Abstract
Bioactive natural products containing a C-P bond act as mimics of phosphate esters and carboxylic acids, thereby competing with these compounds for active sites of enzymes. Dehydrophos (DHP), a broad-spectrum antibiotic, is a phosphonotripeptide produced by Streptomyces luridus, in which glycine and leucine are linked to an aminophosphonate analog of dehydroalanine, ΔAla(P). This unique feature, in combination with the monomethylation of the phosphonic acid, renders DHP a Trojan horse type antibiotic because peptidase-mediated hydrolysis will release methyl acetylphosphonate, a potent inhibitor of pyruvate dehydrogenase. Bioinformatic analysis of the biosynthetic gene cluster suggested that ΔAla(P) would be generated from Ser(P), the phosphonate analog of Ser, by phosphorylation and subsequent elimination, and that ΔAla(P) would be condensed with Leu-tRNALeu. DhpH was anticipated to carry out this elimination/ligation cascade. DhpH is a multidomain protein, in which a pyridoxal phosphate binding domain is fused to an N-acetyltransferase domain related to the general control nonderepressible-5 (GCN5) family. In this work, the activity of DhpH was reconstituted in vitro. The enzyme was able to catalyze the β-elimination reaction of pSer(P) to generate ΔAla(P), but it was unable to condense ΔAla(P) with Leu. Instead, ΔAla(P) is hydrolyzed to acetyl phosphonate, which is converted to Ala(P) by a second pyridoxal phosphate-dependent enzyme, DhpD. Ala(P) is the substrate for the condensation with Leu-tRNALeu catalyzed by the C-terminal domain of DhpH. DhpJ, a 2-oxoglutarate/Fe(II)-dependent enzyme, introduces the vinyl functionality into Leu-Ala(P) acting as a desaturase, and addition of Gly by DhpK in a Gly-tRNAGly-dependent manner completes the in vitro biosynthesis of dehydrophos.
Phosphonopeptides containing a P-terminal aminophosphonate moiety are well-documented bioactive agents. In 1971, incorporation of l-1-aminoethylphosphonic acid, l-Ala(P), in the dipeptide mimetic l-Ala-l-Ala(P) lead Roche researchers to the discovery of alaphosphin (Fig. 1A), a potent inhibitor of cell-wall biosynthesis (1). Since then, numerous synthetic phosphonopeptides have been screened for their antimicrobial action (2), and several naturally occurring oligopeptides harboring a C-P bond have been isolated (3) (SI Appendix, Fig. S1). Two unique modes of action have been recognized: (i) the intact phosphonopeptide may directly inhibit an enzymatic activity, such as K-26 (SI Appendix, Fig. S1), which exhibits antihypertensive properties by inhibiting angiotensin converting enzyme (4), or (ii) the bioactive phosphonate warhead is released after cellular uptake of the peptide mimic by the target organism and subsequent peptidase-mediated hydrolysis, as in the case of alaphosphin (5), bialaphos (6), and rhizocticin (7) (SI Appendix, Fig. S1). The affinity of the C-P compounds for their targets is usually high owing to their structural similarity to analogous phosphate esters or carboxylic acids that are the substrates of the target enzymes (8, 9).
Fig. 1.
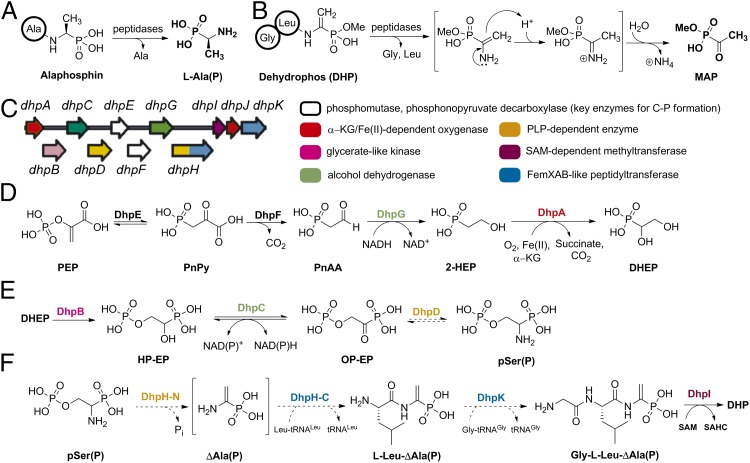
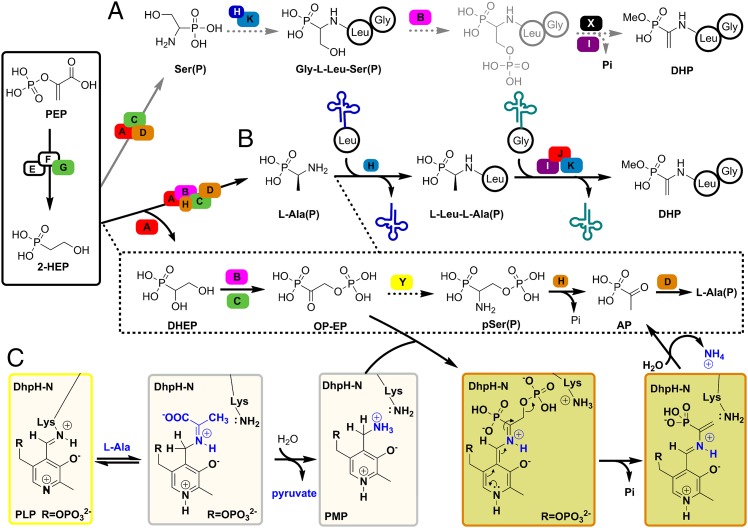
Proposed biosynthesis of dehydrophos. (A) Alaphosphin: a man-made, Trojan horse type, phosphonopeptide. (B) Conversion of DHP into the active compound MAP. (C) Organization of the DHP biosynthetic genes and functional assignment of putative proteins. The dhpLMNOP genes that encode putative transcriptional regulatory proteins and/or secondary transporters are not shown. (D) Confirmed early steps in DHP biosynthesis. (E) Proposed biosynthesis of pSer(P) based on single-gene deletion experiments. (F) Proposed late steps in DHP biosynthesis. Abbreviations not defined in the text: PEP, phosphoenolpyruvic acid; PnAA, phosphonoacetaldehyde; PnPy: phosphonopyruvic acid; SAHC, S-adenosylhomocysteine; SAM, S-adenosylmethionine; 2-HEP, 2-hydroxyethylphosphonate. Dashed reaction arrows are hypothetical.
