Abstract
Photosynthetic organisms flourish under low light intensities by converting photoenergy to chemical energy with near unity quantum efficiency and under high light intensities by safely dissipating excess photoenergy and deleterious photoproducts. The molecular mechanisms balancing these two functions remain incompletely described. One critical barrier to characterizing the mechanisms responsible for these processes is that they occur within proteins whose excited-state properties vary drastically among individual proteins and even within a single protein over time. In ensemble measurements, these excited-state properties appear only as the average value. To overcome this averaging, we investigate the purple bacterial antenna protein light harvesting complex 2 (LH2) from Rhodopseudomonas acidophila at the single-protein level. We use a room-temperature, single-molecule technique, the anti-Brownian electrokinetic trap, to study LH2 in a solution-phase (nonperturbative) environment. By performing simultaneous measurements of fluorescence intensity, lifetime, and spectra of single LH2 complexes, we identify three distinct states and observe transitions occurring among them on a timescale of seconds. Our results reveal that LH2 complexes undergo photoactivated switching to a quenched state, likely by a conformational change, and thermally revert to the ground state. This is a previously unobserved, reversible quenching pathway, and is one mechanism through which photosynthetic organisms can adapt to changes in light intensities.
Keywords: photosynthesis, purple bacteria, fluorescence spectroscopy
In photosynthetic light harvesting, networks of antenna pigment–protein complexes (PPCs) absorb sunlight, then efficiently transport the excitation through these networks to the reaction center, a PPC dedicated to charge separation (1, 2). Photosynthetic systems can complete this process both with near unity quantum efficiency and also function at light levels that provide photoenergy in excess of the capacity of downstream photochemistry. This is accomplished through mechanisms that dissipate harmful by-products produced by unused photoenergy (2, 3). The absorption, energy transport, and dissipation properties are governed by the balance of pigment–pigment and pigment–protein couplings (4). However, the molecular machinery responsible for this balance, and for the couplings themselves, is still poorly understood. This is because the couplings are highly sensitive to intermolecular distances. As a result of this sensitivity, the light-harvesting properties vary drastically among individual PPCs, because of small differences in protein conformation, and vary drastically even within a single PPC over time, because of protein fluctuations (2). In ensemble measurements, these drastic variations appear solely as a static, average value. Therefore, only through single-molecule spectroscopy can we investigate the photodynamics of individual PPCs, specifically how the absorption, energy transport, and dissipation of each change with time. We apply a room-temperature, single-molecule technique, the anti-Brownian electrokinetic (ABEL) trap (5, 6), to reveal static and dynamic heterogeneity for the primary antenna PPC of purple bacteria, light-harvesting complex 2 (LH2), without surface attachment or encapsulation.
Single-molecule techniques are particularly important in describing the electronic structure of LH2, because the complex-to-complex heterogeneity in excited-state energies is an order of magnitude larger for LH2 than for most other PPCs (7). Previous single-molecule experiments provided an incisive tool for elucidating variation in emissive spectra (8) and for switching between states with different emissive spectra (9) and lifetimes (10). These experiments, however, relied on immobilization by attachment chemistry or nonspecific surface association that may alter protein structure significantly or introduce an artificial near-environment (11–13), producing greater heterogeneity in fluorescence intensities and lifetimes. Indeed, modeling of these two sets of previous room temperature experiments yielded conflicting results, which the authors suggest may arise from differing immobilization schemes (14). Here, we remove this uncertainty by studying single LH2 complexes in solution to access the native heterogeneity and photodynamics (6, 15).
LH2 from Rhodopseudomonas acidophila strain 10050 exhibits a ring-like architecture, made up of nine subunits (Fig. S1A). Each subunit noncovalently binds three bacteriochlorophylls (BChl) and a carotenoid (16). The BChl are organized into two concentric rings, B800 and B850 (named after their absorption maxima). Pigment–pigment couplings drive rapid (<1 ps) and efficient transfer of singlet excitations from the carotenoid and B800 to B850 (Fig. S1C) (17–20). Fluorescence occurs out of B850. The singlet state also may undergo intersystem crossing to a triplet state, which, in principle, can generate deleterious singlet oxygen. One mechanism of photoprotection is dissipation of the triplet excitations through efficient transfer from the BChl to the carotenoid (3), which cannot sensitize singlet oxygen species (3).
