Abstract
Toll-like receptor (TLR) signaling is critical in innate response against invading pathogens. However, the molecular mechanisms for full activation of TLR-triggered innate immunity need to be fully elucidated. The broad complex tramtrack bric-a-brac/poxvirus and zinc finger (BTB/POZ) family is a class of transcription factors involved in many biological processes. However, few BTB/POZ proteins were reported to function in innate immune response. Zinc finger and BTB domain-containing 20 (ZBTB20), a member of BTB/POZ family, functions in neurogenesis and represses α-fetoprotein gene transcription in liver. However, the immunological functions of ZBTB20 remain unknown. Here, we found that myeloid cell-specific ZBTB20 KO mice were resistant to endotoxin shock and Escherichia coli-caused sepsis. ZBTB20 deficiency attenuated TLR-triggered production of proinflammatory cytokines and type I IFN in macrophages, which attributed to higher abundance of IκBα protein and impaired activity of NF-κB. Furthermore, ChIP and next generation high-throughput DNA sequencing assay showed that ZBTB20 specifically bound to IκBα gene promoter (+1 to +60 region) after TLR activation. ZBTB20 could inhibit IκBα gene transcription, govern IκBα protein expression, and then promote NF-κB activation. Therefore, transcriptional repressor ZBTB20 is needed to promote full activation of TLR signaling and TLR-triggered innate immune response by selectively suppressing the suppressor IκBα gene transcription.
Toll-like receptors (TLRs) are important pattern recognition receptors that can detect the conserved structures in pathogens and initiate protective innate immune responses (1). After the recognition of pathogen-associated molecule patterns, TLRs recruit adaptor molecules, such as myeloid differentiation protein 88 and toll-interleukin 1 receptor domain-containing adaptor protein-inducing IFN-β (TRIF), to activate subsequent signaling pathways and induce the production of proinflammatory cytokines and type I IFN (2). On stimulation, most TLRs recruit myeloid differentiation protein 88 and use the downstream molecules, such as IL-1 receptor-associated kinases, TNF receptor-associated factor 6, and TGF-β–activated kinase 1, to activate inhibitor of κB kinases (IKKs). The IKK complex can phosphorylate IκBα and lead to the degradation of IκBα by ubiquitination. Then, NF-κB, which has been inhibited by IκBα, is released to translocate into nucleus and initiate the transcription of genes encoding various proinflammatory mediators (3). Apart from that, TLR3 and TLR4 can signal through a TRIF-dependent pathway, which activates TNF receptor-associated factor family member-associated NF-κB activator binding kinase 1 to phosphorylate IFN regulatory factor 3 (IRF3), leading to the production of type I IFN. In addition, TRIF can also interact with TNF receptor-associated factor 6 and mediate NF-κB activation, leading to the proinflammatory cytokine production and enhanced type I IFN production (4).
Although full activation of TLR signaling is critical for host defense to eliminate the invading microbial pathogens, excessive activation of TLRs pathways may contribute to the pathogenesis of inflammatory and autoimmune diseases (5, 6). Thus, TLR signaling is tightly regulated to maintain the immunological homeostasis. Previous studies have shown that the TLR pathway is positively or negatively regulated by various signaling molecules (7–9). However, the underlying molecular mechanism through which TLR response is properly initiated and fully activated still needs to be further clarified.
The broad complex tramtrack bric-a-brac/poxvirus and zinc finger (BTB/POZ) family is a class of nuclear DNA binding transcription factors that is involved in a variety of biological processes, including development, differentiation, tumorigenesis, and chromatin remodeling (10, 11). However, up to now, few BTB/POZ zinc finger proteins were reported to function in innate immune response. Zinc finger and BTB domain-containing 20 (ZBTB20), also known as HOF and zinc finger protein 288 (12, 13), is identified in human dendritic cells (DCs) and named as dendritic cell-derived BTB/POZ zinc finger (14). ZBTB20 is a unique member of BTB/POZ family and functions primarily as a transcriptional repressor by the N-terminal BTB/POZ domain and conserved C-terminal C2H2 Krüppel-type zinc finger. By generating ZBTB20 KO mice, we show that ZBTB20 is a unique regulator that directly binds to the α-fetoprotein (Afp) promoter and controls postnatal AFP repression (15). Moreover, it can regulate β-cell function through transcriptional suppression of fructose-1,6-bisphosphatase 1 and thus, becomes crucial for postnatal survival and glucose homeostasis in mice (16, 17). In addition, ZBTB20 is essential for the specification of CA1 field identity in the developing hippocampus (18).
Our previous studies show that ZBTB20 is widely expressed in human hematopoietic tissues, including DCs, monocytes, B cells, and T cells (14). However, the precise role of ZBTB20 in immune response, especially in TLR-triggered innate response, has not been established yet. Here, we found that myeloid cell-specific ZBTB20 KO mice were resistant to endotoxin shock and Escherichia coli-caused sepsis, which was caused by the attenuated production of proinflammatory cytokines and type I IFN in TLR-triggered or bacterial-challenged macrophages. Furthermore, ZBTB20 specifically bound to IκBα gene promoter, repressed IκBα gene transcription, and then inhibited IκBα protein expression, thus promoting NF-κB activation and leading to the enhanced TLR-triggered cytokine production. Therefore, ZBTB20 can act as a transcriptional repressor of the inhibitory gene IκBα, promoting full activation of TLR-triggered innate immune responses.
Results
Generation of Myeloid Cell-Specific ZBTB20 KO Mice.
