Abstract
Cells of Flavobacterium johnsoniae and of many other members of the phylum Bacteroidetes exhibit rapid gliding motility over surfaces by a unique mechanism. These cells do not have flagella or pili; instead, they rely on a novel motility apparatus composed of Gld and Spr proteins. SprB, a 669-kDa cell-surface adhesin, is required for efficient gliding. SprB was visualized by electron microscopy as thin 150-nm-long filaments extending from the cell surface. Fluorescence microscopy revealed movement of SprB proteins toward the poles of the cell at ∼2 μm/s. The fluorescent signals appeared to migrate around the pole and continue at the same speed toward the opposite pole along an apparent left-handed helical closed loop. Movement of SprB, and of cells, was rapidly and reversibly blocked by the addition of carbonyl cyanide m-chlorophenylhydrazone, which dissipates the proton gradient across the cytoplasmic membrane. In a gliding cell, some of the SprB protein appeared to attach to the substratum. The cell body moved forward and rotated with respect to this point of attachment. Upon reaching the rear of the cell, the attached SprB often was released from the substratum, and apparently recirculated to the front of the cell along a helical path. The results suggest a model for Flavobacterium gliding, supported by mathematical analysis, in which adhesins such as SprB are propelled along a closed helical loop track, generating rotation and translation of the cell body.
Keywords: cell motility, proton motive force, immunofluorescence microscopy, continuous track, left-handed helix
Cells of Flavobacterium johnsoniae, and of many other members of the phylum Bacteroidetes, move rapidly over surfaces at speeds of 1–3 μm/s by gliding motility (1). These cells lack flagella and pili, and the mechanism of cell movement is poorly understood. Flavobacterium gliding is thought to rely on motors embedded in the cell envelope that propel large cell-surface adhesins such as SprB and related proteins (2). Deletion of sprB results in dramatic reduction in motility. Twelve Gld proteins also are required for gliding (3). Some of the Gld proteins are components of a Bacteroidetes-specific protein secretion system, the porphyromonas secretion system (PorSS), required for transport of SprB to the cell surface (4). The PorSS originally was identified in the nonmotile periodontal pathogen Porphyromonas gingivalis. Comparative genome analysis revealed the widespread occurrence of PorSS genes in members of the phylum Bacteroidetes and their absence in members of other bacterial phyla (5). The PorSS is not related to the well-studied bacterial type I–VIII secretion systems (6, 7), and for this reason it recently has been referred to as the type IX secretion system (T9SS) (5, 8).
Gliding motility has been described in many other phyla of bacteria and has been studied at the molecular level for the δ proteobacterium Myxococcus xanthus (9) and for mollicutes belonging to the genus Mycoplasma (10). Cells of F. johnsoniae, M. xanthus, and Mycoplasma mobile each crawl over surfaces, but the proteins known to be involved in cell movement in each are unrelated, suggesting that gliding motility may have evolved independently in different bacterial phyla (5, 11). M. mobile gliding is powered by ATP hydrolysis and is thought to rely on conformational changes to cell surface adhesins that allow the cell essentially to “walk” over surfaces (10). M. xanthus has two motility systems, referred to as Social (S) and Adventurous (A) motility systems. S-motility involves type IV pilus extension and retraction, as does bacterial twitching motility (12). A-motility appears to be driven by motors in the cell envelope that harvest the proton gradient and exert force on proteins in the periplasm. One model envisions cargo proteins propelled within the periplasm along a helical track, causing localized deformation of the peptidoglycan layer and outer membrane, resulting in cell movement (13). A second model postulates a connection to a cell-surface adhesin, not yet identified, that is propelled by the motors, resulting in cell movement (9, 14). A difference between the models describing M. xanthus and F. johnsoniae motility is that in the former, motor proteins have been proposed to move long distances within the cell, whereas in the latter they are thought to be anchored within the cell envelope to be able to propel cell-surface adhesins such as SprB.
Genetic and molecular analyses indicate that SprB is the primary cell-surface motility adhesin of F. johnsoniae (2). SprB is a huge (669-kDa) cell-surface protein, but structures formed by SprB have not previously been identified. Rapid lateral movement of SprB along the cell surface is supported by the observation that 500-nm Protein G-coated polystyrene spheres carrying antibodies against SprB bound specifically to cells that expressed SprB and were propelled at 2 μm/s. The large size and multivalency of the Protein G-coated spheres prevented high-resolution analyses of SprB movements and left open the possibility that spheres might be passed from one molecule of SprB to another. Here, using electron microscopy and fluorescence microscopy, we demonstrate that SprB forms filaments that extend from the cell surface, and that SprB moves rapidly from pole to pole along what appears to be a closed helical loop track.
Results
Filamentous Surface Protein SprB.
