Abstract
Many fungi restructured their proteomes through incorporation of serine (Ser) at thousands of protein sites coded by the leucine (Leu) CUG codon. How these fungi survived this potentially lethal genetic code alteration and its relevance for their biology are not understood. Interestingly, the human pathogen Candida albicans maintains variable Ser and Leu incorporation levels at CUG sites, suggesting that this atypical codon assignment flexibility provided an effective mechanism to alter the genetic code. To test this hypothesis, we have engineered C. albicans strains to misincorporate increasing levels of Leu at protein CUG sites. Tolerance to the misincorporations was very high, and one strain accommodated the complete reversion of CUG identity from Ser back to Leu. Increasing levels of Leu misincorporation decreased growth rate, but production of phenotypic diversity on a phenotypic array probing various metabolic networks, drug resistance, and host immune cell responses was impressive. Genome resequencing revealed an increasing number of genotype changes at polymorphic sites compared with the control strain, and 80% of Leu misincorporation resulted in complete loss of heterozygosity in a large region of chromosome V. The data unveil unanticipated links between gene translational fidelity, proteome instability and variability, genome diversification, and adaptive phenotypic diversity. They also explain the high heterozygosity of the C. albicans genome and open the door to produce microorganisms with genetic code alterations for basic and applied research.
Keywords: codon reassignment, evolution, tRNA
Natural alterations to the standard genetic code have been discovered in Mycoplasma (1, 2), Micrococci (3), ciliates (4), fungi (5, 6), and mitochondria (7), modifying the hypothesis of a universal genetic code (8). Both neutral (9) and nonneutral theories (10) have been proposed to explain codon reassignments; however, experimental data to support or refute them are scarce, and genetic code alterations remain an intriguing biological puzzle. Despite this fact, it is becoming clear that genetic code alterations are associated with mutations in tRNAs and translation release factors that expand or restrict codon decoding capacity (7). In other words, alterations of translational factors have the potential to release the genetic code from its frozen state. This hypothesis is strongly supported by the widespread cotranslational incorporation of selenocysteine into the active site of selenoprotein (11) and pyrrolysine in the active site of the methyltransferases of several Metanosarcina species (12), Desulfitobacterium hafniense (13), and the gutless worm Olavius algarvensis (14). The selective advantages produced by these two amino acids are associated with evolution of proteins with unique catalytic properties.
The flexibility of the genetic code is further highlighted by the in vivo incorporation of artificial amino acids into recombinant proteins of Escherichia coli, yeast, and mammalian cells using orthogonal pairs of tRNAs and aminoacyl-tRNA synthetases (aaRSs) (15, 16). These tRNA-aaRS pairs recognize, activate, and incorporate fluorescent and other amino acid analogs into proteins in response to reprogrammed stop codons (17). The UAG stop codon was also reassigned in a modified E. coli strain, where it was selectively replaced by the UAA stop codon in seven essential genes (coaD, hda, hemA, mreC, murF, lolA, and lpxK), and the release factor 1 (RF-1) was mutated to abolish recognition of the UAG codon (18). However, these codon reassignments decreased fitness, and the evolution of genetic code alterations has been very difficult to rationalize under such negative selection.
Fungi of the Candida, Debaryomyces, and Lodderomyces genera (CTG clade) (19) reassigned the CUG codon from leucine (Leu) to serine (Ser) through CUG decoding ambiguity (20, 21) and are good experimental model systems to investigate sense codon reassignments (22). Remarkably, these fungi still incorporate Ser (∼97%) and Leu (∼3%) at thousands of CUG sites present in over 60% of their genes (23). Protein X-ray crystallography and molecular modeling showed that the Leu and Ser residues encoded by CUGs are partially exposed to the solvent or on the interface of subunits of protein complexes where they can be better tolerated (24). These structural data explain the high tolerance to CUG ambiguity (23) and are fundamental to rationalize the evolution of CUG reassignment in the fungal CTG clade. However, the dynamic range of Leu misincorporation at CUGs and the selective advantages that it spawns are not fully understood. Here, we push CUG ambiguity to its limit to show that codon ambiguity provides an effective mechanism to alter the genetic code.
Results
Reversion of CUG Identity from Ser Back to Leu.
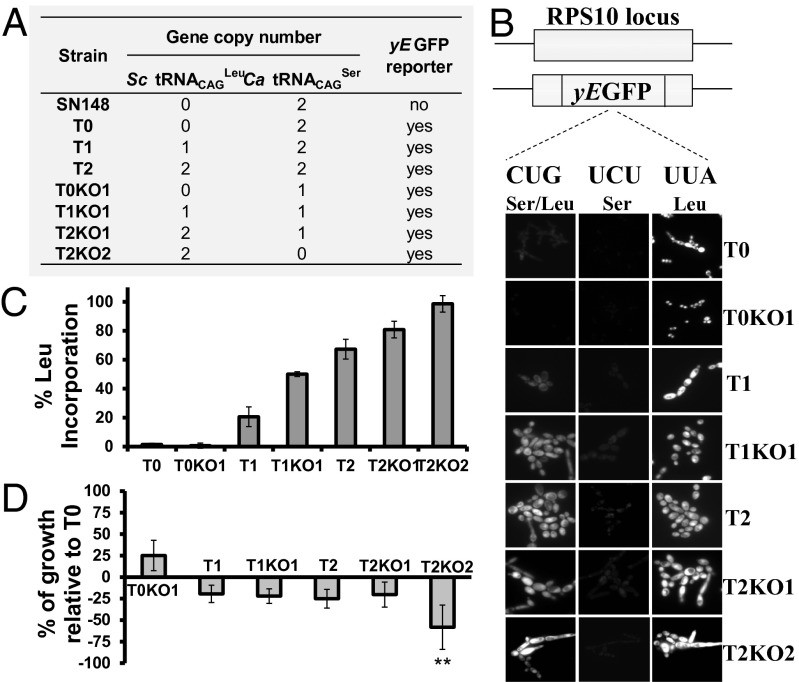