Dehydrophos (formerly A53868 factor A), a broad-spectrum antibiotic isolated originally from Streptomyces luridus (10), has been of interest because the reassignment of its structure revealed a unique O-methylated dehydroaminophosphonate connected to a glycine-l-leucine dipeptide by an amide bond (11) (Fig. 1B). Structure–activity relationship studies and screening of Salmonella mutants for dehydrophos sensitivity have provided strong evidence for a Trojan horse type mechanism (12, 13). Thus, whereas hydrolysis of alaphosphin releases l-Ala(P) (Fig. 1A), a competitive inhibitor for alanine racemase (14), enzymatic hydrolysis of dehydrophos will unmask 1-aminovinylphosphonate, ΔAla(P), in its monomethylated form (Fig. 1B). The enamine of ΔAla(P) will tautomerize to the corresponding imine and hydrolyze to afford methyl acetylphosphonate (MAP), a 125 times stronger inhibitor of pyruvate dehydrogenase compared with nonesterified acetylphosphonate (15) and a potent inhibitor of bacterial 1-deoxy-d-xylulose 5-phosphate synthase (16, 17).
As part of a program focused on elucidating phosphonate biosynthetic pathways, the dehydrophos gene cluster was integrated into the chromosome of S. lividans and heterologous production of dehydrophos (DHP) was accomplished (18). Bioinformatic analysis of the proteins involved revealed the apparent duplication of several enzymatic functions (Fig. 1C) including two putative 2-oxoglutarate/Fe(II)-dependent oxygenases, DhpA and DhpJ, two putative alcohol dehydrogenases, DhpC and DhpG, two putative pyridoxal 5′-phosphate (PLP)-dependent enzymes, DhpD and DhpH (N-terminal domain), and two putative nonribosomal peptidyl transferases, DhpH (C-terminal domain) and DhpK (18). Single-gene deletions and characterization of the accumulated intermediates by 31P-NMR spectroscopy delineated the order of the first four biosynthetic steps (Fig. 1D). With the exception of the DhpA-catalyzed reaction, these steps are all similar to reactions described for other phosphonate natural products (19–21). Furthermore, the accumulation of intermediates such as 1, 2-dihydroxyethyl phosphonate (DHEP), 1-hydroxy-2-phosphorylethyl phosphonate (HP-EP), and 1-amino-2-phosphorylethyl phosphonate [pSer(P)] suggested a pathway that resembles that of serine biosynthesis in Escherichia coli (Fig. 1E and SI Appendix, Fig. S2). Based on that scheme, the two PLP-dependent enzymes (DhpD and DphH) would act in tandem and the transient product of the N-terminal domain of DhpH, ΔAla(P), would be the substrate of the factors essential for methicillin (Fem) XAB-like C-terminal domain of DhpH for amide bond formation (Fig. 1F). The fact that the two functional domains are fused and encoded by one gene (dhpH) implies a physical proximity for the two active sites and could compensate for the expected very short life of the reactive enamine in ΔAla(P). Thus, it was anticipated that both the carbon-carbon double bond and the amide bond to Leu would be formed by the dual action of DhpH. Formation of the second amide bond was assigned to DphK, whereas DhpI, a phosphonate O-methyltransferase, would furnish dehydrophos (Fig. 1F) (22).
In this study, we reconstituted the activity of DhpH in vitro and discovered that it is not responsible for the formation of the vinyl group in dehydrophos. Instead, we propose that DhpD and the PLP domain of DhpH convert the phosphorylated intermediate 1-oxo-2-phosphorylethylphosphonate (OP-EP) into l-Ala(P), and that the C-terminal domain of DhpH subsequently forms the dipeptide l-Leu-l-Ala(P) in a Leu-tRNALeu–dependent manner. Subsequent methylation by DhpI affords the substrate for the 2-oxoglutarate/Fe(II)-dependent enzyme DhpJ, which is shown to be a desaturase. DhpK, a Gly-tRNAGly-dependent peptidyl transferase, is shown to furnish the final tripeptide.
Results
In Vitro Reconstitution of PLP-Dependent Activity of DhpD and DhpH.
The dhpD and dhpH genes were PCR amplified from the fosmid 17E11-4 harboring the dehydrophos gene cluster (18) and ligated into the expression vector pET-15b. Both proteins were expressed in E. coli as N-terminal hexahistidine-tagged constructs and purified by immobilized metal affinity chromatography (IMAC). DhpH, a multidomain protein (SI Appendix, Fig. S3) with an expected molecular mass of 75 kDa, was copurified with a second protein of approximately 45 kDa after IMAC, based on SDS/PAGE analysis. When the polyacrylamide protein gel was stained with a polyhistidine sequence-specific dye, the lower molecular mass band was also stained (SI Appendix, Fig. S4). We suspected therefore that the second band was the product of partial proteolytic degradation of DhpH. Therefore, we generated a second His-tagged construct (His6-DhpH-N) in which only the first 356 amino acid residues of DhpH were included. His6-DhpH-N was also successfully expressed in E. coli in soluble form.
Based on the previously proposed biosynthetic scheme, DhpD was expected to reversibly convert the phosphonate analog of phosphoserine, pSer(P), and an amino-acceptor molecule to OP-EP (Fig. 1E). In addition, pSer(P) was expected to be the substrate of the PLP domain of DhpH and undergo a β-elimination reaction (Fig. 1F). To test these hypotheses, we prepared pSer(P) (SI Appendix, Fig. S5) by using phosphorylchloride (POCl3) as a phosphorylating reagent under conditions similar to those reported for the phosphorylation of serine (23). Unexpectedly, when DhpD was incubated with pSer(P) in the presence of pyruvate, oxaloacetate, or α-ketoglutarate (α-KG) as amino acceptors, only the starting material was recovered. However, l-Ala(P) and Ser(P) were converted to acetylphosphonate (AP) and 1-oxo-2-hydroxyethylphosphonate (OH-EP), respectively, in the presence of pyruvate (Fig. 2 A and B and SI Appendix, Fig. S6), demonstrating that DhpD has aminotransferase activity. Therefore, the lack of transaminase activity with pSer(P) suggests that pSer(P) is not the physiological substrate of DphD.
