Abstract
Theoretical studies have focused on the environmental temperature of the universal common ancestor of life with conflicting conclusions. Here we provide experimental support for the existence of a thermophilic universal common ancestor. We present the thermal stabilities and catalytic efficiencies of nucleoside diphosphate kinases (NDK), designed using the information contained in predictive phylogenetic trees, that seem to represent the last common ancestors of Archaea and of Bacteria. These enzymes display extreme thermal stabilities, suggesting thermophilic ancestries for Archaea and Bacteria. The results are robust to the uncertainties associated with the sequence predictions and to the tree topologies used to infer the ancestral sequences. Moreover, mutagenesis experiments suggest that the universal ancestor also possessed a very thermostable NDK. Because, as we show, the stability of an NDK is directly related to the environmental temperature of its host organism, our results indicate that the last common ancestor of extant life was a thermophile that flourished at a very high temperature.
Keywords: ancient protein, crystal structure, last universal common ancestor, molecular resurrection, phylogenetic analysis
Elucidation of the origin and early evolution of life is fundamental to our understanding of ancient living systems and their environment(s). One debate about the last universal common ancestor, which has been denoted “LUCA,” “LCA,” or “senancestor,” and which we call “Commonote” (1), is its environmental temperature. In a well-referenced phylogenetic tree containing small-subunit rRNA sequences, those of hyperthermophilic archaea and bacteria are found at the deepest and shortest branches (2), and therefore it has been proposed that the common ancestors of Archaea and Bacteria were hyperthermophilic (3, 4). Given the apparent hyperthermophilic ancestry for both lineages, Occam’s razor suggests that the Commonote was a thermophilic organism. However, although some theoretical studies that focused on the environmental temperature of the Commonote support the thermophilic common-ancestry hypothesis (5–7), other theoretical studies have concluded that the universal ancestor was not (hyper)thermophilic (8–10). Therefore, the available theoretical studies disagree among themselves. More seriously, these theoretical studies have not been tested empirically.
Information concerning the properties of ancient proteins is embedded in the sequences of their descendants, so the ancestral sequence of a protein can be inferred by comparing a large number of extant homologous sequences (11–14). A powerful method for experimentally studying the properties of ancient life resurrects ancestral protein sequences, thereby allowing characterization of their features (15–23). Such resurrections have been used to understand the evolution of ethanol production and consumption in yeast (16), the evolutionary trajectory of changes in ligand specificity of hormone receptors (17, 18, 20), and the evolution of the increased complexity of vacuolar H+-ATPases (22). This empirical technique has provided new ways to elucidate the environmental temperatures experienced by ancient bacteria (15, 19). For the study reported here, we chose archaeal and bacterial ancestral nucleoside diphosphate kinase (NDK) sequences as the resurrection targets. NDK catalyzes the transfer of a phosphate from a nucleoside triphosphate to a nucleoside diphosphate. The ancestral NDK may have arisen early, because most extant organisms contain at least one gene that encodes NDK. The sequences of extant NDKs are relatively well conserved, allowing inference of the ancestral sequence with confidence. In addition, a number of 3D structures are available for the NDK family of proteins isolated from a wide variety of organisms, including bacteria, archaea, and eukaryotes. Therefore, NDK is an ideal model for studying the physical characteristics of ancient proteins. By resurrecting ancestral NDK sequences and characterizing their properties, which should reflect the ancestor’s characteristics and its environment, we experimentally estimated the environmental temperature of the Commonote that would have existed about 3,800 million years ago (24).
Results and Discussion
Ancestral Sequence Reconstruction Using a Small NDK Sequence Set.
The first step in the reconstruction of an ancestral sequence is to prepare a multiple amino acid sequence alignment, using the sequences of a given protein from extant species, which then is used to build a phylogenetic tree (12). Two methods, the maximum-likelihood (ML) method (25) and the Bayesian method (26), have been used commonly for tree building and reconstructing the ancestral sequence. In the ML method, the likelihood of each type of amino acid at each position in the sequence associated with the deepest node of the tree is computed using a statistical model of evolution. The ancestral sequence is defined as the set of residues in which each residue has the greatest likelihood of existing at its associated position. The Bayesian method integrates uncertainties associated with the tree topology, branch lengths, and substitution models into the ancestral sequence calculation, whereas only the most likely estimate of the tree and substitution models are assumed with the ML method.
As a suboptimal test case, we first constructed an ML tree from 66 extant NDK amino acid sequences (Fig. S1A). Then 10 sequences representing the main tree branches were selected to build an ML phylogenetic tree by CODEML in PAML (27) and a Bayesian tree by nhPhybloBayes (28) to identify the deepest archaeal and bacterial nodes (Fig. S1B). The CODEML-derived tree and the nhPhyloBayes-built tree have the same topology. The ancestral sequences predicted by CODEML are designated Arc1 (archaeal) and Bac1 (bacterial); those predicted by nhPhylobayes are designated Arc2 and Bac2 (Fig. S2). These reconstructed proteins are extremely thermally stable (Table 1 and Fig. S3 A and B), a property that is compatible with the proposal that the ancestors of bacteria and archaea were thermophiles. However, the 10 sequences used to build the trees seemed not to represent NDK sequence space accurately. Therefore, the thermal stability of the reconstructed proteins may be an artifact of the reconstruction methods (29).
Table 1.
Tm (°C) for A. fulgidus (Afu), T. thermophilus (Tth), and the resurrected ancestral NDKs
Ancestral Sequences Inferred from 204 NDK Sequences.