To increase Leu misincorporation at CUG sites, we have inserted one (strain T1) or two (strain T2) copies of a yeast Leu tDNACAGLeu gene (25) into the RPS10 genome locus of the Candida albicans SN148 strain (26). This heterologous tRNACAGLeu misincorporates Leu only at the atypical C. albicans Ser CUGs (23, 25). We also knocked out one or two copies of the chromosomal C. albicans Ser tRNACAGSer gene in strains T1 and T2, producing strains T1KO1 and T2KO1 or T2KO2 (Fig. 1A, Figs. S1, S2, and S3, and Dataset S1, Tables S1–S3). Leu misincorporation at CUG sites was monitored in each strain using a gain-of-function reporter system that expresses active GFP if Leu is incorporated at codon 201 (Leu201) and inactive GFP if Ser is incorporated at the same site (Ser201) (Fig. 1B and Fig. S4). Western blot analysis showed that Ser incorporation at residue 201 destabilizes GFP, leading to its rapid degradation (Fig. S4). Quantification of fluorescence in each of the constructed strains showed relative increase of Leu misincorporation from 1.45% in strain T0 up to 98.46% in strain T2KO2 (Fig. 1C and Dataset S1, Table S3). Surprisingly, a C. albicans strain harboring a KO in one copy of the Ser tRNACAGSer gene (strain T0KO1) produced a lower level of fluorescence than the control strain (T0), corresponding to relative Leu misincorporation levels of 0.6% (Fig. 1 A–C). This result suggested that the cellular abundance of this tRNACAGSer is a determinant of CUG ambiguity levels. Transformation efficiency of strains T1, T2, T1KO1, and T2KO1 was lower than the T0 control, and growth rate decreased ∼20% (Fig. 1D and Fig. S5). Interestingly, the T0KO1 strain (0.6% Leu) grew faster (∼25%) than the control (T0) strain (Fig. 1D), indicating that the natural ambiguity is detrimental to C. albicans fitness in rich medium. The transformation efficiency of the reverted T2KO2 strain was low, and growth rate was strongly affected (∼60%) (Fig. 1D); however, similarly to the other strains, it adapted readily to the genetic code alteration and recovered viability to nearby WT levels (Fig. S5).
Fig. 1.
Construction of highly mistranslating and C. albicans reverted strains. (A) Table depicting the main characteristics of the mistranslating strains. (B) The reporter system is based on the yEGFP gene. The Leu UUA codon at position 201 (positive control) was mutated to a UCU Ser codon (negative control) and the ambiguous CUG codon. Incorporation of Ser at this position inactivates yEGFP, whereas Leu incorporation produces functional yEGFP. Fluorescence measured by epifluorescence microscopy is directly proportional to the level of Leu inserted at the CUG-201 site. (C) In T0, T1, and T2 cells, ambiguity is 1.45 ± 0.85%, 20.61 ± 1.81%, and 67.29 ± 6.83%, respectively. Strains T0KO1, T1KO1, and T2KO1 show 0.64 ± 0.82%, 50.04 ± 1.64%, and 80.84 ± 5.79% of Leu incorporation, whereas KO cells (T2KO2) incorporate 98.46 ± 1.75% of Leu. Data represent the mean ± SD of five independent experiments. (D) Growth rate analysis of mistranslating strains. Data represent the mean ± SD of triplicates of three independent clones. **P < 0.01, one-way ANOVA post-Dunnett comparison test with 95% confidence interval relative to T0 cells.
Phenomics of Mistranslating and Reverted Strains.
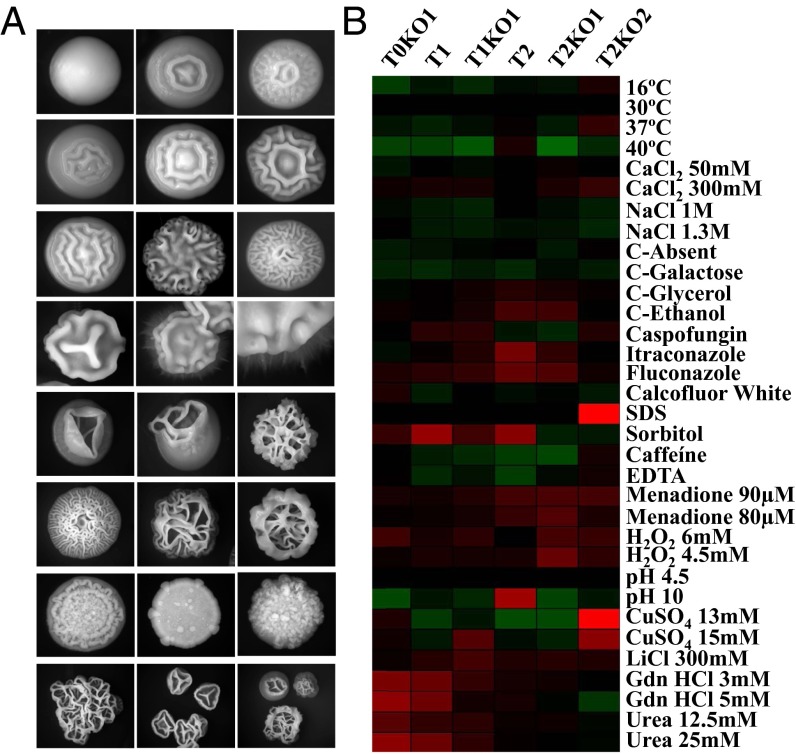
Strains with GFP fluorescence that indicated >50% of Leu misincorporation produced colonies with highly variable morphologies on agar plates, variable cell sizes, and differentiation heterogeneity in liquid media (Fig. 2A). Colony and cell morphology variation increased with increasing levels of Leu misincorporation, and the T2KO2 reverted strain produced morphotypes unrelated to those morphotypes previously observed in the control strain, which produced smooth and ring colonies only. The smooth phenotype was characterized by dome-shaped colonies consisting of yeast cells, and the ring phenotype had a wrinkled center with a smooth periphery (27). Clones T0KO1 and T1 produced a small proportion of regular-wrinkled colonies (Fig. 2A), but strains with GFP fluorescence that corresponded to >50% of Leu misincorporation produced colonies with irregular-wrinkled and jagged morphologies (Fig. 2A). These phenotypes were characterized by heavily rippled colonies composed of a mixture of yeast, pseudohyphal, and hyphal cells (27). Colonies obtained in this primary screen were replated onto fresh agar plates to assess phenotypic stability, because switching among these different phenotypes is associated with the capacity of cells within colonies to undergo the yeast-to-hypha transition (28). The strains with lower levels of GFP fluorescence (<50% of Leu misincorporation at CUGs), namely T0, T0KO1, and T1, switched from smooth to ring at high frequency, but switching between smooth and regular-wrinkled was less frequent. The strains with higher levels of GFP fluorescence (>50% of Leu misincorporation: T1KO1, T1KO1, and T2KO2) switched frequently between wrinkled and jagged morphologies, but switching to the smooth phenotype was not observed. Therefore, increased CUG ambiguity increases morphological plasticity beyond the levels already described in the literature (29), indicating that C. albicans phenotypic variation is more extensive and flexible than previously reported.
