Abstract
Tunas are migratory fishes in offshore habitats and top predators with unique features. Despite their ecological importance and high market values, the open-ocean lifestyle of tuna, in which effective sensing systems such as color vision are required for capture of prey, has been poorly understood. To elucidate the genetic and evolutionary basis of optic adaptation of tuna, we determined the genome sequence of the Pacific bluefin tuna (Thunnus orientalis), using next-generation sequencing technology. A total of 26,433 protein-coding genes were predicted from 16,802 assembled scaffolds. From these, we identified five common fish visual pigment genes: red-sensitive (middle/long-wavelength sensitive; M/LWS), UV-sensitive (short-wavelength sensitive 1; SWS1), blue-sensitive (SWS2), rhodopsin (RH1), and green-sensitive (RH2) opsin genes. Sequence comparison revealed that tuna's RH1 gene has an amino acid substitution that causes a short-wave shift in the absorption spectrum (i.e., blue shift). Pacific bluefin tuna has at least five RH2 paralogs, the most among studied fishes; four of the proteins encoded may be tuned to blue light at the amino acid level. Moreover, phylogenetic analysis suggested that gene conversions have occurred in each of the SWS2 and RH2 loci in a short period. Thus, Pacific bluefin tuna has undergone evolutionary changes in three genes (RH1, RH2, and SWS2), which may have contributed to detecting blue-green contrast and measuring the distance to prey in the blue-pelagic ocean. These findings provide basic information on behavioral traits of predatory fish and, thereby, could help to improve the technology to culture such fish in captivity for resource management.
Keywords: tuna genome, visual system, animal opsin
Tunas are considered “the ultimate fish,” because they are top predators in ocean ecosystems, in addition to their unique morphology and physiology (1, 2). Worldwide, tunas have been exploited because of their high market values, causing concern about the state of the wild stocks (3). The investigation of the open-ocean lifestyle of tuna is therefore a major issue from the view of ocean-wide resource management and conservation policies.
In general, a predatory lifestyle requires efficient sensing systems such as color vision. One of the important proteins for the animal visual system is opsin (∼350 aa) of the retinylidene protein family in pigment cells (4–7). The fish visual system includes five opsin genes attributed to gene duplication: rhodopsin (RH1) in rod cells and four in cone cells: red-sensitive (middle/long-wavelength sensitive; M/LWS), UV-sensitive (short-wavelength sensitive 1; SWS1), blue-sensitive (SWS2), and green-sensitive (RH2). Fish species often have multiple copies of opsin genes (6, 7); for example, medaka has three RH2 genes (8), and guppy has four M/LWS genes (9). In addition, many fish species have two copies of SWS2 genes, consistent with the concept that an ancient duplication occurred and each copy has been retained for a long time. Previous studies have accumulated molecular information on opsins, focusing on the residues surrounding the retinal-binding pocket (10–12).
Many physiological studies have been conducted on fish visual system underwater (13, 14). As for tuna, spectrophotometric analyses have demonstrated several wavelengths of maximal absorbance (λmax) in the visual pigments of yellowfin tuna (Thunnus albacares) (15) and Pacific bluefin tuna (Thunnus orientalis) (16), respectively. However, little is known about the genetic basis and evolutionary history of tuna’s optic adaptation to an open-ocean predatory lifestyle. In this study, we have sequenced the draft genome of Pacific bluefin tuna and analyzed the opsin genes in a phylogenetic framework to look for evidence of optic adaptation at the molecular level. The origins of tuna’s opsin paralogs were dated by genome-wide comparison among the teleosts for which genomic information is available, and the relationship between the evolutionary pathway of opsin genes and adaptation to ocean environment is discussed.
Results
Genome Sequencing and Gene Prediction.
The diploid genome of a wild-caught male Pacific bluefin tuna (T. orientalis) was sequenced. A whole-genome shotgun sequencing and assembling strategy with a combination of Roche 454 FLX Titanium (Roche Diagnostics) and Illumina GaIIx platforms provided 192,169 contigs (>500 bp) and 16,802 scaffolds (>2 kb) totaling 740.3 Mb, corresponding to 92.5% of the estimated genome size (∼800 Mb) (ref. 17; Table 1). The scaffolds were obtained by assembling 31.9 million 454 reads, including 4.9 million paired ends (11.9-fold coverage) and 229.7 million Illumina paired-end reads (43-fold coverage) (Tables S1 and S2 and Fig. S1). Sequence accuracy of the scaffolds was improved by crosschecking those 454 and Illumina reads whose error profiles were different from each other (18). cDNA of 10 different tissues from an adult Pacific bluefin tuna were sequenced to identify coding gene regions in the scaffolds, recovering 5,741 full-length cDNA sequences out of 180,512 contigs obtained from 3.8 million 454 reads (Table S3). A tuna-specific training model (19) was then constructed with cDNA of this study and ESTs of the Atlantic bluefin tuna (Thunnus thynnus) (20), as well as protein sequence alignments from Ensembl fish genes (21). As a result, 26,433 protein-coding gene candidates were predicted in the scaffolds (Table 2 and Table S4).