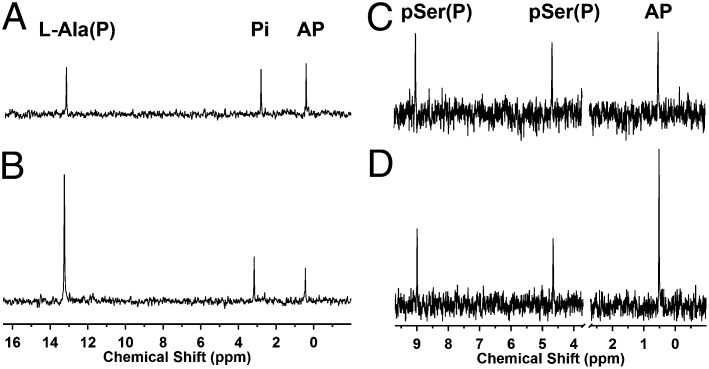
Fig. 2.
31P-NMR analysis of DhpD activity converting l-Ala(P) to AP and vice versa and DhpH activity with rac-pSer(P). (A) 31P-NMR spectrum after conversion of l-Ala(P) to AP by DhpD. Reaction mixture contained 10 mM l-Ala(P), 10 mM pyruvate, and 40 µM DhpD in 50 mM Na-Hepes at pH 8.0. (B) 31P-NMR spectrum after conversion of AP to l-Ala(P) by DhpD. Reaction mixture contained 10 mM l-Ala, 10 mM AP, and 40 µM DhpD in 50 mM Na-Hepes at pH 8.0. (C) 31P NMR spectrum after conversion of rac-pSer(P) to AP by DhpH. (D) 31P-NMR spectrum of C after spiking with authentic standard of AP.
We determined the apparent steady-state kinetic parameters for the conversion of AP and l-Ala to l-Ala(P) and pyruvate by coupling the formation of pyruvate with the oxidation of β-NADH, in the presence of lactate dehydrogenase (LDH) and l-Ala (kcat = 1.9 s−1, Km = 0.02 mM, kcat/Km = 1.0 × 105 M−1⋅s−1) (SI Appendix, Fig. S7A). The high catalytic efficiency observed suggested that this transformation could be the physiological reaction catalyzed by DhpD. The enzyme also converted MAP to the corresponding aminophosphonate in the presence of alanine, albeit with 100-fold lower catalytic efficiency (i.e., kcat = 2.1 s−1, Km = 2.1 mM; kcat/Km =1.0 × 103 M−1⋅s−1) (SI Appendix, Fig. S7B), suggesting that MAP is not the physiological substrate of DhpD.
When His6-DhpH or His6-DhpH-N were incubated with rac-pSer(P) in the absence or presence of typical amino-recipient keto acids such as pyruvate, oxaloacetate, or α-ketoglutarate (α-KG), AP was observed as the only product by 31P-NMR spectroscopy (Fig. 2 C and D and SI Appendix, Fig. S8). Thus, like DhpD, DhpH did not convert pSer(P) to OP-EP. AP is the expected product for β-elimination of the phosphate group followed by tautomerization (Fig. 3A). Interestingly, the (S)-enantiomer of the carboxylic acid analog of pSer(P), phosphoserine (l-pSer), was also a substrate for DhpH-catalyzed β-elimination. By coupling the observed formation of pyruvate with the oxidation of β-NADH in the presence of LDH, we determined the apparent steady-state kinetic parameters for both DhpH (kcat = 0.06 s−1, Km = 0.22 mM, kcat/Km = 0.3 × 103 M−1⋅s−1) and DhpH-N (kcat = 1.10 s−1, Km = 0.28 mM, kcat/Km = 0.4 × 104 M−1⋅s−1) (SI Appendix, Fig. S9). We note that DhpH exhibited transamination activity with unphosphorylated compounds such as SerP and l-Ala(P) in the presence of pyruvate as amino acceptor (SI Appendix, Fig. S10). In summary, the in vitro studies on the PLP-dependent enzymes DhpD and DhpH-N did not provide confirmation of the proposed pathway in Fig. 1 E and F, but DhpH generated AP from pSer(P) and DhpD converted AP to l-Ala(P) (Fig. 3A), prompting further evaluation whether these reactions could be consecutive steps in the DHP biosynthetic pathway.
Fig. 3.
Radioactive TLC analysis of the conversion of rac-pSer(P) to l-[14C(U)]-Leu-Ala(P) by DhpH and DhpD in a one-pot reaction. (A) Reaction scheme. (B) Reaction progress and scanned phosphorimaging plate of silica TLC sheets spotted with: lane 1, aliquot of a reaction containing DhpH, rac-pSer(P), l-[14C(U)]-Leu, tRNA and (re)generation components of Leu-tRNALeu; lane 2, aliquot of a reaction containing l-Ala in addition to the components in lane 1; lane 3, aliquot of a reaction containing l-Ala and DhpD in addition to the components in lane 1.
In Vitro Reconstitution of DhpH tRNA-Dependent Activity.
A PSI-BLAST search using the last 346 amino acids of DhpH as query against the UniProtKB database at the PredictProtein server (24) revealed that this part of the protein shares similarity with numerous uncharacterized proteins that belong to the domain of unknown function 482 (25). The latter is a member of the acetyltransferase clan, which consist of 30 families including the Fem resistance family members FemXAB (26, 27) and the leucyl/phenylalanyl-tRNA protein transferase family (28). Indeed, a BLAST search of the Protein Data Bank database (29) pinpointed FemX from Weissella viridescens (FemXWv) as the closest 3D structural match to the C-terminal domain of DhpH (sequence identity: 17%; similarity: 29%). When we purified the full-length DhpH protein, we observed a high A260/A280 ratio (approximately 1.3) after IMAC and desalting purification steps, a strong indicator of nucleic acid copurification. We also cloned and overexpressed in E. coli the C-terminal domain of DhpH as a histidine-tagged fusion protein (His6-DhpH-C) and observed the same high A260/A280 ratio. In contrast, purified His6-DhpH-N exhibited a A260/A280 ratio of 0.7 (SI Appendix, Fig. S11A). To identify the nature of the nucleic acids bound to DhpH, we treated samples of DhpH-C separately with DNase, RNase, or Proteinase K and analyzed them on a 1% agarose gel. In the RNase-treated sample, nucleic acids were no longer visible under UV light when the gel was stained with ethidium bromide, whereas nucleic acids were still visible in all other samples, suggesting that it was RNA that copurified with DhpH-C. Subsequently, we isolated the RNA by phenol extraction and ethanol precipitation. The isolated RNA sample was incubated either with l-[14C(U)]-leucine in the presence of ATP and purified leucyl-tRNA synthetase from E. coli (LeuRS) or with [14C(U)]-glycine in the presence of ATP and purified glycyl-tRNA synthetase from E. coli (GlyRS) following a standard aminoacylation assay protocol (30). Only the pair l-[14C(U)]-leucine/LeuRS was able to radioactively label the RNA sample with carbon-14 (SI Appendix, Fig. S11B).