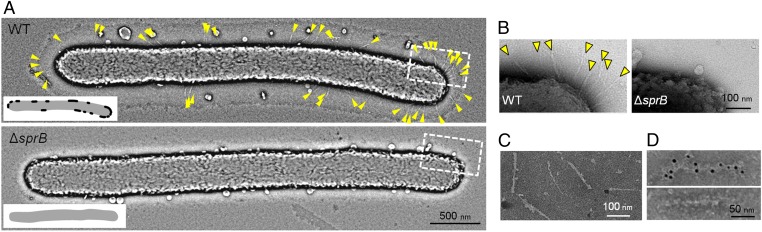
SprB is thought to be one of the outermost components of the motility machinery, but details regarding the structures formed by SprB are lacking. Previous work showed that wild-type cells of F. johnsoniae have thin filaments extending from the cell surface that appear to be anchored in the periplasm, and that these filaments were absent in a nonmotile mutant (15). Similar structures were observed on negatively stained wild-type cells, and these were absent from cells of an sprB mutant (Fig. 1 and Fig. S1). The filaments were distributed unevenly on the cell and typically emerged from the cell surface at angles of 30–90° in negative stained samples. A similar patchy distribution of SprB on wild-type cells was observed by immunofluorescent and immunoelectron microscopic localization of SprB on intact cells (Figs. S2 and S3), suggesting that SprB was a component of the filaments, and that the filaments have a role in gliding. In addition to the filaments, a capsule-like material was observed on the surface of wild-type cells but not on cells of the sprB mutant (Fig. 1 and Fig. S1). SprB may influence the production or distribution of this capsule-like material.
Fig. 1.
SprB forms cell-surface filaments. (A) Negative staining of cells of wild-type F. johnsoniae UW101 and of the sprB deletion mutant CJ1922. Images were treated with a bandpass filter to visualize the surface features clearly. (Inset) Surface regions where the filaments were found are drawn schematically as black dots. (B) Magnified image of polar regions (dashed boxes in A) of wild-type and sprB deletion mutant cells. Yellow arrowheads indicate filamentous structures extending from the cell surface. (C) Negative staining of the SprB fraction partially purified from wild-type cells. The SprB fraction is the same as that used in Fig. S4A, lane 5. (D) Immunogold EM on the SprB fraction treated with antisera against SprB (Upper) and GldJ (Lower).
To confirm that SprB forms filaments, a cell fraction enriched for SprB was obtained by detergent extraction followed by ammonium sulfate precipitation (Fig S4); 150-nm–long filaments were observed in the SprB-enriched fraction (Fig. 1C and Fig. S1). The filaments reacted specifically with anti-SprB antiserum (Fig. 1D and Fig. S2). The 669-kDa SprB may be large enough to form 150-nm filaments, because the M. mobile gliding motility protein Gli521 is 521 kDa and has been shown to form 110-nm–long structures (16). The SprB-enriched cell fraction obtained by detergent solubilization followed by ammonium sulfate precipitation contained several motility proteins in addition to SprB, including GldJ, GldK, and GldN (Fig. S4). GldK and GldN are known to be required for secretion of SprB to the cell surface and are part of the PorSS (4, 17). GldJ is required for gliding, but its exact function is not known (18). GldJ, GldK, and GldN may be associated with the SprB filaments; however, antibodies against GldJ did not label the SprB filaments, but rather labeled separate nonfilamentous structures in the cell fraction (Fig. S2D).
Rapid Movement of SprB Along the Outer Membrane.
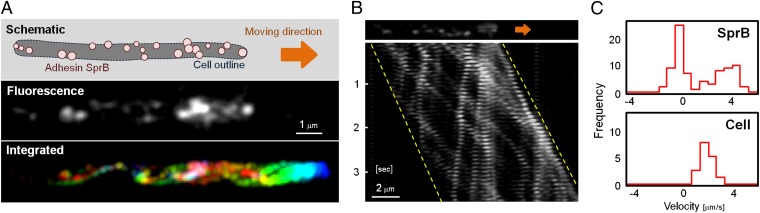
Wild-type cells moved rapidly on glass surfaces, whereas cells of an sprB deletion mutant exhibited little net movement, as previously observed (19), suggesting that SprB has an important role in motility. Addition of antisera against SprB weakened the ability of wild-type cells to bind to and move on glass (Fig. S5), suggesting that SprB may be involved in adhesion to glass. The inhibitory effects depended on the concentration of anti-SprB, and most of the cells were detached from glass at a concentration of 1/10 dilution of the antiserum. A 1/100 dilution of anti-SprB was used to analyze the dynamic movements of SprB, because this allowed most cells to bind and glide (Fig. S5). To observe the dynamic movements of SprB in a living cell, antiserum against SprB and fluorescent secondary antibody were added to cells gliding on glass. Immunolabeling of living cells revealed that the SprB signals, which clearly were separated from one another, moved rapidly along the long axes of gliding cells (Movies S1 and S2). When the apparent velocities of SprB signals were determined with respect to the glass substratum in a translocating cell (Fig. 2 A and B and Movie S2), about half the SprB signals (those moving toward the front of the cell) moved with velocities of 3.4 ± 1.1 μm/s and the other half (those moving toward the rear of the cell) moved with velocities of −0.5 ± 0.5 μm/s (Fig. 2C). The kymographs representing the spatial position of the SprB signals along the x-axis over time depict examples of SprB signals of both types (Fig. 2B). Taking the velocity of cells (1.9 ± 0.6 μm/s) into consideration, SprB signals migrating in opposite directions appeared to move at similar speeds with respect to the cell. When SprB reached the cell pole of a translocating cell, it changed direction, apparently by looping around the pole, and continued migrating toward the opposite pole (Fig. S6). As an SprB signal moving toward the forward pole at about 3.4 μm/s navigated the pole and returned toward the opposite end, its velocity with respect to the substratum decreased to about −0.5 μm/s, which indicates that the velocity of the SprB signal with respect to the cell surface was constant before and after moving around the cell pole. Signals moving toward the rear of the cell often remained stationary or nearly stationary with respect to the substratum, as though they were firmly attached to it. The rapid movement of SprB observed in translocating cells is in sharp contrast to what has been observed for other bacterial outer membrane or cell-surface proteins, such as Escherichia coli LamB, which traveled less than 0.3 μm during 5 min (20).