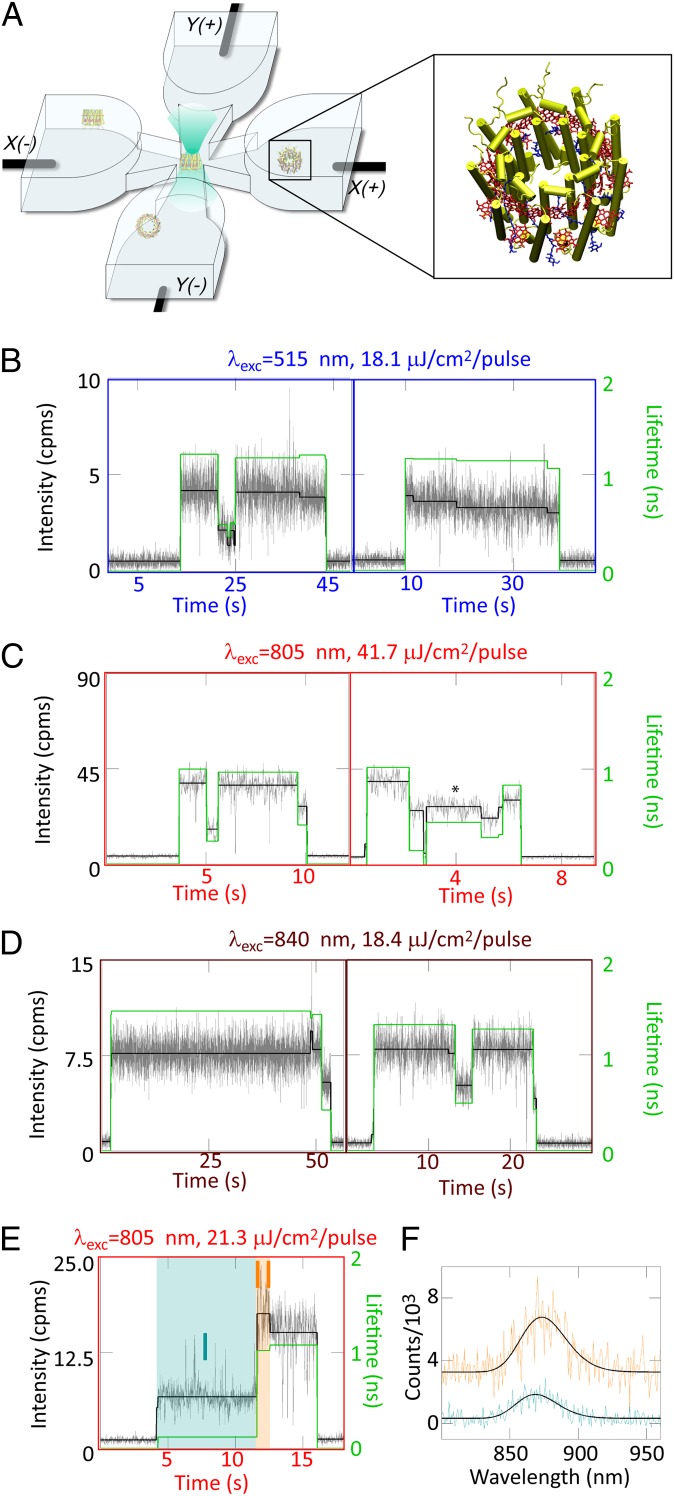
Here, we implement a solution-phase, single-molecule approach to investigate LH2, by taking advantage of the ABEL trap, which maintains individual LH2 complexes in the center of a microfluidic cell (Fig. 1A) using a closed-loop feedback system (21). To evaluate the contribution of each pigment group, we have performed three sets of experiments to individually excite the carotenoids with 515 nm (unique for single-molecule studies), B800 with 805 nm, and B850 with 840 nm. Furthermore, we record simultaneous measurements of fluorescence intensity, emission lifetime, and spectrum (22) and investigate correlated changes in all three variables, a measurement not possible with ensemble-averaged methods. This approach uncovers previously unknown states of LH2 and protein conformational dynamics—accessible across the solar spectrum—by which LH2 switches between these states. Notably, these results reveal that individual LH2 complexes undergo photoactivated, reversible switching to a quenched state, which serves as a previously unknown mechanism for dissipation of excitation energy under high light conditions.
Fig. 1.
