fig. 3), which apparently facilitates association of the G protein and further stabilization of the receptor active conformation.
Relocation of Trp4006.48 toward Phe1955.47 and Val1995.51 is found to be a key conformational change during activation of the M2 receptor (Fig. 5). This finding is consistent with previous structural studies on rhodopsin and the A2A adenosine receptor (A2AAR) that suggest the conserved Trp6.48 to be a transmission switch, which links agonist binding to the movement of the intracellular domains of TM5 and TM6 during GPCR activation (27). Another key conformational change involves relocation of the Tyr4307.43 side chain, whose hydrogen-bonding interaction with Asp1033.32 can be replaced by Tyr4267.39. This change resembles breaking of a Lys7.43–Glu3.28 salt bridge in the activation of rhodopsin (5, 6), as well as relocation of Ser7.42 and His7.43 coordinated by Thr3.36 during agonist binding of A2AAR (27, 28). These residue interactions and conformational changes play important roles in GPCR activation.
With generalized cross-correlation analysis, residue motions in the ligand-binding and G-protein-coupling sites are found to be correlated during activation of the M2 receptor. Notably, the intracellular domain of TM6 that undergoes large-scale outward movement during receptor activation is highly correlated with the extracellular domains of TM5, TM6, and TM7 surrounding the ligand-binding site (Fig. 4). Such correlations can be justified by the conformational changes triggered by the Trp4006.48 transmission switch discussed above. The intracellular domain of TM7 is also correlated to its extracellular counterpart. Tyr4307.43 flips from the ligand-binding cavity to the TM7–TM2 interface at ∼80 ns, which coincides with displacement of the NPxxY motif in the intracellular domain of TM7. These correlated motions between the ligand-binding and G-protein–coupling sites may provide a coherent picture for allosteric regulation of GPCR activation.
Apart from the active conformation that resembles opsin, another different M2 conformation is observed in one of the four 200-ns dual-boost aMD simulations (Sim11). Relative to the inactive X-ray conformation, this conformation depicts shear motion of intracellular domains of TM6 and TM7 toward TM1 by ∼3 Å and outward movement of the TM5 cytoplasmic end by ∼6 Å (SI Appendix, Fig. S11). It may be relevant for coupling of the M2 receptor with different signaling effectors other than the G protein, e.g., protein kinases and arrestins. Site-directed mutagenesis experiments identified two clusters of Ser/Thr residues (Ser286–Ser290 and Thr307–Ser311) in ICL3 as agonist-dependent phosphorylation sites for arrestin binding (29, 30). This finding may justify the large opening between the intracellular domains of TM5 and TM6 in this different conformation, because the opening appears to be necessary for exposure of the two Ser/Thr clusters in ICL3 for phosphorylation and arrestin binding. However, further validation is still required, ideally with a high-resolution arrestin-coupled GPCR structure. Nevertheless, the activation-associated conformational states of the M2 receptor and its highly dynamic network identified in the present aMD simulations may allow us to perform structural screening to search for allosteric drugs.
Methods
Both cMD and aMD simulations have been performed by using NAMD2 (31, 32) on the M2 receptor that is embedded in a palmitoyl-oleoyl-phosphatidyl-choline (POPC) lipid bilayer and solvated an aqueous medium of 0.15 M NaCl with all atoms represented explicitly. The CHARMM27 parameter set was used for the protein (with CMAP terms included) (33, 34), CHARMM36 for POPC lipids (35), and TIP3P model for water molecules (36). Force-field parameters for QNB were obtained from the CHARMM ParamChem web server (37). Details of the simulation protocols and analyses are provided in SI Appendix.
Supplementary Material
Acknowledgments
We thank Irina Tikhonova, Jeff Wereszcynski, Yi Wang, Cesar de Oliveira, and William Sinko for valuable discussions; and Ron Dror, Jodi Hezky, and Albert Pan from D. E. Shaw’s research group for generously providing the Anton simulation trajectories. Computing time was provided on the Gordon supercomputer by the Extreme Science and Engineering Discovery Environment (XSEDE) Award TG-MCA93S013. This work was supported by National Science Foundation Grant MCB1020765), National Institutes of Health Grant GM31749, the Howard Hughes Medical Institute, the Center for Theoretical Biological Physics, and the National Biomedical Computation Resource.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1309755110/-/DCSupplemental.