Fig. 2.
Helical loop-like motion of SprB. (A) Localization of SprB observed by epifluorescence microscopy. SprB was immunolabeled by antisera against SprB and fluorescent secondary antibody (Materials and Methods). In a translocating cell, the fluorescent signals were recorded at 0.1-s intervals for 2 s, colored from red (time 0) to blue (2 s), and integrated into a single image (Bottom). The images come from Movie S2. (B) Kymograph of SprB signals. The same cell as shown in A was used. The x-axis is the position of SprB signals with respect to the substratum (glass), and the y-axis is time. The orange arrow and dashed lines indicate the translocating direction of the cell and the approximate positions of cell poles, respectively. (C) Velocity of SprB movement and cell motility along the x-axis. SprB signals displaying well-separated foci were used for calculations. The velocities of 80 signals (Upper) and 13 cells (Lower) were integrated into the histograms.
Left-Handed Helical Flow of SprB.
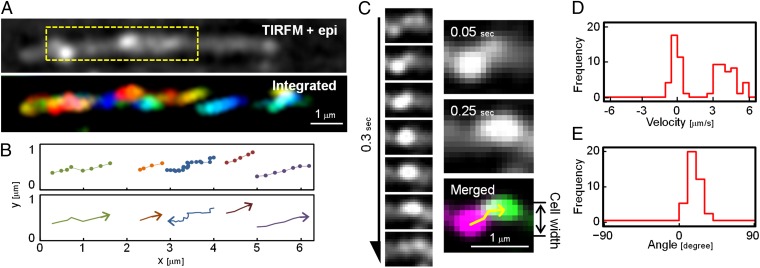
Movements near the substratum were examined in more detail by total internal reflection fluorescence (TIRF) microscopy, which revealed the movement of SprB located at the bottom of the cell. Under TIRF microscopy, many SprB signals entered and exited the narrow field of view adjacent to the substratum (Movie S3), suggesting that SprB moved across the width of the cell body in addition to moving along the length of the cell. In a translocating cell (Fig. 3A and Movie S4), when the apparent velocities of SprB signals were determined with respect to the glass substratum, about half the SprB signals (those moving toward the front of the cell) moved with velocities of 4.1 ± 0.8 μm/s and the other half (those moving toward the rear of the cell) moved with velocities of −0.1 ± 0.4 μm/s (Fig. 3D), consistent with the data obtained by epifluorescence microscopy. All the SprB signals moving toward the front of the cell appeared at the lower left side of the image and moved to the upper right side before disappearing from the field of view, whereas all the signals moving toward the rear of the cell moved backward slowly or appeared not to move with respect to the substratum (Fig. 3 B and C and Movie S4). The traces of SprB toward the front of the cell always had the same apparent pitch angles with respect to the long axis of the cell (17.6 ± 6.7°) (Fig. 3 B and E). This may suggest that SprB follows a left-handed helical path as it migrates along the cell surface.
Fig. 3.
Left-handed helical flow of SprB on the cell surface. (A) Location of SprB observed by TIRF microscopy (TIRFM). A cell translocating to the right was analyzed. Cell outline was visualized by simultaneous weak illumination using a halogen lamp. The SprB signals were colored from red to blue at 0.05-s intervals for 1.25 s and integrated into one image (Lower). The image is the view from the glass side. The images come from Movie S4. (B) Traces of typical SprB signals (Upper). Each signal was dotted with 0.05-s intervals. The same cell as shown in A was used. The moving direction of each trace is indicated by the arrows (Lower). (C) Tracking of typical signals. (Left) Montage of signals at 0.05-s intervals for 0.3 s. (Right) The images at 0.05 s and 0.25 s were colored magenta and green, respectively, and merged into a single image. Tracking of the signal is represented by the yellow arrow. (D) Velocity of SprB signals. The velocities of 72 signals were calculated from the x-axes of the fitting line and integrated into a histogram. (E) Angle of SprB signal traces. The angles of the fitting lines of the traces of 39 SprB signals moving from lower left to upper right with respect to the x-axis were measured and integrated into a histogram. SprB signals displaying well-separated foci were used for calculations.