The accuracy of a predicted ancestral sequence depends on sequence sampling, which can be improved by including as many extant sequences as possible in the reconstruction. Since we performed the aforementioned experiment, improvements in computational power have allowed the use of a larger dataset. Therefore we built two ML phylogenic trees from 204 extant NDK sequences. [The Bayesian approach was not used for this tree-building exercise because Thornton and colleagues (30) recently reported that the ML phylogenetic algorithm accurately reconstructs ancestral sequences.] One tree was built without constraints (Fig. 1A and Fig. S4A), and one was built with the constraint that Archaea and Bacteria each represent a monophyletic group (Fig. 1B and Fig. S4B). For both trees, the major phyla are well grouped into their own monophyletic groups. However, the relationship among the phyla is slightly different for the two trees. In the tree built without constraint, certain archaeal sequences (those of Desulfurococcales and Thermoplasmatales) are paraphyletic and are found among the bacterial sequences, positionings that are inappropriate for a nearly universal phylogenetic tree. In the constrained tree, the Desulfurococcales and Thermoplasmatales sequences also are paraphyletic, but they are positioned near the root of the Archaea domain. Because the difference in the likelihood values of the two trees is not substantial, the sequences at the deepest archaeal and bacterial nodes were inferred from both trees. The resulting ancestral sequences are named, using the nomenclature given above, as Arc3 (nonconstrained) and Arc4 (constrained) and Bac3 (nonconstrained), and Bac4 (constrained) (Fig. S2). The amino acid sequences of Arc3 and Arc4 are very similar (131 of 139 residues are identical), and the sequences of Bac3 and Bac4 also are very similar (129 residues are identical).
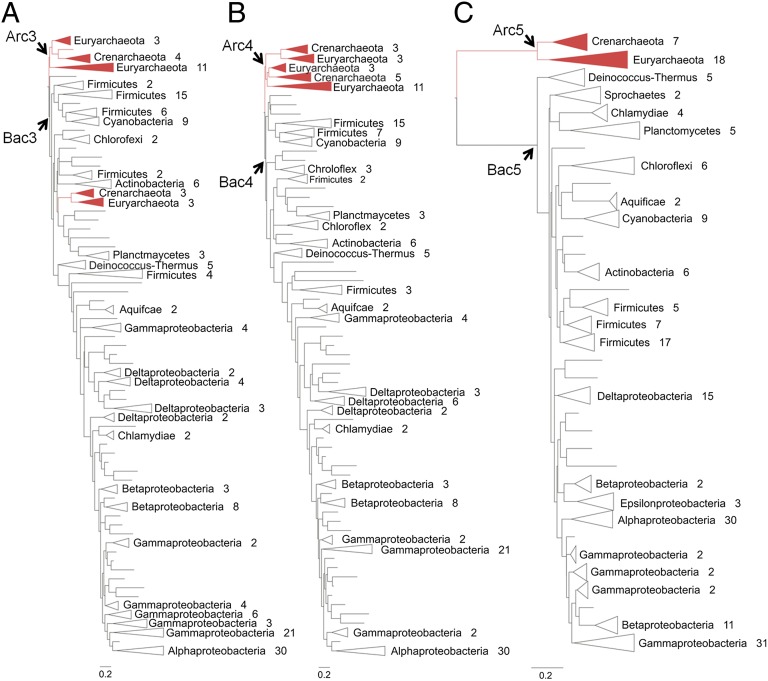
Fig. 1.
Phylogenetic trees used to infer ancestral NDK sequences. (A) The ML phylogenetic tree built from 204 NDK sequences without constraints. (B) The tree built from the same sequences as in A with the constraint that Archaea and Bacteria form separate monophyletic groups. (C) The ML rRNA tree containing the small-subunit rRNA sequences from the same species. Arrows mark the nodes corresponding to the positions of the ancestral proteins. The number of sequences in each clade is shown. Red branches indicate archaeal sequences; black-outlined branches indicate bacterial sequences. For complete trees, see Fig. S4.
The genes encoding the inferred ancestral proteins were PCR constructed, and the encoded proteins were expressed individually in Escherichia coli and purified. CD spectral changes at 222 nm as a function of temperature were acquired to assess the thermostability of the proteins at pH 6.0 and 7.6 (Table 1 and Fig. S3 C–F). The Arc3 and Arc4 unfolding midpoint temperatures (Tm, ∼110 °C for both proteins) show that the two proteins are more stable than is the NDK from the hyperthermophilic archaeon Archaeoglobus fulgidus at pH 6.0. The bacterial ancestors are less stable than the archaeal ancestors but are as thermostable as, or are more stable than, NDK from the thermophile Thermus thermophilus under both pH conditions.
Robustness of the Accuracy of the Sequence Predictions.
Of the 139 reconstructed residues, 115 are identical in Arc3, Arc4, Bac3, and Bac4, and the inclusion of these residues generally seems to be correct [the a posteriori probability (p) for each residue >0.9] (Fig. S2B). In contrast, the correctness of the other inferred ancestral residues is weakly supported, and therefore such residues may not represent the ancestral residues. Williams et al. (29) argued that erroneously inferred ancestral sequences are likely to be biased toward high stability. To characterize the effects of “incorrect” residues in the ancestral sequences empirically, “second-best” residues were substituted for the weakly supported residues (p <0.8) to create Arc3sec, Arc4sec, Bac3sec, and Bac4sec (Fig. S5A), and then their effects on the stability of the corresponding proteins were assessed. Temperature-induced unfolding studies showed that the proteins containing the second-best residues had reduced Tms of 1–10 °C (Table 1 and Fig. S5B). Nevertheless, they still are highly thermostable because their Tms are similar to that of T. thermophilus NDK. Thus, the predicted thermal stabilities of the ancestral NDKs are most likely valid even if some residues that do not represent those of the ancestral sequences are present in the proteins.
Robustness of the Tree Topologies Used to Infer the Ancestral Sequences.
