Significance
A mechanism for regulating production of the antibiotic planosporicin by Planomonospora alba is described. A low level of planosporicin biosynthesis, probably stimulated by nutrient limitation, functions in a feed-forward mechanism to release a key regulatory protein required for high level production. Planosporicin also functions as an extracellular signaling molecule to induce its own synchronous production, presumably ensuring ecologically effective levels of the antibiotic by coordinating biosynthesis in the multicellular colony, regions of which have reached different levels of maturity. This information was used to increase the level of planosporicin production, with important implications for strain improvement.
Abstract
Planosporicin is a ribosomally synthesized, posttranslationally modified peptide lantibiotic produced by the actinomycete Planomonospora alba. It contains one methyl-lanthionine and four lanthionine bridges and inhibits cell wall biosynthesis in other Gram-positive bacteria probably by binding to lipid II, the immediate precursor for cell wall biosynthesis. Planosporicin production, which is encoded by a cluster of 15 genes, is confined to stationary phase in liquid culture and to the onset of morphological differentiation when P. alba is grown on agar. This growth phase-dependent gene expression is controlled transcriptionally by three pathway-specific regulatory proteins: an extracytoplasmic function σ factor (PspX), its cognate anti-σ factor (PspW), and a transcriptional activator (PspR) with a C-terminal helix-turn-helix DNA-binding domain. Using mutational analysis, S1 nuclease mapping, quantitative RT-PCR, and transcriptional fusions, we have determined the direct regulatory dependencies within the planosporicin gene cluster and present a model in which subinhibitory concentrations of the lantibiotic function in a feed-forward mechanism to elicit high levels of planosporicin production. We show that in addition to acting as an antibiotic, planosporicin can function as an extracellular signaling molecule to elicit precocious production of the lantibiotic, presumably ensuring synchronous and concerted lantibiotic biosynthesis in the wider population and, thus, the production of ecologically effective concentrations of the antibiotic.
More than 60 y have passed since the first natural product antibiotic was used in the clinic, and yet infectious disease remains a global problem, exacerbated by the acquisition by a broad range of human pathogens of multiple antibiotic resistance determinants (1). Consequently, there is a growing need for the discovery and development of new anti-infective agents. The majority of clinically used antibiotics are natural products or their derivatives (2), and many of these are derived from actinomycetes. These Gram-positive mycelial multicellular bacteria remain a promising source of potentially useful new compounds (3). Actinomycete antibiotic biosynthetic genes are arranged in clusters, greatly facilitating the isolation and analysis of all of the genes required to make a particular compound. In general, the expression of these gene clusters is not constitutive but occurs in a growth phase-dependent manner; in liquid culture, it usually begins at the onset of stationary phase, whereas in agar grown cultures it occurs at the onset of morphological development. Indeed, there are pleiotropic regulatory genes that are required for both processes (4). This growth phase-dependent gene expression is often exerted, at least in part, by pathway-specific regulatory genes located in the biosynthetic gene cluster, the expression of which is influenced by a variety of environmental and developmental cues (4, 5). In some cases, at least, the small intracellular signaling molecule guanosine tetraphosphate (ppGpp), produced in response to nutrient limitation, appears to activate the transcription of antibiotic biosynthetic genes (6).
Lantibiotics are ribosomally synthesized, posttranslationally modified peptides (RiPPs) that contain characteristic lanthionine and/or methyl-lanthionine bridges that confer structural rigidity, protease resistance, and antimicrobial activity on the resulting compounds (7, 8). Although actinomycetes have not generally been recognized as prolific producers of potentially useful lantibiotics, two actinomycete compounds, NVB302 [an actagardine derivative (9)] and NAI-107 [also known as microbisporicin (10)] are currently in clinical development as anti-infectives, whereas a third, Moli1901 (also known as lancovutide or duramycin, a structural analog of cinnamycin), promises to be a useful adjunct for the treatment of cystic fibrosis (11, 12). Actinomycete RiPPs that have been analyzed genetically include cinnamycin (13), the spore-associated proteins SapB (14) and SapT (15), michiganin (16), actagardine (17), deoxyactagardine (18), microbisporicin (19, 20), cypemycin (21), venezuelin (22), and grisemycin (23).
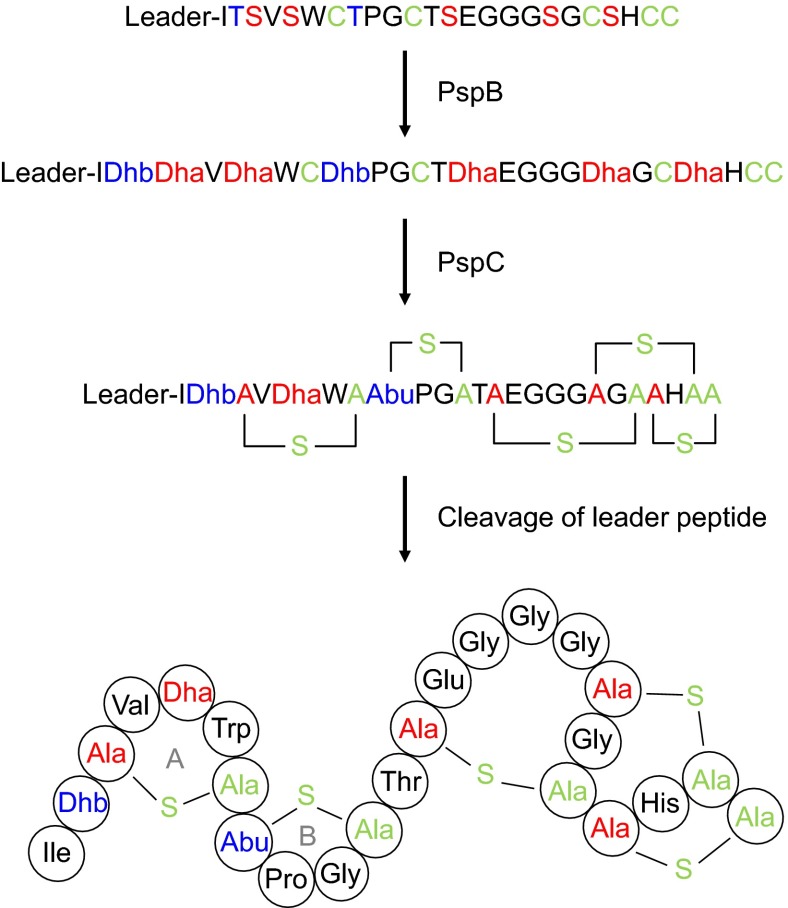
Planosporicin is another actinomycete-derived lantibiotic produced by Planomonospora sp. DSM 14920 (24) and Planomonospora alba (25) and contains one methyl-lanthionine and four lanthionine bridges (Fig. 1). The lantibiotic prevents the growth of other Gram-positive bacteria by inhibiting cell wall biosynthesis (24), and its N-terminal similarity to nisin (26) and microbisporicin (27) suggests an ability to bind to lipid II, the immediate precursor for cell wall biosynthesis (28). Recently, we identified and analyzed the gene cluster from P. alba responsible for the stationary phase production of planosporicin (25). The core biosynthetic genes are pspA (encoding the precursor peptide), pspB [encoding the dehydratase responsible for the dehydration of five serines and two threonines to dehydroalanine (Dha) and dehydrobutyrine (Dhb), respectively], and pspC (encoding the cyclase responsible for the coupling of five cysteines to four Dha and one Dhb to form four lanthionines and one methyl-lanthionine, respectively) (Fig. 1). The cluster also contains three regulatory genes. pspR encodes a protein with a DNA-binding domain also found in the LuxR family of transcriptional activators, whereas pspX and pspW encode an extracytoplasmic function (ECF) family σ factor and putative anti-σ factor, respectively. Both pspR and pspX are essential for planosporicin biosynthesis (25). ECF σ factors are frequently involved in sensing and responding to cell envelope stress (29) and occur in a range of bacterial genera, but are particularly prevalent in actinomycetes (30). Nevertheless, pspX is only the second example of the use of this family of regulatory proteins to directly control antibiotic production in actinomycetes [the first was microbisporicin (20)].
Fig. 1.
Schematic representation of the posttranslational modification of planosporicin. The planosporicin precursor peptide is shown at the top. PspB catalyzes the dehydration of Ser and Thr to Dha and Dhb, respectively. PspC installs the four Lan bridges and one MeLan bridge by regioselective cyclization. Removal of the leader sequence by an unidentified protease liberates the active lantibiotic. The sequence of the planosporicin leader peptide is MGISSPALPQNTADLFQLDLEIGVEQSLASPA.
In this paper, we analyze the transcriptional regulation of the planosporicin biosynthetic gene cluster and show that the lantibiotic can induce its own synthesis at subinhibitory concentrations. This likely occurs by disrupting the interaction between σPspX and PspW that serves to sequester the ECF σ factor, thus preventing premature antibiotic production. We propose a detailed model for the regulation of planosporicin production in which the lantibiotic serves as an extracellular signaling molecule to coordinate the onset of its own biosynthesis throughout the substrate mycelium and potentially in the P. alba population as a whole.
Results
Mapping Transcriptional Start Sites in the Planosporicin Biosynthetic Gene Cluster.