Fig. 2.
Phenotypic diversity produced by mistranslating C. albicans strains. (A) The repertoire of colony morphology phenotypes includes smooth, ring, wrinkled, and hyphae. KO mutants do not show the smooth or ring phenotypes, and colonies consist primarily of pseudohyphal and hyphal cell types. (B) A phenotypic screen carried out in stress media is shown on the panel where growth scores are represented by the color of the indicated square. Black represents a phenotype that is indistinguishable from the control (T0); green and red represent a reduction and enhancement phenotype, respectively. The growth score represents a ratio between growth in normal 2% glucose, 1% yeast extract, and 1% peptone (YPD) medium and growth in YPD supplemented with stressors. Triplicates of three independent clones from each strain were tested and compared with the T0 control cells.
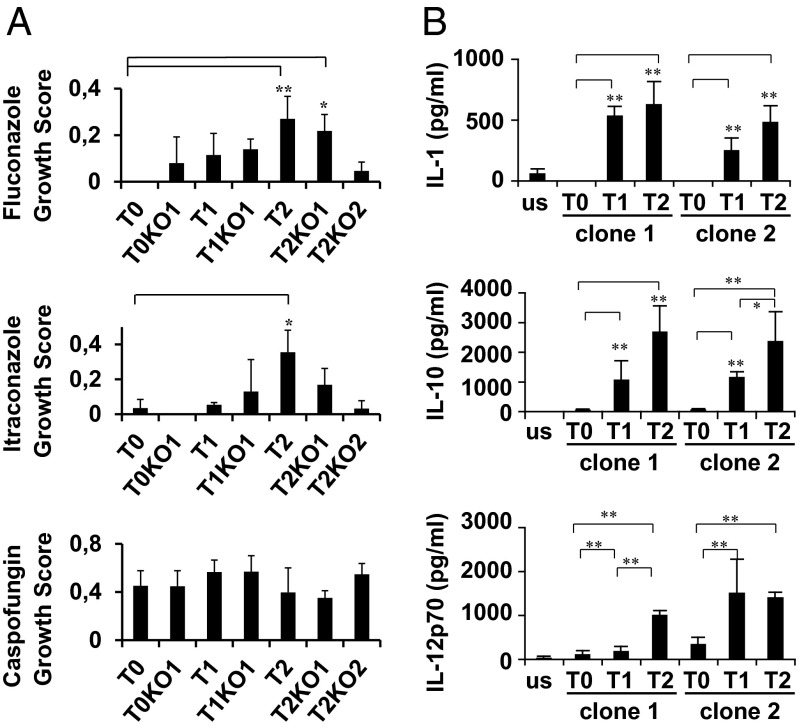
We have also compared the growth characteristics of the misincorporating strains in more than 30 phenotypic assays, including growth at different temperatures, pH, and carbon sources and in the presence of salts, metals, antifungal drugs, and inhibitors of diverse cellular processes (Dataset S1, Table S4). Midlog phase cells were serially diluted and spotted onto test plates, where colony growth was assessed and normalized relative to the control T0. Although the changes in growth were idiosyncratic, with each strain exhibiting a unique pattern of phenotypes, there were clear growth advantages under many conditions (Fig. 2B). For example, all misincorporating strains grew faster than the control in the presence of the oxidative stressors menadione and H2O2, particularly the strains T2, T2KO1, and T2KO2 (Fig. 2B). Strains T0KO1 and T1 grew faster than the control in media supplemented with protein misfolding agents (urea and guanidine hydrochloride) (Fig. 2B). However, all mistranslating strains grew slower than the control in the presence of salts, caffeine, and EDTA and also at 37 °C and 40 °C (Fig. 2B). The most surprising result was the resistance of the T2K02 strain to SDS and CuSO4 (Fig. 2B and Dataset S1, Table S5), which could indicate major alterations in the cell wall of this strain. The behavior of the misincorporating strains in the presence of the clinical antifungals fluconazole, itraconazole, and caspofungin is also noteworthy. Strains T2 and T2KO1 grew faster than the control strain in the presence of fluconazole and itraconazole but poorly in the presence of caspofungin (Fig. 3A), indicating that mistranslation may be linked with tolerance against azoles but not against echinocandins.
Fig. 3.
Mistranslation modulates antifungal and immune responses. (A) Growth of C. albicans strains in YPD medium supplemented with antifungals. The growth score represents a ratio between growth in normal YPD medium and growth in YPD supplemented with fluconazole, itraconazole, and caspofungin, respectively. Data represent the mean ± SD of triplicates of three independent clones (**P < 0.01, *P < 0.1, one-way ANOVA post-Dunnett comparison test with 95% confidence interval relative to the T0 control cells). (B) In vitro immune reactivity to C. albicans mistranslating strains. Monocyte-derived DCs were exposed for 24 h to C. albicans cells or no stimuli (us in the graph) in a concentration (expressed as stimuli:DC ratio) of 5:1. Cytokine production in supernatants was evaluated by Milliplex technology. Data represent the mean ± SD (n = 6). Statistical comparisons were performed using a Kruskal–Wallis test with a posthoc paired comparison. Differences between samples are represented as horizontal bars. *P < 0.05, **P < 0.01.