Table 1.
Overall metrics of the genome assembly and annotation of T. orientalis
| Assembly | Number | N50, bp | Average, bp | Longest, bp | Total length, Mb |
| Contigs (>500 bp) | 192,169 | 7,588 | 3,813 | 79,054 | 732.9 |
| Scaffolds (>2,000 bp) | 16,802 | 136,950 | 44,064 | 1,021,118 | 740.3 |
Table 2.
Gene comparison among teleost genomes
| Sequenced teleost | Number | Protein identity (%) |
|
| Mean | Median | ||
| Protein-coding genes of T. orientalis | 26,433 | ||
| Orthologs | |||
| vs. zebrafish | 13,474 | 69.3 | 70.4 |
| vs. cod | 13,023 | 74.0 | 75.7 |
| vs. medaka | 13,528 | 77.8 | 79.8 |
| vs. greenpuffer | 13,428 | 77.4 | 79.3 |
| vs. fugu | 13,899 | 78.6 | 80.6 |
| vs. stickleback | 14,542 | 80.2 | 82.5 |
| Among 7 genomes | 6,170 | ||
Opsin Gene and Synteny.
A total of 13,023–14,542 orthologous genes were identified by comparing the tuna data with the genomes of six teleosts (Table 2) (21, 22). The largest number of orthologs (14,542) was obtained by comparison with stickleback, which showed the highest protein identity (80.2% in the mean, 82.5% in the median), suggesting that it may be the most closely related fish to tuna among the sequenced teleosts. This observation is consistent with that in the previous study (23) in which stickleback has been shown to be close to Perciformes. Expanding the pairwise ortholog condition (i.e., reciprocal best hit) for multiple genomes, we obtained 6,170 core genes conserved among the seven teleosts. From the core gene set, all types of opsin genes, M/LWS, SWS1, SWS2, RH1, and RH2 were identified in the Pacific bluefin tuna genome. The orthologous relationship was strongly supported by syntenies with other genomes, in which the neighboring genes are also conserved as orthologous gene pairs (Fig. 1A and Fig. S2A). We found that the Pacific bluefin tuna genome possessed 10 visual pigment genes: 5 for RH2, 2 for SWS2, and 1 each for SWS1, M/LWS, and RH1. All of the opsin genes were expressed in the whole eye of a juvenile Pacific bluefin tuna (Fig. 1B). In a previous study, only two RH2, one RH1, and one SWS2 genes were identified in Pacific bluefin tuna (24); thus, three more RH2, one more SWS2, and single SWS1 and M/LWS genes were identified in this study. Of five RH2 genes, three (RH2B, g6738, and g6740) are transcribed in the same direction, whereas the others (RH2A1 and RH2A2) are on the opposite strand. One of the neighboring genes of the RH2 locus [solute carrier family 6, member 13 (slc6a13) in zebrafish] was not identified at the scaffold end but was detected in another scaffold end. Therefore, there might be more than five RH2 copies, although we did not find any significant RH2 homologs from other scaffolds or read sequences.
Fig. 1.
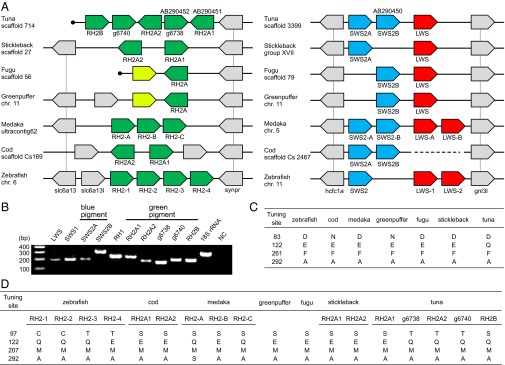
Synteny of opsin gene loci and spectral tuning sites. (A) Synteny of green (Left), blue, and red (Right) visual pigment genes compared with six teleosts whose genomes have been sequenced. Colors indicate different gene families (e.g., green for green pigment genes, blue for blue pigments, and red for red pigments). Gray-colored genes flank the pigment genes, and vertical bars indicate orthology. Previously identified genes in tuna are denoted with their GenBank accession nos. (AB290450–AB290452). The light green-colored green pigment genes of two pufferfishes are pseudogenes (30). The gene names in zebrafish and medaka comply with previous studies, and others are named in this study (g6738/g6740 of tuna are named in the order of genes predicted in scaffolds). The cod scaffold containing blue pigment genes has a gap. Filled circles indicate the scaffold ends. Each gene is arranged in consideration of transcriptional direction, and the physical distance is ignored. (B) Expression of all opsin genes in a juvenile tuna. The 18S ribosomal RNA gene was used as a positive control. (C and D) Representative amino acid sites involved in the light sensitivity of rhodopsin (C) and green opsin (D) (44) in comparison with seven teleosts. The site numbers are standardized to those of bovine rhodopsin (45). The residues of other tuning sites are summarized in Fig. S2C.