Previous RT-PCR studies (25) suggested that the psp gene cluster was transcribed in at least three operons, pspABCTUV, pspXWJYZQR, and pspEF. To identify the transcriptional start sites of these putative operons, and to assess whether pspR and pspJ, which are preceded by intergenic regions of 89 and 231 bp, respectively, were also transcribed from additional promoters, high-resolution nuclease S1 protection analyses were carried out using 32P-5′ end-labeled probes complementary to the predicted 5′ ends of the transcripts of pspA, pspX, pspJ, pspE, and pspR. The probes were hybridized with RNA isolated from wild-type P. alba and the ΔpspW mutant M1304 after 162 h of incubation, when planosporicin was detectable in culture supernatants (Fig. 2). Nonhomologous sequences (15 nt) at the 5′ end of each probe were used to distinguish between probe:probe reannealing and transcriptional read-through from upstream genes; the latter would result in a protected segment 15 nt shorter than the probe fragment. The resulting nuclease S1-protected fragments were subjected to gel electrophoresis adjacent to GA-sequencing ladders derived from the corresponding labeled probe to determine the precise 5′ ends of the transcripts. With the exception of pspR, each probe identified a single upstream promoter (Fig. S1); in contrast, the pspR probes (R1, extending 237 nt into pspQ, and R2, extending 519 nt upstream of the pspR TTG start codon and incorporating all of pspQ), revealed only transcriptional read-through (Fig. S1). Intergenic sequences upstream of pspA, pspX, pspJ, and pspE were then analyzed by multiple expectation maximization for motif elicitation (MEME)* to identify conserved regulatory motifs. Three of these regions (those for pspX, pspJ, and pspE) contain a conserved motif (GAACC–17 NT–TACGTAC) similar to that recognized by ECF σ factors [AAC in the −35 region and CGT in the −10 region (31)] and positioned appropriately upstream of the experimentally determined transcriptional start sites (Fig. 3 A and C). Thus, all three of these promoters were predicted to be regulated directly by the ECF σ factor PspX. In contrast, the transcriptional start site of pspA is preceded by −35 and −10 sequences (Fig. 3B) that resemble the consensus sequence for the major vegetative σ factor of the most studied actinomycete, Streptomyces coelicolor (32). Thus, the psp biosynthetic gene cluster appears to be transcribed in four operons: pspABCTUV, pspXWJYZQR, pspJYZQR, and pspEF, with the last three regulated directly by σPspX.
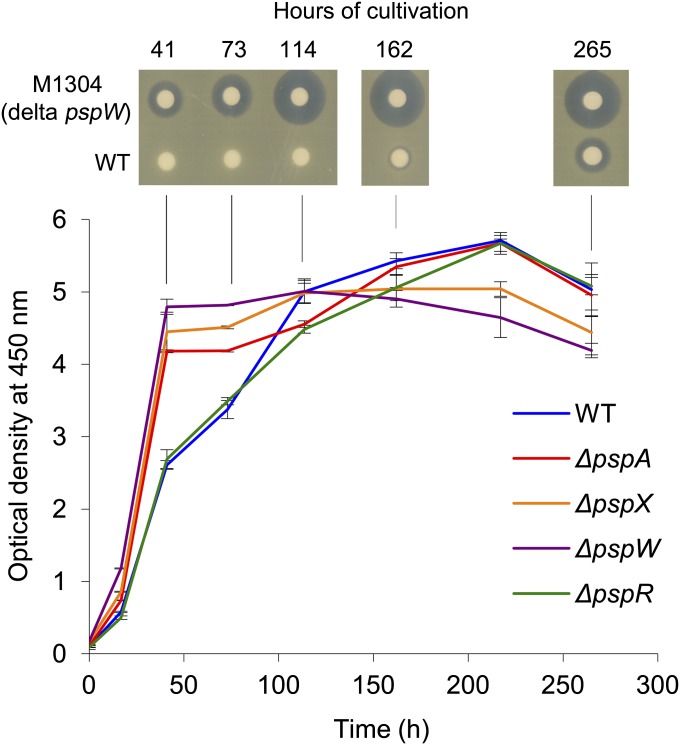
Fig. 2.
Growth and planosporicin production in P. alba. Growth was measured by optical density at 450 nm and averaged for two independent cultures of each strain at each time point (the range and average of each pair of measurements for each strain is shown): P. alba wild type (blue), M1302 ΔpspA (red), M1303 ΔpspX (orange), M1304 ΔpspW (purple), and M1308 ΔpspR (green). Mycelium was collected after 41, 73, 114, and 162 h for RT-PCR analysis. Supernatants were collected at the same time points and at 265 h and assayed for antibacterial activity on a lawn of M. luteus.
Fig. 3.
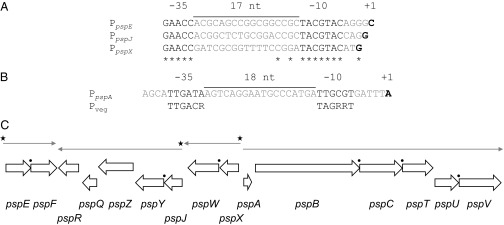
Summary of the results of high-resolution S1 nuclease mapping of the 5′ ends of the four psp transcripts using PCR-generated probes and RNA from P. alba. In A and B, predicted −35 and −10 elements are in black type, and transcriptional start sites are highlighted in bold. (A) Alignment of the promoter regions of pspE, pspJ, and pspX. (B) Promoter region of pspA aligned with the consensus motif for promoters recognized by the major vegetative σ factor of streptomycetes (31). (C) Transcriptional organization of the psp gene cluster. Occurrences of translational coupling are indicated by black circles. Starred promoters contain an ECF σ factor consensus motif.
Quantitative RT-PCR Analyses Reveal Likely Regulatory Dependences in the psp Gene Cluster.
The level of transcription of representative psp genes was analyzed in five P. alba strains: wild type, M1302 (ΔpspA::oriT-hyg), M1303 (ΔpspX::oriT-hyg), M1304 (ΔpspW::oriT-hyg), and M1308 (ΔpspR::oriT-hyg). The strains were grown in AF/MS liquid medium (Fig. 2); although the wild type and M1308 (ΔpspR::oriT-hyg) exhibited lower initial growth rates, all five strains reached the same final biomass. RNA was extracted from mycelium harvested after 41, 73, 113, and 162 h. Culture supernatants from each time point were assayed for planosporicin production (Fig. 2), which could be detected in M1304 and the wild-type strain after 41 and 162 h, respectively; M1302, M1303 and M1308 failed to produce the lantibiotic, as observed previously (25). Interestingly, M1304 (ΔpspW::oriT-hyg) exhibited precocious and increased levels of antibiotic activity compared with the wild-type strain (see later for further discussion).
Reverse transcriptase (RT)-PCR analysis was used initially to assess psp gene expression at all four time points in all five strains (Fig. S2). As a positive control for each cDNA template, a gene with 87% nt identity across 1036 nt of 3′ sequence of hrdB from S. coelicolor was identified in the P. alba 454 sequence data (25). hrdB encodes the major vegetative σ factor of S. coelicolor (σ70) and is commonly used as a control in RT-PCR and quantitative (q)RT- PCR experiments in Streptomyces spp (33). Expression of the putative P. alba hrdB occurred at approximately equal levels in all five strains at each time point (Fig. S2) (the apparently lower levels of expression in later time points may represent decreased transcription of the hrdB homolog during stationary phase). To obtain an overview of transcription within the cluster in the different strains, primer pairs were used that annealed within each of the psp operons (Table S1). Transcription of essentially all of the psp genes was markedly reduced in the ΔpspR, ΔpspX, and surprisingly ΔpspA mutants and markedly elevated in the ΔpspW mutant (Fig. S2), with little difference between the different time points.
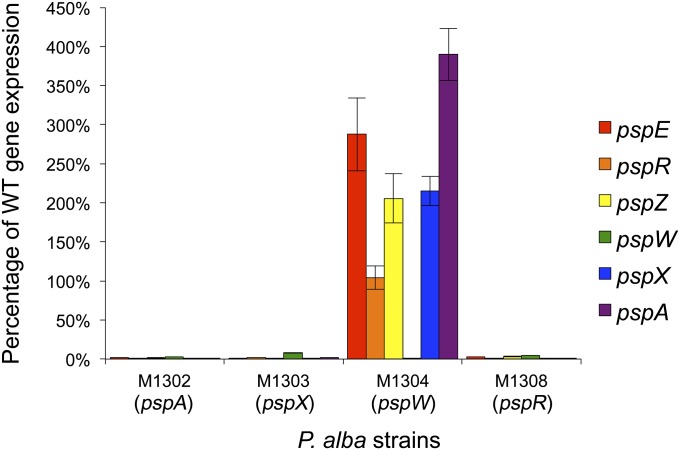
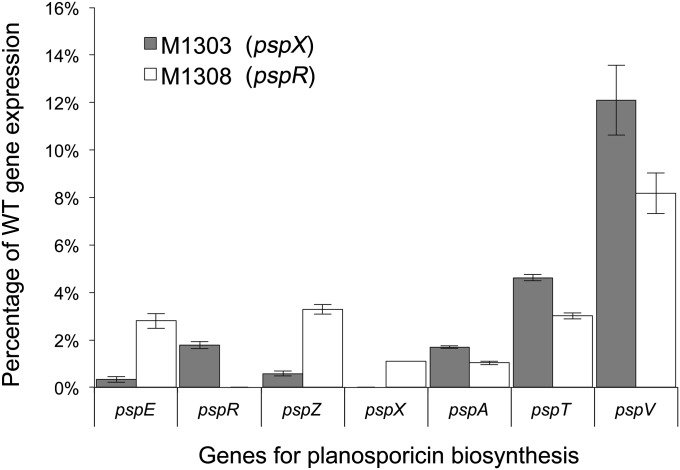
To obtain a more quantitative view of the effect of the regulatory mutations on psp gene expression, RNA isolated from all five strains after 162 h of growth was subjected to qRT-PCR analysis (Fig. 4). The copy number of each transcript was first normalized to the expression of P. alba hrdB and then expressed as a percentage of the wild-type level of expression. Expression of all four psp operons was markedly increased in the ΔpspW mutant M1304, and markedly decreased in the ΔpspA (M1302), ΔpspX (M1303), and ΔpspR (M1308) mutants. To compare the relative effect of deleting the two positive regulatory genes, expression of psp genes was compared in M1303 (ΔpspX) and M1308 (ΔpspR) (Fig. 5). The levels of expression of pspZ and pspE were reduced 5.70- and 8.37-fold, respectively, more in M1303 (ΔpspX) than in M1308 (ΔpspR), potentially reflecting direct regulation by σPspX. In contrast, the levels of expression of pspA, pspT, and pspV were reduced 1.64-, 1.54-, and 1.48-fold, respectively, more in M1308 than in M1303, potentially reflecting direct regulation by PspR.