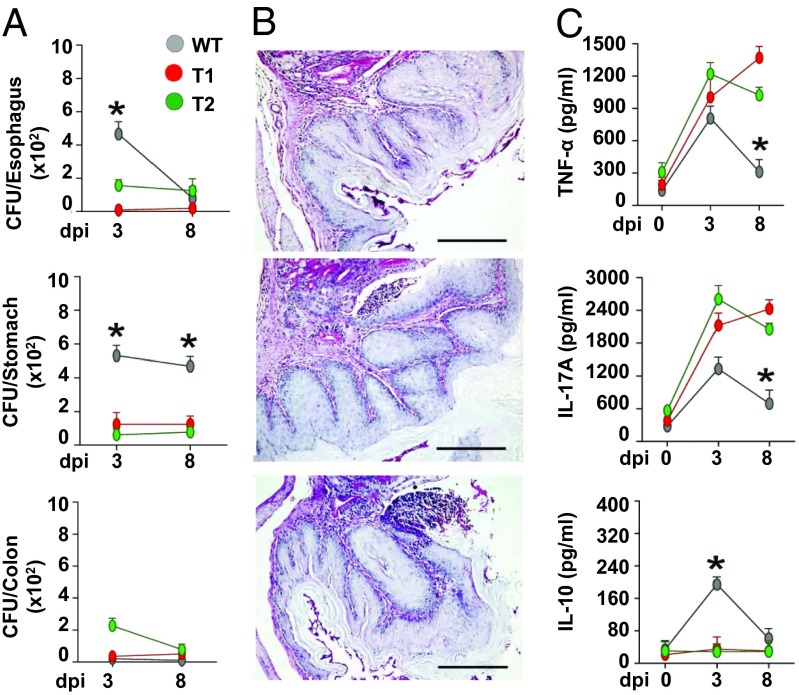
The remarkable cell and colony morphotypes observed (Fig. 2) likely reflect changes in the cell wall influencing fungal recognition by human immune cells. Indeed, part of the fungal cell wall is decorated with various pathogen-associated molecular patterns, which are the main target for recognition by host innate immune cells (30). To test this hypothesis, we have exposed T1 and T2 strains to human monocyte-derived dendritic cells (DCs). There was increased in vitro release of the inflammatory ILs IL-1β, IL-10, and IL-12p70 by human DCs (Fig. 3B), suggesting that CUG ambiguity modulates the immune responses to the fungus. To assess the pathogenic potential of the mistranslating strains in vivo, C57BL/6 mice were inoculated intragastrically with WT, T1, and T2 strains. Despite higher fungal growth in mice infected with the WT strain (Fig. 4A), colonization by the T1 and T2 misincorporating strains was associated with a noticeable inflammatory pathology in the stomach of infected mice, which was indicated by the increased mucin content of goblet cells and the number of infiltrating cells (Fig. 4B). The levels of the proinflammatory cytokines TNFα and IL-17A were also higher, and the antiinflammatory IL-10 levels were lower relative to the control strain (Fig. 4C), indicating that high inflammatory responses are likely elicited by dynamic Leu misincorporation at CUGs.
Fig. 4.
In vivo immune responses to C. albicans mistranslating strains. (A) C57BL/6 mice were infected intragastrically with control T0, T1, or T2 strains (six mice/group). Fungal growth (mean log10 CFU ± SE, n = 3) in the stomach, exophagus, and colon of infected mice was assessed at different days postinfection (dpi). (B) Stomach histology (periodic acid–Schiff staining) and mononuclear or polymorphonuclear cells staining were done at 3 dpi. Representative images of two independent experiments were depicted; bars indicate magnifications. (C) Stomach homogenates at 3 dpi time points were tested for levels of TNFα, IL-17A, and IL-10 by specific ELISA (mean values ± SD, n = 3). *P ≤ 0.01, WT strain.
CUG Ambiguity Is Associated with Genome Diversification.
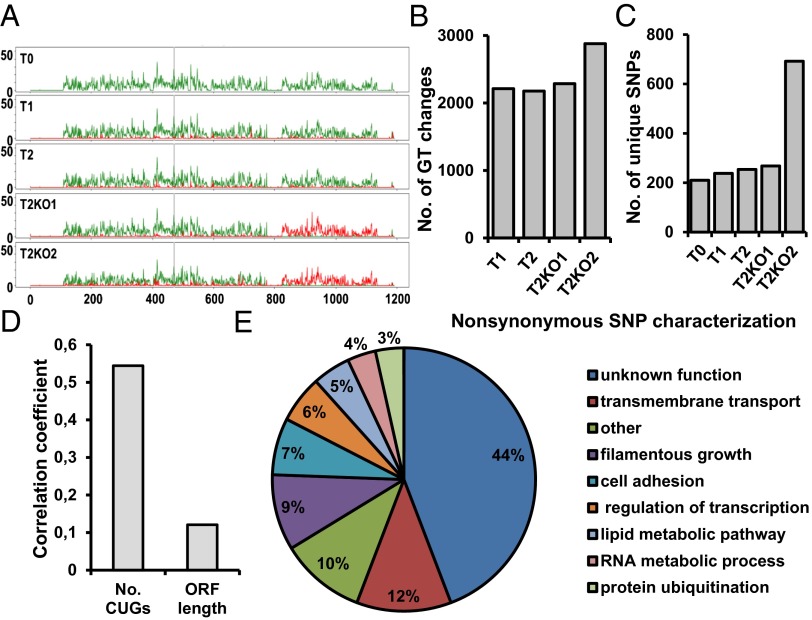
To clarify the molecular nature of the observed phenotypes, we have carried out an extensive analysis of the genome of the control and misincorporating strains. Genome sequencing showed that increasing CUG ambiguity leads to rapid genome evolution through mutation and loss of heterozygosity (LOH) relative to the T0 control. Although read depth was relatively uniform across the genome in all strains (Fig. S6 and Dataset S1, Tables S7 and S8), there was a near-complete LOH in a 300-kb region on chromosome V (825,000–1,136,000) (Fig. 5A) in strains T2KO1 and T2KO2 and the entire chromosome R in T2KO1 (Fig. S7 and Dataset S1, Tables S10–S12). The affected region on chromosome V contains 178 ORFs (24, 31). Most of these ORFs have no known function, but the known genes are involved in the regulation of biological processes (21.3%), organelle organization (13.5%), stress response (9.6%), antifungal drug resistance (6.2%), filamentous growth (9%), pathogenesis (2.8%), and conjugation (2.2%), which is in line with the phenotypic diversity observed.