We also were concerned that the NDK phylogenies differ somewhat from the species phylogenies of Bacteria and Archaea. The most commonly referenced species tree contains small-subunit rRNA sequences (2). Therefore we created a phylogenetic tree containing the small-subunit rRNA sequences of the species that were used to infer the sequences of Arc3/4 and Bac3/4 (Fig. 1C and Fig. S4C) to infer additional ancestral NDK sequences (Arc5 and Bac5) (Fig. S2). In the NDK trees, Euryarchaeota diverged into two groups near the root of the trees (Fig. 1 A and B). Crenarchaeota formed a monophyletic group but appeared with some of the Euryarchaeota in the unconstrained tree (Fig. 1A). However, Euryarchaeota and Crenarchaeota formed unique monophyletic clusters in the rRNA-derived tree (Fig. 1C). The relationship among the phyla within the Bacterial domain also is different in the NDK- and rRNA-derived trees. Although the Firmucutes diverged closest to the root of the Bacterial domain in the NDK trees, the Deinocosccus-Thermus group diverged closest to the root of the rRNA-derived tree. The minor differences between the NDK and the rRNA-derived trees suggest that horizontal gene transfer played only a small role in NDK evolution. Arc5 has 131 and 129 identical residues found at the same positions in Arc3 and Arc4, respectively, and 132 and 130 residues of Bac5 are found at the same positions in Bac3 and Bac4, respectively. The large degree of sequence similarity indicates that horizontal gene-transfer events did not affect the inference of the ancestral sequences substantially.
Thermal denaturations of Arc5 and Bac5 were monitored by measuring their θ222nm (ellipticity at 222 nm) values as a function of temperature. The Tms of Arc5 are 108 °C (pH 6.0) and 107 °C (pH 7.6) (Table 1 and Fig. S3 C and D). Bac5, with Tms of 107 °C (pH 6.0) and 105 °C (pH 7.6), is more thermally stable than T. thermophilus NDK (Table 1 and Fig. S3 E and F). Therefore the inferred thermal stabilities of the ancestral NDKs were not affected substantially by the topologies of the trees used to reconstruct the ancestral sequences.
Comparison with the Consensus NDK.
The consensus approach has been used in conjunction with multiple amino acid sequence alignments to design proteins with enhanced stability (31, 32). Notably, a consensus residue at a given position often is the same as that found in the ancestral protein. Indeed, the consensus sequence for the 204 NDKs that were used to infer the ancestral sequences contains more than 100 residues (∼72%) that are identical to those in the same positions in the ancestral sequences. We therefore constructed a consensus NDK (ConsNDK) that contains the amino acid most frequently found at each position (Fig. S2A). The Tm for ConsNDK is 84 °C, which is substantially lower than the Tms of the ancestral NDKs (Fig. S3G). However, the ConsNDK sequence may not accurately reflect the sequence with the highest Tm. Consensus residues do not always improve protein stability. Sullivan et al. (33) reported that removal of consensus mutations at positions for which their effect is statistically coupled to a residue(s) at another site(s) tends to improve stability. Therefore, the stability of ConsNDK might be increased if its sequence was optimized according to the finding of Sullivan et al.
Implications for the Environment Temperatures of Ancient Organisms.
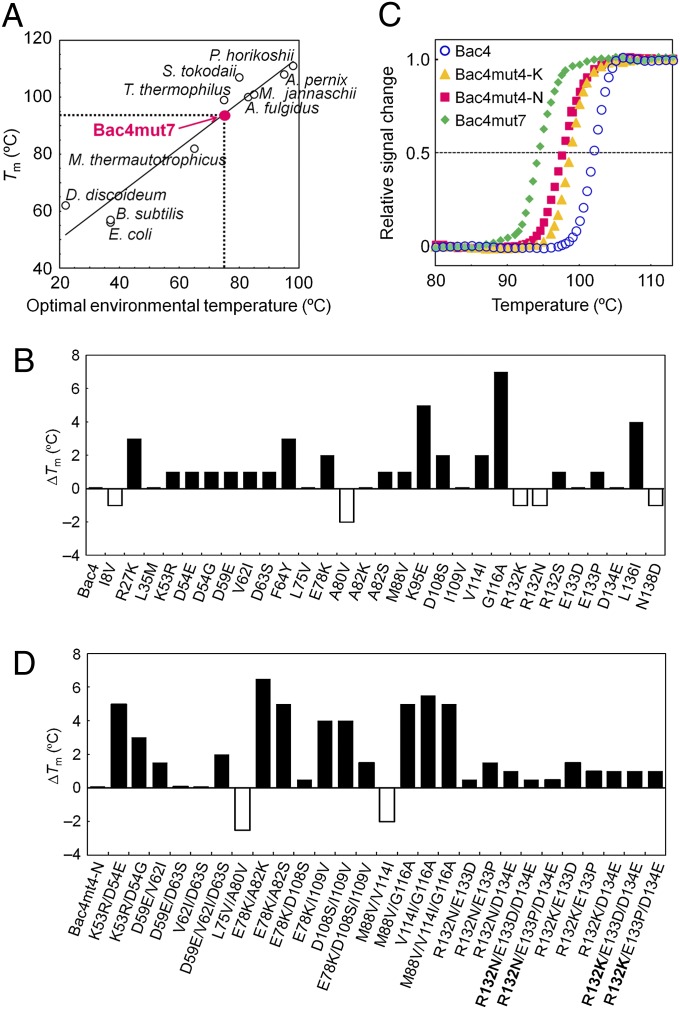
The measured Tms of the resurrected NDKs provide experimental evidence for the existence of extremely thermally stable ancestral archaea and bacteria, if one assumes that, in general, the denaturation temperature of a protein reflects the environmental temperature of its host organism. Because this assumption is the key premise of our study, we determined the thermal stabilities of extant NDKs from organisms that thrive at various temperatures. A plot of the Tms of the extant NDKs as a function of the optimal environmental temperatures of their host organisms is shown in Fig. 2A. The Tms correlate strongly with the optimal environmental temperatures [correlation coefficient = 0.96, which is nearly the same as a value (0.91) reported for 56 globular proteins from 16 different families (34)]. Therefore, the stability of each NDK appears to be related directly to its host’s natural environmental temperature.
Fig. 2.