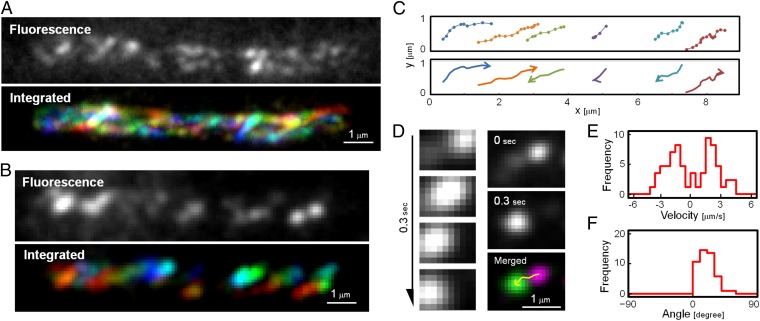
SprB moved rapidly on gliding cells, as described above. Most cells of F. johnsoniae moved rapidly on glass surfaces, but a few (∼1 in 100) did not move during the observation period. SprB also moved rapidly along these nontranslocating cells (Fig. 4A and Movie S5). SprB moved from pole to pole, traced an apparent loop around the end of the cell, and returned toward the opposite pole (Fig. S6). To observe the SprB movements in greater detail, we used TIRF microscopy to reveal the movement of SprB located near the substratum (Fig. 4B and Movie S6). At any one time, approximately half the SprB signals moved toward one cell pole with a velocity of ∼2 μm/s (2.1 ± 1.0 μm/s), and the rest moved toward the opposite pole with similar speed (−1.9. ± 0.8 μm/s) (Fig. 4E). As SprB migrated from pole to pole, it also traversed the width of the cell several times, as shown in Fig. 4 C and D. In a nontranslocating cell, about half the signals moved from the lower left side to the upper right side (with “lower” and “upper” not corresponding to the top and bottom of the cell, but rather to the relative positions in the recorded images), whereas the rest moved from the upper right to the lower left side (Fig. 4 C and D and Movie S6). The traces of SprB always have the same apparent pitch angles with respect to the long axis of the cell (19.0 ± 11.5°) (Fig. 4F). These observations suggest that SprB follows a left-handed closed helical loop path as it migrates along the cell surface.
Fig. 4.
Helical flow of SprB in a nontranslocating cell. (A) Localization of SprB observed by epifluorescence microscopy. Cells were immunolabeled with anti-SprB antiserum and fluorescent secondary antibody (Materials and Methods). The fluorescent signals were traced at 0.1-s intervals for 2 s and integrated into one image (Lower), with red indicating time 0 and blue indicating 2 s. The images come from Movie S5. (B) Localization of SprB observed by TIRFM. The SprB signals were colored from red to blue at 0.05-s intervals for 1 s and integrated into one image (Lower). The image is the view from the glass side. The images come from Movie S6. (C) Traces of typical SprB signals (Upper). Each signal was dotted with 0.05-s intervals for 1 s. The same cell as shown in B was used. The moving direction of each trace is indicated by the arrows (Lower). (D) Tracking of typical signals. Montage of signals at 0.1-s intervals for 0.3 s (Left). The images at 0 and 0.3 s were colored magenta and green, respectively, and merged into a single image (Right). Tracking of the signal is represented by the yellow arrow. (E) Velocity of SprB signals. The velocities of 61 signals were calculated from the x-axes of the fitting lines and integrated into a histogram. + and −, right and left directions of SprB motion, respectively. (F) Angle of SprB signal traces. The angles of the fitting lines with respect to the x-axis were measured and integrated into a histogram. SprB signals displaying well-separated foci were used for calculations.
Energy Source of Helical Flow.
Previous studies indicated that proton motive force (PMF) powers gliding of Flavobacterium and related gliding bacteria (21–24). To determine whether the SprB motion is powered by PMF, we examined the effects of carbonyl cyanide m-chlorophenylhydrazone (CCCP), a PMF uncoupler, on SprB motion. The SprB molecules, as well as the bacterial cell itself, stopped moving within 3 s after addition of CCCP, and the cells started moving again within 6 s after removal of CCCP (Fig. S7 and Movies S7 and S8). Addition of arsenate (which inhibits synthesis of ATP) had no immediate effect on gliding or on SprB movements, whereas it had a significant effect on intracellular ATP levels (Fig. S7D). These results suggest that movement of SprB is powered by PMF.