Fig. 5.
Genome sequencing of mistranslating and reverted C. albicans strains. (A) Genomic analysis of the control (T0) and misincorporating strains shows LOH in a region of chromosome 5 in T2KO1 and T2KO2. SNPs per kilobase are shaded green; density of LOH SNPs is in red, and gray vertical lines indicate the major repeat sequence. (B) Discounting LOH regions, the number of genotype (GT) changes compared with the control strain increases with the degree of mistranslation in the four altered strains. (C) The number of heterozygote SNPs unique to a specific strain increases as the degree of amino acid misincorporation increases. (D) Genes with a higher number of CUGs accumulated higher levels of unique nonsynonymous mutations in strain T2KO2. Pearson correlation was used as a measure of relationship between the number of nonsynonymous mutations and the number of CUG codons. This correlation was stronger than the relation between ORF length and nonsynonymous genotype changes. (E) Gene ontology process terms for the unique genes containing nonsynonymous genotype changes identified in T2KO2 compared with control T0. The graph represents the percentage of genes from the total of 86 containing nonsynonymous mutations.
In the five sequenced strains, we have observed between 45,297 and 55,429 SNPs from a unique set of 61,312 locations (Dataset S1, Table S13). In the part of the genome not affected by LOH, we have observed an increasing number of heterozygote SNPs for strains with increasing Ser-for-Leu misincorporation (Fig. 5B). Of 49,922 SNPs observed outside the regions affected by LOH in any of the strains, each strain is a heterozygote between 44,660 and 46,250 of these positions, with the fewest SNPs in the control strain and the most SNPs in T2KO2 (Fig. S8A and Dataset S1, Table S13). Across the polymorphic locations outside LOH regions, T2KO2 changed genotype compared with the T0 control at 2,879 SNPs, nearly 600 SNPs more than the other three mistranslating strains (Fig. S8B and Dataset S1, Table S14). Interestingly, the T2KO2 strain has 692 unique SNPs not found in any other strain (nearly three times as many as the other strains) (Fig. 5C). A number of genes with important biological functions accumulated more than just a single new nonsynonymous SNP per gene in the T2KO2 strain. For example, the filamentous growth regulators (FGRs) FGR23, FGR6-4, and FGR6-1 had more than three mutations, and members of the ALS family of cell surface glycoproteins, namely ALS4 and ALS9, had two nonsynonymous mutations (Dataset S1, Table S14). The increased rate of genotype changes from T0 control is accumulated in coding regions at the same rate for all strains (between 31.9% and 34.6% of the SNPs), and analogously, there was no shift in nonsynonymous mutation rate (13.6–14.1%) in any of the strains (Dataset S1, Table S14). In C. albicans, 63.0% of the genome constitutes coding regions (19, 32), and the lower proportion of SNPs in those coding regions expected by a uniform random rate can be explained by the increased risk for a functionally disruptive mutation in those regions. We did not observe any shift in codon use between the strains (between 12,514 and 12,576 CTG codons detected) (Dataset S1, Tables S15 and S16), but genes with a higher number of CUGs accumulated higher levels of unique SNPs in strain T2KO2 (Fig. 5D), which is consistent with the higher rate of genotype changes in the most strongly misincorporating strain. These changes accumulated mostly in genes with unknown functions (44%), filamentous growth (9%), cell adhesion (7%), metabolic processes (9%), and transcription regulation (6%) (Fig. 5E). This result may indicate that the increased degree of mistranslation produces a strong pressure on the fungal population to genotype changes at polymorphic locations in genes conferring survivability in the altered proteome landscape, where most proteins have Ser replaced by Leu.
Discussion
Evolutionary Implications.
We have altered the identity of a sense codon of an organism. This alteration is a significant shift from previous studies, where stop codons were partially reassigned to both canonical and artificial amino acids (15, 18). Our data show that the genetic code is evolvable and that codon ambiguity provides the mechanism for such evolvability. The data presented here and previous discoveries showing that codon ambiguity allows for growth of ambiguous cells in ecological niches, where WT cells are not able to grow or grow poorly (33, 34), show unequivocally that the proteome disruption caused by codon ambiguity is not an impediment to the evolution of genetic code alterations, providing that the phenotypic diversity produced by such ambiguity allows the organism to explore available ecological landscapes. These data are in line and strongly support genetic code theories postulating that ambiguous codons played fundamental roles in the evolution of the genetic code (10, 35, 36), particularly during the initial stages of the code development when amino acids were gradually assigned to codons (36) and during the late evolution of alterations to the standard genetic code (35). Because natural codon ambiguities produced through defects in the editing domain of aaRSs have also been discovered in various species of Mycoplasma (31) and because E. coli, yeast, and mammalian cells are highly tolerant to such codon ambiguities (20, 34, 37), we postulate that they are more common in nature than expected. We also postulate that such ambiguities can be used to alter the genetic code for evolutionary biology, synthetic biology, biotechnological, and biomedical applications.
Insights for C. albicans Biology.
C. albicans has a parasexual lifecycle but reproduces mainly mitotically (19, 38). It also has one of the most polymorphic genomes sequenced to date, with >48,000 SNPs in strain SC5314 and >41,000 SNPs in strain WO1, giving 1 SNP per 330 and 390 bases, respectively (19, 32, 39). These numbers match our sequencing-based predictions for SN148 and the four derived mistranslating strains well. Our data show that codon ambiguity increases the rate of genotype changes compared with the control strain at polymorphic locations likely to suppress or minimize protein structural instability generated by amino acid misincorporations. The selection of alternative amino acids at nonambiguous sites may retune protein–protein, protein–RNA, and protein–DNA interactions and networks destabilized by misincorporations. This result is consistent with known effects of aminoglycosidic antibiotics, mutant tRNAs, and editing defective aminoacyl-tRNA synthetases on mutation rate in bacteria (40) and genome destabilization induced by codon ambiguity in yeast (25).