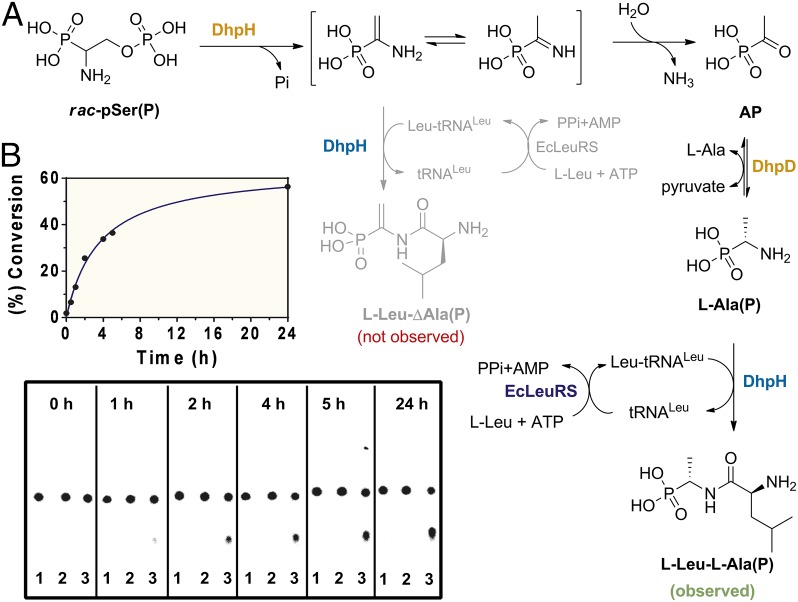
To evaluate whether the C-terminal domain of DhpH could generate an amide bond, the expected product l-Leu-ΔAla(P) was chemically synthesized (SI Appendix, Figs. S12A and S13). This synthetic standard was first used to assess whether pSer(P) could be converted to l-Leu-ΔAla(P) by the coordinated action of the two domains of DhpH in the presence of Leu-tRNALeu as expected by the pathway in Fig. 1F. We set up a sensitive TLC assay in which aminoacylated tRNALeu was (re)generated in situ by LeuRS in the presence of total tRNA from E. coli, ATP, l-[14C(U)]-leucine, and thermostable inorganic pyrophosphatase (TIPP). Incubation of this regeneration system with DhpH and rac-pSer(P) did not result in formation of the desired product (Fig. 3). Thus, once again, the model in Fig. 1F could not be confirmed.
The results described thus far showed that the PLP domain of DhpH converts pSer(P) to AP and that DhpD can generate Ala(P) from AP. We thus decided to investigate whether the general control nonderepressible-5 (GCN5)-related N-acetyltransferase domain of DhpH could form l-Leu-Ala(P), which was also chemically synthesized (SI Appendix, Figs. S12B and S14 A and B). Indeed, when DhpH and DhpD were incubated with rac-pSer(P) and l-Ala in the presence of the tRNALeu regeneration system, an additional radioactive spot appeared on the TLC (Fig. 3). The Rf value of the product was identical to that of synthetic l-Leu-l-Ala(P) (SI Appendix, Fig. S15). To confirm the chemical structure of the radioactive product, l-Ala(P) was incubated on larger scale with DhpH-C or DhpH in the presence of leucine and the Leu-tRNALeu regeneration components. We purified the product by high performance liquid chromatography (HPLC) and showed it exhibited spectral data identical to that of synthetic l-Leu-l-Ala(P) (SI Appendix, Fig. S14 C and D). Combined, these results suggest that Ala(P) is the physiological substrate for the C-terminal domain of DhpH, but it raises the question of how the alkene is then installed in dehydrophos because phosphate elimination is no longer possible. We address this question in the next section.
A series of control experiments confirmed that amide bond formation absolutely depends on tRNA (SI Appendix, Figs. S16 and S17). When using l-Ala(P) as substrate, only partial conversion to the corresponding dipeptide was observed in the absence of externally added nucleic acids. Furthermore, the dipeptide l-Leu-l-Ala(P) was not formed when DhpH was preincubated with RNase (SI Appendix, Fig. S18). Thus, the observed partial activity in the absence of exogenous tRNA was the result of the RNA content of the DhpH preparation as described above. Lastly, by coupling the continuous formation of AMP, owing to the combined action of LeuRS and DhpH, with NADH oxidation in the presence of phosphoenolpyruvate (PEP), myokinase, PEP kinase, and LDH (31), we were able to demonstrate the dependence of DhpH and DhpH-C activity on l-Ala(P) concentration (SI Appendix, Fig. S19).
In Vitro Reconstitution of the Fe(II)/α-KG/O2–Dependent DhpJ Activity.