Fig. 4.
qRT-PCR analysis of psp gene expression in P. alba M1302 (ΔpspA), M1303 (ΔpspX), M1304 (ΔpspW), and M1308 (ΔpspR) compared with the wild-type strain. cDNA was synthesized from RNA extracted after 162 h of culture. psp gene expression was normalized to that of the putative hrdB homolog of P. alba and expressed as a percentage of wild-type gene expression. Error bars depict SD between three technical replicates.
Fig. 5.
Relative effects of deletion of pspX and pspR on psp gene expression measured by qRT-PCR analysis. cDNA was synthesized from RNA extracted from P. alba wild type, M1303 (pspX), and M1308 (pspR) after 162 h of culture. psp gene expression was normalized to that of the putative hrdB homolog of P. alba and expressed as a percentage of wild-type gene expression. Error bars depict SD between three technical replicates.
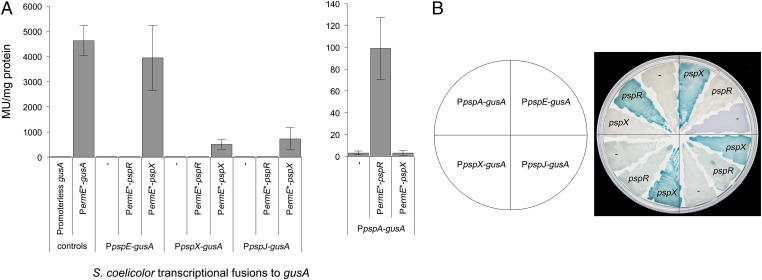
gusA Transcriptional Fusions in S. coelicolor Confirm the Direct Regulatory Targets of σPspX and PspR.
To confirm the likely direct regulatory targets of PspR and σPspX, a series of promoter fusions were made to a version of gusA encoding β-glucuronidase (GUS) that had been codon-optimized for expression in streptomycetes (34). The constitutive promoter of ermE (PermE*) (35) was cloned upstream of gusA in pGUS (34), yielding pIJ10740, which was integrated into the ΦC31 chromosomal attachment site (attB) of S. coelicolor M1152 to provide a positive control strain. M1152 containing pGUS served as a negative control, displaying basal levels of GUS activity (3.77 Miller units per milligram of protein). Expression of gusA from PermE* gave readily detectable GUS activity in both spectrophotometric (4628 Miller units per milligram of protein; Fig. 6A) and agar-based assays (Fig. 7B). The pspA, pspX, pspJ, and pspE promoters were then cloned separately upstream of gusA in pGUS, and the resulting plasmids were integrated into the ΦC31 attB site of S. coelicolor M1152. In parallel, the coding sequences of the regulatory genes pspR, pspX, and pspXW were cloned downstream of PermE* in pIJ10257, and the resulting constructs were integrated into the ΦBT1 attB site of the S. coelicolor M1152 derivatives containing the four different reporter constructs (Table S2).
Fig. 6.
GUS activity in S. coelicolor derivatives containing psp promoter-gusA fusions in the absence or presence of either pspX or pspR transcribed from PermE*. (A) Spectrophotometric assay using p-nitrophenyl-β-d-glucoronide as substrate. Miller units are shown per milligram of protein present in cell lysate. Error bars depict SD between three biological replicates. (B) Chromogenic assay on R2 agar containing the substrate 5-bromo-4-chloro-3-indolyl-β-d-glucoronide.
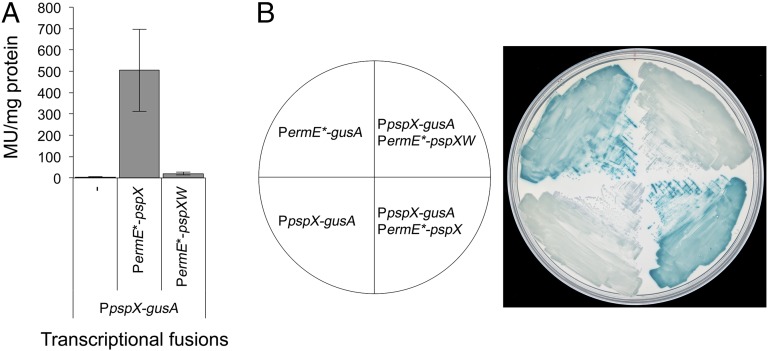
Fig. 7.
GUS activity in S. coelicolor M1152 derivatives containing the pspX promoter fused to gusA in the absence or presence of either pspX or pspXW transcribed from PermE*. PermE* fused to gusA was used as a positive control and the pspX promoter fused to gusA with no additional activator present as the negative control. (A) Spectrophotometric assay using the substrate p-nitrophenyl-β-d-glucoronide. Miller units are displayed per milligram of protein present in cell lysate. See Fig. 6 for the PermE*-gusA data. Error bars depict SD between three biological replicates. (B) Chromogenic assay on R2 agar containing the substrate 5-bromo-4-chloro-3-indolyl-β-d-glucoronide.
Initially, gusA transcriptional fusions were assessed in agar-based chromogenic assays using 5-bromo-4-chloro-3-indolyl-β-d-glucuronide as substrate (Fig. 6B). Subsequently, quantitative data were obtained using p-nitrophenyl-β-d-glucuronide as substrate in spectrophotometric kinetic assays (Fig. 6A). None of the strains containing psp promoter::gusA promoter fusions that lacked constitutively expressed pspR or pspX gave GUS activity levels (3.15–9.83 Miller units per milligram of protein) significantly above those of the negative control strain carrying pGUS alone (3.77 Miller units per milligram of protein; Fig. 6A). Constitutive expression of pspX resulted in readily detected transcription of gusA from the pspX, pspJ and pspE promoters, all of which contain an ECF σ factor motif, but no detectable transcription from the pspA promoter (Fig. 6). In contrast, transcription from the pspA promoter, which lacks an ECF σ factor motif, was only activated in the strain containing constitutively expressed pspR (99 Miller units per milligram of protein; Fig. 6A). Thus, in S. coelicolor, σPspX activates transcription of pspX, pspJ, and pspE, while PspR activates transcription of pspA, consistent with the qRT-PCR analyses in P. alba (Fig. 5).
pspW is predicted to encode an anti-σ factor that sequesters σPspX at the membrane [PspW appears to possess six transmembrane helices (25)], preventing expression of the psp gene cluster until the appropriate signals are sensed. Consistent with this, constitutive expression of pspXW from the ermE* promoter suppressed the activation of gusA transcription observed upon constitutive expression of pspX alone (Fig. 7B), reducing GUS activity levels from 505 to 20 Miller units per milligram of protein; Fig. 7A), further supporting the proposed negative role of PspW in regulating planosporicin production.
Deletion of pspW and Overexpression of pspR or pspX Results in Precocious Overproduction of Planosporicin in P. alba.
In previous studies (25), deletion of pspW appeared to result in increased levels of planosporicin biosynthesis. To characterize this phenotype further, P. alba wild type and three independent ΔpspW mutants were grown both on agar and in liquid culture and assayed for planosporicin production using bioassays with the indicator organism Micrococcus luteus and, for the liquid cultures, MALDI-TOF mass spectrometry. There was no perceptible difference in growth rate or sporulation of the wild-type and mutant strains on agar plates. After 2 d of growth on AF/MS agar, planosporicin production was readily detected in the ΔpspW mutant but not in the wild-type strain (Fig. S3 shows the results for one of the three independent ΔpspW mutants, M1304). In liquid culture, planosporicin production was readily observed after 41 h of growth and at a level equivalent to that seen in the wild-type strain after 265 h of incubation (Fig. 2); production in the wild-type strain was not observed until 162 h of incubation (in each case, planosporicin production was confirmed by MALDI-TOF analysis). To assess the effect of increasing the copy number of pspR and pspX on planosporicin production, pspX under the control of its own promoter, and pspR under the control of the P. alba hrdB promoter, were cloned in pSET152, yielding pIJ12539 and pIJ12709, respectively (Table S2). Each plasmid was integrated into the ΦC31 attB site of the P. alba wild-type strain. Both strains started producing planosporicin (confirmed by MALDI-TOF analysis) earlier than the wild-type strain and at much higher levels (Fig. 8).
Fig. 8.
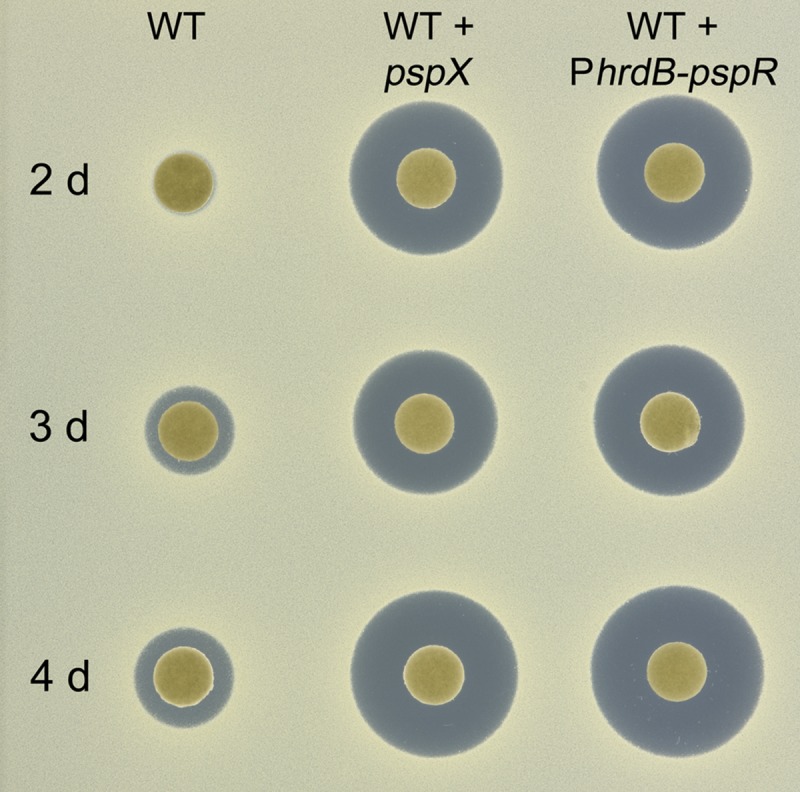
Planosporicin production in P. alba derivatives containing a second copy of pspX or pspR. Strains were cultured in AF/MS medium and supernatants assayed on a lawn of M. luteus after 2, 3, and 4 d of incubation.