The remarkable diversity and plasticity of the phenotypes produced by CUG ambiguity complicate significantly the clarification of how they are produced. Nevertheless, our discovery that ambiguous cells are tolerant to commonly used antifungals, namely fluconazole and itraconazole, indicates that CUG ambiguity is relevant to evolution of antifungal drug resistance, particularly in the case of the azoles. C. albicans drug tolerance involves mutations in antifungal target genes that block drug binding (41, 42) and up-regulation of multidrug transporters that remove the drug from the cell (43, 44). For example, mutations in the lanosterol 14-alpha-demethylase gene (ERG11) are associated with resistance to azoles (41, 42), and up-regulation of genes from the major facilitator superfamily of transporters (MDR1) (43) and the ATP binding cassette transporter superfamily (CDR1 and CDR2) are associated with general drug resistance (44). Our genome sequencing data did not show accumulation of mutations in ergosterol pathway genes, suggesting that mistranslation-induced drug tolerance is independent of mutations in known drug target genes. Beyond the above mechanisms, metabolic alterations, activation of chaperones, and signal transduction cascades that sense and respond to the stress induced by antifungal drugs also contribute to drug tolerance (45). Indeed, the signaling regulator Ca2+-calmodulin–activated protein phosphatase calcineurin, which is a heterodimeric phosphatase composed of catalytic (CNA1) and regulatory (encoded by CNB1) subunits, is activated by external stimuli. Membrane stress activates this pathway by increasing the levels of intracellular Ca2+ through either its release from intracellular stores or an increased influx of calcium ions from the extracellular environment (46). One of the main targets of this phosphatase is the transcription factor Crz1 that relocalizes to the nucleus upon dephosphorylation and activates expression of genes involved in cell wall-related functions (46, 47). Our data are insufficient to clarify the direct involvement of this pathway in the observed resistance to the azoles; however, the near-complete LOH on chromosome V of the highly azole-tolerant strains (T2KO1 and T2KO2), which affected the regulatory subunit of calcineurin (CNB1) and the calcium channel (MID1), supports the hypothesis that such tolerance may be mediated through the calcineurin pathway. Furthermore, genes associated with the plasma membrane and cell wall processes are enriched in CUGs (19, 24, 48), suggesting that increased Leu misincorporation levels may have a stronger impact on the activation of the calcineurin pathway than on other known drug resistance pathways.
The genome sequencing data also showed that unique SNPs accumulated mostly in genes related to filamentous growth and cell adhesion (Fig. 5E), which are important virulence traits. Members of the ALS adhesion family of cell surface glycoproteins, namely ALS4 and ALS9, and various genes involved in regulation of filamentous growth, including FGR23, FGR47, FGR6-10, and FGR6-1, contained SNPs unique to the most mistranslating strain (Dataset S1, Table S14), indicating that CUG ambiguity diversifies the genes involved in pathogenesis-related processes. The different inflammatory behavior of our mistranslating strains observed both in vitro and in vivo suggests that mistranslation may induce phenotypic changes that determine the way the fungus is recognized and cleared by the immune system. The increased resistance to oxidative stress of mistranslating strains could negatively influence killing by phagocytes, determining high fungal burden at the sites of infection and stronger inflammatory responses.
Conclusions
Genetic code ambiguity provides a powerful mechanism to alter the genetic code using experimental evolution. Our data show that mutations introduced directly into proteins during mRNA decoding by the ribosome flow back to DNA as compensatory mutations and mitigate proteome destabilization and loss of proteostasis. The data provide strong coevolutionary links between the genetic code and the genome and add dimensions to the study of genome evolution, phenotypic variation, and ecological adaptation by showing unanticipated power of codon ambiguity in shaping gene evolution. Our data are too preliminary to establish direct links between CUG ambiguity and C. albicans pathogenesis; however, the phenotypes uncovered support the hypothesis that CUG ambiguity modulates immune responses and accelerates the evolution of drug resistance. It will be fascinating to identify gain-of-function mutations and the molecular mechanisms that spawn C. albicans pathogenesis, drug resistance, and ecological adaptation phenotypes.
Methods
The experimental procedures are briefly described here, and a detailed description is provided in SI Text. C. albicans strain SN148 (26) was used to construct the highly ambiguous strains using homologous recombination methodologies. For this construction, tRNA genes and the yeast-enhanced GFP (yEGFP) reporter gene were cloned into the integration vector CIp10 (49), and linearized DNA was integrated into the RPS10 locus of the C. albicans SN148 genome as described in SI Text (Figs. S1, S2, S3, and S4). yEGFP fluorescence was measured using a Zeiss MC80 Axioplan 2 microscope, and images were analyzed using ImageJ software. At least 1,000 cells of each strain were analyzed in each case. For whole-genome sequencing, genomic DNA was prepared using the Genomic-tip 100/G kit (Qiagen), and one paired-end library was prepared for each sample using Illumina DNA sample prep protocols. Libraries were sequenced using the Illumina Genome Analyzer IIx. Reads were mapped to the reference C. albicans genome (www.candidagenome.org) using BWA (version 0.5.9); processing, filtering, and SNP detection/genotype calling were carried out using Samtools (version 0.1.17) (50, 51). Sequencing data were archived in the European Nucleotide Archive under accession no. E-SYBR-7. Phenotypic assays were carried out on solid agar plates supplemented with stressors with the help of a robot equipped with a 96-pin bolt replicator (Caliper). Colony growth was monitored using an AxioCam HRc camera and Axio Vision Software (Zeiss). Images were processed using ImageJ software. Human monocyte-derived DC responses to the mistranslated strains were scored to assess cytokines levels on challenges using the Milliplex technology. The experimental plan was approved by the local Ethical Committee of Azienda Universitaria Ospedaliera Careggi (Careggi Hospital, Florence, Italy), and written informed consent was obtained from all donors (approval document no. 87/10). The study conforms to the ethical guidelines of the 1975 Declaration of Helsinki, the international recommendation Dir. EU 2001/20/EC, and its Italian counterpart (DM 15 Luglio 1997; D.Lvo 211/2003; D.L.vo 200/2007). For infection, female C57BL6 mice were injected with a suspension of 2 × 107 viable conidia/20 μL saline intragastrically. Mice were monitored for fungal growth in esophagus, stomach, and colon (CFU/organ, mean ± SE), histopathology (Periodic acid–Schiff staining of sections of paraffin-embedded tissues), and patterns of cytokine production. Total cell counts were done by staining with May–Grünwald Giemsa reagents (Sigma) before analysis. Photographs were taken using a high-resolution microscopy Olympus DP71 (Olympus).