ZBTB20 null mice, generated by homologous recombination, displayed a stark phenotype characterized by postnatal growth retardation, metabolic dysfunction, and lethality (16). To specifically investigate the function of ZBTB20 in immune cell, especially in macrophages, we created myeloid cell-specific ZBTB20 KO mice (MZB20KO mice) using the LysM-Cre/loxP recombination system, which allowed for specific and highly efficient Cre-mediated deletion of loxP-flanked target genes in myeloid cells, mainly including mature macrophages and granulocytes. MZB20KO mice were generated by repeated mating of ZBTB20flox/flox mice (15) with LysM-cre mice (19). MZB20KO (ZBTB20flox/flox/LysM-Cre) and control mice (ZBTB20flox/flox) were viable, fertile, and normal in size, and they displayed no gross physical or behavioral abnormalities. Efficient deletion of Zbtb20 gene in MZB20KO peritoneal macrophages was confirmed at the mRNA and protein levels (Fig. S1). No deletion was detected in T cells, and slight deletion was observed in splenic DCs from MZB20KO mice (Fig. S1). MZB20KO and control mice also had similar numbers of splenic or bone marrow-derived macrophages, DCs, and peritoneal macrophages (Fig. S2). Therefore, MZB20KO mice have normal myeloid development and macrophage differentiation.
ZBTB20 Deficiency Protects Mice from LPS and Bacterial Challenge.
To investigate the role of ZBTB20 in the TLR-triggered innate immune response, we challenged MZB20KO mice with TLR ligands LPS, the synthetic RNA duplex poly(I:C), or the CpG oligodeoxynucleotide (ODN). MZB20KO mice produced significantly less TNF, IL-6, and IFN-β than control littermates in response to LPS, poly(I:C), or CpG ODN challenge (Fig. 1A). Accordingly, MZB20KO mice exhibited prolonged survival compared with control littermates after lethal LPS challenge (Fig. 1B). MZB20KO mice were also more resistant to lethal challenge with high-dose poly(I:C) (Fig. S3). To further confirm the effects of ZBTB20 deficiency on TLR-triggered cytokine production in vivo, bone marrow cells from MZB20KO or control mice were transplanted into lethally irradiated WT mice. The LPS-induced in vivo production of TNF, IL-6, and IFN-β was much less in the reconstituted MZB20KO chimeras than control chimeras (Fig. 1C). These data show that MZB20KO mice exhibit impaired TLR-triggered inflammatory responses and are more resistant to endotoxin shock, indicating that ZBTB20 plays an important role in the full activation of TLR-triggered innate immune responses.
Fig. 1.
ZBTB20 deficiency protects mice from challenge with TLR ligands. (A) ELISA of TNF, IL-6, and IFN-β in the serum of MZB20KO or control mice (n = 3 per genotype) 2 h after i.p. administration of PBS and LPS, poly(I:C), or CpG ODN (CpG; at a dose of 10, 15, or 20 mg/kg body weight, respectively). *P < 0.05, **P < 0.01 (Student t test). (B) Survival of MZB20KO or control mice (n = 10 per genotype) given i.p. injection of LPS (15 mg/kg body weight). P < 0.01 (Wilcoxon test). (C) ELISA of TNF, IL-6, and IFN-β in serum from control mice (n = 3 per genotype) lethally irradiated and reconstituted for 8 wk with bone marrow cells from MZB20KO or control mice followed by challenge with PBS or LPS and analysis 2 h later. **P < 0.01 (Student t test). Data are from three independent experiments (mean ± SEM).
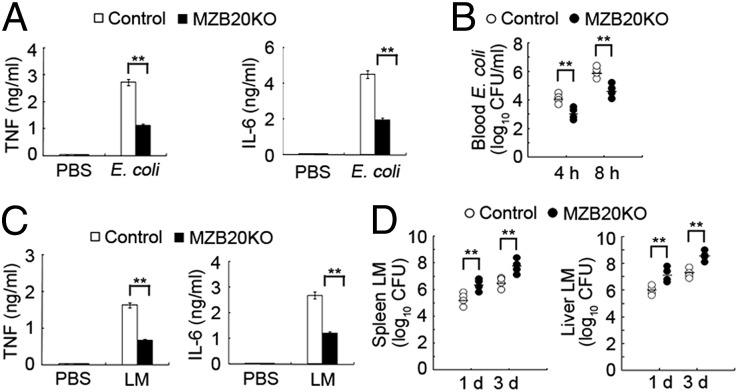
To assess the role of ZBTB20 in the host innate response to pathogen infection, MZB20KO mice were challenged with intact Gram-negative E. coli or Gram-positive Listeria monocytogenes. After infection with E. coli, the production of TNF and IL-6 in serum of MZB20KO mice was significantly less than the production in serum of their control littermates (Fig. 2A). Accordingly, MZB20KO mice had a smaller load of E. coli bacteria in the blood (Fig. 2B), consistent with published finding that proinflammatory cytokines promote the dissemination of E. coli (20). After infection with L. monocytogenes, MZB20KO mice also produced fewer proinflammatory cytokines and had a greater bacterial load in the spleen and liver (Fig. 2 C and D). These data indicate that ZBTB20 deficiency attenuates the inflammatory innate response of host and protects mice from lethal challenge by endotoxin shock and bacteria-caused sepsis.
Fig. 2.
ZBTB20-deficient mice are more resistant to E. coli infection but more susceptible to L. monocytogenes infection. (A) ELISA of TNF and IL-6 in the serum and (B) analysis of bacterial loads in the blood of MZB20KO or control mice (n = 4 per genotype) 2, 4, and 8 h after i.p. infection with 1 × 107 E. coli 0111:B4. (C) ELISA of TNF and IL-6 in the serum and (D) analysis of bacterial loads in the spleen and liver of MZB20KO or control mice (n = 4 per genotype) after treatment with PBS or infection with 1 × 104 L. monocytogenes. Serum was obtained 4 h after infection. In B and D, each symbol represents an individual mouse; small horizontal lines indicate the mean. **P < 0.01 (Student t test). Data are from three independent experiments with similar results (mean ± SEM).
Impaired Cytokine Production in TLR-Triggered ZBTB20-Deficient Macrophages.