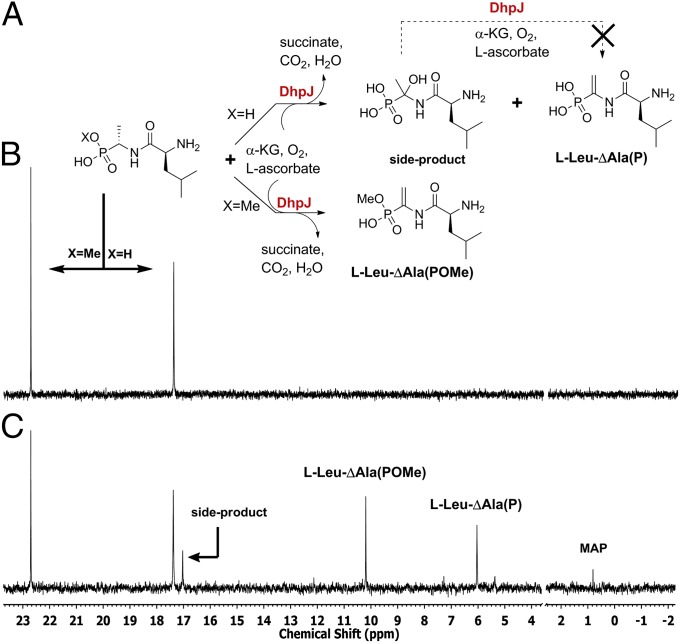
As mentioned above, the formation of l-Leu-l-Ala(P) raises the question how the alkene is formed in dehydrophos. Based on bioinformatic analysis, DhpJ exhibits up to 40% sequence similarity with previously characterized aspartyl/asparaginyl β-hydroxylases (32). The prediction that DhpJ might modify peptides prompted us to overexpress the protein in E. coli. His-tagged DhpJ possessed limited solubility and, thus, we elected to work with a maltose binding protein (MBP) fusion protein (MBP-DhpJ). After excluding the possibility that MBP-DhpJ acted on other intermediates of dehydrophos biosynthesis, such as 2-HEP, l-Ala(P), and DHEP, we tested its activity against a panel of dipeptides and tripeptides listed in SI Appendix, Fig. S20. Interestingly, l-Leu-l-Ala(P) was converted to l-Leu-ΔAla(P), whereas for all other substrates, only the starting material was recovered. The formation of the unsaturated dipeptide was accompanied by the formation of a second major product (Fig. 4C and SI Appendix, Figs. S21 and S22). The 1H-NMR spectrum of the HPLC-purified side product suggested that the dipeptide had been hydroxylated at the α-carbon of l-Ala(P) (Fig. 4A and SI Appendix, Figs. S22 and S24); attempts to confirm this assignment by mass spectrometry were unsuccessful (SI Appendix, Fig. S24). When the isolated side product was incubated with a fresh solution of DhpJ in the presence of Fe(II)/α-KG/O2 and l-ascorbic acid, no conversion to l-Leu-ΔAla(P) was observed (SI Appendix, Figs. S25–S27). Together, these observations suggest that the putative hydroxylated dipeptide is not an intermediate in l-Leu-ΔAla(P) formation and imply that perhaps the l-Leu-l-Ala(P) is not the true physiological substrate of DhpJ. Indeed, incubation of DhpJ with the monomethylated version of l-Leu-l-Ala(P) (SI Appendix, Fig. S28) afforded the corresponding unsaturated dipeptide as the only product (Fig. 4C and SI Appendix, Figs. S29–S32), suggesting that methylation of l-Leu-l-Ala(P) by DhpI occurs before desaturation. In support of this hypothesis, when nearly equimolar amounts of l-Leu-l-Ala(POMe) and l-Leu-l-Ala(P) were allowed to compete for MBP-DhpJ in the same reaction mixture, the methylated substrate was cleanly desaturated, whereas the nonmethylated substrate once again was partially hydroxylated (Fig. 4 B and C).
Fig. 4.
31P-NMR analysis of DhpJ activity with a 1:1.2 mixture of monomethylated and unmethylated l- Leu-l-Ala(P). (A) Reaction scheme of DhpJ activity. (B) 31P-NMR spectrum of the a solution of l-Leu-l-Ala(POMe) and l-Leu-l-Ala(P). (C) 31P-NMR spectrum taken 6 h after addition of Fe(II)-reconstituted DhpJ, α-KG, O2, and l-ascorbic acid. MAP was presumably the side product of l-Leu-ΔAla(POMe) hydrolysis. The phosphate peak (between 2 and 4 ppm) was omitted for clarity.
In Vitro Reconstitution of DhpK Activity.
The observed selectivity of DhpJ for l-Leu-l-Ala(P)-OMe as substrate suggested that addition of the N-terminal Gly would be the last step of DHP biosynthesis. DhpK possesses only 16% identity to the C-terminal domain of DhpH, yet both amino acid sequences have the FemXWv peptidyl transferase as closest 3D structural match. His-tagged DhpK was soluble in E. coli only when coexpressed with the chaperones GroEL/S; therefore, we decided to work with a MBP-fused construct. MBP-DhpK catalyzed the addition of Gly to the N terminus of synthetic l-Leu-Ala(P), l-Leu-ΔAla(P), and methylated l-Leu-ΔAla(P) in assays similar to those described for DhpH-C (SI Appendix, Figs. S33–S35). DhpK could not ligate Gly to l-Ala(P).
Discussion
Based on nature’s strategy to introduce dehydroalanine residues into peptides (33) and the small natural product valanimycin (34), the biosynthesis of the phosphonate analog of dehydroalanine in dehydrophos could have featured generation of the tripeptide Gly-Leu-Ser(P) and phosphorylation of the alcohol of Ser(P) by a kinase. Elimination across the Cα-Cβ bond would then generate the ΔAla(P) group (Fig. 5A). However, although a gene encoding a putative kinase, dhpB, is present in the dehydrophos biosynthetic gene cluster, a gene encoding a pSer lyase-like enzyme is not. pSer(P) was the Ariadne’s thread in our previous efforts to resolve the puzzle of dehydrophos biosynthesis. The appearance of phosphorylated intermediates such as HP-EP and pSer(P) in spent supernatant of the blocked mutants ΔdhpC and/or ΔdhpH (18, 35) suggested that the phosphorylation event occurs early in DHP biosynthesis, and that the phosphate would be eliminated subsequently to install the vinyl group. However, DhpH was unable to incorporate pSer(P) into a peptide in our in vitro studies. Instead, DphH eliminated the phosphate from pSer(P) to generate AP, DhpD converted AP to Ala(P), and Ala(P) was efficiently condensed with Leu by the C-terminal domain of DhpH. DhpJ then installed the olefin in a desaturation reaction (Fig. 5B). This pathway is attractive because it provides a role for DhpJ, which had no function in the previous proposal. It also provides further evidence that methylation occurs late in the biosynthetic pathway, because otherwise the active compound MAP would be formed inside the producing organism. The 100-fold lower activity of DhpD on MAP than on AP also agrees with methylation in a subsequent step. However, by using DhpD to convert AP to Ala(P), a PLP-dependent enzyme appears to be missing from the dehydrophos gene cluster to convert OP-EP to pSer(P). Indeed, when DhpD or DhpH was incubated with pSer(P) to look at the reverse reaction (conversion to OP-EP), no activity was observed. We cannot rule out that an aminotransferase that is not encoded in the biosynthetic gene cluster may generate pSer(P) from OP-EP (e.g., protein Y in Fig. 5B); in fact, the formation of pSer(P) by the dhpH blocked mutant in combination with our in vitro characterization of DhpD/DhpH support this notion. However, an additional aminotransferase is not absolutely necessary if OP-EP encounters DhpH in the pyridoxamine phosphate (PMP) form (SI Appendix, Fig S36). If so, the N-terminal domain of DhpH could catalyze phosphate elimination to AP, followed by the regeneration of the PMP form with an appropriate amine donor such as Ala (Fig. 5 B and C); indeed, we show that DhpH has aminotransferase activity. In this explanation, pSer(P) would never appear as an intermediate during the in vivo biosynthesis of DHP; it would have accumulated in the dhpH mutant (35) because of a fortuitous action of a nonspecific aminotransferase on OP-EP. Indeed, a similar phenomenon has been observed in the case of a dhpG blocked mutant, which accumulates 2-aminoethylphosphonate instead of phosphonoacetaldehyde (18) and in the biosynthesis of phosphinothricin (19). We intended to test this possibility by accessing OP-EP enzymatically from DHEP by the consecutive actions of DhpB and DhpC (or vice versa), but attempts to obtain the kinase DhpB by expression in E. coli were unsuccessful.