Estimation of the environmental temperature of the Commonote. (A) Relationship between the thermostabilities of extant microbial NDKs and the optimal environmental temperatures of their hosts. The Tms for E. coli and Dictyostelium discoideum NDKs were obtained from Giartosio et al. (45). For the other NDKs, Tms were obtained from thermal unfolding experiments as described for Fig. S3, with the pH values of the solutions being 6.0. The lower limit of the estimate for the environmental temperature of the Commonote was found using the calibration curve and the Tm of Bac4mut7. (B) Effects of single amino acid substitutions on the Tm of Bac4 monitored at θ222nm. (C) Temperature-induced unfolding of Bac4, Bac4mut4-K, Bac4mut4-N, and Bac4mut7 monitored at θ222nm. Duplicated measurements are identical within experimental error. The plots were normalized with respect to the baselines of the native and denatured states. (D) Effect of combined amino acid substitutions on the Tm of Bac4mut4-N monitored at θ222nm. The mutants are named to identify the nonconserved residues within 5 Å of each other, although for the triple mutations only one of the mutations needs to be within 5 Å of the others. In those cases, the residue within 5 Å of the others is highlighted in bold type.
Gaucher et al. (15, 19) reproduced the ancestor of bacterial elongation factor Tu, and, by virtue of its properties, suggested that the common bacterial ancestor was thermophilic. The ancestral archaeon also is thought to be a thermophile or a hyperthermophile. In the most commonly referenced phylogenetic tree composed of small-subunit rRNA sequences, those of hyperthermophilic archaea are located near the node of their ancestor (2, 3). Theoretical studies also support the thermophilic origin of archaea (10, 35). According to the Tms of the reconstructed NDKs (Table 1) and the calibration curve shown in Fig. 2A, the optimal environmental temperatures of the common ancestors of Bacteria and Archaea are ∼80–93 °C and ∼81–97 °C, respectively.
Catalytic Properties of Ancestral NDKs.
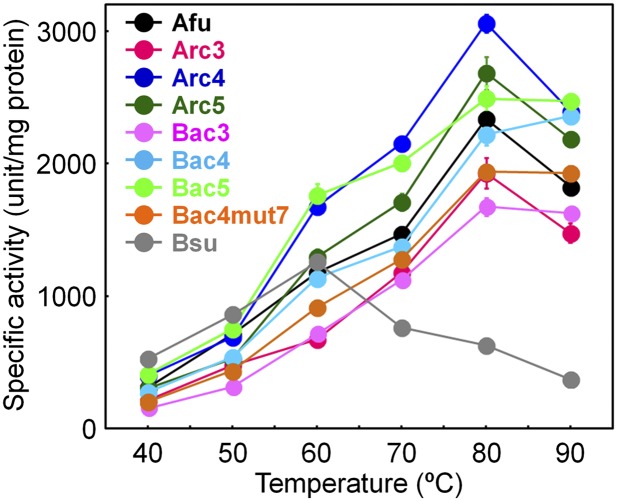
We next measured the enzymatic activities of the ancestral NDKs by assessing the extent of γ-phosphate transfer from GTP to ADP to produce GDP and ATP. Fig. 3 depicts the specific activities of the ancestral NDKs as a function of temperature. The specific activities of the hyperthermophilic A. fulgidus and mesophilic Bacillus subtilis NDKs also were measured for comparison. The temperature dependence of the specific activities of the ancestral NDKs more closely resembles that of A. fulgidus NDK than that of the B. subtilis protein. Although B. subtilis NDK functions optimally at 60 °C, the specific activities of the ancestral NDKs and the A. fulgidus NDK all increase as the temperature increases up to 80 °C. A high optimal temperature also has been found for other naturally occurring thermophilic enzymes.
Fig. 3.
Temperature dependence of the specific activities of B. subtilis, A. fulgidus, and ancestral NDKs. The specific activities were measured using a reaction mixture containing 5 mM each of ADP and GTP. Each value is the average of at least four replicas. Afu, A. fulgidus; Bsu, B. subtilis.
The kinetic parameters of the ancestral, A. fulgidus, and T. thermophilus NDKs were obtained from initial velocity experiments at 70 °C that used various concentrations of ADP and 2.5 mM GTP (Table 2). Arc4 has the most unfavorable Km for ADP but the best kcat. Conversely, Bac1 has the best Km for ADP but the worst kcat. Overall, however, the resurrected ancestral and contemporary enzymes have similar kinetic parameters.
Table 2.
Kinetic parameters for the resurrected ancestral, A. fulgidus (Afu), and T. thermophilus (Tth) NDKs at 70 °C
| Protein | KmADP, μM* | kcat, s−1* | kcat/KmADP, s−1·μM−1 |
| Afu NDK | 1,480 ± 300 | 1,800 ± 240 | 1.22 |
| Arc1 | 411 ± 45 | 1,720 ± 100 | 4.18 |
| Arc2 | 205 ± 11 | 1,030 ± 20 | 5.02 |
| Arc3 | 630 ± 89 | 1,690 ± 130 | 2.68 |
| Arc4 | 850 ± 101 | 2,100 ± 146 | 2.47 |
| Arc5 | 312 ± 17 | 1,260 ± 30 | 4.04 |
| Tth NDK | 248 ± 29 | 1,120 ± 60 | 4.52 |
| Bac1 | 114 ± 17 | 241 ± 12 | 2.11 |
| Bac2 | 320 ± 31 | 780 ± 35 | 2.44 |
| Bac3 | 246 ± 26 | 1,210 ± 50 | 4.92 |
| Bac4 | 411 ± 49 | 1,520 ± 90 | 3.70 |
| Bac5 | 480 ± 33 | 1.920 ± 70 | 4.00 |
The values and SEs of the kinetic constants were obtained by nonlinear least-square fitting of the steady-state velocity data to the Michaelis–Menten equation.
Crystal Structures of Ancestral NDKs.