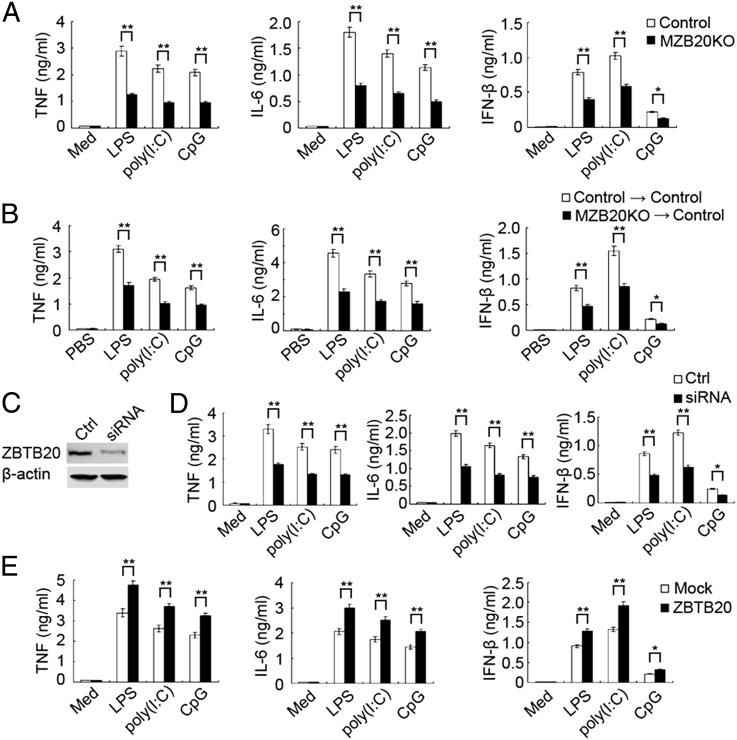
Macrophages are the main mediator of TLR-triggered inflammatory responses in vivo (1). Next, we assessed whether ZBTB20 deficiency attenuated the production of proinflammatory cytokines and type I IFN in TLR-triggered macrophages. In accordance with the in vivo observations, peritoneal macrophages from MZB20KO mice had lower expression of TNF, IL-6, and IFN-β mRNA and protein than macrophages from control littermate mice in response to LPS, poly(I:C), or CpG ODN (Fig. 3A and Fig. S4). In addition, there was no substantial difference between MZB20KO and control mice in the expression level of TLR4, TLR3, or TLR9 protein in peritoneal macrophages (Fig. S5), which excluded the possibility that the attenuation of TLR response achieved by ZBTB20 deficiency was simply because of lower expression of TLRs. To further confirm that the impaired TLR-triggered inflammatory response in vivo was caused by ZBTB20 deficiency in macrophages, we adoptively transferred bone marrow-derived macrophages from MZB20KO or control mice into WT mice depleted of endogenous macrophages by pretreatment with clodronate liposomes. Mice reconstituted with MZB20KO macrophages produced less proinflammatory cytokines and IFN-β in response to LPS, poly(I:C), or CpG ODN challenge than mice reconstituted with macrophages from control mice (Fig. 3B).
Fig. 3.
ZBTB20 promotes TLR-triggered production of proinflammatory cytokines and type I IFN in macrophages. (A) ELISA of cytokines in supernatants of MZB20KO or control macrophages left unstimulated (Med) or stimulated for 6 h with LPS (100 ng/mL), poly(I:C) (10 μg/mL), or CpG ODN (CpG; 0.3 μM). (B) ELISA of cytokines in serum from control mice first depleted of endogenous macrophages and then transplanted with 1 × 107 MZB20KO or control bone marrow-derived macrophages 6 h before challenge with LPS, poly(I:C), or CpG ODN (measured 2 h after challenge). (C) Immunoblot analysis of the expression of ZBTB20 and β-actin in lysates of macrophages 48 h after transfected with control siRNA (Ctrl) or siRNA specific for Zbtb20 (siRNA). (D) ELISA of cytokines in supernatants of macrophages transfected as in C and 48 h later, left unstimulated or stimulated for 6 h with LPS, poly(I:C), or CpG ODN. (E) ELISA of cytokines in supernatants of WT macrophages given mock transfection (mock) or transfected with vectors for the expression of ZBTB20 and 36 h later, stimulated for 6 h with LPS, poly(I:C), or CpG ODN. *P < 0.05, **P < 0.01. Data are from (A, B, D, and E) three independent experiments (mean ± SEM) or (C) representative of three independent experiments with similar results.
We further observed the effect of ZBTB20 knockdown on cytokine production by TLR-activated macrophages. The endogenous expression of ZBTB20 in peritoneal macrophages was markedly diminished by Zbtb20-specific siRNA (Fig. 3C). Zbtb20-specific siRNA resulted in significantly less production of TNF, IL-6, and IFN-β in peritoneal macrophages stimulated with LPS, poly(I:C), or CpG ODN (Fig. 3D). Consistently, overexpression of ZBTB20 significantly promoted the production of TNF, IL-6, and IFN-β in macrophages induced by LPS, poly(I:C), or CpG ODN, respectively (Fig. 3E). Therefore, ZBTB20 significantly promotes TLR-triggered production of proinflammatory cytokines and type I IFN in macrophages.
Impaired TLR-Induced NF-κB Activation in ZBTB20-Deficient Macrophages.