Fig. 5.
Revised biosynthesis of dehydrophos. (A) Proposed model for the biosynthesis of dehydrophos based on precedent in dehydroalanine formation. (B) Proposed model for the biosynthesis of dehydrophos based on the in vitro biochemical analysis of DhpD, DhpH, DhpK, and DhpJ. (C) Alternative mechanism for the PLP domain of DhpH. A single capital letter denotes proteins participating in the DHP biosynthesis. Capital letters X and Y denote hypothetical proteins that are not present in the DHP biosynthetic gene cluster.
The evolutionary advantage of the fused domains of DhpH (PLP domain and GCN5-related N-acetyltransferase domain) is unknown but, based on our data, is unrelated to channeling of a reactive intermediate from one active site to another. DhpH and DhpK are two examples of a small but growing list of peptide ligases that are not directly ATP-dependent but instead rely on the cell’s primary metabolism and use aminoacyl-tRNAs, normally dedicated to protein biosynthesis, to accomplish their functions in natural product biosynthesis. Parry and coworkers initially reported the involvement of Ser-tRNASer in the biosynthesis of a natural product (valanimycin) (36), and more recently Walsh and coworkers showed that PacB, which also has similarity to the FemXWv peptidyltransferase, transfers l-Ala from l-Ala-tRNAAla to the N terminus of a tetrapeptide intermediate during biosynthesis of pacidamycin (37). The molecular and structural basis of peptide formation by this emerging class of ligases is unexplored and most likely distinct from a second recently introduced class of tRNA-dependent cyclodipeptide synthetases (38, 39). Our results strengthen the notion that the role of amino acylated-tRNAs goes well beyond their use in protein and cell wall biosynthesis (40, 41).
The introduction of the vinyl moiety into dehydrophos is accomplished in an unusual way: A Fe(II)/α-KG/O2-dependent hydroxylase evolved into a desaturase. The apparent dilemma (hydroxylation vs. desaturation) is well-documented for this class of enzymes (42–44), but the ability of an α-KG–dependent oxygenase to desaturate linear peptides has been demonstrated only very recently (45) and only as a promiscuous activity. It remains to be seen how general this function is.
In conclusion, the biosynthetic pathway to dehydrophos can be divided into three parts. The first part consists of three steps involved in the biosynthesis of many of the natural phosphonates investigated thus far: installation of the C-P bond, decarboxylation, and reduction of phosphonoacetaldehyde to 2-HEP (Figs. 1D and 5). From 2-HEP onwards, the reactions are at present unique to dehydrophos biosynthesis. The middle part of the pathway involves the introduction of an amine functionality at the α-carbon and complete removal of functional groups at the β-carbon of 2-HEP to generate Ala(P), which heretofore was considered to be only a manmade compound (Fig. 5B). Hence, although late methylation prevents formation of the active molecule MAP during DHP biosynthyesis, the biosynthetic pathway delineated herein requires the producing organism to protect itself from inhibition of Ala racemase by l-Ala(P) in a manner that is unknown. The final part includes four steps: ligation of the proteinogenic amino acid leucine to Ala(P) by the C-terminal domain of DhpH, monomethylation of the phosphonic acid by DhpI, introduction of the C-C double bond by DhpJ, and attachment of glycine to the unsaturated dipeptide by DhpK. These four steps are executed by a set of quite unusual enzymes in the context of biosynthesis of natural products (Fig. 5B).
Materials and Methods
All experimental techniques are described in detail in SI Appendix.
Radioactive-TLC Assay.
All three reaction mixtures (50 µL) contained 0.3 mg of tRNA, 10 mM rac-pSer(P), 5 mM l-Leu, 0.85 µCi l-[14C(U)]-Leu, 5 mM ATP, 5 µM DhpH, 5 µM LeuRS and 2 U TIPP, 100 mM Hepes, 10 mM KCl, and 20 mM MgCl2 at pH 7.5. The reaction mixture corresponding to TLC lane 2 was supplemented with 10 mM l-Ala. The reaction mixture corresponding to TLC lane 3 was supplemented with 10 mM l-Ala and 10 µM DhpD. The reactions were incubated at room temperature and 0.5-µL aliquots were removed periodically and spotted on a 5 × 10 cm TLC aluminum sheet Si 60. The TLC plates were developed by using butanol/water/acetic acid at a ratio of 48/12/20 as developing solvent and were dried before they were placed in the phosphorimaging cassette. Rf values of radioactive spots were compared with those obtained from a TLC plate spotted with standards of leucine, l-Leu-l-Ala(P), and l-Leu-l-ΔAla(P) developed identically and visualized with ninhydrin solution (SI Appendix, Fig. S15).