Interestingly, complementation of the ΔpspR mutant M1308 with pspR expressed from the constitutive PhrdB promoter resulted in precocious planosporicin production that, in contrast to the wild-type strain, appeared to decrease upon prolonged incubation (Fig. S4). Failure to complement the mutant with pspR preceded by the pspQ-pspR intergenic region (Fig. S4, Lower) corroborated the S1 nuclease protection experiments that failed to identify a promoter that transcribed just pspR (Fig. S1), further suggesting that pspR is expressed only as part of a polycistronic transcript.
Subinhibitory Concentrations of Planosporicin Induce Planosporicin Production in P. alba.
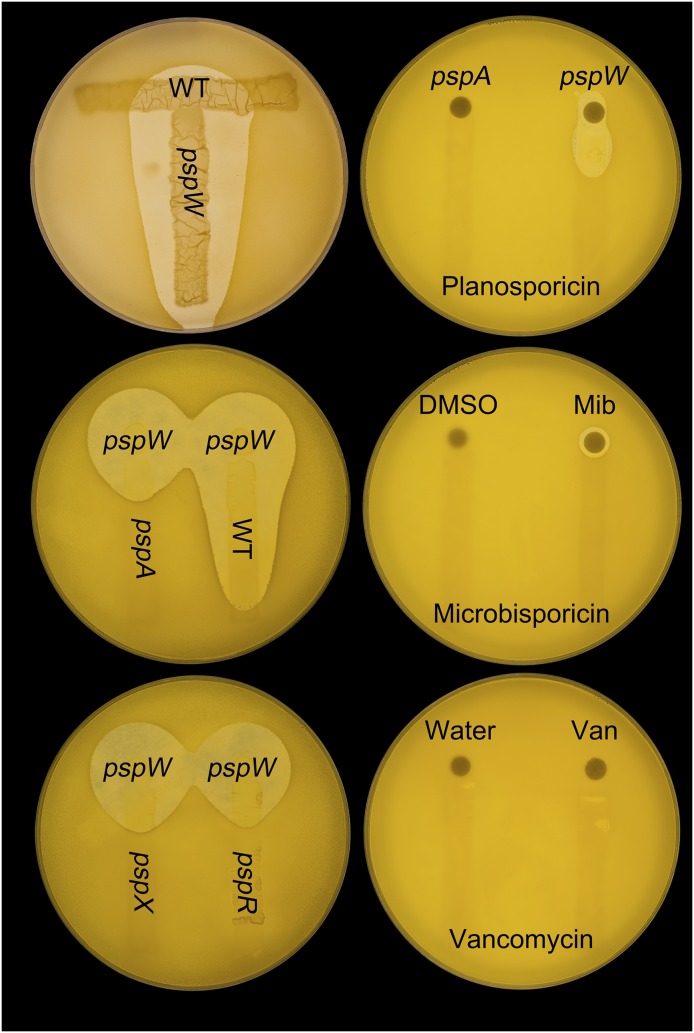
The requirement of pspA for transcription of the psp gene cluster (Fig. 4 and Fig. S2) is consistent with a role for planosporicin in inducing its own biosynthesis. To address this possibility, M1304 (ΔpspW), which appears to produce planosporicin constitutively, was grown on AF/MS agar plates in close proximity to the wild-type strain and the ΔpspA, ΔpspX, and ΔpspR mutants. After 3 d of incubation, when the wild-type strain had not commenced planosporicin biosynthesis (usually detected only after 6 d of incubation), the plates were overlaid with soft nutrient agar (SNA) containing M. luteus. The resulting zones of inhibition clearly indicated induction of planosporicin biosynthesis in the wild-type strain by the ΔpspW mutant (without any detectable inhibition of growth of the wild-type strain) but not by the other three mutants (Fig. 9, left plates). In an alternative approach, concentrated culture supernatants of M1302 (ΔpspA), M1304 (ΔpspW), and 20 μg of microbisporicin or 10 μg of vancomycin were spotted onto antibiotic assay discs adjacent to streaks of the wild-type P. alba strain. After incubation for 3 d, the plates were overlaid with SNA containing M. luteus. Again, the resulting zones of inhibition indicated autoinduction of planosporicin production (Fig. 9, right plates) in the absence of any detectable inhibition of the wild-type strain; neither of the other two antibiotics induced biosynthesis. A range of other antibiotics were also tested: the cell wall biosynthesis inhibitors deoxy-actagardine B, bacitracin, and tunicamycin; the protein synthesis inhibitor apramycin; and the RNA synthesis inhibitor rifampicin. A range of concentrations were used, many of which inhibited the growth of P. alba, but none induced precocious planosporicin production. These results suggest that planosporicin induces its own synthesis in a highly specific manner and at subinhibitory concentrations.
Fig. 9.
Autoinduction of planosporicin production in P. alba. Top left plate: P. alba wild-type and M1304 (ΔpspW) spores were streaked at right angles to each other on AF/MS agar. Middle and bottom left plates: P. alba wild-type, M1302 (ΔpspA), M1303 (ΔpspX), and M1308 (ΔpspR) spores were streaked on AF/MS agar adjacent to a spot of M1304 (ΔpspW) spores. After 3 d at 30 °C, all three plates were overlaid with SNA containing the indicator organism M. luteus. Right plates: wild-type P. alba spores were streaked on AF/MS agar. Antibiotic assay discs impregnated with concentrated culture supernatants of M1302 (ΔpspA) or M1304 (ΔpspW), 20 μg of microbisporicin (Mib) in DMSO, or 10 μg of vancomycin (Van) in water were placed at one end of each streak. After 3 d at 30 °C, the plates were overlaid with SNA containing M. luteus.
Manipulating the Regulatory Genes of the psp Gene Cluster Results in Heterologous Production of Planosporicin in Streptomyces coelicolor.
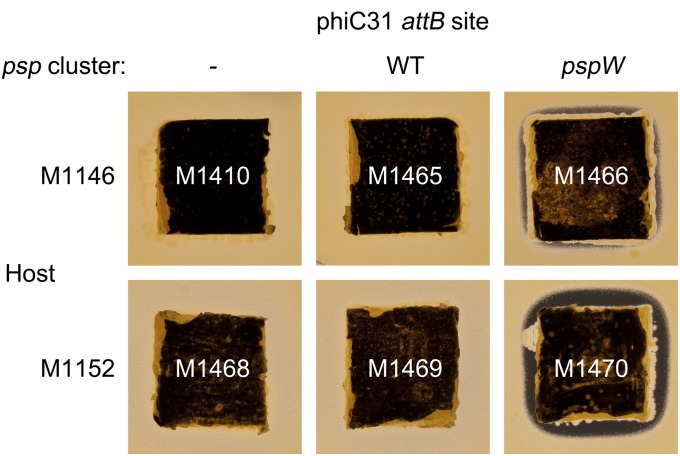
Previous attempts to detect planosporicin production from the psp gene cluster when transferred to the genetically tractable S. coelicolor M1146 had failed (ref. 25 and Fig. 10). Attempts in the course of this work to achieve expression in S. coelicolor M1152 (36), specifically engineered for enhanced expression of heterologous gene clusters, also failed (Fig. 10). Given the marked stimulatory effect on planosporicin production of deleting pspW or of increasing the copy number of pspR and pspX in P. alba, further attempts were made to achieve heterologous production in S. coelicolor by manipulating each of these regulatory genes in the heterologous host. pIJ12719, containing the psp gene cluster but with an in-frame deletion of pspW, was introduced into S. coelicolor M1146 and M1152 by conjugation, creating M1466 and M1470, respectively. After 5 d of growth on R5 agar medium both strains, unlike the parental strains, inhibited the growth of M. luteus (Fig. 10); planosporicin production was confirmed by MALDI-TOF analysis of 5% (vol/vol) formic acid extracts of agar plugs supporting M1466 or M1470 (Fig. S5). As expected (36), the M1152 derivative gave higher levels of planosporicin production than the M1146 exconjugant.
Fig. 10.
Heterologous expression of the psp gene cluster integrated into the S. coelicolor M1146 and M1152 chromosomes. S. coelicolor M1146 derivatives M1410 (pIJ10702, vector control), M1465 (pIJ12328, wild-type psp gene cluster), and M1466 (pIJ12718, psp gene cluster with ΔpspW) and S. coelicolor M1152 derivatives M1468 (pIJ10702), M1469 (pIJ12328), and M1470 (pIJ12718) were grown on 2 × 2 cm squares of R5 agar for 5 d before being placed on a lawn of M. luteus.
To assess whether overexpression of pspR or pspX would activate planosporicin production in S. coelicolor derivatives containing the wild-type psp gene cluster, pspX and pspR were PCR-amplified to incorporate 5′ NdeI and 3′ PacI sites and cloned individually into NdeI plus PacI cleaved pIJ10257, creating pIJ12715 and pIJ12716, respectively, with each gene transcribed from the strong constitutive ermE* promoter. These plasmids, along with the empty vector pIJ10257, were integrated individually at the ΦBT1 site of M1469 (S. coelicolor M1152 with pIJ12328, containing the psp minimal gene set, integrated at the ΦC31 attB site; Table 1). After 6 d of growth on R5 agar medium, a cork borer was used to embed agar plugs containing each of the strains into a lawn of SNA containing M. luteus; after 1 d of further incubation, a small zone of inhibition was visible around the strains containing the PermE*::pspX and PermE*::pspR constructs (M1475 and M1476, respectively) but not the negative control strain (M1469) (Fig. 11); MALDI-TOF mass spectrometry confirmed the production of planosporicin (Fig. S5).