We further investigated the effect of ZBTB20 deficiency on TLR-activated downstream signal pathways in macrophages, including the MAPK, NF-κB, and IRF3 pathways. There was no substantial difference of phosphorylation of ERK, JNK, and p38 induced by LPS, poly(I:C), or CpG ODN between macrophages from MZB20KO or control mice (Fig. 4A and Fig. S6), suggesting that ZBTB20 did not affect TLR-triggered activation of the MAPK pathway. In the NF-κB pathway, LPS-induced phosphorylation of IKKα/β remain unchanged in MZB20KO macrophages, whereas the protein expression level of IκBα was substantially higher in MZB20KO macrophages than control macrophages at 45 and 60 min after LPS stimulation (Fig. 4A). A high abundance of IκBα can result in less translocation of NF-κB to the nucleus and impaired activation of transcription (21). As shown in Fig. 4, LPS-induced phosphorylation and nuclear translocation of the NF-κB subunit p65 were lower in MZB20KO macrophages than control macrophages. We obtained similar results in MZB20KO macrophages stimulated with poly(I:C) or CpG ODN (Fig. S6). Both NF-κB and IRF3 activations contribute to TLR-mediated transcriptional activation of type I IFN gene (22, 23). We then detected the impact of ZBTB20 on TLR-triggered activation of IRF3. ZBTB20 deficiency did not affect TLR-induced phosphorylation of IRF3 (Fig. 4A). Thus, these data indicate that ZBTB20 deficiency-impaired production of TNF and IL-6 was caused by the high abundance of IκBα and the attenuated activation of NF-κB, whereas the impaired production of IFN-β was also caused by the attenuated activation of NF-κB but not IRF3, suggesting that ZBTB20 promotes TLR-triggered innate immune response by inhibiting IκBα expression and subsequently enhancing NF-κB activation.
Fig. 4.
ZBTB20 deficiency impairs TLR-triggered activation of NF-κB but not MAPK and IRF3 pathway in macrophages. (A) Immunoblot analysis of phosphorylated (p-) or total protein in lysates of MZB20KO or control macrophages stimulated for 0–60 min with LPS (100 ng/mL). (B) Immunoblot analysis of p65 among nuclear proteins from macrophages stimulated with LPS; lamin A served as a loading control. Data are representative of three independent experiments with similar results.
ZBTB20 Binds to IκBα Promoter.
Considering that ZBTB20 could bind to the Afp promoter and repress Afp gene transcription (15), we investigated the innate gene promoter ZBTB20 to which it could potentially bind. ChIP followed by the next generation high-throughput DNA sequencing (ChIP-Seq) assay was performed with antibody to ZBTB20 in LPS-stimulated or untreated macrophages. Among the target genes that we detected, IκBα (encoding by Nfkbia) attracted our attention, because our above results had indicated that ZBTB20 controlled IκBα protein expression. The ChIP-Seq results showed that the binding site (peak region) of ZBTB20 on the IκBα gene was located in the chr12:56593075–56593640 region. Another ChIP assay with specific primers for the binding site confirmed that ZBTB20 was recruited to the IκBα gene promoter after stimulation of macrophages with LPS, poly(I:C), or CpG ODN (Fig. 5 A and B). We also explored the signal mechanism underlying the recruitment of ZBTB20 to the IκBα promoter after TLR activation. As shown in Fig. 5C, inhibition of NF-κB efficiently impaired the recruitment of ZBTB20 to the IκBα promoter, whereas inhibition of MAPK kinase 1/ERK, JNK, or p38 had little effect. These data indicate that TLR activation causes recruitment of ZBTB20 to the IκBα promoter, which is mainly dependent on the NF-κB pathway.
Fig. 5.
ZBTB20 directly binds to IκBα promoter. (A and B) ChIP analysis of the recruitment of ZBTB20 to IκBα promoter in macrophages left unstimulated or stimulated for 30 min with LPS, poly(I:C), or CpG ODN; IκBα promoter sequences in input DNA and DNA recovered from antibody-bound chromatin segments were detected by (A) semiquantitive PCR or (B) quantitative PCR. The graph represents fold enrichment of ZBTB20 bound at the IκBα promoter region. (C) ChIP analysis of the recruitment of ZBTB20 to the IκBα promoter in macrophages pretreated with DMSO, PD98059 (10 μM), SP600125 (10 μM), SB203580 (10 μM), or pyrrolidine dithiocarbamate (100 μM) for 30 min and then left unstimulated or stimulated for 30 min with LPS, poly(I:C), or CpG ODN. (B and C) All values generated by quantitative PCR analysis were normalized to control IgG precipitations. **P < 0.01. Data are (A) representative of three independent experiments with similar results or (B and C) from three independent experiments (mean ± SEM).
ZBTB20 Represses IκBα Gene Transcription.
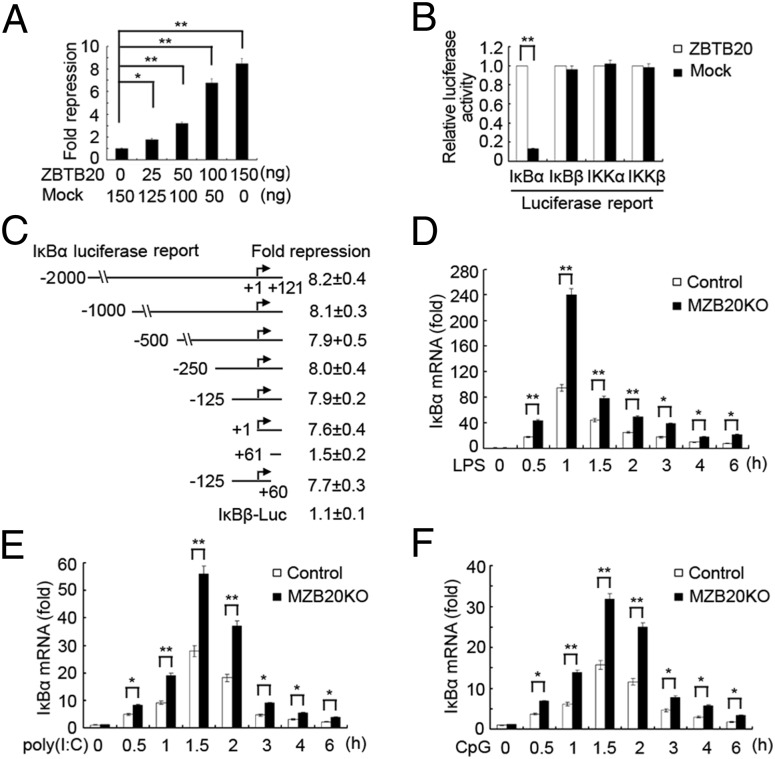