Discussion
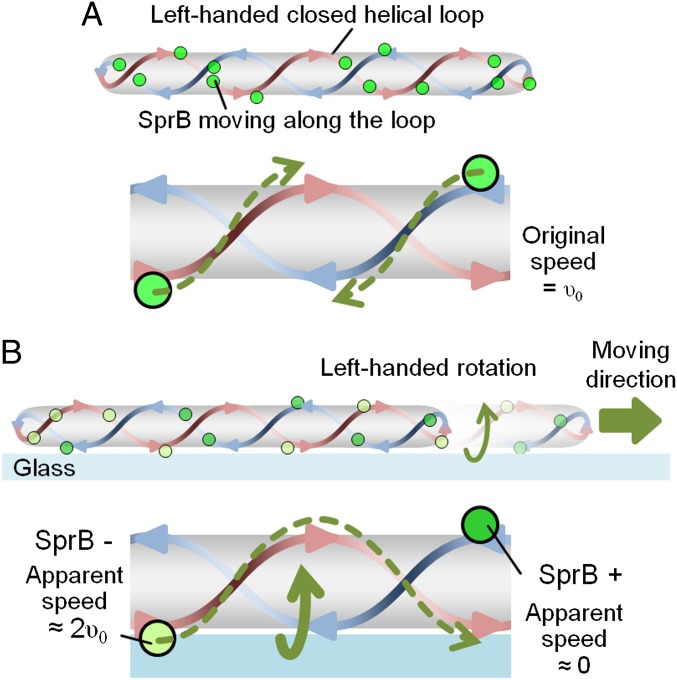
Here, we show the dynamic movement of SprB adhesin, which mediates the gliding motility of F. johnsoniae. SprB appears to form cell-surface filaments that move rapidly along the length of the cell. The observations of SprB movement on gliding and nongliding cells suggest that SprB moves in a left-handed helical manner in both cases. The results that (i) the pitch angles of the traces of SprB toward one direction were almost the same as those toward the other direction in a nontranslocating cell, (ii) the apparent pitch angles of the traces of SprB toward the front of a translocating cell were almost the same as those in a nontranslocating cell, (iii) the velocity of translocating cells was almost the same as that of SprB in a nontranslocating cell, and (iv) SprB moving toward the front of a translocating cell moved about twice as fast as the cell body, suggest a “helical loop track” model (Fig. 5 and Movie S9). The behavior of SprB in a translocating cell may be explained by adherence to the glass of SprB proteins moving toward the rear of the cell. Firm adherence of SprB to the glass would prevent its movement with respect to the substratum, with the result that the cell would rotate and translocate forward. This model is consistent with previous reports implying that cells of F. johnsoniae and related bacteria rotate as they glide (25–27).
Fig. 5.
Model of Flavobacterium gliding motility. (A) A nontranslocating cell. Adhesin SprB moves along the left-handed helical loop with a speed of υ0. (B) A translocating cell on glass. SprB has two different states: SprB moving toward the front of the cell and SprB moving toward the rear of the cell. In a translocating cell, SprB moving toward the rear of the cell adheres to the surface, generating left-handed rotation and right-directed translocation of the cell. SprB moving toward the front of the cell apparently runs twice as fast with respect to the glass surface than SprB on a nontranslocating cell.
To understand the cell motion observed in our experiments, we consider a simple mechanical model that describes force and torque balance of a cell body based on the left-handed helical geometry of a looped track and the bidirectional SprB motions found in our experiments (SI Text). The fundamental assumption of our model is that all SprB proteins are propelled uniformly along a rigid track on the cell surface with a constant speed. We also assume that the frictional interactions between SprB proteins and the substratum provide the major driving force to push the cell body forward. To obtain the speed of a cell body comparable with the experimentally measured values, our model requires the condition that only the SprB proteins moving toward the rear of the cell exert forces on the substrate. Given this, our mechanical model predicts the following features that are consistent with our experimental observations (1). As the cell moves forward, the cell body rotates counterclockwise about its axis when viewed from the rear of the cell (2). The apparent speed of SprB proteins moving toward the front of the cell is roughly twice the speed of the cell body (3). The apparent pitch angle of the trajectory of SprB moving toward the front of the cell is almost the same as that of SprB moving toward the rear of the cell, which is about 20°. The last result explains why the bacterial pitch angle appears to be insensitive to the translational motion of the cell body in our experiments (Fig. S8).
We propose a helical loop track model for Flavobacterium motility in which the filamentous protein SprB is propelled along a left-handed helical loop on the bacterial cell surface. When SprB adheres to a solid surface and thus can no longer move with respect to that surface, the cell is propelled helically in the opposite direction. This model has some similarity to models recently proposed for M. xanthus, in which motility motors are proposed to travel along an endless closed helical loop in the cytoplasm or cytoplasmic membrane (9, 13). We do not know the nature of the F. johnsoniae gliding motor, and we have no evidence indicating that it migrates rapidly within the cell. Because the peptidoglycan layer would provide a barrier to long-range movement of a protein complex spanning the periplasm, we envision a cytoplasmic membrane-spanning motor that is anchored to the cell wall and propels cell-surface adhesins such as SprB. F. johnsoniae and M. xanthus belong to different phyla of bacteria, and there are many differences between their motility systems. Flavobacterium cells move 50–100 times faster than myxobacterial cells, and there is little if any overlap between the proteins known to be involved in Flavobacterium gliding and those involved in myxobacterial gliding (2, 3, 28). The common features of the models may reflect convergent evolution of analogous systems to move similarly shaped cells over surfaces.