Opsin Sequence Comparison and Phylogenetic Analysis.
Fish opsin sequences were extracted from the public databases and compared with the sequences of Pacific bluefin tuna. Based on multiple alignments, we surveyed the amino acid sites involved in light sensitivity (Fig. 1 C and D and Fig. S2C). Spectral tuning sites of RH1 are almost identical among sequenced fish species; however, a tuna-specific amino acid was observed at site 122 (glutamine; Q), whereas other teleost genomes have glutamic acid (E). Glutamine at site 122 was widely conserved among the suborder Scombroidei containing the family Scombridae (e.g., tuna, mackerel, and bonito) (Fig. S2B), as well as coelacanth and lampfish (25, 26). Moreover, in four of five RH2 genes, we found amino acid substitutions at sites known for spectral tuning (11): E to Q at site 122.
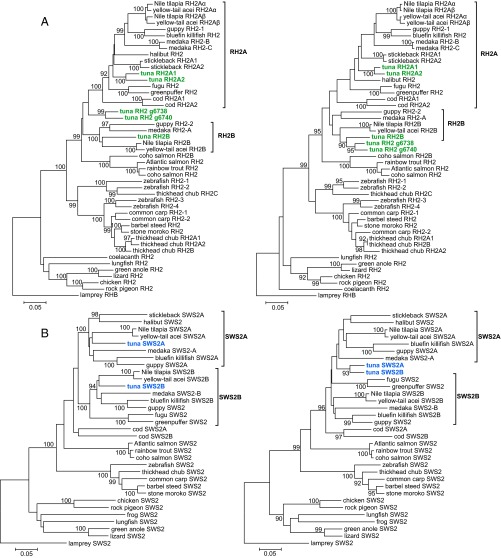
Phylogenetic analysis demonstrated that two pairs of green pigment genes, RH2A1/A2 and g6738/g6740, were recently duplicated, and that RH2A1/A2 and RH2B fall into distinct clusters, namely, RH2A and RH2B, respectively (Fig. 2 A, Left). The RH2 genes of medaka, guppy, and cichlid are in each cluster; therefore, the origin of RH2A1/A2 and RH2B is probably common to these fishes, before the appearance of the superorder Acanthopterygii. In particular, considering the position of the cod RH2 genes, the origin of RH2A and RH2B might predate the divergence of the Acanthopterygii and the Paracanthopterygii, and, subsequently, RH2B was lost in cod. A recently duplicated pair, g6738/g6740, seems to be a lone group in the phylogenetic tree, and a robust position for this pair was not found. Interestingly, high local similarities were found at the C-terminal regions between RH2B and g6738/g6740 sequences by a window analysis (Fig. S3 A, Left). This observation implies a sequence homogenization by gene conversion, which can also explain the unstable position of g6738/g6740. Therefore, the phylogenetic tree of RH2 genes was reconstructed by using only C-terminal sequences (average length: 112 aa), which placed g6738/g6740 significantly closer to RH2B2 (Fig. 2 A, Right), whereas the tree of N-terminal sequences was similar to that of the full sequences (Fig. S3B). All pairs of tuna RH2 genes exhibited nonsynonymous/synonymous substitution ratios (dN/dS; a measure for selection) of <1; therefore, these genes have maintained their functions under purifying selection (Table S5).
Fig. 2.
Phylogenetic trees of green and blue pigment genes. Numbers at branches indicate the bootstrap probabilities (≥90%) with 1,000 replicates. (A) Trees of green pigment genes. Tuna RH2 genes are shown in green. Note that the previously named RH2-A/B genes of medaka do not correspond to cichlid RH2A/B, and the tuna RH2A/B correspond to the cichlid genes. The trees were constructed by using full sequences (Left) and C-terminal sequences (Right), respectively. (B) Trees of blue pigment genes: full sequences (Left) and C-terminal sequences (Right). Tuna SWS2 genes are shown in blue.
The sequences of tuna SWS2 were found to be different from other teleosts among the predicted spectral tuning sites (sites 94 and 116) (ref. 27; Fig. S2C). Phylogenetic analysis showed that two copies of the tuna SWS2 were predicted to fall into distinct and large clades like those of other fishes (e.g., medaka and cichlid) (Fig. 2 B, Left), indicating that both tuna SWS2 genes come from an ancient gene duplication. The dN/dS between the two SWS2 copies was also <1 (0.1964) (Table S5). Moreover, we found that tuna SWS2 genes were highly similar to each other at C-terminal regions (average length: 123 aa), such as RH2B and g6738/g6740 (Fig. S3 A, Right). We then constructed the phylogenetic trees for the subsequences (Fig. 2 B, Right) and obtained the topologies suggestive of a gene conversion in the tuna SWS2 locus.