Given that ZBTB20 was identified as a transcriptional repressor (15, 17) and that ZBTB20 was found to bind to the IκBα promoter and control IκBα protein expression, we wanted to know whether ZBTB20 repressed IκBα gene transcription. We first determined whether ZBTB20 could repress IκBα-driven transcriptional activity by using a transiently transfected luciferase reporter assay in HEK293 cells. ZBTB20 exerted up to an eightfold repression of the reporter IκBα-2000Luc, driven by −2,000 to +121 of the IκBα gene, in a dose-dependent manner (Fig. 6A). However, it had no significant effects on IκBβ-, IKKα-, or IKKβ-driven reporters (Fig. 6B). In addition, ZBTB20 deficiency did not impair the mRNA expression of IFN-stimulated gene 15 and inflammatory protein 10 induced by IFN-β as well as IL-10 mRNA expression induced by TGF-β1 in macrophages (Fig. S7). These results suggest that ZBTB20-mediated repression of gene transcription and expression is gene-specific but not global and extensive. To localize the potential region in the IκBα gene mediating transcriptional repression by ZBTB20, we made serial 5′ deletions of the IκBα gene promoter in the IκBα reporter constructs and analyzed the effects of ZBTB20 on their activity (Fig. 6C). Serial deletion of the fragment from −2,000 to −1 or from +61 to +121 of the IκBα gene did not significantly alter the repressive effects of ZBTB20 on the resultant IκBα reporters, whereas the +1 to +60-region deletion significantly impaired the repressive effects of ZBTB20 on the resultant IκBα reporters, suggesting that the ZBTB20-responsive region might be located in the +1 to +60 region of the IκBα promoter (Fig. 6C). Our ChIP-Seq results also showed that the binding site (peak region) of ZBTB20 on IκBα gene was located in the chr12:56593075–56593640 region, which contained 5′ UTR (+1 to +121 region), the first exon, and part of first intron. Together, these data indicate that the minimal IκBα promoter (+1 to +60 region) contains a cis-acting element mediating ZBTB20 repressive activity. We further observed differences of the mRNA expression kinetics of IκBα in TLR-activated macrophages from MZB20KO and control mice. The IκBα mRNA level in MZB20KO macrophages was significant higher than the level in control macrophages stimulated with LPS, poly(I:C), or CpG ODN, whereas the basal level of IκBα mRNA had no differences between MZB20KO and control macrophages (Fig. 6 D–F), which was consistent with the difference of the IκBα protein level in TLR-activated MZB20KO and control macrophages (Fig. 4A and Fig. S6). Furthermore, the mRNA expression levels of TNF, IL-6, and IFN-β induced by LPS in MZB20KO macrophages were significantly lower than the levels in control macrophages (Fig. S8). These data indicate that ZBTB20 is recruited to the IκBα gene promoter after TLR signaling activation and gene-specifically represses IκBα gene transcription, which results in the lower abundance of IκBα protein, the enhanced NF-κB activation, and the more production of proinflammatory cytokines and type I IFN in macrophages.
Fig. 6.
ZBTB20 represses IκBα gene transcription. (A) Luciferase activity assay in lysates of HEK293 cells cotransfected with different doses of ZBTB20 expressing plasmids, IκBα-2000 Luc reporter plasmid (−2,000 to +121 region of the IκBα promoter), and pTK-Renilla-luciferase. (B) Luciferase activity assay in lysates of HEK293 cells cotransfected with ZBTB20 expressing plasmids, IκBα, IκBβ, IKKα, or IKKβ gene promoter-driven luciferase reporter plasmid, and pTK-Renilla-luciferase. (C) Luciferase activity assay in lysates of HEK293 cells cotransfected with ZBTB20 expressing plasmids, IκBα reporter plasmid driven by IκBα promoter −2,000 to +121, 5′ truncations at −1,000, −500, −250, −125, −1, +61 to +121, or −125 to +60, and pTK-Renilla-luciferase. (D–F) Quantitative PCR analysis of IκBα mRNA level in macrophages from MZB20KO or control mice stimulated with LPS, poly(I:C), or CpG ODN for the indicated times. Results are presented as fold induction relative to IκBα mRNA level in control cells without agonist stimulation. *P < 0.05, **P < 0.01. Data are from three independent experiments (mean ± SEM).
Discussion
Various signaling molecules are involved in the tight positive or negative regulation of the TLR pathway to maintain the immunological balance. However, the great majority of regulatory machinery for TLR signaling was focused on the changes of signaling protein function, the interaction of proteins, and the degradation of protein. Little attention was paid to the transcriptional regulation of TLR signaling molecules. Here, we have provided evidence that deficiency in zinc finger protein ZBTB20 impairs TLR-triggered production of proinflammatory cytokines and type I IFN in macrophages, which attributed to the high abundance of IκBα protein and attenuated NF-κB activation, and protects mice from lethal challenge with TLR ligands and live Gram-negative bacteria. ZBTB20 was also found to bind IκBα gene promoter on TLR activation and inhibit IκBα gene transcription, governing IκBα protein expression. Therefore, we show that the transcriptional repressor ZBTB20 is required for the full activation of TLR signaling and present a model for the activation and regulation of TLR-triggered innate immune responses.