We solved the Arc1 and Bac1 structures to 2.4-Å resolution. A protein’s stability generally originates not from a single factor but rather from a combination of many different types of structural features, with the contribution of each being small and often strongly dependent on the structural context. However, examination of the two crystal structures reveals several features that likely contribute to their high thermal stabilities: reduced nonpolar accessible surface areas and increased numbers of intersubunit ion pairs and hydrogen bonds (Figs. S6 and S7, Tables S1 and S2, and SI Text).
Thermostability of the Commonote NDK.
There has been substantial hypothetical debate regarding the environmental temperature of the Commonote, but no experimental evidence for the temperature is available (5–10). Although we expected that the node for the Commonote would be located between the roots of the bacterial and archaeal NDKs, we could not determine the precise position of the Commonote, because the trees are not rooted; therefore we also could not its precise sequence. However, of the 139 reconstructed residues in the 10 NDK ancestral sequences, 115 are identical (Fig. S2A). These residues therefore probably are present at the same positions in the NDK sequence of the Commonote. For the 24 remaining positions, the Commonote’s NDK sequence likely would contain residues found in at least one of the reconstructed ancestral NDK sequences because Occam’s razor suggests that the sequence of the Commonote NDK had residues present in either the last common bacterial or archaeal ancestor sequence. To investigate the effects on stability of the amino acid composition of the remaining 24 positions, we individually replaced the 24 nonconserved residues of Bac4 with the residues found in the same positions in Arc3–5, Bac3–5, Arc3/4sec, and Bac3/4sec to generate 29 NDKs. We used the sequence of Bac4 as the starting point because it had the lowest Tm of the ancestral NDKs that had been constructed up until that point. Only five of the amino acid substitutions, I8V, A80V, R132K, R132N, and N138D, reduced the Tm (Fig. 2B). We then prepared the Bac4 variants Bac4mut4-K and Bac4mut4-N that contain V8, V80, D138, and K132 or N132, respectively. The Tms of Bac4mut4-K and Bac4mut4-N are 99 °C and 98 °C, respectively (Fig. 2C).
The effect of a mutation on the stability of the protein may be affected by its surroundings, so the presence of one mutation may negatively impact the stabilizing effect of a second mutation. Therefore, an amino acid substitution that enhances the thermostability when individually introduced into Bac4 may be destabilizing in combination with other mutations. However, expressing all possible combinations of the 29 amino acid substitutions (5.4 × 108 combinations) was not practical. Instead, using the Bac1 structure as the guide, we identified the combinations of the nonconserved residues whose side chains are located within 5 Å of each other, because residues that are near each other are more likely to affect stability than those that are far apart. Then we introduced the 27 identified combinations of such substitutions into Bac4mut4-N and assessed the thermal stabilities of the resulting proteins (Fig. 2D). The Tms of two proteins decreased compared with that of Bac4mut4-N. L75 of Bac4 is buried within the core and interacts with A80. When alanine is present at position 80, the L75 → V mutation does not affect the thermal stability of Bac4. However, when A80 is replaced by valine (i.e., Bac4mut4-N), valine at position 75 decreases the Tm. Similarly, the simultaneous replacements M88 → V and V114 → I decrease the Tm of Bac4mut4-N. We then mutated Bac4mut4-N so that the resulting protein Bac4mut7 contains V75, V88, and I114. The Tm of Bac4mut7 (94 °C) (Fig. 2C) probably represents the lowest estimate of Tm for the Commonote NDK and, consequently, the lowest estimate for the Commonote’s environment temperature. In addition, Bac4mut7 functions optimally between 80 and 90 °C (Fig. 3). This finding strongly supports the hypothesis that the Commonote was a thermophile that flourished at a temperature above 75 °C (Fig. 2A).
Limitations of Ancestral Sequence Reconstruction in Estimating Ancestral Environment Temperatures.
In this study, we found that the ancestral NDKs are extremely thermostable and functioned optimally at high temperatures, and therefore we concluded that the ancestral organisms lived in high-temperature environments. However, the environmental temperatures that were estimated using the reconstructed proteins’ Tms and the calibration curve (Fig. 2A) may be too high. In extant organisms, the stability of their cytoplasmic proteins is affected by cellular osmolytes and molecular chaperons, which might have been absent from primordial cells. Oxidative stress and low fidelity of the transcription–translation machinery also could be driving forces for the development of high intrinsic stability in primitive proteins (36).
Williams et al. (29) reported that use of the ML method for reconstruction of an ancestral sequence may result in an overestimation of its thermal stability. The basis for their assertion is that ancestral reconstruction using the ML method tends to incorporate amino acids that are found frequently in extant sequences and often are stabilizing. Therefore, the method tends to exclude variants at a position that have deleterious effects on structural stability because such deleterious variants are found less frequently. However, this caveat does not seem to apply to the ancestral NDKs, because all of them are more thermally stable than ConsNDK. If we assume that the differences between the residues of the ancestral and consensus NDKs are responsible for the differences in their thermal stability, then less frequently used amino acids would contribute to the greater stability of the ancestral NDKs.
Even so, the high thermal stability of the ancestral proteins possibly is related to the inherent nature of the ancestral sequence reconstructions, specifically to the presence of identical residues at many positions in the consensus and ancestral sequences. Maintenance of a protein’s tertiary structure relies on a delicate balance of intramolecular interactions, and simultaneously replacing many residues risks introducing deleterious substitutions because it is never certain that all stabilizing consensus residues have been identified correctly. Therefore, it is possible that the lower stability of ConsNDK is caused simply by its nonoptimized sequence. Moreover, we previously found that the method of ancestral reconstruction can correct, at least partially, for potentially erroneous predictions of consensus residues that would be caused by selection bias in the sequence collection (37). Similar interpretations may be applied to the ancestral NDK sequences reconstructed in this study. Currently, there is no way to discriminate between effects that can be ascribed to the “antiquity” of the residues and ones associated with consensus residues. However, the ancestral NDKs functioning optimally at high temperatures (Fig. 3) provides additional evidence that the ancestral NDKs would be compatible with a thermophilic environment.
Methods
Phylogenetic Tree Building Using a Small NDK Sequence Set.