With reference to M/LWS, in which five sites are major determinants of optimum light sensitivity (λmax) (12), tuna has the same amino acids as medaka and stickleback (S, H, Y, T, and A) (Fig. S2C). Thus, following the five-sites rule, the λmax of tuna M/LWS is inferred to be similar to that of medaka, at ∼560 nm. Unlike M/LWS, significant insights into the tuning sites of SWS1 are not gained (Fig. S2C); there are no simple rules, although a previous study has shown that site-directed mutations cause wavelength shifts (28).
To date the duplication events of opsin genes, the divergence time of seven teleost species in Table 2 was initially calibrated by using nucleotide substitution rates between orthologous genes. Here, we used the synonymous substitution rate (dS), which reflects the molecular evolution in a neutral manner (29). First, 4,991 of 6,170 core genes common to the seven teleost genomes (Table 2) were selected according to the criterion that the dS of any orthologous pair was computable. The mean dS values were then computed among the seven genomes and plotted vs. the divergence times (Fig. 3A and Table S6). A linear correlation was observed between the divergence time and mean dS, and the clock was estimated as 2.5 × 10−9 per site per year. The dS values for each of RH2A/B pairs in tuna, guppy, medaka, and cichlids were estimated, and the mean was 0.92 (Table S5). The dS values of recent duplicate pairs were 0.11 for RH2A1/A2 and 0.16 for g6738/g6740. Similarly, the mean dS of SWS2A/B was 0.95. Based on the regression formula, the estimated duplication times (with a 95% confidence interval) were 183 (163–203) million years ago (Mya) for RH2A/B, 22 Mya (13–31 Mya) for RH2A1/A2, 32 Mya (21–43 Mya) for g6738/g6740, and 189 Mya (168–211 Mya) for SWS2A/B (Fig. 3B). We also applied the formula for estimating the time of gene conversion in each of the RH2 and SWS2 genes, using the C-terminal alignments; the average dS value between RH2B and g6738/g6740 was 0.29, corresponding to 58 Mya (38–79 Mya). In the case of SWS2A/B, the dS value was 0.13 (12–41) Mya. Thus, the time of gene conversion between SWS2 genes may be close to the time of birth of recent RH2 duplicates.
Fig. 3.
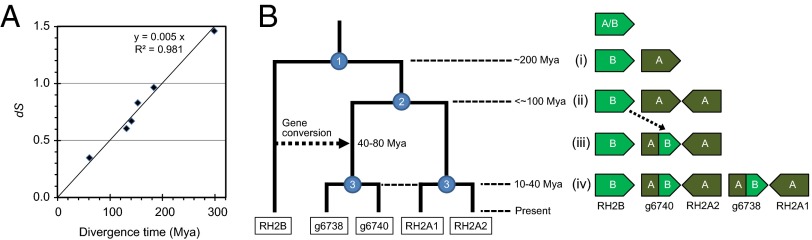
Calibration of divergence time among teleosts and evolutionary scenario of green pigment genes in T. orientalis. (A) The relationship between divergence time and the synonymous substitution rate (dS). The divergence times are based on previous studies (Table S6). The mean dS values were computed by using 4,991 of 6,170 core genes among seven teleost genomes. (B) A proposed evolutionary pathway of tuna RH2 genes. Each node with a number in the dendrogram (Left) indicates a duplication event. Right shows a diagram of the changes to the RH2 locus structure during the course of evolution. The duplication times were estimated by the regression formula computed in A.
Discussion
Genetic Feature of the Tuna Visual System.
In this study, all five opsins (M/LWS, SWS1, SWS2, RH1, and RH2) were identified in the Pacific bluefin tuna genome. Recently, three light absorption peaks (λmax: 423–436, 473, and 512–515 nm) were estimated for visual pigments of juvenile Pacific bluefin tuna, corresponding to at least one SWS2 and two RH2 genes, respectively (16), but such a color sensitivity in tuna may be explained by the combinatorial function of more opsin genes. In particular, five green pigment genes are present in the tuna genome, whereas the other six teleost genomes sequenced to date have only one to four genes (8, 21, 22, 30). This number is the highest among the sequenced fish species, and three of the five genes are identified in this study. Thus, one can speculate that the abundance of RH2 genes is involved in the tuna-specific visual system. Previous studies demonstrated that the expression pattern of fish opsins might be regulated spatially, as well as temporally, in the retina (31–33). In the case of zebrafish, shorter-wavelength RH2 proteins are first expressed in the central part of the retina, whereas longer wavelength proteins are expressed later in the peripheral part (34). Similarly, the five RH2 genes of tuna might be differentially expressed in retina, which would contribute to the visual diversification of tuna. Although the two known RH2 genes are expressed in the retina of juvenile Pacific bluefin tuna (24), we found that the other three were also expressed in the eye. It should be noted that tuna has been shown to change the light sensitivity with growth in the spectrophotometric studies (15, 35). Thus, the expression control of tuna’s opsin genes at different developmental stages might contribute to the fine tuning of color vision in the bluish ocean.