Several observations require further comment in relation to this model. In our observations of moving cells, only the SprB proteins moving toward the rear of the cell appeared to attach to the substratum. It is not known how this is orchestrated or whether the cell has a mechanism for regulating binding and release of individual SprB proteins to facilitate movement in one direction or another. When SprB attached to the substratum reaches the trailing end of the cell, it must release from the substratum if the cell is to continue moving forward. This event was observed repeatedly and may be facilitated by the geometry at the pole of the cell, with SprB being “peeled off” the substratum at the lagging pole by the action of the gliding motor. Alternative “binding” and “releasing” states of the SprB filaments may be generated by conformation of the SprB filaments. Occasional failure to release from the substratum upon reaching the trailing end of the cell would result in the cell flipping end over end and continuing to glide but making little net progress. This behavior has been observed often for F. johnsonaie and related bacteria (29, 30) and is easily explained by the model.
Addition of CCCP completely suppressed both the movement of SprB and gliding of cells, suggesting that the energy source for SprB movement and cell gliding is PMF. PMF presumably is harvested by cytoplasmic membrane proteins. Of the proteins essential for F. johnsoniae gliding, only GldF, GldG, GldL, and GldM are thought to span the cytoplasmic membrane. GldF and GldG are components of an ATP-binding cassette transporter and thus are likely to rely on ATP rather than PMF to perform their functions (31). Furthermore, several relatives of F. johnsoniae that lack GldA, GldF, and GldG exhibit rapid gliding motility (5). This suggests that GldF and GldG may not have a central role in gliding, leaving GldL and GldM as known gliding motility proteins that might harvest PMF, resulting in cell movement. GldL and GldM are components of the PorSS (T9SS) required for secretion of cell-surface motility adhesins such as SprB. They also may be involved directly in movement of SprB along the cell surface, and thus constitute components of the gliding “motor.” Alternatively, some other proteins not yet identified may perform this function. The PorSS originally was identified in the nonmotile periodontal pathogen P. gingivalis, which, like F. johnsoniae, belongs to the phylum Bacteroidetes. Comparative genome analysis reveals the widespread occurrence of PorSS genes in members of the phylum Bacteroidetes and their absence in members of other bacterial phyla (5). The secretion mechanism and protein components of PorSSs are distinct from those of the type I–VIII secretion systems (4, 5, 32), resulting in the use of T9SS to describe this Bacteroidetes-specific secretion system.
Several mysteries regarding Flavobacterium gliding remain, including the identity of the motor components that propel SprB along the cell surface, the association of the motor with the PorSS (T9SS), and the mechanism by which the direction of cell movement is controlled. The cell-surface adhesin SprB, which migrates rapidly along a helical track, provides a handle to explore each of these mysteries associated with the functioning of the Flavobacterium gliding motility machinery.
Materials and Methods
Bacterial Strains.
F. johnsoniae ATCC 17061 (UW101), CJ1827 (a streptomycin-resistant rpsl2 derivative of UW101 used for construction of deletion mutations), and CJ1922 (sprB deletion mutant) were used in this study (17, 33, 34). The cells were grown in casitone yeast extract (CYE) medium at 25 °C with shaking (35) to an optical density of around 1.0 at 600 nm. For selection and maintenance of the antibiotic-resistant strains, antibiotics were added to the medium at the following concentrations: erythromycin, 100 μg/mL; streptomycin, 100 μg/mL; tetracycline, 20 μg/mL (17, 18).
Optical Microscopy.
For epifluorescence microscopy, the cells were irradiated through CY3-4040C, FITC-5050A, and DAPI-5060C filters (Semrock) for red, green, and blue fluorescence, respectively, by using a USH-102D mercury lamp (USHIO). Cell behavior was observed under an Axiovert 200 inverted microscope (Carl Zeiss). Images were recorded with a CoolSNAP EZ camera (Roper Scientific) and a 100×/1.4 DLL Plan-Apochromat objective using MetaVue software (Molecular Devices) (see SI Materials and Methods for details).
Immunolabeling of SprB on Cells.
For immunofluorescence microscopy of live cells, cultures were poured into a tunnel slide assembled by taping a coverslip onto a glass slide (36). After incubation for 3 min at room temperature (RT), the medium containing floating cells was removed and replaced with fresh medium, and subsequently with CYE medium with 1/100 dilution of antisera against SprB, incubated for 5 min, and washed three times with CYE medium. The cells were treated with 1/100 dilution of Cy3-labeled secondary antibody in CYE, incubated for 5 min, and washed three times with CYE medium. Fluorescence observation was performed within 10 min (see SI Materials and Methods for details).
Electron Microscopy.
Samples bound to grids were stained with ammonium molybdate and observed by transmission electron microscopy as previously described (37). Carbon-coated EM grids were glow-discharged by the hydrophilic treatment device HDT-400 (JEOL). F. johnsoniae cells were put on the grid and incubated for 5 min at RT. The cells were chemically fixed with 3% (vol/vol) paraformaldehyde and 0.1% glutaraldehyde in PBS for 10 min at RT and were washed three times with PBS. After removing PBS, the cells were stained with1% ammonium molybdate (wt/vol) and air-dried. To examine cell fractions, protein samples in PBS buffer (100 μg/mL) were applied to the grid and incubated for 5 min at RT. After PBS was removed, the cells were stained with 2% ammonium molybdate (wt/vol) and air-dried. Samples were observed using a JEM-1210 transmission electron microscope (JEOL) at 80 kV. Whole micrographs were digitized as 16-bit images using a GT-X970 scanner (Epson) and analyzed by ImageJ 1.45s (http://rsb.info.nih.gov/ij/) (see SI Materials and Methods for details).