(A) The microfluidic sample cell used for ABEL trap experiments in which LH2 complexes diffuse into the focal region in the center of the cell and are trapped. (Inset) Structure of a diffusing LH2 complex. (B–E) Representative fluorescence intensity–lifetime traces for seven single LH2 complexes. The dynamics of the emission intensity levels (black, left axis) are identified by a change-point–finding algorithm (38) on the data binned at 10 ms (gray, left axis), and the lifetime (green, right axis) is calculated by binning all photons for a given intensity level. Correlated changes in intensity/lifetime are observed for (B) carotenoid, (C) B800, and (D) B850 excitation. Periods of stability also are observed at lower excitation fluences for carotenoid excitation (B, Right) and B850 excitation (D, Left). For higher excitation fluence, similar intensities have different concomitant lifetimes (e.g. C, Right, *). Occasionally, spectral shifts accompany intensity shifts. (E) B800 excitation and (F) the corresponding spectra for intensity intervals I and II, with II offset by 3,000 counts for clarity. The spectral maxima with 95% confidence intervals are found by a nonlinear least-squares fit to an asymmetric Gaussian (black), and are 873.3 (871.1,875.6) nm for interval I (turquoise) and 868.6 (866.6,879.7) nm for interval II (orange).
Results
Fluorescence intensity–lifetime traces from representative, single LH2 complexes are displayed in Fig. 1 B–D for excitation of the carotenoids, B800, and B850. Under all three excitation wavelengths, LH2 complexes show correlated changes in intensity/lifetime (Fig. 1 B, Left; C, Left and Right; and D, Left and Right). The rate of the photodynamics increases as a function of excitation fluence. Thus, the lower fluence for carotenoid excitation and B850 excitation produces periods of stability tens of seconds long (Fig. 1 B and D, Left), which is particularly remarkable under the relatively high photon energy of carotenoid excitation. Because of the higher fluence, the B800 excitation experiments (Fig. 1C) show faster photodynamics. Additionally, a higher intensity/shorter lifetime state (Fig. 1 C, Right, asterisk) is observed. Here, the intensity/lifetime changes no longer are only correlated, as may be seen from the asterisked state contrasted with the final intensity level in the trace, where a similar intensity exhibits a longer lifetime.
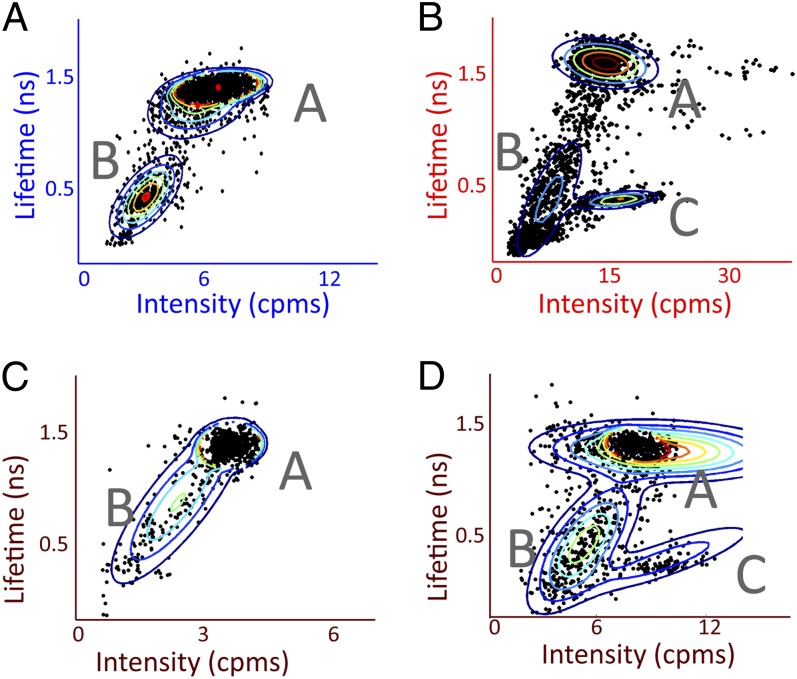
The distribution of LH2 behaviors is displayed by representing each period of constant intensity with its concomitant lifetime as a dot on a 2D intensity–lifetime plot, shown in Fig. 2 for four different combinations of excitation wavelength and fluence. Additional combinations are shown in Fig. S2. Three groups of correlated intensity–lifetime values are visible: states A, B, and C. All excitation conditions give rise to state A, the higher intensity/longer lifetime state, and state B, the lower intensity/shorter lifetime state. Individual complexes switch back and forth between these two states (Fig. 1 B, Left; C; and D). Under the higher excitation fluences (Fig. 2 B and D), state C, a higher intensity/shorter lifetime state, appears (Fig. 1 C, Right, asterisk).
Fig. 2.
Correlations between intensity and lifetime reveal three states of LH2. Fluorescence lifetime-vs.-intensity plots in which each period of constant intensity is represented as a ● and clusters are identified by a Gaussian mixture model, shown by the rainbow lines at 10% contour intervals for four different combinations of excitation wavelength and fluence: (A) 515 nm, 18.1 μJ/cm2 per pulse; (B) 805 nm, 41.7 μJ/cm2 per pulse; (C) 840 nm, 2.4 μJ/cm2 per pulse; and (D) 840 nm, 18.4 μJ/cm2 per pulse. All combinations show states A and B, and state C appears with higher excitation fluence.