At the molecular level, specific amino acid patterns were found at spectral tuning sites for both of RH1 and RH2 genes. In RH1, the amino acid at site 122 was glutamine, whereas other teleosts possess glutamic acid. A previous study using site-directed mutagenesis demonstrated that the E122Q replacement in coelacanth RH1 causes a short-wave shift of the pigments (i.e., blue shift) (25). This pattern is often observed in deep-sea fishes irrespective of lineage, involving an adaptation to dark environments (26, 36). The present study demonstrated that this pattern is well conserved in the suborder, Scombroidei, suggesting that the amino acid was substituted to Q from E in an ancestor of this lineage. Many of the species in Scombroidei are not deep-sea dwellers but are open water predators. Regarding the RH2 genes, in which Q122E replacement shifts the absorption toward red (11), four of the five genes in tuna might be shifted to a shorter wavelength than the other because of Q at site 122. Therefore, we propose that these E122Q substitutions in tuna RH1 and RH2 may contribute to effective detection of prey in the bluish ocean.
Evolution of Tuna Blue-Green Genes.
The phylogenetic analysis showed that an ancient duplication of fish green pigment gene into RH2A and RH2B occurred before the divergence of the Acanthopterygii. This duplication was estimated to have occurred ∼180 Mya (163–203 Mya), based on the synonymous substitution rates. Thus, the emergence of RH2A and RH2B may predate the divergence of the Acanthopterygii and the Paracanthopterygii (∼160 Mya) (23, 37). Here, the duplication of the SWS2 genes is also estimated to have occurred ∼190 Mya (168–211 Mya), near the time of the RH2 duplication. These estimates are consistent with those from published reviews (6, 7), suggesting that the calibration by synonymous substitution rates using genome-wide ortholog pairs is effective. Regarding recent duplicate pairs of tuna RH2 genes, the dS values are similar for each pair (RH2A1/RH2A2, 0.11; g6738/g6740, 0.16). Taking into account the gene order and corresponding directions of transcription, it is likely that these pairs originated simultaneously from a single tandem duplication (∼10–40 Mya). The duplication of RH2A was estimated to have occurred ∼92 Mya (79–105 Mya), with a mean dS = 0.46 (Table S5); however, this date (92 Mya) is probably an overestimate, because the C-terminal sequences of g6738/g6740 were replaced by that of RH2B through gene conversion (38–79 Mya). In addition to the RH2 genes, we obtained evidence of a recent gene conversion between SWS2 genes (12–41 Mya). Because the alignments examined were partial and short (∼110–120 aa), further analysis is needed to accurately estimate the times of gene conversion. Interestingly, g6738/g6740 is still a lone group even in the tree of N-terminal regions (Fig. S3B), suggesting that the duplication of RH2A1/A2 and g6738/g6740 might be specific to the Scombridae/Scombroidei or the Perciformes.
In the present study, we propose the following evolutionary pathway of RH2 genes in tuna based on parsimony, in which gene duplication and conversion events are minimized to three and one, respectively (Fig. 3B). (i) An RH2 gene was duplicated into RH2A and RH2B before the divergence of the Paracanthopterygii and the Acanthopterygii (∼200 Mya). (ii) An inverted duplication then occurred in the RH2A lineage (<∼100 Mya). (iii) Gene conversion partially replaced one of the RH2A copies by a counterpart from the RH2B lineage (40–80 Mya). (iv) A tandem duplication doubled two copies to four (10–40 Mya). If the gene inversion and duplication in stage ii occurred separately, a gene fusion by deletion could also account for the RH2 evolution by three duplications (Fig. S3C). Under any scenario, we can argue that RH2 increased from three to five copies in stage iv in a relatively short period. In a previous study, the divergence time of Thunnus and Scomber (mackerel) is inferred to be ∼80 Mya (37), suggesting that RH2 copy number variation and both conversions in RH2 and SWS2 may have arisen within the Scombridae/Scombroidei and may be different between tuna and other closely related species, such as mackerel and bonito.