Supplementary Material
Acknowledgments
We thank Alexander Johnson for providing the C. albicans strains and plasmids and Judith Berman, Csaba Pál, and Dieter Söll for their useful comments and suggestions on the manuscript. The study was funded by the European Union Framework Program 7 (EU-FP7) Sybaris Consortium Project 242220 and the Portuguese Science Foundation through Fundo Europeu de Desenvolvimento Regional (FEDER/FCT) Project PTDC/BIA-MIC/099826/2008.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the European Nucleotide Archive (accession no. E-SYBR-7).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1302094110/-/DCSupplemental.
References
- 1.Yamao F, et al. UGA is read as tryptophan in Mycoplasma capricolum. Proc Natl Acad Sci USA. 1985;82(8):2306–2309. doi: 10.1073/pnas.82.8.2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oba T, Andachi Y, Muto A, Osawa S. CGG: An unassigned or nonsense codon in Mycoplasma capricolum. Proc Natl Acad Sci USA. 1991;88(3):921–925. doi: 10.1073/pnas.88.3.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ohama T, Muto A, Osawa S. Role of GC-biased mutation pressure on synonymous codon choice in Micrococcus luteus, a bacterium with a high genomic GC-content. Nucleic Acids Res. 1990;18(6):1565–1569. doi: 10.1093/nar/18.6.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meyer F, et al. UGA is translated as cysteine in pheromone 3 of Euplotes octocarinatus. Proc Natl Acad Sci USA. 1991;88(9):3758–3761. doi: 10.1073/pnas.88.9.3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ohama T, et al. Non-universal decoding of the leucine codon CUG in several Candida species. Nucleic Acids Res. 1993;21(17):4039–4045. doi: 10.1093/nar/21.17.4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Santos MA, Tuite MF. The CUG codon is decoded in vivo as serine and not leucine in Candida albicans. Nucleic Acids Res. 1995;23(9):1481–1486. doi: 10.1093/nar/23.9.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yokobori S, Suzuki T, Watanabe K. Genetic code variations in mitochondria: tRNA as a major determinant of genetic code plasticity. J Mol Evol. 2001;53(4–5):314–326. doi: 10.1007/s002390010221. [DOI] [PubMed] [Google Scholar]
- 8.Crick FH. The origin of the genetic code. J Mol Biol. 1968;38(3):367–379. doi: 10.1016/0022-2836(68)90392-6. [DOI] [PubMed] [Google Scholar]
- 9.Osawa S, Jukes TH. Codon reassignment (codon capture) in evolution. J Mol Evol. 1989;28(4):271–278. doi: 10.1007/BF02103422. [DOI] [PubMed] [Google Scholar]
- 10.Schultz DW, Yarus M. Transfer RNA mutation and the malleability of the genetic code. J Mol Biol. 1994;235(5):1377–1380. doi: 10.1006/jmbi.1994.1094. [DOI] [PubMed] [Google Scholar]
- 11.Böck A, et al. Selenocysteine: The 21st amino acid. Mol Microbiol. 1991;5(3):515–520. doi: 10.1111/j.1365-2958.1991.tb00722.x. [DOI] [PubMed] [Google Scholar]
- 12.Srinivasan G, James CM, Krzycki JA. Pyrrolysine encoded by UAG in Archaea: Charging of a UAG-decoding specialized tRNA. Science. 2002;296(5572):1459–1462. doi: 10.1126/science.1069588. [DOI] [PubMed] [Google Scholar]
- 13.Herring S, Ambrogelly A, Polycarpo CR, Söll D. Recognition of pyrrolysine tRNA by the Desulfitobacterium hafniense pyrrolysyl-tRNA synthetase. Nucleic Acids Res. 2007;35(4):1270–1278. doi: 10.1093/nar/gkl1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y, Gladyshev VN. High content of proteins containing 21st and 22nd amino acids, selenocysteine and pyrrolysine, in a symbiotic deltaproteobacterium of gutless worm Olavius algarvensis. Nucleic Acids Res. 2007;35(15):4952–4963. doi: 10.1093/nar/gkm514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chatterjee A, Xiao H, Schultz PG. Evolution of multiple, mutually orthogonal prolyl-tRNA synthetase/tRNA pairs for unnatural amino acid mutagenesis in Escherichia coli. Proc Natl Acad Sci USA. 2012;109(37):14841–14846. doi: 10.1073/pnas.1212454109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chin JW, et al. An expanded eukaryotic genetic code. Science. 2003;301(5635):964–967. doi: 10.1126/science.1084772. [DOI] [PubMed] [Google Scholar]
- 17.Wang L, Schultz PG. A general approach for the generation of orthogonal tRNAs. Chem Biol. 2001;8(9):883–890. doi: 10.1016/s1074-5521(01)00063-1. [DOI] [PubMed] [Google Scholar]
- 18.Mukai T, et al. Codon reassignment in the Escherichia coli genetic code. Nucleic Acids Res. 2010;38(22):8188–8195. doi: 10.1093/nar/gkq707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Butler G, et al. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature. 2009;459(7247):657–662. doi: 10.1038/nature08064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Santos MA, Cheesman C, Costa V, Moradas-Ferreira P, Tuite MF. Selective advantages created by codon ambiguity allowed for the evolution of an alternative genetic code in Candida spp. Mol Microbiol. 1999;31(3):937–947. doi: 10.1046/j.1365-2958.1999.01233.x. [DOI] [PubMed] [Google Scholar]
- 21.Massey SE, et al. Comparative evolutionary genomics unveils the molecular mechanism of reassignment of the CTG codon in Candida spp. Genome Res. 2003;13(4):544–557. doi: 10.1101/gr.811003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Santos MA, Moura G, Massey SE, Tuite MF. Driving change: The evolution of alternative genetic codes. Trends Genet. 2004;20(2):95–102. doi: 10.1016/j.tig.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 23.Gomes AC, et al. A genetic code alteration generates a proteome of high diversity in the human pathogen Candida albicans. Genome Biol. 2007;8(10):R206. doi: 10.1186/gb-2007-8-10-r206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rocha R, Pereira PJ, Santos MA, Macedo-Ribeiro S. Unveiling the structural basis for translational ambiguity tolerance in a human fungal pathogen. Proc Natl Acad Sci USA. 2011;108(34):14091–14096. doi: 10.1073/pnas.1102835108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miranda I, et al. A genetic code alteration is a phenotype diversity generator in the human pathogen Candida albicans. PLoS One. 2007;2(10):e996. doi: 10.1371/journal.pone.0000996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Noble SM, Johnson AD. Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans. Eukaryot Cell. 2005;4(2):298–309. doi: 10.1128/EC.4.2.298-309.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Slutsky B, Buffo J, Soll DR. High-frequency switching of colony morphology in Candida albicans. Science. 1985;230(4726):666–669. doi: 10.1126/science.3901258. [DOI] [PubMed] [Google Scholar]