Table 1.
Strains used in this study
| Strain | Description | Source |
| P. alba | ||
| NRLL18924 | WT strain | Ref. 51 |
| M1302 | ∆pspA::(oriT-hyg) | Ref. 25 |
| M1303 | ∆pspX::(oriT-hyg) | Ref. 25 |
| M1304 | ∆pspW::(oriT-hyg) | Ref. 25 |
| M1308 | ∆pspR::(oriT-hyg) | Ref. 25 |
| M1574 | pIJ12539 in ΦC31 attB (PpspX-pspX) | Ref. 25 |
| M1575 | pIJ12709 in ΦC31 attB (PhrdB-pspR) | Ref. 25 |
| S. coelicolor M1146 and derivatives | ||
| M1146 | M145 ∆act ∆red ∆cpk ∆cda | Ref. 36 |
| M1410 | pIJ10702 in ΦC31 attB | Ref. 22 |
| M1288 | pIJ12323 in ΦC31 attB (cosmid B4-1) | Ref. 25 |
| M1465 | pIJ12328 in ΦC31 attB (minimal psp gene set) | This work |
| M1466 | pIJ12719 in ΦC31 attB (minimal psp gene set; ∆pspW::scar) | This work |
| S. coelicolor M1152 and derivatives: heterologous expression | ||
| M1152 | M145 ∆act ∆red ∆cpk ∆cda rpoB[C1298T] | Ref. 36 |
| M1468 | pIJ10702 in ΦC31 attB | Ref. 22 |
| M1469 | pIJ12328 in ΦC31 attB (minimal psp gene set) | This work |
| M1470 | pIJ12719 in ΦC31 attB (minimal psp gene set; ∆pspW::scar) | This work |
| M1472 | pIJ12719 in ΦC31 attB and pIJ12715 in ΦBT1 attB (PermE*-pspX) | This work |
| M1473 | pIJ12719 in ΦC31 attB and pIJ12716 in ΦBT1 attB (PermE*-pspR) | This work |
| M1474 | pIJ12719 in ΦC31 attB and pIJ12717 in ΦBT1 attB (PermE*-pspEF) | This work |
| M1475 | pIJ12328 in ΦC31 attB and pIJ12715 in ΦBT1 attB | This work |
| M1476 | pIJ12328 in ΦC31 attB and pIJ12716 in ΦBT1 attB | This work |
| M1477 | pIJ12328 in ΦC31 attB and pIJ12717 in ΦBT1 attB | This work |
| M1478 | pIJ12721 in ΦC31 attB (minimal psp gene set; ∆pspEF; ∆pspW::scar) | This work |
| M1479 | pIJ12721 in ΦC31 attB and pIJ12716 in ΦBT1 attB (PermE*-pspR) | This work |
| M1480 | pIJ12723 in ΦC31 attB | |
| S. coelicolor M1152 and derivatives: transcriptional fusions | ||
| M1551 | pIJ12715 in ΦBT1 attB (PermE*-pspX) | This work |
| M1552 | pIJ12716 in ΦBT1 attB (PermE*-pspR) | This work |
| M1553 | pIJ12717 in ΦBT1 attB (PermE*-pspEF) | Ref. 25 |
| M1554 | pIJ10740 | This work |
| M1555 | pIJ12724 in ΦC31 attB (PpspA-gusA) | This work |
| M1556 | pIJ12724 in ΦC31 attB and pIJ12715 in ΦBT1 attB (PermE*-pspX) | This work |
| M1557 | pIJ12724 in ΦC31 attB and pIJ12716 in ΦBT1 attB (PermE*-pspR) | This work |
| M1561 | pIJ12726 in ΦC31 attB (PpspX-gusA) | This work |
| M1562 | pIJ12726 in ΦC31 attB and pIJ12715 in ΦBT1 attB (PermE*-pspX) | This work |
| M1563 | pIJ12726 in ΦC31 attB and pIJ12716 in ΦBT1 attB (PermE*-pspR) | This work |
| M1564 | pIJ12726 in ΦC31 attB and pIJ12718 in ΦBT1 attB (PermE*-pspXW) | This work |
| M1565 | pIJ12727 in ΦC31 attB (PpspJ-gusA) | This work |
| M1566 | pIJ12727 in ΦC31 attB and pIJ12715 in ΦBT1 attB (PermE*-pspX) | This work |
| M1567 | pIJ12727 in ΦC31 attB and pIJ12716 in ΦBT1 attB (PermE*-pspR) | This work |
| M1568 | pIJ12728 in ΦC31 attB (PpspE-gusA) | This work |
| M1569 | pIJ12728 in ΦC31 attB and pIJ12715 in ΦBT1 attB (PermE*-pspX) | This work |
| M1570 | pIJ12728 in ΦC31 attB and pIJ12716 in ΦBT1 attB (PermE*-pspR) | This work |
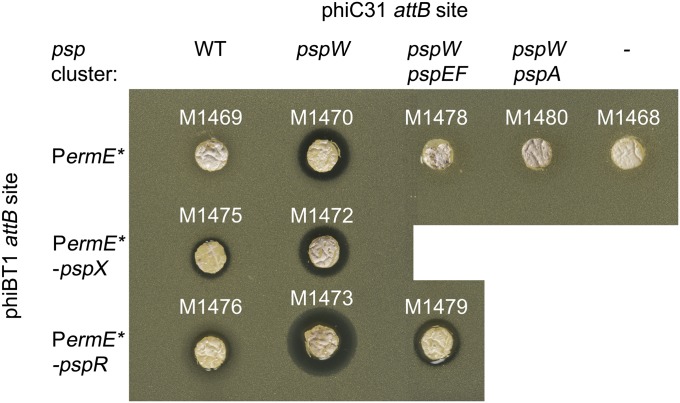
Fig. 11.
Heterologous expression of the psp gene cluster in S. coelicolor M1152. Each strain was grown as a lawn on R5, and after 6 d, a cork borer was used to embed an agar plug in a lawn of M. luteus. Genes deleted from the wild type gene cluster are indicated at the top of the figure.
To assess whether overexpression of pspR or pspX in the ΔpspW S. coelicolor M1152 derivative would enhance planosporicin production further, pIJ12715 (pIJ10257::pspX), pIJ12716 (pIJ10257::pspR) and pIJ10257 were transferred by conjugation into M1470 [M1152 with pIJ12718 (containing the psp minimal gene set, but with pspW deleted) integrated at the ΦC31 attB] to create M1472, M1473 and M1470, respectively (Table 1). The strains were grown on R5 agar medium as above and assayed for planosporicin production using M. luteus as indicator. Whereas overexpression of pspR further increased production in the ΔpspW mutant, overexpression of pspX did not (Fig. 11), presumably reflecting constitutive activity of σPspX in the anti-σ factor mutant. In each case, the production of planosporicin was confirmed by MALDI-TOF mass spectrometry (Fig. S5).
Earlier attempts to delete pspEF, one of three gene pairs encoding ATP-binding cassette (ABC) transporters in the psp gene cluster, in P. alba had failed (25), raising the possibility that PspEF played an essential role in immunity in the producing organism and that their deletion would be lethal. With expression now achieved in S. coelicolor, pIJ12718 containing the psp gene cluster with pspW deleted was PCR-targeted to remove pspEF. The resulting construct, pIJ12721, was transferred by conjugation to M1152 to yield M1478 (Table 1), which failed to inhibit M. luteus in agar plug bioassays (Fig. 11); MALDI-TOF mass spectrometry also failed to detect the presence of planosporicin (Fig. S5). However, introduction of pIJ12716 (pIJ10257::pspR), creating M1479, restored planosporicin production to the level observed before deleting pspEF (Fig. 11 and Fig. S5). Thus, if PspEF play a role in immunity, they are not essential for planosporicin biosynthesis in S. coelicolor, although they may be in P. alba. It is conceivable that the putative lipoprotein encoded by pspQ also functions to mediate resistance in the producing strains by sequestering the lantibiotic, preventing interaction with lipid II. A similar function has been proposed for other lipoproteins encoded in lantibiotic biosynthetic gene clusters (37, 38).