Competition Experiment Between Monomethylated and Unmethylated l-Leu-l-Ala(P) in the Presence of MBP-DhpJ.
l-Leu-l-Ala(POMe) was prepared by reacting 3 mM l-Leu-LAla(P) with 5 mM SAM in the presence of 50 μM DhpI and 20 μM SAHC nucleosidase in 50 mM Na-Hepes at pH 7.5. After 6-h incubation at ambient temperature, the enzymes were removed by passing the reaction mixture through an Amicon spin column (10-kDa molecular weight cutoff; MWCO). The conversion of the reaction (60%) was calculated by integrating the signals of starting material and product by 31P-NMR spectroscopy. To this solution, l-Leu-l-Ala(P) was added such that the final ratio of monomethylated/unmethylated dipeptide was 1:1.2. After lyophilization, the material was dissolved in 0.5 mL of a solution containing 6 mM α-KG, 3 mM l-ascorbic acid, 0.2 mM (NH4)2Fe(SO4)2, and 40 µM MBP-DhpJ reconstituted on ice in an anaerobic chamber with 1.2 equivalent of Fe(II). The reaction mixture was incubated for 5 h at room temperature. Then, approximately 100 µL of Chelex 100 resin was added to the reaction tube to remove iron ions, and the solution was passed through an Amicon spin column (30 kDa MWCO). Before NMR analysis, 150 µL of D2O was added into the sample. Based on integration of the signals of Fig. 4C, about 21% was side-product, about 37% l-Leu-ΔAla(POMe), about 37% l-Leu-ΔAla(P), and about 5% belonged to MAP.
Supplementary Material
Acknowledgments
We thank Susan A. Martinis (Department of Biochemistry, University of Illinois at Urbana–Champaign) for providing the E. coli strain that expressed the leucyl-tRNA synthetase. This work was supported by National Institutes of Health (NIH) Grant P01 GM077596. NMR spectra were recorded on a 600-MHz instrument purchased with support from NIH Grant S10 RR028833.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1303568110/-/DCSupplemental.
References
- 1.Allen JG, et al. Phosphonopeptides, a new class of synthetic antibacterial agents. Nature. 1978;272(5648):56–58. doi: 10.1038/272056a0. [DOI] [PubMed] [Google Scholar]
- 2.Atherton FR, Hassall CH, Lambert RW. Synthesis and structure-activity relationships of antibacterial phosphonopeptides incorporating (1-aminoethyl)phosphonic acid and (aminomethyl)phosphonic acid. J Med Chem. 1986;29(1):29–40. doi: 10.1021/jm00151a005. [DOI] [PubMed] [Google Scholar]
- 3.Mastalerz P, Kafarski P. Naturally Occurring Aminophosphonic and Aminophosphinic Acids. 1st Ed. Wiley, Chichester, NY; 2000. [Google Scholar]
- 4.Ntai I, Bachmann BO. Identification of ACE pharmacophore in the phosphonopeptide metabolite K-26. Bioorg Med Chem Lett. 2008;18(10):3068–3071. doi: 10.1016/j.bmcl.2007.11.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Atherton FR, et al. Phosphonopeptides as antibacterial agents: Mechanism of action of alaphosphin. Antimicrob Agents Chemother. 1979;15(5):696–705. doi: 10.1128/aac.15.5.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lea PJ, Joy KW, Ramos JL, Guerrero MG. The action of 2-amino-4-(methylphosphinyl)-butanoic acid (phosphinothricin) and its 2-oxo-derivative on the metabolism of cyanobacteria and higher-plants. Phytochemistry. 1984;23(1):1–6. [Google Scholar]
- 7.Kugler M, Loeffler W, Rapp C, Kern A, Jung G. Rhizocticin A, an antifungal phosphono-oligopeptide of Bacillus subtilis ATCC 6633: Biological properties. Arch Microbiol. 1990;153(3):276–281. doi: 10.1007/BF00249082. [DOI] [PubMed] [Google Scholar]
- 8.Metcalf WW, van der Donk WA. Biosynthesis of phosphonic and phosphinic acid natural products. Annu Rev Biochem. 2009;78:65–94. doi: 10.1146/annurev.biochem.78.091707.100215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Demmer CS, Krogsgaard-Larsen N, Bunch L. Review on modern advances of chemical methods for the introduction of a phosphonic acid group. Chem Rev. 2011;111(12):7981–8006. doi: 10.1021/cr2002646. [DOI] [PubMed] [Google Scholar]
- 10.Hunt AH, Elzey TK. Revised structure of A53868A. J Antibiot (Tokyo) 1988;41(6):802. doi: 10.7164/antibiotics.41.802. [DOI] [PubMed] [Google Scholar]
- 11.Whitteck JT, et al. Reassignment of the structure of the antibiotic A53868 reveals an unusual amino dehydrophosphonic acid. Angew Chem Int Ed Engl. 2007;46(47):9089–9092. doi: 10.1002/anie.200703810. [DOI] [PubMed] [Google Scholar]
- 12.Kuemin M, van der Donk WA. Structure-activity relationships of the phosphonate antibiotic dehydrophos. Chem Commun. 2010;46(41):7694–7696. doi: 10.1039/c0cc02958k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Circello BT, Miller CG, Lee JH, van der Donk WA, Metcalf WW. The antibiotic dehydrophos is converted to a toxic pyruvate analog by peptide bond cleavage in Salmonella enterica. Antimicrob Agents Chemother. 2011;55(7):3357–3362. doi: 10.1128/AAC.01483-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conti P, et al. Drug discovery targeting amino acid racemases. Chem Rev. 2011;111(11):6919–6946. doi: 10.1021/cr2000702. [DOI] [PubMed] [Google Scholar]
- 15.O’Brien TA, Kluger R, Pike DC, Gennis RB. Phosphonate analogues of pyruvate. Probes of substrate binding to pyruvate oxidase and other thiamin pyrophosphate-dependent decarboxylases. Biochim Biophys Acta. 1980;613(1):10–17. doi: 10.1016/0005-2744(80)90186-2. [DOI] [PubMed] [Google Scholar]