NF-κB signaling is one of the particularly crucial pathways activated when cells are exposed to a variety of stimuli, especially including pathogen-associated molecular patterns recognized by TLRs (24). Under rested conditions, the short-lived inhibitor IκBα binds to NF-κB in the cytoplasm, which prevents translocation of NF-κB to the nucleus and subsequent activation of transcription. After infection with a pathogen or stimulation with TLR ligand, IκBα is phosphorylated by IKKα/β, which results in ubiquitination and degradation of IκBα. NF-κB then translocates into the nucleus and facilitates transcription of NF-κB–regulated genes, including Il6, Tnf, and Ifnb1 (24, 25). NF-κB signaling is eventually terminated through cytoplasmic resequestration of NF-κB, which depends on newly synthesized and expressed IκBα. Newly synthesized IκBα also enters the nucleus and binds NF-κB, thereby enhancing its dissociation from the DNA (the affinity of NF-κB to IκBα seems to be higher than its affinity to κB sites on DNA) and causing its reexportation to the cytoplasm by means of a nuclear export sequence present on IκBα (21, 26). The IκBα promoter contains six NF-κB binding sequences; thus, IκBα gene transcription and synthesis are controlled by autoregulation of NF-κB signaling (26). However, except for NF-κB, no other molecule was reported to regulate IκBα gene transcription. Our result identified ZBTB20 as a transcriptional repressor of the IκBα gene, the important suppressor of NF-κB pathway, which presents a positive regulatory machinery for NF-κB signaling. The aberrant activation of NF-κB is involved in many kinds of inflammatory or autoimmune diseases (27); therefore, ZBTB20 may act as a potential target for drug discovery or therapeutic approach for NF-κB–associated human diseases.
The BTB/POZ domain is an evolutionary conserved protein–protein interaction domain. In most cases, this domain is associated with the C2H2 Krüppel-type zinc finger domain in transcription factors involved in transcriptional regulation. The proteins comprising these two domains are defined as members of the so-called POK (POZ and Krüppel) family, also known as the ZBTB (Zinc finger and BTB domain) family (28). Both in human and mouse genomes, about 43 genes encode POK proteins, including the cancer-associated proteins promyelocytic leukemia zinc finger, ZBTB7, and ZBTB20 (10, 29). POK proteins have been implicated in many biological processes, including B-cell fate determination and cell cycle progression. Consequently, dysfunction of POK proteins has been involved in tumorigenesis and developmental disorders (10, 30). However, few POK proteins were reported to function in immune response, especially in innate immune response. Only promyelocytic leukemia zinc finger was found to activate transcription of a subset of IFN-stimulated genes and contribute to the antiviral activity of IFN (31). Our results show that ZBTB20 promotes TLR-triggered innate immunity by repressing IκBα gene transcription, providing insight into the roles of POK proteins in innate immune response. POK transcription factors generally interact with DNA through their zinc finger motifs to bring about chromatin modification and/or restructuring and localized transcriptional activation or repression (the majority of POK proteins are transcription repressors) (10, 29). The binding sites of most POK proteins in target genes promoters are sequence-nonspecific, whereas Kaiso binds the sequence-specific element (TCCTGCnA) and methylated CpG dinucleotides in target gene promoters, although the mechanism underlying transcription repression through the sequence-specific sites is unclear (32). Our current study identified that the binding site of ZBTB20 was located in the +1 to +60 region of the IκBα promoter, and our previous study identified that ZBTB20 could bind to the AFP promoter directly at the −108 to −53 region (15). However, the DNA sequences of these two binding regions in the IκBα and Afp promoters had nothing in common, indicating that ZBTB20 may bind the target gene promoter through the sequence-nonspecific sites to regulate gene expression. In addition, our data indicate that TLR activation causes recruitment of ZBTB20 to the IκBα promoter mainly through the NF-κB pathway. Activation of the NF-κB pathway by TLR and other unknown machinery may be involved in the induction of chromatin structure changes of the IκBα promoter, which is remodeled into an open and accessible conformation that allows the specific binding of ZBTB20. In addition, it is possible that other unknown intermediates mediating the specific binding of ZBTB20 to the IκBα promoter exist that need to be investigated in the future.
Except for C2H2-type POK zinc finger proteins, the zinc finger protein family comprises many other subfamily members, including C4 type and C6 type (28). They possess zinc finger motifs and play important roles in many physiological and pathological processes. In recent years, some attention was given to the function of zinc finger proteins in innate immune response. Growth factor independent 1 was found to control TLR-triggered inflammatory response by antagonizing NF-κB p65 and inhibit p65-mediated transcriptional transactivation of cytokine gene (33, 34).The CCCH-zinc finger protein family members Zc3h12b, Zc3h12c, and Zc3h12d negatively regulate LPS-induced inflammatory cytokine production by inhibiting cytokine target gene promoter activation (35). In addition, Zc3h12a is also an RNase essential for controlling immune responses by regulating IL-6 and IL-12p40 mRNA decay (36). These studies focused on transcription regulation of inflammatory cytokine genes or mRNA decay, whereas our current work provides evidence that ZBTB20 binds the promoter of suppressor gene IκBα and represses its transcription; then, it facilitates TLR-triggered cytokine production, thus presenting a different regulatory mechanism for zinc finger protein to function in innate immune response.
In conclusion, our results have shown that ZBTB20 specifically binds to the IκBα gene promoter and represses IκBα gene transcription; then, it inhibits IκBα protein expression and enhances NF-κB activation, finally leading to the enhanced TLR-triggered innate immune response. Therefore, transcriptional repressor ZBTB20 is needed to promote the full activation of TLR signaling. Our findings provide regulatory machinery with suppressing suppressor for zinc finger proteins in the innate immune responses.
Materials and Methods
Mice.
The LysM-cre mice (B6.129P2-Lyzstm1(cre)Ifo/J ; 004781) were from Jackson Laboratories and bred in specific pathogen-free conditions; 6- to 8-wk-old littermate mice were used in the experiments (body weight and sex balanced). All animal experiments were performed in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals, with the approval of the Scientific Investigation Board of the Second Military Medical University.
RNA Interfering.
The sequence of siRNA targeting Zbtb20 was 5′-GAATCTACTCCGCACTCTA-3′. The control small RNA sequence was 5′-TTCTCCGAACGTGTCACGT-3′. siRNA duplexes were transfected into mouse peritoneal macrophages using INTERFERin reagent (Polyplus) (37).
Establishment of the Endotoxin Shock Model and the Bacterial Sepsis Model.
The establishment of the endotoxin shock and bacterial sepsis models was performed as described previously (38). Details are described in SI Materials and Methods.