Sixty-six amino acid sequences of single-domain NDKs, retrieved from GenBank, were aligned by ClustalX 1.83 (38) using its default settings before tree building. The alignment was adjusted manually to correct for gaps. Regions of uncertain alignment were removed by Gblocks 0.91b (39). A total of 120 residue positions were selected to build an ML tree using TREEFINDER ver. December 2005 (40) with the JTT+F+G (four-class) model (Fig. S1A).
Using the structure of the ML tree as a guide, we then selected and aligned 10 relatively slowly evolving sequences for further tree building and prediction of the ancestral bacterial and archaeal sequences. After poorly aligned regions were excluded by a Gblocks run under default conditions, the remaining regions were used for ML tree building. The 105 best ML trees were selected by PROTML in Molphy 2.3b (41) using the JTT-F model. Log-likelihoods of the 105 trees generated were compared by CODEML in PAML (27) under the JTT-F-G substitution model (an eight-class discrete model). The shape parameter alpha of a gamma distribution was calculated from the data. The most likely ML tree is shown in Fig. S1B.
Starting with the tree shown in Fig. S1B, prediction of ancestral sequences for Arc1 and Bac1 was performed by CODEML in PAML and by GASP (42) to predict the indel positions. Ancestral sequence predictions also were made by nhPhyloBayes (28). The 10 Gblocks-treated sequences were used again for tree building by nhPhyloBayes. The nhPhyloBayes-built tree has the same topology as that of the CODEML-derived tree. nhPhyloBayes then was used to predict the ancestral sequences for Arc2 and Bac2 in conjunction with the 10 sequences including their indels.
Phylogenetic Tree Building Using 204 NDK Sequences.
A prerequisite for the resurrection of accurate ancestral sequences is the construction of a highly reliable phylogenetic tree. Keeping in mind that lateral transfer of NDK genes between Bacteria and Archaea may have occurred frequently, we carefully selected NDK sequences on the basis of their entries in GenBank as follows. NDKs with multiple domains and eukaryotic sequences were excluded. The remaining 204 sequences (179 bacterial and 25 archaeal sequences) were aligned by ClustalX 2.1 (38) and then were adjusted manually to correct for gap positions to generate the multiple sequence alignment MSA-204.
Starting with MSA-204, TREEFINDER ver. Oct. 2008 (40), in conjunction with the LG + Gamma (eight-class) + F amino acid substitution model, was used to compute an ML tree. However, the archaeal sequences did not form a monophyletic group, because some were found within the bacterial branches (Fig. 1A and Fig. S4A). Therefore we built a second tree (Fig. 1B and Fig. S4B with the constraint that Archaea and Bacteria formed separate monophyletic groups. The difference in log likelihood between the two trees is 13 (−24,826 for the tree built without constraints; −24,839 for the tree built with the constraint). Both trees then were used for ancestral sequence prediction by CODEML after it had been used to build the ML tree. The inferred ancestral sequences are designated Arc3 and Bac3 (nonconstrained tree), and Arc4 and Bac4 (constrained tree) (Fig. S2).
Ancestral Sequence Prediction Using a Small-Subunit rRNA Tree.
Ancestral sequences also were inferred from a small-subunit rRNA tree. Using Gblocks 0.91B run under its default settings, we constructed a multiple small-subunit rRNA sequence alignment (708 positions) using sequences from the 204 species whose NDK sequences were used to construct MSA-204. A phylogenetic tree then was built by TREEFINDER ver. Oct. 2008 using the ML algorithm in conjunction with the GTR + Gamma Invariant (eight-class) model (Fig. 1C and Fig. S4C). Prediction of the best-fit evolutionary model was performed by Kakusan4 (www.fifthdimension.jp/products/kakusan/). By using the rRNA tree topology and replacing each rRNA sequence with its corresponding species NDK sequence, the sequences for the last common archaeal and bacterial ancestors were inferred by CODEML with the sequences named Arc5 and Bac5, respectively (Fig. S2).
Construction of the Ancestral Genes.
The inferred ancestral amino acid sequences were reverse translated to obtain their gene sequences, which then were PCR synthesized using complementary synthetic oligonucleotides with 18–22 bp overlaps (12). For PCR amplification, the reaction mixture contained 1 × PCR buffer for KOD-plus-polymerization (Toyobo), 1 mM MgSO4, 0.2 mM each of the dNTPs, 0.2 μM each of the synthetic oligonucleotides, and 1.0 unit KOD-plus-DNA polymerase (Toyobo). The temperature/time program was step 1, 95 °C, 3 min; step 2, 95 °C, 30 s; step 3, 55 °C, 30 s; and step 4, 68 °C, 1 min; steps 2–4 were repeated 25 times. Each amplified product was treated with NdeI and BamHI (New England Biolabs) and then was purified by agarose gel electrophoresis. Each purified gene was ligated into pET21c or pET23a (Merck).
Protein Expression and Purification.
To prepare and purify each of the resurrected enzymes, an E. coli Rosetta2 (DE3) inoculum (Merck) harboring the respective expression plasmid was cultivated in Luria–Bertani medium supplemented with ampicillin (150 μg/mL). Heterologous gene expression was induced using Overnight Express Autoinduction system reagents (Novagen). After overnight culture at 37 °C, cells were harvested and disrupted by sonication. The soluble fractions were each isolated by centrifugation at 60,000 × g for 20 min. The supernatants were individually heated at 70 °C for 20 min and centrifuged at 60,000 × g for 20 min to precipitate proteins other than NDK. To purify each enzyme, its respective supernatant was chromatographed successively through HiTrapQ, ResourceQ, and Superdex 200 resins (GE Healthcare Biosciences). SDS-PAGE was used to verify the homogeneity of each enzyme. The extant NDKs were prepared in a similar manner.
Analytical Methods.
Protein concentrations were determined using the OD280 of the solutions as described by Pace and colleagues (43), who followed the procedure of Gill and von Hippel (44).