References
- 1.Kow RL, Nathanson NM. Structural biology: Muscarinic receptors become crystal clear. Nature. 2012;482(7386):480–481. doi: 10.1038/482480a. [DOI] [PubMed] [Google Scholar]
- 2.Spalding TA, Burstein ES. Constitutive activity of muscarinic acetylcholine receptors. J Recept Signal Transduct Res. 2006;26(1–2):61–85. doi: 10.1080/10799890600567349. [DOI] [PubMed] [Google Scholar]
- 3.Vogel WK, Sheehan DM, Schimerlik MI. Site-directed mutagenesis on the m2 muscarinic acetylcholine receptor: The significance of Tyr403 in the binding of agonists and functional coupling. Mol Pharmacol. 1997;52(6):1087–1094. doi: 10.1124/mol.52.6.1087. [DOI] [PubMed] [Google Scholar]
- 4.Haga K, et al. Structure of the human M2 muscarinic acetylcholine receptor bound to an antagonist. Nature. 2012;482(7386):547–551. doi: 10.1038/nature10753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scheerer P, et al. Crystal structure of opsin in its G-protein-interacting conformation. Nature. 2008;455(7212):497–502. doi: 10.1038/nature07330. [DOI] [PubMed] [Google Scholar]
- 6.Park JH, Scheerer P, Hofmann KP, Choe H-W, Ernst OP. Crystal structure of the ligand-free G-protein-coupled receptor opsin. Nature. 2008;454(7201):183–187. doi: 10.1038/nature07063. [DOI] [PubMed] [Google Scholar]
- 7.Rasmussen SGF, et al. Crystal structure of the β2 adrenergic receptor-Gs protein complex. Nature. 2011;477(7366):549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rasmussen SGF, et al. Structure of a nanobody-stabilized active state of the β(2) adrenoceptor. Nature. 2011;469(7329):175–180. doi: 10.1038/nature09648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ballesteros JA, Weinstein H (1995) Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. Methods in Neurosciences, ed Stuart CS (Academic, New York), Vol 25, pp 366–428.
- 10.Dror RO, et al. Activation mechanism of the β2-adrenergic receptor. Proc Natl Acad Sci USA. 2011;108(46):18684–18689. doi: 10.1073/pnas.1110499108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Niesen MJM, Bhattacharya S, Vaidehi N. The role of conformational ensembles in ligand recognition in G-protein coupled receptors. J Am Chem Soc. 2011;133(33):13197–13204. doi: 10.1021/ja205313h. [DOI] [PubMed] [Google Scholar]
- 12.Provasi D, Artacho MC, Negri A, Mobarec JC, Filizola M. Ligand-induced modulation of the free-energy landscape of G protein-coupled receptors explored by adaptive biasing techniques. PLOS Comput Biol. 2011;7(10):e1002193. doi: 10.1371/journal.pcbi.1002193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kruse AC, et al. Structure and dynamics of the M3 muscarinic acetylcholine receptor. Nature. 2012;482(7386):552–556. doi: 10.1038/nature10867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vilardaga J-P, Bünemann M, Krasel C, Castro M, Lohse MJ. Measurement of the millisecond activation switch of G protein-coupled receptors in living cells. Nat Biotechnol. 2003;21(7):807–812. doi: 10.1038/nbt838. [DOI] [PubMed] [Google Scholar]
- 15.Markwick PRL, McCammon JA. Studying functional dynamics in bio-molecules using accelerated molecular dynamics. Phys Chem Chem Phys. 2011;13(45):20053–20065. doi: 10.1039/c1cp22100k. [DOI] [PubMed] [Google Scholar]
- 16.Hamelberg D, Mongan J, McCammon JA. Accelerated molecular dynamics: A promising and efficient simulation method for biomolecules. J Chem Phys. 2004;120(24):11919–11929. doi: 10.1063/1.1755656. [DOI] [PubMed] [Google Scholar]
- 17.Hamelberg D, de Oliveira CAF, McCammon JA. Sampling of slow diffusive conformational transitions with accelerated molecular dynamics. J Chem Phys. 2007;127(15):155102. doi: 10.1063/1.2789432. [DOI] [PubMed] [Google Scholar]