Supplementary Material
Acknowledgments
We thank M. Miyata for discussion; S. Suematsu and Y. Tahara for technical help; Y. Kamasaki and the cell function laboratories at Osaka City University for the use of their facilities; and members of the K.N. laboratories for valuable comments. This work was supported by the Global Center of Excellence Program at Nagasaki University (K.N.); Grants-in-Aid for Scientific Research on the priority area “Harmonized Supramolecular Motility Machinery and Its Diversity (to K.S., H.W., and K.N.); Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to K.S. and K.N.); National Science Foundation Grant MCB-1021721 (to M.J.M.); and a grant from the Japan Society for the Promotion of Science Research Fellowship for Young Scientists (to D.N.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1219753110/-/DCSupplemental.
References
- 1.McBride MJ. Bacterial gliding motility: Multiple mechanisms for cell movement over surfaces. Annu Rev Microbiol. 2001;55:49–75. doi: 10.1146/annurev.micro.55.1.49. [DOI] [PubMed] [Google Scholar]
- 2.Nelson SS, Bollampalli S, McBride MJ. SprB is a cell surface component of the Flavobacterium johnsoniae gliding motility machinery. J Bacteriol. 2008;190(8):2851–2857. doi: 10.1128/JB.01904-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braun TF, Khubbar MK, Saffarini DA, McBride MJ. Flavobacterium johnsoniae gliding motility genes identified by mariner mutagenesis. J Bacteriol. 2005;187(20):6943–6952. doi: 10.1128/JB.187.20.6943-6952.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sato K, et al. A protein secretion system linked to bacteroidete gliding motility and pathogenesis. Proc Natl Acad Sci USA. 2010;107(1):276–281. doi: 10.1073/pnas.0912010107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McBride MJ, Zhu Y. Gliding motility and Por secretion system genes are widespread among members of the phylum bacteroidetes. J Bacteriol. 2013;195(2):270–278. doi: 10.1128/JB.01962-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Desvaux M, Hébraud M, Talon R, Henderson IR. Secretion and subcellular localizations of bacterial proteins: A semantic awareness issue. Trends Microbiol. 2009;17(4):139–145. doi: 10.1016/j.tim.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 7.Holland IB. The extraordinary diversity of bacterial protein secretion mechanisms. Methods Mol Biol. 2010;619:1–20. doi: 10.1007/978-1-60327-412-8_1. [DOI] [PubMed] [Google Scholar]
- 8.Sato K, et al. Identification of Porphyromonas gingivalis proteins secreted by the Por secretion system. FEMS Microbiol Lett. 2013;338(1):68–76. doi: 10.1111/1574-6968.12028. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Y, Ducret A, Shaevitz J, Mignot T. From individual cell motility to collective behaviors: Insights from a prokaryote, Myxococcus xanthus. FEMS Microbiol Rev. 2012;36(1):149–164. doi: 10.1111/j.1574-6976.2011.00307.x. [DOI] [PubMed] [Google Scholar]
- 10.Miyata M. Unique centipede mechanism of Mycoplasma gliding. Annu Rev Microbiol. 2010;64:519–537. doi: 10.1146/annurev.micro.112408.134116. [DOI] [PubMed] [Google Scholar]
- 11.Jarrell KF, McBride MJ. The surprisingly diverse ways that prokaryotes move. Nat Rev Microbiol. 2008;6(6):466–476. doi: 10.1038/nrmicro1900. [DOI] [PubMed] [Google Scholar]
- 12.Wall D, Kaiser D. Type IV pili and cell motility. Mol Microbiol. 1999;32(1):1–10. doi: 10.1046/j.1365-2958.1999.01339.x. [DOI] [PubMed] [Google Scholar]
- 13.Nan B, et al. Myxobacteria gliding motility requires cytoskeleton rotation powered by proton motive force. Proc Natl Acad Sci USA. 2011;108(6):2498–2503. doi: 10.1073/pnas.1018556108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun M, Wartel M, Cascales E, Shaevitz JW, Mignot T. Motor-driven intracellular transport powers bacterial gliding motility. Proc Natl Acad Sci USA. 2011;108(18):7559–7564. doi: 10.1073/pnas.1101101108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu J, McBride MJ, Subramaniam S. Cell surface filaments of the gliding bacterium Flavobacterium johnsoniae revealed by cryo-electron tomography. J Bacteriol. 2007;189(20):7503–7506. doi: 10.1128/JB.00957-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nonaka T, Adan-Kubo J, Miyata M. Triskelion structure of the Gli521 protein, involved in the gliding mechanism of Mycoplasma mobile. J Bacteriol. 2010;192(3):636–642. doi: 10.1128/JB.01143-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rhodes RG, Nelson SS, Pochiraju S, McBride MJ. Flavobacterium johnsoniae sprB is part of an operon spanning the additional gliding motility genes sprC, sprD, and sprF. J Bacteriol. 