State C exhibits a nonlinear dependence on excitation fluence and, as a result, appears only at high excitation fluences. Over an order-of-magnitude increase in fluence for B850 excitation (0.51–2.4–18.4 μJ per cm2 per pulse), the ratio of total time in state C to state A increased from 0 to 5.4 × 10−4 to 0.1. This suggests a multistep process, either via a simultaneous double excitation of the protein or two sequential steps, such as state A to B to C. A likely mechanism for this process is exciton–exciton annihilation after excited-state absorption, which is possible at these fluences, as shown in Table 1 (23).
Table 1.
Intensity dependence of switching rates between states A and B
| Excitation wavelength, nm | Fluence, μJ/cm2 per pulse | Absorption probability per pulse | kA→B, s−1 | kB→A, s−1 |
| 515 | 26.9 | 0.32 | 0.94 | 0.50 |
| 11.4 | 0.14 | 0.077 | 0.33 | |
| 4.1 | 0.049 | 0.011 | 0.31 | |
| 840 | 18.4 | 0.41 | 0.086 | 0.26 |
| 8.1 | 0.18 | 0.11 | 0.27 | |
| 2.4 | 0.054 | 0.016 | 0.098 | |
| 1.3 | 0.029 | 0.0079 | 0.26 | |
| 0.51 | 0.011 | 0.0068 | 0.11 |
The switching rates kA→B and kB→A were determined for carotenoid and B850 excitation. The rates (ki→j) are calculated by dividing the number of events (i→j) by the total time in state i. The orders-of-magnitude increase in kA→B upon orders-of-magnitude increase in excitation fluence suggests this process is photoactivated. In contrast, kB→A remains the same order of magnitude, suggesting a thermal process. The linear dependence on excitation fluence, as well as the per pulse absorption probabilities for a single LH2 complex (23, 39), shows that the dynamics of states A and B are not a result of nonlinear effects.
To explore the molecular mechanism behind the changing emissive state, Fig. 1 shows an intensity/lifetime trace (Fig. 1D) and the simultaneously measured spectra for two of the intensity levels (Fig. 1E). As illustrated for this molecule, intensity changes occasionally are accompanied by a spectral shift. This occurred ∼3% of the time over a total of 1,073 intensity transitions analyzed. The spectral changes most likely arise from fluctuations of the protein structure, as has been well described (9, 14). Remarkably, however, the dominant switches between states (Fig. 2) were not correlated with any specific spectral change, and the average spectral maxima for states A, B, and C are the same within the 1-nm resolution of the spectrometer. Consequently, the states defined here do not correspond to the previously reported correlated spectral and intensity shifts of immobilized single LH2 complexes.
We quantify the rate of switching between states A and B as a function of excitation fluence and absorption probability per pulse for carotenoid (515 nm, Table 1, Upper) and B850 (840 nm, Table 1, Lower) excitations. As shown by the results, kA→B increases with increasing excitation fluence, indicating a photoactivated transition. In contrast, kB→A remains the same order of magnitude, indicating a thermal transition.
Discussion
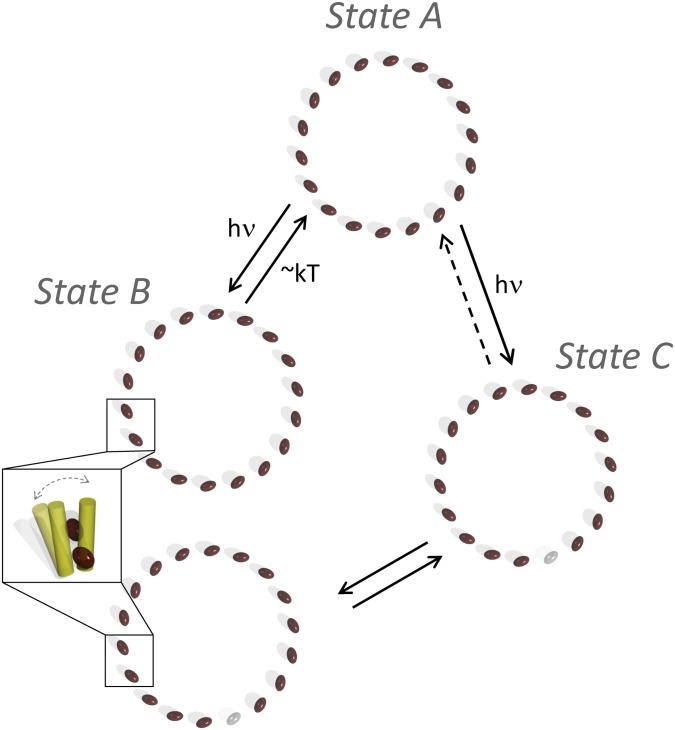
The most likely molecular mechanism of switching from state A to B is a local conformational change (Fig. 3, Inset). We propose a local conformational change as opposed to a large distortion of the ring, because a large distortion would be accompanied by a spectral signature (24, 25), which is not observed here. We assign this mechanism for two other major reasons: (i) We can eliminate other states of LH2 (triplets, cations) as the source of state B because of the dwell times of seconds. Previous experiments determined that triplet states relax on a microsecond timescale (19) and cations recombine on a picosecond timescale (26). (ii) Each photon deposits sufficient energy into the PPC (∼50–100 kT) to easily move it out of the ground conformational state (27). Furthermore, the reverse (kB→A) rate of hundreds of milliseconds suggests that state B is a metastable conformation that returns to the ground conformation via thermal fluctuations, as shown by the intensity independence of kB→A.