Bone Marrow Transplantation.
Bone marrow cells (1 × 107) from MZB20KO or control mice were transplanted into lethally irradiated WT C57BL/6J mice (cumulative dose of 10 Gy) through tail vein injection. After 8 wk, ZBTB20 expression in macrophages was analyzed by immunoblot (39).
ChIP and ChIP-Seq Assay.
Chromatin was immunoprecipitated according to the instructions of the EZ ChIP Kit (Millipore) with modifications. Cells (1 × 107) were treated for 30 min with LPS, poly(I:C), or CpG ODN and then fixed for 10 min at room temperature with formaldehyde. Cells were then collected and lysed in SDS lysis buffer. Equal amounts of lysates were used for immunoprecipitation of chromatin with anti-ZBTB20 or IgG (Santa Cruz). DNA was purified with a DNA purification column according to the manufacturer’s instructions (Qiagen). Occupancy was assessed by semiquantitive or quantitiative PCR for samples precipitated with a specific antibody vs. samples precipitated with the control IgG (40). The primers were 5′-AAAGTTCCCTGTGCATGACC-3′ (sense) and 5′-CTGGCAGGGGATTTCTCAG-3′ (antisense). The ChIP DNA was also sequenced to saturation with ChIP-Seq, and data processing was performed as described previously (41).
Statistical Analysis.
The statistical significance of comparisons between two groups was determined with Student t test. The statistical significance of survival curves was estimated according to the method of Kaplan and Meier, and curves were compared with the generalized Wilcoxon test. P values of less than 0.05 were considered statistically significant.
Additional methods are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Panpan Ma, Mei Jin, and Yan Li for technical assistance and Chaofeng Han for helpful discussion. This work was supported by National Key Basic Research Program of China Grants 2013CB530500 and 2013CB530603 and National Natural Science Foundation of China Grants 81230074, 81130084, 31070790, 31100635, and 31270943.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. G.C. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1301257110/-/DCSupplemental.
References
- 1.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: Update on Toll-like receptors. Nat Immunol. 2010;11(5):373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 2.Iwasaki A, Medzhitov R. Regulation of adaptive immunity by the innate immune system. Science. 2010;327(5963):291–295. doi: 10.1126/science.1183021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O’Neill LA, Bowie AG. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat Rev Immunol. 2007;7(5):353–364. doi: 10.1038/nri2079. [DOI] [PubMed] [Google Scholar]
- 4.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140(6):805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 5.Marshak-Rothstein A, Rifkin IR. Immunologically active autoantigens: The role of toll-like receptors in the development of chronic inflammatory disease. Annu Rev Immunol. 2007;25:419–441. doi: 10.1146/annurev.immunol.22.012703.104514. [DOI] [PubMed] [Google Scholar]
- 6.Netea MG, Wijmenga C, O’Neill LA. Genetic variation in Toll-like receptors and disease susceptibility. Nat Immunol. 2012;13(6):535–542. doi: 10.1038/ni.2284. [DOI] [PubMed] [Google Scholar]
- 7.Liu J, Liu S, Cao X. Highlights of the advances in basic immunology in 2011. Cell Mol Immunol. 2012;9(3):197–207. doi: 10.1038/cmi.2012.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murray PJ, Smale ST. Restraint of inflammatory signaling by interdependent strata of negative regulatory pathways. Nat Immunol. 2012;13(10):916–924. doi: 10.1038/ni.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schneider M, et al. The innate immune sensor NLRC3 attenuates Toll-like receptor signaling via modification of the signaling adaptor TRAF6 and transcription factor NF-κB. Nat Immunol. 2012;13(9):823–831. doi: 10.1038/ni.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kelly KF, Daniel JM. POZ for effect—POZ-ZF transcription factors in cancer and development. Trends Cell Biol. 2006;16(11):578–587. doi: 10.1016/j.tcb.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 11.Lee SU, Maeda T. POK/ZBTB proteins: An emerging family of proteins that regulate lymphoid development and function. Immunol Rev. 2012;247(1):107–119. doi: 10.1111/j.1600-065X.2012.01116.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mitchelmore C, et al. Characterization of two novel nuclear BTB/POZ domain zinc finger isoforms. Association with differentiation of hippocampal neurons, cerebellar granule cells, and macroglia. J Biol Chem. 2002;277(9):7598–7609. doi: 10.1074/jbc.M110023200. [DOI] [PubMed] [Google Scholar]
- 13.Nielsen JV, Nielsen FH, Ismail R, Noraberg J, Jensen NA. Hippocampus-like corticoneurogenesis induced by two isoforms of the BTB-zinc finger gene Zbtb20 in mice. Development. 2007;134(6):1133–1140. doi: 10.1242/dev.000265. [DOI] [PubMed] [Google Scholar]
- 14.Zhang W, et al. Identification and characterization of DPZF, a novel human BTB/POZ zinc finger protein sharing homology to BCL-6. Biochem Biophys Res Commun. 2001;282(4):1067–1073. doi: 10.1006/bbrc.2001.4689. [DOI] [PubMed] [Google Scholar]
- 15.Xie Z, et al. Zinc finger protein ZBTB20 is a key repressor of alpha-fetoprotein gene transcription in liver. Proc Natl Acad Sci USA. 2008;105(31):10859–10864. doi: 10.1073/pnas.0800647105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sutherland AP, et al. Zinc finger protein Zbtb20 is essential for postnatal survival and glucose homeostasis. Mol Cell Biol. 2009;29(10):2804–2815. doi: 10.1128/MCB.01667-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y, et al. The zinc finger protein ZBTB20 regulates transcription of fructose-1,6-bisphosphatase 1 and β cell function in mice. Gastroenterology. 2012;142(7):1571–1580. doi: 10.1053/j.gastro.2012.02.043. [DOI] [PubMed] [Google Scholar]