Discussion
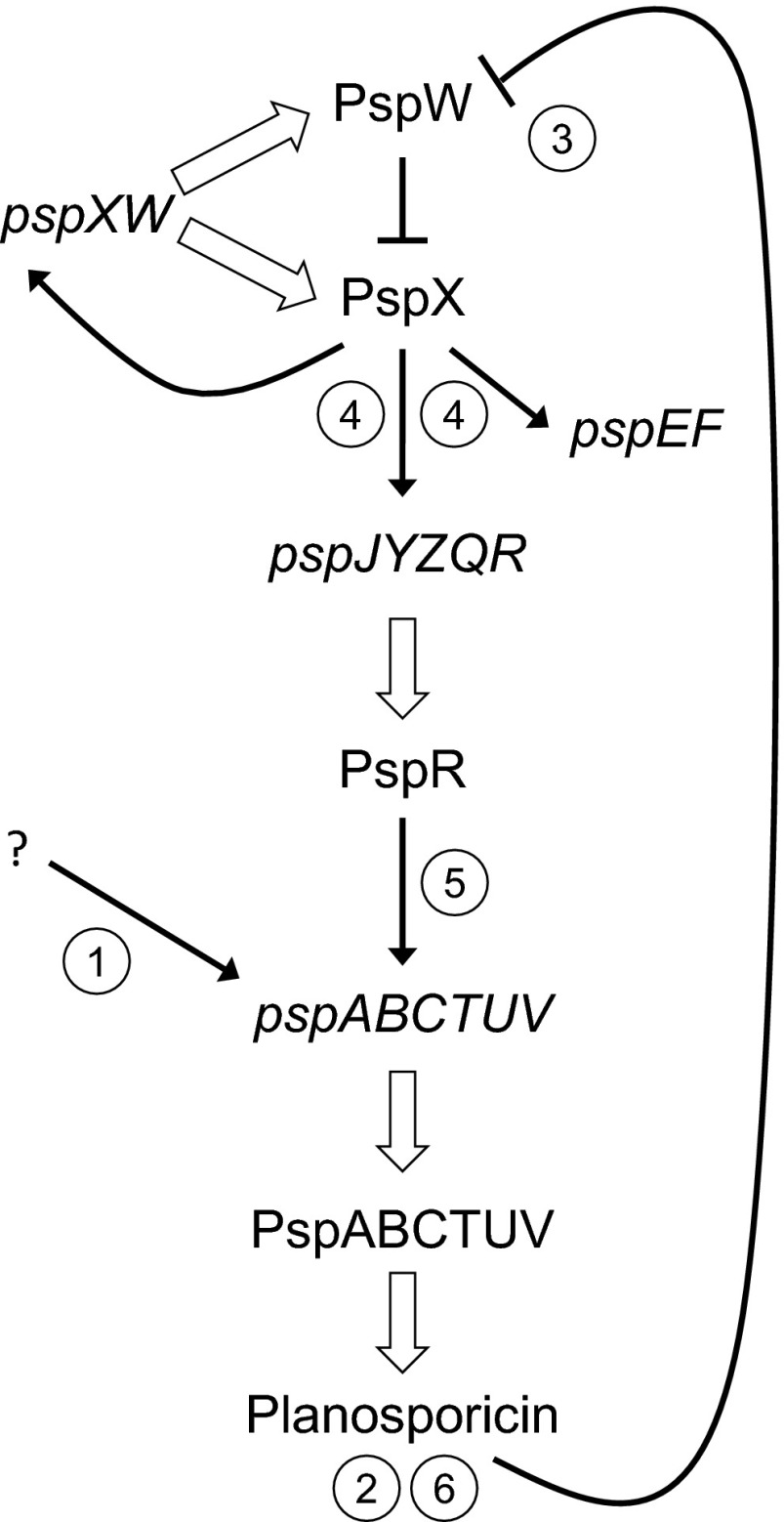
The results described in this paper show that an actinomycete antibiotic with potent antibacterial activity can induce its own synthesis and allow us to present a detailed model for the regulation of planosporicin biosynthesis in P. alba that involves three pathway-specific regulatory genes (Fig. 12). We propose that before the onset of planosporicin production, a basal level of expression of pspXW leaves the system poised for activation, with σPspX sequestered at the membrane by PspW. At the onset of nutrient limitation a low level of transcriptional activation of the pspABCTUV operon occurs, possibly mediated by the intracellular signaling molecule ppGpp (7), resulting in the production of a small amount of the lantibiotic (the pspABCTUV operon encodes the precursor peptide and enzymatic machinery required for the production of planosporicin and an ABC transporter for its initial export from the cell). The resulting extracellular lantibiotic then interacts either directly with PspW or with lipid II causing a low level of inhibition of peptidoglycan biosynthesis that is perceived by the anti-σ factor; in either case, σPspX is released from PspW, resulting in high-level expression of the pspXWJYZQR, pspJYZQR, and pspEF operons [note that the possible functions of pspJYZQ, and their potential role in regulating planosporicin production, were discussed previously (25)]. This, in turn, results in high level expression of pspR and further activation of transcription of the pspABCTUV operon and, consequently, high levels of planosporicin production. Thus, there is a feed-forward mechanism of regulation in which the production of a small, apparently subinhibitory concentration of the antibiotic results in activation of planosporicin biosynthesis throughout the substrate mycelium, much of which has not reached the stage of maturity at which normal, stationary phase biosynthesis of the antibiotic would occur and thus ensuring that sufficient quantities of the lantibiotic are made to provide P. alba with a competitive advantage. Feed-forward regulation of antibiotic biosynthesis by an immature form of the antibiotic was proposed (but not demonstrated) for microbisporicin biosynthesis (19), and biosynthetic pathway intermediates, as well as the final product, have been postulated to activate export and potentially immunity mechanisms in streptomycetes (39–42). Here we have demonstrated the ability of a fully mature actinomycete antibiotic to function as an extracellular signaling molecule to induce its own synthesis in the wider population. The inability of even closely related lantibiotics, such as microbisporicin, and of other classes of antibiotics that have markedly different mechanisms of action (for example, apramycin and rifampicin) to elicit planosporicin biosynthesis even at inhibitory levels is again consistent with a specific role for the lantibiotic as a signaling molecule capable of triggering its own production. Other inhibitors of cell wall biosynthesis (vancomycin, bacitracin, deoxyactagardine B, and tunicamycin) failed to elicit precocious planosporicin biosynthesis, suggesting that the release of σPspX from PspW likely results from a specific interaction of planosporicin with the anti-σ factor PspW rather than through the latter sensing cell envelope stress induced by the antibiotic.
Fig. 12.
Model for the regulation of planosporicin production in P. alba. A basal level of pspXW transcription results in the presence of σPspX-PspW at the cell membrane and leaves the system poised for activation. Low-level transcription of the pspABCTUV operon (1), triggered by an unknown signal, possibly nutrient limitation, results in the production of a small amount of planosporicin (2), eliciting the release of σPspX from PspW (3). This leads to high-level expression of the pspXW, pspEF, and pspJYZQR operons (4). The PspR produced then further activates transcription of pspABCTUV (5), leading to high-levels of planosporicin production (6). White-filled arrows represent transcription, translation, and posttranslational modification. Black arrows represent transcriptional activation. Inhibition is indicated by a blocked line.
All of the data presented here are consistent with this model, most notably the ability of subinhibitory concentrations of planosporicin to induce production in the wild-type strain (Fig. 9), the apparently constitutive production of the lantibiotic in the ΔpspW mutant (M1304; Fig. 2), the precocious production of planosporicin observed upon early and/or overexpression of either pspX (presumably resulting in a molar excess of σPspX over PspW) or pspR (Fig. 8), and the transcriptional dependence of the entire psp gene cluster on, strikingly, pspA, as well as on pspX and pspR (Fig. 4).
Initial bioinformatic analysis revealed highly conserved ECF-like promoter sequences upstream of the experimentally determined transcriptional start sites for pspX, pspJ, and pspE, suggesting that the pspXWJYZQR, pspJYZQR, and pspEF operons were regulated directly by σPspX, whereas PspR appeared to be a possible transcriptional activator of the pspABCTUV operon, which lacked the consensus motif. Consistent with this hypothesis, deletion of pspX had a more severe deleterious effect on transcription of pspE and pspZ than deletion of pspR, whereas the converse was true for expression of pspA, pspT, and pspV (Fig. 5). Fusions of the pspA, pspE, pspJ, and pspX promoters to the gusA reporter gene in S. coelicolor subsequently confirmed these direct transcriptional dependences (Fig. 6), further supporting the model.
Complementation of the ΔpspR mutant with pspR expressed from the constitutive PhrdB promoter led to precocious planosporicin production that, in contrast to the wild-type strain, decreased upon further incubation (Fig. S4). This observation is again in accordance with the regulatory model and presumably reflects the lack of a feed-forward mechanism in the complemented mutant, where transcription of pspR is no longer under σPspX control.
Autoinduction of antibiotic biosynthetic gene clusters has also been observed in low-GC Gram-positive bacteria, where nisin, subtilin, and mersacidin may act as quorum sensors to elicit lantibiotic production in Lactococcus lactis, Bacillus subtilis, and Bacillus sp. strain HIL Y-8554728, respectively (43–45). In L. lactis and B. subtilis, a low level of lantibiotic production results in phosphorylation of membrane-bound histidine kinases that then phosphorylate cytoplasmic response regulators that activate expression of the respective biosynthetic operons (46, 47). The planosporicin biosynthetic gene cluster does not encode a comparable two-component regulatory system, and the mechanism of feed-forward regulation described in this paper is, like P. alba itself, of distinct evolutionary origin. Unlike their generally planktonic low-GC counterparts, actinomycetes do not grow as single cells but usually as surface-attached multicellular networks of branched substrate mycelia that give rise to aerial hyphae and eventually spores, enabling dispersal to new environments. As a colony grows, different parts of the substrate mycelium will be in different physiological states and unlikely to encounter simultaneous nutrient limitation. However, synchronous and concerted expression of antibiotic biosynthetic gene clusters throughout the substrate mycelium, and in neighboring colonies, may be needed if effective levels of antibiotic production are to be achieved. For planosporicin, and we suspect other actinomycete antibiotics, this is apparently achieved by a feed-forward mechanism in which the modified peptide plays dual roles as an antibiotic and a signaling molecule, seemingly using a strategy that is distinct from the quorum-sensing mechanisms proposed for low-GC Gram-positive bacteria.
Initial attempts to produce planosporicin in S. coelicolor strains specifically engineered for the expression of heterologous gene clusters failed (Fig. 10), a deficiency that could be overcome either by deleting pspW (Fig. 10) or by overexpressing pspX or pspR from the strong constitutive ermE* promoter (Fig. 11). These observations are consistent with the inability, for whatever reason, of the feed-forward mechanism to function in a distantly related streptomycete host.
A frequent stumbling block in the development of a natural product as it progresses toward commercialization is the provision of sufficient material for preclinical and clinical trials. In addition to providing fundamental insights into the regulation of antibiotic production in actinomycetes, the work described in this paper illustrates how knowledge-based approaches can be used to effect marked increases in productivity, either by deleting negatively acting regulating genes or by overexpressing those encoding transcriptional activators, either in the natural producer or in a heterologous host.
Experimental Procedures
For information on manipulation and expression of the psp gene cluster in S. coelicolor, and for the gusA transcriptional fusions and GUS assays, see SI Experimental Procedures.
Strains and General Methods.
Oligonucleotides are described in Table S1, and plasmids and strains are described in Table S2. Growth and manipulation of P. alba and the detection of planosporicin were performed as described previously (25). Streptomyces and Escherichia coli strains were cultured and manipulated as in (48) and (49), respectively. Restriction and PCR enzymes were purchased from Roche or New England Biolabs. DNA fragments were cloned generally through amplification using the Expand High Fidelity PCR system (Roche) and TA-cloned into pGEM-T (Promega) for Sanger sequencing (TGAC). Error-free fragments were subsequently excised and ligated into the relevant vector. All subcloning was carried out in E. coli DH5α (Invitrogen).