For thermal denaturation measurements, we used a J-720 spectropolarimeter (Jasco) equipped with a programmable temperature controller and a pressure-proof cell compartment that prevented the aqueous solution from bubbling over and evaporating at high temperatures. Thermal denaturations were monitored using θ222 of the protein solutions. Each enzyme was dissolved in 20 mM potassium phosphate (pH 6.0 or 7.6), 50 mM KCl, and 1 mM EDTA to a final concentration of 20 μM. A path-length cell with a path length of 0.1 cm was used. The temperature was increased at a rate of 1.0 °C/min.
The enzymatic activity assay monitored the increase in the amount of product ATP (Kinase-Glo Luminescent Kinase Assay kit; Promega). The assay solution was 50 mM Hepes (pH 8.0), 25 mM KCl, 10 mM (NH4)2SO4, 2.0 mM Mg(CH3COO)2, 1.0 mM DTT, 5.0 mM ADP, and 5.0 mM GTP. One enzyme unit equaled 1 μmol ATP formed per min. For Michaelis–Menten kinetic measurements, the assay mixtures were 50 mM Hepes (pH 8.0), 25 mM KCl, 10 mM (NH4)2SO4, 2.0 mM Mg(CH3COO)2, 1.0 mM DTT, 2.5 mM GTP, with ADP at a concentration between 50 and 1,000 μM. The Michaelis constant (Km, substrate ADP) and the catalytic rate constant (kcat) were obtained by nonlinear-least-squares fitting of the steady-state velocities as functions of ADP concentrations to the Michaelis–Menten equation using the Enzyme Kinetics module in SigmaPlot (Systat Software).
X-Ray Crystallography of Ancestral NDKs.
Procedures for crystallization, data collection, and X-ray crystallography of Arc1 and Bac1 are described in SI Text. The crystal structures have been deposited in the Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank (www.rcsb.org) under the accession numbers 3VVT (Arc1), 3VVU (Bac1).
Supplementary Material
Acknowledgments
This work was supported in part by Grants-in-Aid from the Japanese Society for the Promotion of Science.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The crystal structures of Arc1 and Bac1 have been deposited in the Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank (www.rcsb.org) [accession numbers 3VVT (Arc1), and 3VVU (Bac1)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1308215110/-/DCSupplemental.
References
- 1.Yamagishi A, Kon T, Takahashi G, Oshima T. In: Thermophiles: The keys to molecular evolution and the origin of life? Wiegel J, Adams MWW, editors. London: Taylor & Francis; 1998. pp. 287–295. [Google Scholar]
- 2.Woese CR. Bacterial evolution. Microbiol Rev. 1987;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pace NR. Origin of life—facing up to the physical setting. Cell. 1991;65(4):531–533. doi: 10.1016/0092-8674(91)90082-a. [DOI] [PubMed] [Google Scholar]
- 4.Stetter KO. Hyperthermophiles in the history of life. Philos Trans R Soc Lond B Biol Sci. 2006;361(1474):1837–1842, discussion 1842–1843. doi: 10.1098/rstb.2006.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Di Giulio M. The universal ancestor lived in a thermophilic or hyperthermophilic environment. J Theor Biol. 2000;203(3):203–213. doi: 10.1006/jtbi.2000.1086. [DOI] [PubMed] [Google Scholar]
- 6.Di Giulio M. The universal ancestor was a thermophile or a hyperthermophile: Tests and further evidence. J Theor Biol. 2003;221(3):425–436. doi: 10.1006/jtbi.2003.3197. [DOI] [PubMed] [Google Scholar]
- 7.Brooks DJ, Fresco JR, Singh M. A novel method for estimating ancestral amino acid composition and its application to proteins of the Last Universal Ancestor. Bioinformatics. 2004;20(14):2251–2257. doi: 10.1093/bioinformatics/bth235. [DOI] [PubMed] [Google Scholar]
- 8.Galtier N, Tourasse N, Gouy M. A nonhyperthermophilic common ancestor to extant life forms. Science. 1999;283(5399):220–221. doi: 10.1126/science.283.5399.220. [DOI] [PubMed] [Google Scholar]
- 9.Becerra A, Delaye L, Lazcano A, Orgel LE. Protein disulfide oxidoreductases and the evolution of thermophily: Was the last common ancestor a heat-loving microbe? J Mol Evol. 2007;65(3):296–303. doi: 10.1007/s00239-007-9005-0. [DOI] [PubMed] [Google Scholar]
- 10.Boussau B, Blanquart S, Necsulea A, Lartillot N, Gouy M. Parallel adaptations to high temperatures in the Archaean eon. Nature. 2008;456(7224):942–945. doi: 10.1038/nature07393. [DOI] [PubMed] [Google Scholar]
- 11.Messier W, Stewart CB. Episodic adaptive evolution of primate lysozymes. Nature. 1997;385(6612):151–154. doi: 10.1038/385151a0. [DOI] [PubMed] [Google Scholar]
- 12.Thornton JW. Resurrecting ancient genes: Experimental analysis of extinct molecules. Nat Rev Genet. 2004;5(5):366–375. doi: 10.1038/nrg1324. [DOI] [PubMed] [Google Scholar]
- 13.Richter M, et al. Computational and experimental evidence for the evolution of a (β α)8-barrel protein from an ancestral quarter-barrel stabilised by disulfide bonds. J Mol Biol. 2010;398(5):763–773. doi: 10.1016/j.jmb.2010.03.057. [DOI] [PubMed] [Google Scholar]
- 14.Perez-Jimenez R, et al. Single-molecule paleoenzymology probes the chemistry of resurrected enzymes. Nat Struct Mol Biol. 2011;18(5):592–596. doi: 10.1038/nsmb.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaucher EA, Thomson JM, Burgan MF, Benner SA. Inferring the palaeoenvironment of ancient bacteria on the basis of resurrected proteins. Nature. 2003;425(6955):285–288. doi: 10.1038/nature01977. [DOI] [PubMed] [Google Scholar]
- 16.Thomson JM, et al. Resurrecting ancestral alcohol dehydrogenases from yeast. Nat Genet. 2005;37(6):630–635. doi: 10.1038/ng1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bridgham JT, Carroll SM, Thornton JW. Evolution of hormone-receptor complexity by molecular exploitation. Science. 2006;312(5770):97–101. doi: 10.1126/science.1123348. [DOI] [PubMed] [Google Scholar]