- 18.Xie Z, et al. Zbtb20 is essential for the specification of CA1 field identity in the developing hippocampus. Proc Natl Acad Sci USA. 2010;107(14):6510–6515. doi: 10.1073/pnas.0912315107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clausen BE, Burkhardt C, Reith W, Renkawitz R, Förster I. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 1999;8(4):265–277. doi: 10.1023/a:1008942828960. [DOI] [PubMed] [Google Scholar]
- 20.Haziot A, Hijiya N, Gangloff SC, Silver J, Goyert SM. Induction of a novel mechanism of accelerated bacterial clearance by lipopolysaccharide in CD14-deficient and Toll-like receptor 4-deficient mice. J Immunol. 2001;166(2):1075–1078. doi: 10.4049/jimmunol.166.2.1075. [DOI] [PubMed] [Google Scholar]
- 21.Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132(3):344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 22.Hiscott J. Convergence of the NF-kappaB and IRF pathways in the regulation of the innate antiviral response. Cytokine Growth Factor Rev. 2007;18(5-6):483–490. doi: 10.1016/j.cytogfr.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 23.Apostolou E, Thanos D. Virus infection induces NF-kappaB-dependent interchromosomal associations mediating monoallelic IFN-beta gene expression. Cell. 2008;134(1):85–96. doi: 10.1016/j.cell.2008.05.052. [DOI] [PubMed] [Google Scholar]
- 24.Dev A, Iyer S, Razani B, Cheng G. NF-κB and innate immunity. Curr Top Microbiol Immunol. 2011;349(2011):115–143. doi: 10.1007/82_2010_102. [DOI] [PubMed] [Google Scholar]
- 25.Karin M, Ben-Neriah Y. Phosphorylation meets ubiquitination: The control of NF-[kappa]B activity. Annu Rev Immunol. 2000;18(2000):621–663. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- 26.Hoffmann A, Levchenko A, Scott ML, Baltimore D. The IkappaB-NF-kappaB signaling module: Temporal control and selective gene activation. Science. 2002;298(5596):1241–1245. doi: 10.1126/science.1071914. [DOI] [PubMed] [Google Scholar]
- 27.Egan LJ, Toruner M. NF-kappaB signaling: Pros and cons of altering NF-kappaB as a therapeutic approach. Ann N Y Acad Sci. 2006;1072(2006):114–122. doi: 10.1196/annals.1326.009. [DOI] [PubMed] [Google Scholar]
- 28.Krishna SS, Majumdar I, Grishin NV. Structural classification of zinc fingers: Survey and summary. Nucleic Acids Res. 2003;31(2):532–550. doi: 10.1093/nar/gkg161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Costoya JA. Functional analysis of the role of POK transcriptional repressors. Brief Funct Genomics Proteomics. 2007;6(1):8–18. doi: 10.1093/bfgp/elm002. [DOI] [PubMed] [Google Scholar]
- 30.Beaulieu AM, Sant’Angelo DB. The BTB-ZF family of transcription factors: Key regulators of lineage commitment and effector function development in the immune system. J Immunol. 2011;187(6):2841–2847. doi: 10.4049/jimmunol.1004006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu D, et al. Promyelocytic leukemia zinc finger protein regulates interferon-mediated innate immunity. Immunity. 2009;30(6):802–816. doi: 10.1016/j.immuni.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Daniel JM, Spring CM, Crawford HC, Reynolds AB, Baig A. The p120(ctn)-binding partner Kaiso is a bi-modal DNA-binding protein that recognizes both a sequence-specific consensus and methylated CpG dinucleotides. Nucleic Acids Res. 2002;30(13):2911–2919. doi: 10.1093/nar/gkf398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karsunky H, et al. Inflammatory reactions and severe neutropenia in mice lacking the transcriptional repressor Gfi1. Nat Genet. 2002;30(3):295–300. doi: 10.1038/ng831. [DOI] [PubMed] [Google Scholar]
- 34.Sharif-Askari E, et al. Zinc finger protein Gfi1 controls the endotoxin-mediated Toll-like receptor inflammatory response by antagonizing NF-kappaB p65. Mol Cell Biol. 2010;30(16):3929–3942. doi: 10.1128/MCB.00087-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liang J, et al. A novel CCCH-zinc finger protein family regulates proinflammatory activation of macrophages. J Biol Chem. 2008;283(10):6337–6346. doi: 10.1074/jbc.M707861200. [DOI] [PubMed] [Google Scholar]
- 36.Matsushita K, et al. Zc3h12a is an RNase essential for controlling immune responses by regulating mRNA decay. Nature. 2009;458(7242):1185–1190. doi: 10.1038/nature07924. [DOI] [PubMed] [Google Scholar]
- 37.Chen W, et al. Induction of Siglec-G by RNA viruses inhibits the innate immune response by promoting RIG-I degradation. Cell. 2013;152(3):467–478. doi: 10.1016/j.cell.2013.01.011. [DOI] [PubMed] [Google Scholar]
- 38.Liu X, et al. Intracellular MHC class II molecules promote TLR-triggered innate immune responses by maintaining activation of the kinase Btk. Nat Immunol. 2011;12(5):416–424. doi: 10.1038/ni.2015. [DOI] [PubMed] [Google Scholar]
- 39.Xu S, et al. Constitutive MHC class I molecules negatively regulate TLR-triggered inflammatory responses via the Fps-SHP-2 pathway. Nat Immunol. 2012;13(6):551–559. doi: 10.1038/ni.2283. [DOI] [PubMed] [Google Scholar]
- 40.Hou J, et al. Identification of miRNomes in human liver and hepatocellular carcinoma reveals miR-199a/b-3p as therapeutic target for hepatocellular carcinoma. Cancer Cell. 2011;19(2):232–243. doi: 10.1016/j.ccr.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 41.Wei L, et al. Discrete roles of STAT4 and STAT6 transcription factors in tuning epigenetic modifications and transcription during T helper cell differentiation. Immunity. 2010;32(6):840–851. doi: 10.1016/j.immuni.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.