- 18.Wereszczynski J, McCammon JA. Nucleotide-dependent mechanism of Get3 as elucidated from free energy calculations. Proc Natl Acad Sci USA. 2012;109(20):7759–7764. doi: 10.1073/pnas.1117441109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gasper PM, Fuglestad B, Komives EA, Markwick PRL, McCammon JA. Allosteric networks in thrombin distinguish procoagulant vs. anticoagulant activities. Proc Natl Acad Sci USA. 2012;109(52):21216–21222. doi: 10.1073/pnas.1218414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pierce LCT, Salomon-Ferrer R, Augusto F de Oliveira C, McCammon JA, Walker RC. Routine access to millisecond time scale events with accelerated Molecular Dynamics. J Chem Theory Comput. 2012;8(9):2997–3002. doi: 10.1021/ct300284c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sethi A, Eargle J, Black AA, Luthey-Schulten Z. Dynamical networks in tRNA:protein complexes. Proc Natl Acad Sci USA. 2009;106(16):6620–6625. doi: 10.1073/pnas.0810961106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eargle J, Luthey-Schulten Z. NetworkView: 3D display and analysis of protein·RNA interaction networks. Bioinformatics. 2012;28(22):3000–3001. doi: 10.1093/bioinformatics/bts546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lange OF, Grubmüller H. Generalized correlation for biomolecular dynamics. Proteins. 2006;62(4):1053–1061. doi: 10.1002/prot.20784. [DOI] [PubMed] [Google Scholar]
- 24.Grant BJ, Rodrigues APC, ElSawy KM, McCammon JA, Caves LSD. Bio3d: An R package for the comparative analysis of protein structures. Bioinformatics. 2006;22(21):2695–2696. doi: 10.1093/bioinformatics/btl461. [DOI] [PubMed] [Google Scholar]
- 25.Spalding TA, Burstein ES, Henderson SC, Ducote KR, Brann MR. Identification of a ligand-dependent switch within a muscarinic receptor. J Biol Chem. 1998;273(34):21563–21568. doi: 10.1074/jbc.273.34.21563. [DOI] [PubMed] [Google Scholar]
- 26.Venkatakrishnan AJ, et al. Molecular signatures of G-protein-coupled receptors. Nature. 2013;494(7436):185–194. doi: 10.1038/nature11896. [DOI] [PubMed] [Google Scholar]
- 27.Deupi X, Standfuss J. Structural insights into agonist-induced activation of G-protein-coupled receptors. Curr Opin Struct Biol. 2011;21(4):541–551. doi: 10.1016/j.sbi.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 28.Xu F, et al. Structure of an agonist-bound human A2A adenosine receptor. Science. 2011;332(6027):322–327. doi: 10.1126/science.1202793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pals-Rylaarsdam R, Hosey MM. Two homologous phosphorylation domains differentially contribute to desensitization and internalization of the m2 muscarinic acetylcholine receptor. J Biol Chem. 1997;272(22):14152–14158. doi: 10.1074/jbc.272.22.14152. [DOI] [PubMed] [Google Scholar]
- 30.Lee KB, Ptasienski JA, Bünemann M, Hosey MM. Acidic amino acids flanking phosphorylation sites in the M2 muscarinic receptor regulate receptor phosphorylation, internalization, and interaction with arrestins. J Biol Chem. 2000;275(46):35767–35777. doi: 10.1074/jbc.M002225200. [DOI] [PubMed] [Google Scholar]
- 31.Phillips JC, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26(16):1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Y, Harrison CB, Schulten K, McCammon JA. Implementation of accelerated molecular dynamics in NAMD. Comput Sci Discov. 2011;4:015002. doi: 10.1088/1749-4699/4/1/015002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.MacKerell AD, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102(18):3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 34.MacKerell AD, Jr, Feig M, Brooks CL., 3rd Improved treatment of the protein backbone in empirical force fields. J Am Chem Soc. 2004;126(3):698–699. doi: 10.1021/ja036959e. [DOI] [PubMed] [Google Scholar]
- 35.Klauda JB, et al. Update of the CHARMM all-atom additive force field for lipids: Validation on six lipid types. J Phys Chem B. 2010;114(23):7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79(2):926–935. [Google Scholar]
- 37.Vanommeslaeghe K, et al. CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J Comput Chem. 2010;31(4):671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.