- 18.Ortlund EA, Bridgham JT, Redinbo MR, Thornton JW. Crystal structure of an ancient protein: Evolution by conformational epistasis. Science. 2007;317(5844):1544–1548. doi: 10.1126/science.1142819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaucher EA, Govindarajan S, Ganesh OK. Palaeotemperature trend for Precambrian life inferred from resurrected proteins. Nature. 2008;451(7179):704–707. doi: 10.1038/nature06510. [DOI] [PubMed] [Google Scholar]
- 20.Bridgham JT, Ortlund EA, Thornton JW. An epistatic ratchet constrains the direction of glucocorticoid receptor evolution. Nature. 2009;461(7263):515–519. doi: 10.1038/nature08249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaucher EA, Kratzer JT, Randall RN. Deep phylogeny—how a tree can help characterize early life on Earth. Cold Spring Harb Perspect Biol. 2010;2(1):a002238. doi: 10.1101/cshperspect.a002238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Finnigan GC, Hanson-Smith V, Stevens TH, Thornton JW. Evolution of increased complexity in a molecular machine. Nature. 2012;481(7381):360–364. doi: 10.1038/nature10724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boussau B, Gouy M. What genomes have to say about the evolution of the Earth. Gondwana Res. 2012;21(2–3):483–494. [Google Scholar]
- 24.Feng DF, Cho G, Doolittle RF. Determining divergence times with a protein clock: Update and reevaluation. Proc Natl Acad Sci USA. 1997;94(24):13028–13033. doi: 10.1073/pnas.94.24.13028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang Z, Kumar S, Nei M. A new method of inference of ancestral nucleotide and amino acid sequences. Genetics. 1995;141(4):1641–1650. doi: 10.1093/genetics/141.4.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang Z, Rannala B. Bayesian phylogenetic inference using DNA sequences: A Markov Chain Monte Carlo Method. Mol Biol Evol. 1997;14(7):717–724. doi: 10.1093/oxfordjournals.molbev.a025811. [DOI] [PubMed] [Google Scholar]
- 27.Yang Z. PAML: A program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13(5):555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- 28.Blanquart S, Lartillot N. A site- and time-heterogeneous model of amino acid replacement. Mol Biol Evol. 2008;25(5):842–858. doi: 10.1093/molbev/msn018. [DOI] [PubMed] [Google Scholar]
- 29.Williams PD, Pollock DD, Blackburne BP, Goldstein RA. Assessing the accuracy of ancestral protein reconstruction methods. PLOS Comput Biol. 2006;2(6):e69. doi: 10.1371/journal.pcbi.0020069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hanson-Smith V, Kolaczkowski B, Thornton JW. Robustness of ancestral sequence reconstruction to phylogenetic uncertainty. Mol Biol Evol. 2010;27(9):1988–1999. doi: 10.1093/molbev/msq081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steipe B. Consensus-based engineering of protein stability: From intrabodies to thermostable enzymes. Methods Enzymol. 2004;388:176–186. doi: 10.1016/S0076-6879(04)88016-9. [DOI] [PubMed] [Google Scholar]
- 32.Nikolova PV, Henckel J, Lane DP, Fersht AR. Semirational design of active tumor suppressor p53 DNA binding domain with enhanced stability. Proc Natl Acad Sci USA. 1998;95(25):14675–14680. doi: 10.1073/pnas.95.25.14675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sullivan BJ, et al. Stabilizing proteins from sequence statistics: The interplay of conservation and correlation in triosephosphate isomerase stability. J Mol Biol. 2012;420(4-5):384–399. doi: 10.1016/j.jmb.2012.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gromiha MM, Oobatake M, Sarai A. Important amino acid properties for enhanced thermostability from mesophilic to thermophilic proteins. Biophys Chem. 1999;82(1):51–67. doi: 10.1016/s0301-4622(99)00103-9. [DOI] [PubMed] [Google Scholar]
- 35.Gribaldo S, Brochier-Armanet C. The origin and evolution of Archaea: A state of the art. Philos Trans R Soc Lond B Biol Sci. 2006;361(1470):1007–1022. doi: 10.1098/rstb.2006.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goldsmith M, Tawfik DS. Potential role of phenotypic mutations in the evolution of protein expression and stability. Proc Natl Acad Sci USA. 2009;106(15):6197–6202. doi: 10.1073/pnas.0809506106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Akanuma S, et al. Phylogeny-based design of a B-subunit of DNA gyrase and its ATPase domain using a small set of homologous amino acid sequences. J Mol Biol. 2011;412(2):212–225. doi: 10.1016/j.jmb.2011.07.042. [DOI] [PubMed] [Google Scholar]
- 38.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25(24):4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17(4):540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 40.Jobb G, von Haeseler A, Strimmer K. TREEFINDER: A powerful graphical analysis environment for molecular phylogenetics. BMC Evol Biol. 2004;4:18. doi: 10.1186/1471-2148-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 41. Adachi J, Hasegawa M (1996) MOLPHY (Tokyo Institute of Statistical Mathematics, Tokyo)
- 42.Edwards RJ, Shields DC. GASP: Gapped Ancestral Sequence Prediction for proteins. BMC Bioinformatics. 2004;5:123. doi: 10.1186/1471-2105-5-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4(11):2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem. 1989;182(2):319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 45.Giartosio A, et al. Thermal stability of hexameric and tetrameric nucleoside diphosphate kinases. Effect of subunit interaction. J Biol Chem. 1996;271(30):17845–17851. doi: 10.1074/jbc.271.30.17845. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.