We have thus observed that molecular evolutionary changes in three opsin genes, RH1, RH2, and SWS2, may have occurred around the time when the ancestor of tuna appeared or diverged. All of the changes are involved in blue-green light sensitivity. In Pacific bluefin tuna, particularly, not only stepwise duplications of RH2 genes but also gene conversions in such multicopy genes (including SWS2) may have facilitated the functional diversification of spectral tuning to the offshore environment, which are useful for detecting bluish contrasts. Moreover, each opsin is in general considered to be expressed in a single pigment cell in the retina; therefore, the difference in blue-green light sensitivity can lead to a lag in focal distances among blue or green pigment cells. Although fish camouflage mechanism underwater is complicated (38), tuna RH2 and SWS2 copies might be used for measuring the distance to blue-backed fishes in the bluish ocean as well as for improving dynamic vision.
Impact of Tuna Genome Sequencing.
Our study represents a tuna genome assembly, which has not been described previously and which provides significant insight into the genetic and evolutionary background of the optic adaptation of Pacific bluefin tuna. These findings will shed light on behavioral traits of predatory fish in the open ocean. In tuna aquaculture, for example, a high mortality due to wall collisions has been problematic (39), which can be considered as an adverse consequence of adaptation to the bluish pelagic environment. The findings we present here are thus applicable for design of tuna farms: perceptible and high-contrast coloring. In particular, because tuna is still difficult to culture and handle for experimental approaches, genomics strategy is widely expected to complement physiological studies on tuna. Genomic information from a wide range of pelagic organisms such as bluefin tunas will provide significant clues for comprehensive understanding of the diversity of those organisms.
Materials and Methods
Detailed methods are described in SI Materials and Methods.
Genome Sequencing.
Genomic DNAs of Pacific bluefin tuna were extracted from a male individual reared in Amami, Japan, and the sequence templates for 454 and Illumina sequencing were prepared followed the manufacturers’ instructions. The de novo genome assembly and gene prediction are described in SI Materials and Methods. The T. orientalis whole-genome assembly sequences have been deposited in the DNA Data Bank of Japan/European Molecular Biology Laboratory/GenBank (accession nos. BADN01000001–BADN01133062).
Gene Comparison.
Genomic data from the six teleosts, zebrafish (Danio rerio), cod (Gadus morhua), medaka (Oryzias latipes), greenpuffer (Tetraodon nigroviridis), fugu (Takifugu rubripes), and stickleback (Gasterosteus aculeatus), were downloaded from the Ensembl database (Release 61). All-vs.-all BLASTP was performed among the seven teleost genes (Ensembl fishes and tuna) with an E-value < 10−10. An orthologous gene pair was defined as one reciprocal best hit. Each of the core genes conserved among seven teleost fishes was defined as the gene set in which any pair was defined as orthologous.
Phylogenetic Analysis.
Opsin gene sequences from other fishes and animals were collected from the GenBank (40) and Ensembl databases (listed in SI Materials and Methods). The detailed methods for multiple alignment (41), computation of synonymous and nonsynonymous substitution rates (42), and molecular phylogenetic tree (43) are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
S. Shiozawa and members of the Tuna Seed Production Group, Seikai National Fisheries Research Institute (Fisheries Research Agency; FRA), kindly provided the tuna tissue specimens for sequencing. We thank K. Furuya, C. Shindo, H. Inaba, E. Iioka, Y. Takayama, E. Ohmori, M. Kikuchi, W. Suda, and Y. Hattori for technical support. S. Song and the Roche Diagnostics technical support team helped us operate the sequencing machine. Drs. Y. Sakaki, H. L. Bart, Jr., M. H. Doosey, and S. Yokoyama provided invaluable comments on the manuscript. This research was partly supported by general research and a facility setting-up funds to the FRA from the Fisheries Agency (Ministry of Agriculture, Forestry and Fisheries, Japan); and Ministry of Education, Culture, Sports, Science and Technology (Japan) Grants-in-Aid for Scientific Research on Priority Areas “Comprehensive Genomics,” on Innovative Areas ”Genome Science,” and the global Center of Excellence Project “Genome Information Big Bang.”
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The T. orientalis whole-genome assembly sequences have been deposited in the DNA Data Bank of Japan, European Molecular Biology Laboratory, and GenBank databases (accession nos. BADN01000001–BADN01133062).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1302051110/-/DCSupplemental.