- 16.Brammer LA, Smith JM, Wade H, Meyers CF. 1-Deoxy-D-xylulose 5-phosphate synthase catalyzes a novel random sequential mechanism. J Biol Chem. 2011;286(42):36522–36531. doi: 10.1074/jbc.M111.259747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith JM, Vierling RJ, Meyers CF. Selective inhibition of E. coli 1-deoxy-D-xylulose-5-phosphate synthase by acetylphosphonates. Medchemcomm. 2012;3:65–67. doi: 10.1039/C1MD00233C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Circello BT, Eliot AC, Lee J-H, van der Donk WA, Metcalf WW. Molecular cloning and heterologous expression of the dehydrophos biosynthetic gene cluster. Chem Biol. 2010;17(4):402–411. doi: 10.1016/j.chembiol.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blodgett JAV, et al. Unusual transformations in the biosynthesis of the antibiotic phosphinothricin tripeptide. Nat Chem Biol. 2007;3(8):480–485. doi: 10.1038/nchembio.2007.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eliot AC, et al. Cloning, expression, and biochemical characterization of Streptomyces rubellomurinus genes required for biosynthesis of antimalarial compound FR900098. Chem Biol. 2008;15(8):765–770. doi: 10.1016/j.chembiol.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shao Z, et al. Biosynthesis of 2-hydroxyethylphosphonate, an unexpected intermediate common to multiple phosphonate biosynthetic pathways. J Biol Chem. 2008;283(34):23161–23168. doi: 10.1074/jbc.M801788200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee J-H, et al. Characterization and structure of DhpI, a phosphonate O-methyltransferase involved in dehydrophos biosynthesis. Proc Natl Acad Sci USA. 2010;107(41):17557–17562. doi: 10.1073/pnas.1006848107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neuhaus FC, Korkes S. Phosphoserine. Biochem Prep. 1958;6:76–79. [Google Scholar]
- 24.Rost B, Yachdav G, Liu J. The PredictProtein server. Nucleic Acids Res. 2004;32(Web Server issue suppl 2):W321-6. doi: 10.1093/nar/gkh377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goonesekere NCW, Shipely K, O’Connor K. The challenge of annotating protein sequences: The tale of eight domains of unknown function in Pfam. Comput Biol Chem. 2010;34(3):210–214. doi: 10.1016/j.compbiolchem.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 26.Hegde SS, Shrader TE. FemABX family members are novel nonribosomal peptidyltransferases and important pathogen-specific drug targets. J Biol Chem. 2001;276(10):6998–7003. doi: 10.1074/jbc.M008591200. [DOI] [PubMed] [Google Scholar]
- 27.Rohrer S, Berger-Bächi B. FemABX peptidyl transferases: A link between branched-chain cell wall peptide formation and beta-lactam resistance in gram-positive cocci. Antimicrob Agents Chemother. 2003;47(3):837–846. doi: 10.1128/AAC.47.3.837-846.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tasaki T, Sriram SM, Park KS, Kwon YT. The N-end rule pathway. Annu Rev Biochem. 2012;81:261–289. doi: 10.1146/annurev-biochem-051710-093308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kelley LA, Sternberg MJE. Protein structure prediction on the Web: A case study using the Phyre server. Nat Protoc. 2009;4(3):363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 30.Splan KE, Musier-Forsyth K, Boniecki MT, Martinis SA. In vitro assays for the determination of aminoacyl-tRNA synthetase editing activity. Methods. 2008;44(2):119–128. doi: 10.1016/j.ymeth.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lloyd AJ, et al. Characterization of tRNA-dependent peptide bond formation by MurM in the synthesis of Streptococcus pneumoniae peptidoglycan. J Biol Chem. 2008;283(10):6402–6417. doi: 10.1074/jbc.M708105200. [DOI] [PubMed] [Google Scholar]
- 32.Jia S, et al. A fully active catalytic domain of bovine aspartyl (asparaginyl) beta-hydroxylase expressed in Escherichia coli: Characterization and evidence for the identification of an active-site region in vertebrate alpha-ketoglutarate-dependent dioxygenases. Proc Natl Acad Sci USA. 1994;91(15):7227–7231. doi: 10.1073/pnas.91.15.7227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Knerr PJ, van der Donk WA. Discovery, biosynthesis, and engineering of lantipeptides. Annu Rev Biochem. 2012;81:479–505. doi: 10.1146/annurev-biochem-060110-113521. [DOI] [PubMed] [Google Scholar]
- 34.Garg RP, Alemany LB, Moran S, Parry RJ. Identification, characterization, and bioconversion of a new intermediate in valanimycin biosynthesis. J Am Chem Soc. 2009;131(28):9608–9609. doi: 10.1021/ja901243p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Circello BT. 2010. Investigations into the biosynthesis and mode of action of the phosphonate antibiotic dehydrophos. PhD thesis (University of Illinois at Urbana–Champaign, Urbana, IL)
- 36.Garg RP, Qian XL, Alemany LB, Moran S, Parry RJ. Investigations of valanimycin biosynthesis: Elucidation of the role of seryl-tRNA. Proc Natl Acad Sci USA. 2008;105(18):6543–6547. doi: 10.1073/pnas.0708957105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang W, Ntai I, Kelleher NL, Walsh CT. tRNA-dependent peptide bond formation by the transferase PacB in biosynthesis of the pacidamycin group of pentapeptidyl nucleoside antibiotics. Proc Natl Acad Sci USA. 2011;108(30):12249–12253. doi: 10.1073/pnas.1109539108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Belin P, et al. The nonribosomal synthesis of diketopiperazines in tRNA-dependent cyclodipeptide synthase pathways. Nat Prod Rep. 2012;29(9):961–979. doi: 10.1039/c2np20010d. [DOI] [PubMed] [Google Scholar]
- 39.Bonnefond L, et al. Structural basis for nonribosomal peptide synthesis by an aminoacyl-tRNA synthetase paralog. Proc Natl Acad Sci USA. 2011;108(10):3912–3917. doi: 10.1073/pnas.1019480108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Francklyn CS, Minajigi A. tRNA as an active chemical scaffold for diverse chemical transformations. FEBS Lett. 2010;584(2):366–375. doi: 10.1016/j.febslet.2009.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Phizicky EM, Hopper AK. tRNA biology charges to the front. Genes Dev. 2010;24(17):1832–1860. doi: 10.1101/gad.1956510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Solomon EI, Decker A, Lehnert N. Non-heme iron enzymes: Contrasts to heme catalysis. Proc Natl Acad Sci USA. 2003;100(7):3589–3594. doi: 10.1073/pnas.0336792100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou J, et al. Spectroscopic studies of substrate interactions with clavaminate synthase 2, a multifunctional alpha-KG-dependent non-heme iron enzyme: Correlation with mechanisms and reactivities. J Am Chem Soc. 2001;123(30):7388–7398. doi: 10.1021/ja004025+. [DOI] [PubMed] [Google Scholar]
- 44.Hausinger RP. FeII/α-ketoglutarate-dependent hydroxylases and related enzymes. Crit Rev Biochem Mol Biol. 2004;39(1):21–68. doi: 10.1080/10409230490440541. [DOI] [PubMed] [Google Scholar]
- 45.Yang M, et al. Substrate selectivity analyses of factor inhibiting hypoxia-inducible factor. Angew Chem Int Ed Engl. 2013;52(6):1700–1704. doi: 10.1002/anie.201208046. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.