RNA Preparation from P. alba.
P. alba was grown in AF/MS liquid medium (25) and after 41, 73, 114, and 162 h, 3–4 mL of the cultures was harvested and the mycelium and supernatant separated. RNA was extracted from mycelium as described previously (25), and RNA concentrations were determined using Nanodrop (Thermo Scientific). PCR (30 cycles) was carried out using 150–300 ng of RNA as template with primers hrdB_RTF and hrdB_RTR (annealing to a gene with 87% nucleotide sequence identity across 1036 nucleotides of 3′ sequence of hrdB from S. coelicolor) to confirm the absence of DNA contamination.
High-Resolution S1 Nuclease Mapping.
S1 nuclease protection analysis used probes amplified by PCR using the oligonucleotides shown in Table S1. T4 polynucleotide kinase (Epicentre) was used to label with [γ-P32]ATP according to the manufacturer’s instructions; 30 μg P. alba RNA harvested from mycelium collected after 162 h growth in liquid AF/MS was denatured at 70 °C for 10 min then hybridized with each probe in sodium trichloroacetic acid buffer at 45 °C overnight (48). G+A sequencing ladders were generated from the end-labeled probes by chemical sequencing (50).
qRT-PCR.
qRT-PCR was carried out in a Chromo4 machine (BioRad) using SYBR GreenER qPCR SuperMix Universal (Invitrogen) with 2.5 μL of freshly diluted (1:100) cDNA, 5% DMSO, 0.2 μL ROX reference dye [Invitrogen; 5-carboxy-X-rhodamine, succinimidyl ester (25 μM) in 20 mM Tris-HCl (pH 8.4), 0.1 mM EDTA, 0.01% Tween 20], and 200 nM each primer in a 25 μL reaction. Primers were initially assessed by RT-PCR (25) and are listed in Table S1. Thermal cycling consisted of 50 °C for 2 min and 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s, 56–58 °C for 60 s, and 72 °C for 1 s. Each primer pair was also used in parallel with dilutions of P. alba genomic DNA in the same 96-well plate to generate a standard curve for each gene analyzed. Reactions were run in triplicate with at least two sets of blank wells (no template) in each run. Negative controls used 1/100 dilutions of reaction mixes that lacked RT with primers hrdB_RTF and hrdB_RTR and gave values comparable to the empty wells. qRT-PCR results were analyzed using Opticon 2 Monitor software (MJ Research). Melting curves confirmed that just one product was amplified with each primer pair. The standard curve was used to calculate cDNA copy numbers, which were then normalized against the cDNA copy number of the presumptive P. alba hrdB homolog (see above). The expression level of each gene was then displayed as a percentage of the wild-type expression level.
Induction of Planosporicin Production in P. alba.
As a source of planosporicin, culture supernatants from P. alba M1304 (ΔpspW) [and from M1302 (ΔpspA) as a negative control] were concentrated using a polystyrene resin (25). The third to sixth elutions (1 mL each) were dried down and combined to give a final volume of 500 μL. To test for induction of planosporicin production in wild-type P. alba, 20 μL of each antibiotic together with solvent controls was applied in duplicate to filter-paper discs, which were allowed to dry and placed on AF/MS agar plates adjacent to streaks of wild-type P. alba; the plates were overlaid with M. luteus after incubation at 30°C for 2, 3, and 4 d. Other antibiotics used were 20 μg of microbisporicin in 10% DMSO, 20 μg of deoxy-actagardine B in water, 20 μg of tunicamycin in 100% ETOH, 20 μg of bacitracin in water, 20 μg of vancomycin in water, 20 μg of apramycin in water, and 2 μg of rifampicin in 100% DMSO.
Overexpression of pspR and pspX in P. alba.
For overexpression of pspX and pspR in wild-type P. alba, an additional copy of each gene was integrated at the chromosomal ΦC31 attB site with exconjugants selected on International Streptomyces Project agar medium 4 (ISP4) containing 0.5 mg of apramycin as described previously (25). For pspX, the pSET152 derivative pIJ12539 (25) was used. Because pspR lies at the 3′ end of an operon (25), it was expressed from the promoter (PhrdB) of the hrdB homolog of P. alba. PhrdB was PCR-amplified using a 3′ primer with 5′ sequences complementary to the entire 5′ primer used to PCR-amplify pspR (Table S1). The 5′ primer for PhrdB and the 3′ primer for pspR both contained XbaI sites. PhrdB and pspR were amplified individually, and the PCR products were combined and PCR-amplified again using the 5′ primer for PhrdB and the 3′ primer for pspR. The resulting PCR product was digested with XbaI, cloned into XbaI-digested pGEM-T (Promega), excised with XbaI, and cloned into XbaI-digested pSET152 to give pIJ12709.
Complementation of the P. alba pspR Deletion Mutant.
Complementation was carried out using pIJ12714 (25) with pspR expressed from the strong constitutive ermE* promoter or with pIJ12709 containing the hrdB promoter. pSET152 and pIJ12540 (pSET152 containing pspR and the preceding 128 bp pspQ-pspR intergenic region that lacks promoter activity; Fig. S1) were used as negative controls, respectively. pIJ12540 was made by amplifying the region between pspQ and pspR together with the open-reading frame of pspR with primers introducing 5′ BamHI and 3′ XbaI sites. The resulting fragment was cloned into BamHI- and XbaI-digested pSET152 to create pIJ12540. The plasmids were transferred into M1308 (ΔpspR) by conjugation from E. coli ET12567/pUZ8002. The resulting four exconjugants, together with the wild-type strain, were grown in AF/MS liquid medium and 40 μL of culture supernatants spotted onto sterile filter-paper discs placed on top of a lawn of SNA seeded with M. luteus to assay for antimicrobial activity.
Supplementary Material
Acknowledgments
We thank Morgan Feeney for the gift of pIJ10740 and for advice on the GUS assays, Maureen Bibb and Mahmoud Al-Bassam for technical advice, Gerhard Saalbach and Mike Naldrett for mass spectrometry, and David Hopwood for comments on the manuscript. This work was supported financially by the United Kingdom Biotechnology and Biological Sciences Research Council Institute Strategic Programme Grant “Understanding and Exploiting Plant and Microbial Secondary Metabolism” (BB/J004561/1) and the John Innes Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
*Bailey TL, Elkan C, Proceedings of the 2nd International Conference on Intelligent Systems for Molecular Biology, Menlo Park, CA, August 14–17, 1994, pp 28–36.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1305392110/-/DCSupplemental.
References
- 1.Nikaido H. Multidrug resistance in bacteria. Annu Rev Biochem. 2009;78:119–146. doi: 10.1146/annurev.biochem.78.082907.145923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Newman DJ, Cragg GM. Natural products as sources of new drugs over the last 25 years. J Nat Prod. 2007;70(3):461–477. doi: 10.1021/np068054v. [DOI] [PubMed] [Google Scholar]
- 3.Nett M, Ikeda H, Moore BS. Genomic basis for natural product biosynthetic diversity in the actinomycetes. Nat Prod Rep. 2009;26(11):1362–1384. doi: 10.1039/b817069j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bibb MJ. Regulation of secondary metabolism in streptomycetes. Curr Opin Microbiol. 2005;8(2):208–215. doi: 10.1016/j.mib.2005.02.016. [DOI] [PubMed] [Google Scholar]
- 5.van Wezel GP, McDowall KJ. The regulation of the secondary metabolism of Streptomyces: New links and experimental advances. Nat Prod Rep. 2011;28(7):1311–1333. doi: 10.1039/c1np00003a. [DOI] [PubMed] [Google Scholar]
- 6.Hesketh A, Chen WJ, Ryding J, Chang S, Bibb M. The global role of ppGpp synthesis in morphological differentiation and antibiotic production in Streptomyces coelicolor A3(2) Genome Biol. 2007;8(8):R161. doi: 10.1186/gb-2007-8-8-r161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bierbaum G, Sahl HG. Lantibiotics: Mode of action, biosynthesis and bioengineering. Curr Pharm Biotechnol. 2009;10(1):2–18. doi: 10.2174/138920109787048616. [DOI] [PubMed] [Google Scholar]
- 8.Arnison PG, et al. Ribosomally synthesized and post-translationally modified peptide natural products: Overview and recommendations for a universal nomenclature. Nat Prod Rep. 2013;30(1):108–160. doi: 10.1039/c2np20085f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crowther GS, et al. Evaluation of NVB302 versus vancomycin activity in an in vitro human gut model of Clostridium difficile infection. J Antimicrob Chemother. 2013;68(1):168–176. doi: 10.1093/jac/dks359. [DOI] [PubMed] [Google Scholar]
- 10.Jabés D, et al. Efficacy of the new lantibiotic NAI-107 in experimental infections induced by multidrug-resistant Gram-positive pathogens. Antimicrob Agents Chemother. 2011;55(4):1671–1676. doi: 10.1128/AAC.01288-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grasemann H, et al. Inhalation of Moli1901 in patients with cystic fibrosis. Chest. 2007;131(5):1461–1466. doi: 10.1378/chest.06-2085. [DOI] [PubMed] [Google Scholar]
- 12.Oliynyk I, Varelogianni G, Roomans GM, Johannesson M. Effect of duramycin on chloride transport and intracellular calcium concentration in cystic fibrosis and non-cystic fibrosis epithelia. APMIS. 2010;118(12):982–990. doi: 10.1111/j.1600-0463.2010.02680.x. [DOI] [PubMed] [Google Scholar]