References
- 1.Hochachka PW. Living Without Oxygen: Closed and Open Systems in Hypoxia Tolerance. Cambridge, MA: Harvard Univ Press; 1980. [Google Scholar]
- 2.Block BA, et al. Migratory movements, depth preferences, and thermal biology of Atlantic bluefin tuna. Science. 2001;293(5533):1310–1314. doi: 10.1126/science.1061197. [DOI] [PubMed] [Google Scholar]
- 3.Collette BB, et al. Conservation. High value and long life—double jeopardy for tunas and billfishes. Science. 2011;333(6040):291–292. doi: 10.1126/science.1208730. [DOI] [PubMed] [Google Scholar]
- 4.Yokoyama S. Molecular evolution of color vision in vertebrates. Gene. 2002;300(1-2):69–78. doi: 10.1016/s0378-1119(02)00845-4. [DOI] [PubMed] [Google Scholar]
- 5.Trezise AE, Collin SP. Opsins: Evolution in waiting. Curr Biol. 2005;15(19):R794–R796. doi: 10.1016/j.cub.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 6.Bowmaker JK. Evolution of vertebrate visual pigments. Vision Res. 2008;48(20):2022–2041. doi: 10.1016/j.visres.2008.03.025. [DOI] [PubMed] [Google Scholar]
- 7.Rennison DJ, Owens GL, Taylor JS. Opsin gene duplication and divergence in ray-finned fish. Mol Phylogenet Evol. 2012;62(3):986–1008. doi: 10.1016/j.ympev.2011.11.030. [DOI] [PubMed] [Google Scholar]
- 8.Matsumoto Y, Fukamachi S, Mitani H, Kawamura S. Functional characterization of visual opsin repertoire in Medaka (Oryzias latipes) Gene. 2006;371(2):268–278. doi: 10.1016/j.gene.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 9.Ward MN, et al. The molecular basis of color vision in colorful fish: Four long wave-sensitive (LWS) opsins in guppies (Poecilia reticulata) are defined by amino acid substitutions at key functional sites. BMC Evol Biol. 2008;8:210. doi: 10.1186/1471-2148-8-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sakmar TP, Franke RR, Khorana HG. Glutamic acid-113 serves as the retinylidene Schiff base counterion in bovine rhodopsin. Proc Natl Acad Sci USA. 1989;86(21):8309–8313. doi: 10.1073/pnas.86.21.8309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chinen A, Matsumoto Y, Kawamura S. Reconstitution of ancestral green visual pigments of zebrafish and molecular mechanism of their spectral differentiation. Mol Biol Evol. 2005;22(4):1001–1010. doi: 10.1093/molbev/msi086. [DOI] [PubMed] [Google Scholar]
- 12.Yokoyama S, Yang H, Starmer WT. Molecular basis of spectral tuning in the red- and green-sensitive (M/LWS) pigments in vertebrates. Genetics. 2008;179(4):2037–2043. doi: 10.1534/genetics.108.090449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clarke GL. On the depth at which fish can see. Ecology. 1936;17(3):452–456. [Google Scholar]
- 14.Lythgoe JN. Visual pigments and visual range underwater. Vision Res. 1968;8(8):997–1011. doi: 10.1016/0042-6989(68)90073-4. [DOI] [PubMed] [Google Scholar]
- 15.Loew ER, McFarland WN, Margulies D. Developmental changes in the visual pigments of the yellowfin tuna, Thunnus albacares. Mar Freshwat Behav Physiol. 2002;35:235–246. [Google Scholar]
- 16.Matsumoto T, Okada T, Sawada Y, Ishibashi Y. Visual spectral sensitivity of photopic juvenile Pacific bluefin tuna (Thunnus orientalis) Fish Physiol Biochem. 2012;38(4):911–917. doi: 10.1007/s10695-011-9574-0. [DOI] [PubMed] [Google Scholar]
- 17.Ida H, Oka N, Terashima H, Hayashizaki K. Karyotypes and cellular DNA contents of three species of the family Scombridae from Japan. Japonais. 1993;59:1319–1323. [Google Scholar]
- 18.Holt RA, Jones SJ. The new paradigm of flow cell sequencing. Genome Res. 2008;18(6):839–846. doi: 10.1101/gr.073262.107. [DOI] [PubMed] [Google Scholar]
- 19.Stanke M, Diekhans M, Baertsch R, Haussler D. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics. 2008;24(5):637–644. doi: 10.1093/bioinformatics/btn013. [DOI] [PubMed] [Google Scholar]
- 20.Chini V, et al. Genes expressed in blue fin tuna (Thunnus thynnus) liver and gonads. Gene. 2008;410(1):207–213. doi: 10.1016/j.gene.2007.12.012. [DOI] [PubMed] [Google Scholar]
- 21.Flicek P, et al. Ensembl 2011. Nucleic Acids Res. 2011;39(Database issue):D800–D806. doi: 10.1093/nar/gkq1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Star B, et al. The genome sequence of Atlantic cod reveals a unique immune system. Nature. 2011;477(7363):207–210. doi: 10.1038/nature10342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matschiner M, Hanel R, Salzburger W. On the origin and trigger of the notothenioid adaptive radiation. PLoS ONE. 2011;6(4):e18911. doi: 10.1371/journal.pone.0018911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miyazaki T, Kohbara J, Takii K, Ishibashi Y, Kumai H. Three cone opsin genes and cone cell arrangement in retina of juvenile Pacific bluefin tuna Thunnus orientalis. Fish Sci. 2008;74(2):314–321. [Google Scholar]