2011;193(3):599–610. doi: 10.1128/JB.01203-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Braun TF, McBride MJ. Flavobacterium johnsoniae GldJ is a lipoprotein that is required for gliding motility. J Bacteriol. 2005;187(8):2628–2637. doi: 10.1128/JB.187.8.2628-2637.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rhodes RG, et al. Flavobacterium johnsoniae gldN and gldO are partially redundant genes required for gliding motility and surface localization of SprB. J Bacteriol. 2010;192(5):1201–1211. doi: 10.1128/JB.01495-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gibbs KA, et al. Complex spatial distribution and dynamics of an abundant Escherichia coli outer membrane protein, LamB. Mol Microbiol. 2004;53(6):1771–1783. doi: 10.1111/j.1365-2958.2004.04242.x. [DOI] [PubMed] [Google Scholar]
- 21.Duxbury T, Humphrey BA, Marshall KC. Continuous observations of bacterial gliding motility in a dialysis microchamber: The effects of inhibitors. Arch Microbiol. 1980;124(2-3):169–175. [Google Scholar]
- 22.Dzink-Fox JL, Leadbetter ER, Godchaux W., 3rd Acetate acts as a protonophore and differentially affects bead movement and cell migration of the gliding bacterium Cytophaga johnsonae (Flavobacterium johnsoniae) Microbiology. 1997;143(Pt 12):3693–3701. doi: 10.1099/00221287-143-12-3693. [DOI] [PubMed] [Google Scholar]
- 23.Pate JL, Chang LE. Evidence that gliding motility in prokaryotic cells is driven by rotary assemblies in the cell envelopes. Curr Microbiol. 1979;2(1):59–64. [Google Scholar]
- 24.Ridgway HF. Source of energy for gliding motility in Flexibacter polymorphus: Effects of metabolic and respiratory inhibitors on gliding movement. J Bacteriol. 1977;131(2):544–556. doi: 10.1128/jb.131.2.544-556.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beatson PJ, Marshall KC. A proposed helical mechanism for gliding motility in three gliding bacteria (order Cytophagales) Can J Microbiol. 1994;40(3):173–183. [Google Scholar]
- 26.Godwin SL, Fletcher M, Burchard RP. Interference reflection microscopic study of sites of association between gliding bacteria and glass substrata. J Bacteriol. 1989;171(9):4589–4594. doi: 10.1128/jb.171.9.4589-4594.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ridgway HF, Lewin RA. Characterization of gliding motility in Flexibacter polymorphus. Cell Motil Cytoskeleton. 1988;11(1):46–63. doi: 10.1002/cm.970110106. [DOI] [PubMed] [Google Scholar]
- 28.Shrivastava A, Rhodes RG, Pochiraju S, Nakane D, McBride MJ. Flavobacterium johnsoniae RemA is a mobile cell surface lectin involved in gliding. J Bacteriol. 2012;194(14):3678–3688. doi: 10.1128/JB.00588-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lapidus IR, Berg HC. Gliding motility of Cytophaga sp. strain U67. J Bacteriol. 1982;151(1):384–398. doi: 10.1128/jb.151.1.384-398.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pate JL. Gliding motility in procaryotic cells. Can J Microbiol. 1988;34(4):459–465. [Google Scholar]
- 31.Hunnicutt DW, Kempf MJ, McBride MJ. Mutations in Flavobacterium johnsoniae gldF and gldG disrupt gliding motility and interfere with membrane localization of GldA. J Bacteriol. 2002;184(9):2370–2378. doi: 10.1128/JB.184.9.2370-2378.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shoji M, et al. Por secretion system-dependent secretion and glycosylation of Porphyromonas gingivalis hemin-binding protein 35. PLoS One. 2011;6(6):e21372. doi: 10.1371/journal.pone.0021372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McBride MJ, et al. Novel features of the polysaccharide-digesting gliding bacterium Flavobacterium johnsoniae as revealed by genome sequence analysis. Appl Environ Microbiol. 2009;75(21):6864–6875. doi: 10.1128/AEM.01495-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rhodes RG, Pucker HG, McBride MJ. Development and use of a gene deletion strategy for Flavobacterium johnsoniae to identify the redundant gliding motility genes remF, remG, remH, and remI. J Bacteriol. 2011;193(10):2418–2428. doi: 10.1128/JB.00117-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Agarwal S, Hunnicutt DW, McBride MJ. Cloning and characterization of the Flavobacterium johnsoniae (Cytophaga johnsonae) gliding motility gene, gldA. Proc Natl Acad Sci USA. 1997;94(22):12139–12144. doi: 10.1073/pnas.94.22.12139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakane D, Miyata M. Mycoplasma mobile cells elongated by detergent and their pivoting movements in gliding. J Bacteriol. 2012;194(1):122–130. doi: 10.1128/JB.05857-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakane D, Miyata M. Cytoskeletal “jellyfish” structure of Mycoplasma mobile. Proc Natl Acad Sci USA. 2007;104(49):19518–19523. doi: 10.1073/pnas.0704280104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.