- 13.Widdick DA, et al. Cloning and engineering of the cinnamycin biosynthetic gene cluster from Streptomyces cinnamoneus cinnamoneus DSM 40005. Proc Natl Acad Sci USA. 2003;100(7):4316–4321. doi: 10.1073/pnas.0230516100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kodani S, et al. The SapB morphogen is a lantibiotic-like peptide derived from the product of the developmental gene ramS in Streptomyces coelicolor. Proc Natl Acad Sci USA. 2004;101(31):11448–11453. doi: 10.1073/pnas.0404220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kodani S, Lodato MA, Durrant MC, Picart F, Willey JM. SapT, a lanthionine-containing peptide involved in aerial hyphae formation in the streptomycetes. Mol Microbiol. 2005;58(5):1368–1380. doi: 10.1111/j.1365-2958.2005.04921.x. [DOI] [PubMed] [Google Scholar]
- 16.Holtsmark I, Mantzilas D, Eijsink VG, Brurberg MB. Purification, characterization, and gene sequence of michiganin A, an actagardine-like lantibiotic produced by the tomato pathogen Clavibacter michiganensis subsp. michiganensis. Appl Environ Microbiol. 2006;72(9):5814–5821. doi: 10.1128/AEM.00639-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boakes S, Cortés J, Appleyard AN, Rudd BA, Dawson MJ. Organization of the genes encoding the biosynthesis of actagardine and engineering of a variant generation system. Mol Microbiol. 2009;72(5):1126–1136. doi: 10.1111/j.1365-2958.2009.06708.x. [DOI] [PubMed] [Google Scholar]
- 18.Boakes S, Appleyard AN, Cortés J, Dawson MJ. Organization of the biosynthetic genes encoding deoxyactagardine B (DAB), a new lantibiotic produced by Actinoplanes liguriae NCIMB41362. J Antibiot (Tokyo) 2010;63(7):351–358. doi: 10.1038/ja.2010.48. [DOI] [PubMed] [Google Scholar]
- 19.Foulston L, Bibb M. Feed-forward regulation of microbisporicin biosynthesis in Microbispora corallina. J Bacteriol. 2011;193(12):3064–3071. doi: 10.1128/JB.00250-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Foulston LC, Bibb MJ. Microbisporicin gene cluster reveals unusual features of lantibiotic biosynthesis in actinomycetes. Proc Natl Acad Sci USA. 2010;107(30):13461–13466. doi: 10.1073/pnas.1008285107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Claesen J, Bibb M. Genome mining and genetic analysis of cypemycin biosynthesis reveal an unusual class of posttranslationally modified peptides. Proc Natl Acad Sci USA. 2010;107(37):16297–16302. doi: 10.1073/pnas.1008608107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goto Y, et al. Discovery of unique lanthionine synthetases reveals new mechanistic and evolutionary insights. PLoS Biol. 2010;8(3):e1000339. doi: 10.1371/journal.pbio.1000339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Claesen J, Bibb MJ. Biosynthesis and regulation of grisemycin, a new member of the linaridin family of ribosomally synthesized peptides produced by Streptomyces griseus IFO 13350. J Bacteriol. 2011;193(10):2510–2516. doi: 10.1128/JB.00171-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Castiglione F, et al. A novel lantibiotic acting on bacterial cell wall synthesis produced by the uncommon actinomycete Planomonospora sp. Biochemistry. 2007;46(20):5884–5895. doi: 10.1021/bi700131x. [DOI] [PubMed] [Google Scholar]
- 25.Sherwood EJ, Hesketh AR, Bibb MJ. Cloning and analysis of the planosporicin lantibiotic biosynthetic gene cluster of Planomonospora alba. J Bacteriol. 2013;195(10):2309–2321. doi: 10.1128/JB.02291-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Vos WM, Mulders JW, Siezen RJ, Hugenholtz J, Kuipers OP. Properties of nisin Z and distribution of its gene, nisZ, in Lactococcus lactis. Appl Environ Microbiol. 1993;59(1):213–218. doi: 10.1128/aem.59.1.213-218.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Castiglione F, et al. Determining the structure and mode of action of microbisporicin, a potent lantibiotic active against multiresistant pathogens. Chem Biol. 2008;15(1):22–31. doi: 10.1016/j.chembiol.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 28.Hsu STD, et al. The nisin-lipid II complex reveals a pyrophosphate cage that provides a blueprint for novel antibiotics. Nat Struct Mol Biol. 2004;11(10):963–967. doi: 10.1038/nsmb830. [DOI] [PubMed] [Google Scholar]
- 29.Missiakas D, Raina S. The extracytoplasmic function sigma factors: Role and regulation. Mol Microbiol. 1998;28(6):1059–1066. doi: 10.1046/j.1365-2958.1998.00865.x. [DOI] [PubMed] [Google Scholar]
- 30.Paget MS, Hong H-J, Bibb MJ, Buttner MJ. The ECF Sigma Factors of Streptomyces coelicolor A3(2) Cambridge, UK: Cambridge Univ Press; 2002. [Google Scholar]
- 31.Staroń A, et al. The third pillar of bacterial signal transduction: Classification of the extracytoplasmic function (ECF) sigma factor protein family. Mol Microbiol. 2009;74(3):557–581. doi: 10.1111/j.1365-2958.2009.06870.x. [DOI] [PubMed] [Google Scholar]
- 32.Strohl WR. Compilation and analysis of DNA sequences associated with apparent streptomycete promoters. Nucleic Acids Res. 1992;20(5):961–974. doi: 10.1093/nar/20.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hesketh A, Kock H, Mootien S, Bibb M. The role of absC, a novel regulatory gene for secondary metabolism, in zinc-dependent antibiotic production in Streptomyces coelicolor A3(2) Mol Microbiol. 2009;74(6):1427–1444. doi: 10.1111/j.1365-2958.2009.06941.x. [DOI] [PubMed] [Google Scholar]
- 34.Myronovskyi M, Welle E, Fedorenko V, Luzhetskyy A. Beta-glucuronidase as a sensitive and versatile reporter in actinomycetes. Appl Environ Microbiol. 2011;77(15):5370–5383. doi: 10.1128/AEM.00434-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bibb MJ, Janssen GR, Ward JM. Cloning and analysis of the promoter region of the erythromycin resistance gene (ermE) of Streptomyces erythraeus. Gene. 1985;38(1-3):215–226. doi: 10.1016/0378-1119(85)90220-3. [DOI] [PubMed] [Google Scholar]
- 36.Gomez-Escribano JP, Bibb MJ. Engineering Streptomyces coelicolor for heterologous expression of secondary metabolite gene clusters. Microb Biotechnol. 2011;4(2):207–215. doi: 10.1111/j.1751-7915.2010.00219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qiao M, Immonen T, Koponen O, Saris PE. The cellular location and effect on nisin immunity of the NisI protein from Lactococcus lactis N8 expressed in Escherichia coli and L. lactis. FEMS Microbiol Lett. 1995;131(1):75–80. doi: 10.1016/0378-1097(95)00238-z. [DOI] [PubMed] [Google Scholar]
- 38.Stein T, Heinzmann S, Solovieva I, Entian KD. Function of Lactococcus lactis nisin immunity genes nisI and nisFEG after coordinated expression in the surrogate host Bacillus subtilis. J Biol Chem. 2003;278(1):89–94. doi: 10.1074/jbc.M207237200. [DOI] [PubMed] [Google Scholar]
- 39.Jiang H, Hutchinson CR. Feedback regulation of doxorubicin biosynthesis in Streptomyces peucetius. Res Microbiol. 2006;157(7):666–674. doi: 10.1016/j.resmic.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 40.Ostash I, et al. Coordination of export and glycosylation of landomycins in Streptomyces cyanogenus S136. FEMS Microbiol Lett. 2008;285(2):195–202. doi: 10.1111/j.1574-6968.2008.01225.x. [DOI] [PubMed] [Google Scholar]
- 41.Otten SL, Ferguson J, Hutchinson CR. Regulation of daunorubicin production in Streptomyces peucetius by the dnrR2 locus. J Bacteriol. 1995;177(5):1216–1224. doi: 10.1128/jb.177.5.1216-1224.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tahlan K, et al. Initiation of actinorhodin export in Streptomyces coelicolor. Mol Microbiol. 2007;63(4):951–961. doi: 10.1111/j.1365-2958.2006.05559.x. [DOI] [PubMed] [Google Scholar]
- 43.Kleerebezem M. Quorum sensing control of lantibiotic production; nisin and subtilin autoregulate their own biosynthesis. Peptides. 2004;25(9):1405–1414. doi: 10.1016/j.peptides.2003.10.021. [DOI] [PubMed] [Google Scholar]
- 44.Stein T, et al. Dual control of subtilin biosynthesis and immunity in Bacillus subtilis. Mol Microbiol. 2002;44(2):403–416. doi: 10.1046/j.1365-2958.2002.02869.x. [DOI] [PubMed] [Google Scholar]
- 45.Schmitz S, Hoffmann A, Szekat C, Rudd B, Bierbaum G. The lantibiotic mersacidin is an autoinducing peptide. Appl Environ Microbiol. 2006;72(11):7270–7277. doi: 10.1128/AEM.00723-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Ruyter PG, Kuipers OP, Beerthuyzen MM, van Alen-Boerrigter I, de Vos WM. Functional analysis of promoters in the nisin gene cluster of Lactococcus lactis. J Bacteriol. 1996;178(12):3434–3439. doi: 10.1128/jb.178.12.3434-3439.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuipers OP, Beerthuyzen MM, de Ruyter PG, Luesink EJ, de Vos WM. Autoregulation of nisin biosynthesis in Lactococcus lactis by signal transduction. J Biol Chem. 1995;270(45):27299–27304. doi: 10.1074/jbc.270.45.27299. [DOI] [PubMed] [Google Scholar]
- 48.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces Genetics. Norwich, UK: John Innes Foundation; 2000. [Google Scholar]
- 49.Sambrook J, Russell DW. Molecular Cloning: A laboratory Manual. 3rd Ed. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2001. [Google Scholar]
- 50.Maxam AM, Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 51.Mertz FP. Planomonospora alba Sp-Nov and Planomonospora sphaerica Sp-Nov, 2 new species isolated from soil by baiting techniques. Int J Syst Bacteriol. 1994;44(2):274–281. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.