- 25.Yokoyama S, Zhang H, Radlwimmer FB, Blow NS. Adaptive evolution of color vision of the Comoran coelacanth (Latimeria chalumnae) Proc Natl Acad Sci USA. 1999;96(11):6279–6284. doi: 10.1073/pnas.96.11.6279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yokoyama S, Tada T, Zhang H, Britt L. Elucidation of phenotypic adaptations: Molecular analyses of dim-light vision proteins in vertebrates. Proc Natl Acad Sci USA. 2008;105(36):13480–13485. doi: 10.1073/pnas.0802426105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chinen A, Matsumoto Y, Kawamura S. Spectral differentiation of blue opsins between phylogenetically close but ecologically distant goldfish and zebrafish. J Biol Chem. 2005;280(10):9460–9466. doi: 10.1074/jbc.M413001200. [DOI] [PubMed] [Google Scholar]
- 28.Shi Y, Radlwimmer FB, Yokoyama S. Molecular genetics and the evolution of ultraviolet vision in vertebrates. Proc Natl Acad Sci USA. 2001;98(20):11731–11736. doi: 10.1073/pnas.201257398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuraku S, Kuratani S. Time scale for cyclostome evolution inferred with a phylogenetic diagnosis of hagfish and lamprey cDNA sequences. Zoolog Sci. 2006;23(12):1053–1064. doi: 10.2108/zsj.23.1053. [DOI] [PubMed] [Google Scholar]
- 30.Neafsey DE, Hartl DL. Convergent loss of an anciently duplicated, functionally divergent RH2 opsin gene in the fugu and Tetraodon pufferfish lineages. Gene. 2005;350(2):161–171. doi: 10.1016/j.gene.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 31.Spady TC, et al. Evolution of the cichlid visual palette through ontogenetic subfunctionalization of the opsin gene arrays. Mol Biol Evol. 2006;23(8):1538–1547. doi: 10.1093/molbev/msl014. [DOI] [PubMed] [Google Scholar]
- 32.Carleton KL, et al. Visual sensitivities tuned by heterochronic shifts in opsin gene expression. BMC Biol. 2008;6:22. doi: 10.1186/1741-7007-6-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shand J, et al. The influence of ontogeny and light environment on the expression of visual pigment opsins in the retina of the black bream, Acanthopagrus butcheri. J Exp Biol. 2008;211(Pt 9):1495–1503. doi: 10.1242/jeb.012047. [DOI] [PubMed] [Google Scholar]
- 34.Takechi M, Kawamura S. Temporal and spatial changes in the expression pattern of multiple red and green subtype opsin genes during zebrafish development. J Exp Biol. 2005;208(Pt 7):1337–1345. doi: 10.1242/jeb.01532. [DOI] [PubMed] [Google Scholar]
- 35.Matsumoto T, Okada T, Sawada Y, Ishibashi Y. Changes in the scotopic vision of juvenile Pacific bluefin tuna (Thunnus orientalis) with growth. Fish Physiol Biochem. 2011;37(3):693–700. doi: 10.1007/s10695-011-9469-0. [DOI] [PubMed] [Google Scholar]
- 36.Hunt DM, Dulai KS, Partridge JC, Cottrill P, Bowmaker JK. The molecular basis for spectral tuning of rod visual pigments in deep-sea fish. J Exp Biol. 2001;204(Pt 19):3333–3344. doi: 10.1242/jeb.204.19.3333. [DOI] [PubMed] [Google Scholar]
- 37.Cantatore P, et al. Evolutionary analysis of cytochrome b sequences in some Perciformes: Evidence for a slower rate of evolution than in mammals. J Mol Evol. 1994;39(6):589–597. doi: 10.1007/BF00160404. [DOI] [PubMed] [Google Scholar]
- 38.Johnsen S. Lifting the cloak of invisibility: The effects of changing optical conditions on pelagic crypsis. Integr Comp Biol. 2003;43(4):580–590. doi: 10.1093/icb/43.4.580. [DOI] [PubMed] [Google Scholar]
- 39.Sawada Y, Okada T, Miyashita S, Murata O, Kumai H. Completion of the Pacific bluefin tuna Thunnus orientalis (Temminck et Schlegel) life cycle. Aquacult Res. 2005;36(5):413–421. [Google Scholar]
- 40.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2011;39(Database issue):D32–D37. doi: 10.1093/nar/gkq1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 2008;9(4):286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- 42.Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3(5):418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 43.Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 44.Yokoyama S. Molecular evolution of vertebrate visual pigments. Prog Retin Eye Res. 2000;19(4):385–419. doi: 10.1016/s1350-9462(00)00002-1. [DOI] [PubMed] [Google Scholar]
- 45.Palczewski K, et al. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289(5480):739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.