- 28.Brown AJ, Gow NA. Regulatory networks controlling Candida albicans morphogenesis. Trends Microbiol. 1999;7(8):333–338. doi: 10.1016/s0966-842x(99)01556-5. [DOI] [PubMed] [Google Scholar]
- 29.Soll DR. High-frequency switching in Candida albicans. Clin Microbiol Rev. 1992;5(2):183–203. doi: 10.1128/cmr.5.2.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bourgeois C, Majer O, Frohner IE, Tierney L, Kuchler K. Fungal attacks on mammalian hosts: Pathogen elimination requires sensing and tasting. Curr Opin Microbiol. 2010;13(4):401–408. doi: 10.1016/j.mib.2010.05.004. [DOI] [PubMed] [Google Scholar]
- 31.Li L, et al. Naturally occurring aminoacyl-tRNA synthetases editing-domain mutations that cause mistranslation in Mycoplasma parasites. Proc Natl Acad Sci USA. 2011;108(23):9378–9383. doi: 10.1073/pnas.1016460108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Braun BR, et al. A human-curated annotation of the Candida albicans genome. PLoS Genet. 2005;1(1):36–57. doi: 10.1371/journal.pgen.0010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pezo V, et al. Artificially ambiguous genetic code confers growth yield advantage. Proc Natl Acad Sci USA. 2004;101(23):8593–8597. doi: 10.1073/pnas.0402893101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ruan B, et al. Quality control despite mistranslation caused by an ambiguous genetic code. Proc Natl Acad Sci USA. 2008;105(43):16502–16507. doi: 10.1073/pnas.0809179105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schultz DW, Yarus M. On malleability in the genetic code. J Mol Evol. 1996;42(5):597–601. doi: 10.1007/BF02352290. [DOI] [PubMed] [Google Scholar]
- 36.Woese CR. On the evolution of the genetic code. Proc Natl Acad Sci USA. 1965;54(6):1546–1552. doi: 10.1073/pnas.54.6.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Netzer N, et al. Innate immune and chemically triggered oxidative stress modifies translational fidelity. Nature. 2009;462(7272):522–526. doi: 10.1038/nature08576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bennett RJ, Johnson AD. Completion of a parasexual cycle in Candida albicans by induced chromosome loss in tetraploid strains. EMBO J. 2003;22(10):2505–2515. doi: 10.1093/emboj/cdg235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jones T, et al. The diploid genome sequence of Candida albicans. Proc Natl Acad Sci USA. 2004;101(19):7329–7334. doi: 10.1073/pnas.0401648101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dorazi R, Lingutla JJ, Humayun MZ. Expression of mutant alanine tRNAs increases spontaneous mutagenesis in Escherichia coli. Mol Microbiol. 2002;44(1):131–141. doi: 10.1046/j.1365-2958.2002.02847.x. [DOI] [PubMed] [Google Scholar]
- 41.Lamb DC, et al. The mutation T315A in Candida albicans sterol 14alpha-demethylase causes reduced enzyme activity and fluconazole resistance through reduced affinity. J Biol Chem. 1997;272(9):5682–5688. doi: 10.1074/jbc.272.9.5682. [DOI] [PubMed] [Google Scholar]
- 42.Sanglard D, Ischer F, Koymans L, Bille J. Amino acid substitutions in the cytochrome P-450 lanosterol 14alpha-demethylase (CYP51A1) from azole-resistant Candida albicans clinical isolates contribute to resistance to azole antifungal agents. Antimicrob Agents Chemother. 1998;42(2):241–253. doi: 10.1128/aac.42.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morschhäuser J, et al. The transcription factor Mrr1p controls expression of the MDR1 efflux pump and mediates multidrug resistance in Candida albicans. PLoS Pathog. 2007;3(11):e164. doi: 10.1371/journal.ppat.0030164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de Micheli M, Bille J, Schueller C, Sanglard D. A common drug-responsive element mediates the upregulation of the Candida albicans ABC transporters CDR1 and CDR2, two genes involved in antifungal drug resistance. Mol Microbiol. 2002;43(5):1197–1214. doi: 10.1046/j.1365-2958.2002.02814.x. [DOI] [PubMed] [Google Scholar]
- 45.Cowen LE, Steinbach WJ. Stress, drugs, and evolution: The role of cellular signaling in fungal drug resistance. Eukaryot Cell. 2008;7(5):747–764. doi: 10.1128/EC.00041-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cruz MC, et al. Calcineurin is essential for survival during membrane stress in Candida albicans. EMBO J. 2002;21(4):546–559. doi: 10.1093/emboj/21.4.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Karababa M, Coste AT, Rognon B, Bille J, Sanglard D. Comparison of gene expression profiles of Candida albicans azole-resistant clinical isolates and laboratory strains exposed to drugs inducing multidrug transporters. Antimicrob Agents Chemother. 2004;48(8):3064–3079. doi: 10.1128/AAC.48.8.3064-3079.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Santos MA, Gomes AC, Santos MC, Carreto LC, Moura GR. The genetic code of the fungal CTG clade. C R Biol. 2011;334(8–9):607–611. doi: 10.1016/j.crvi.2011.05.008. [DOI] [PubMed] [Google Scholar]
- 49.Murad AM, Lee PR, Broadbent ID, Barelle CJ, Brown AJ. CIp10, an efficient and convenient integrating vector for Candida albicans. Yeast. 2000;16(4):325–327. doi: 10.1002/1097-0061(20000315)16:4<325::AID-YEA538>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 50.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26(5):589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.