Fig. 3.
Model of the molecular changes in LH2 upon switching between the observed states. Each BChl in the B850 ring is represented as a brown ellipse. State A shows photoactivated switching to state B. According to the model, this is a conformational change to a quenched form, with increased vibrational dissipation of excitation energy, that thermally reverts to the ground state. State A also shows photoactivated switching to state C, but with a nonlinear dependence on excitation fluence. According to the model, this is a photobleaching of a BChl in the B850 ring, represented by a gray ellipse, lower right. The state C to A transition is represented as a dotted line because of the rarity of this process. Switching between states C and B also is observed and most likely corresponds to a conformational change in an LH2 with a photobleached BChl in the B850 ring. The two forms of state B (with and without a photobleached pigment) overlap spectroscopically and so cannot be separated clearly. Finally, because state C appears only at high excitation fluence, the intensity dependence of the transitions to state B cannot be characterized.
Upon switching between states A and B, a quencher is turned on and off, as determined by the direct proportionality of observed fluorescence lifetime and intensity between states A and B. Direct proportionality between these two variables indicates a change in quenching rate. Generally, in LH2, quenching is performed by intramolecular and protein vibrations. Because LH2 has a fluorescence quantum yield of only 10%, these vibrations dissipate most photoenergy (28, 29). A conformational change might increase vibronic coupling or coupling between intramolecular and protein vibrations to facilitate this dissipation. Furthermore, there might be a distribution of conformations, producing a range of increased couplings that would give rise to the diagonal elongation of state B in Fig. 2.
The most likely molecular mechanism of switching from state A to C is the photobleaching of a pigment. We construct a theoretical model of the electronic structure showing that upon removal of a pigment, the fluorescence spectral maximum barely shifts (SI Text). This is plausible because an analogous effect has been studied experimentally with single-molecule fluorescence excitation spectra on another ring-shaped PPC, light harvesting complex 1 (LH1) (30). A structural gap produced an intensity spike in a narrow wavelength region, which might be the source of the small average intensity increase from state A to C, but did not produce an overall spectral shift. Finally, switching between states C and B is also observed. According to the proposed model, this arises from an LH2 complex with a photobleached pigment (state C) undergoing a conformational change to a quenched state (state B). The diagonal elongation of state B toward C gives rise to the diagonal character of the Gaussian mixture model for state C in Fig. 2D. This provides evidence for the mechanism, as the direct proportionality is a hallmark of a quencher on (state B) and off (state C). The observed state C to state B transitions also suggest heterogeneity in the state B manifold, in which some of the rings have a photobleached pigment and some are intact (Fig. 3).
The nature of the state A to state B quenching mechanism points to its general accessibility to PPCs. Because it is a local conformational change, the process is not specific to a ring-like structure. A conformational change in any PPC may increase the pigment–protein coupling, which in turn increases dissipation of excitation energy into vibrations, similar to what is seen here for LH2. Other PPCs also exhibit vibrational dissipation of excitation energy, as evidenced by their low fluorescence quantum yields. For example, the most abundant membrane protein on Earth, light harvesting complex II from green plants, has a fluorescence quantum yield of only 22% (31). This type of selective tuning of the pigment binding pocket, or more broadly the local environment, is a mechanism available to all light harvesting systems to regulate quenching of excitation energy.
To observe the quenched state on a timescale reasonable for experimental measurements, the experimental incident intensity is ∼102 to 104 larger than actual sunlight (32). During the much longer lifetime of an LH2 complex under sunlight, however, state B is created scores of times yet returns to state A within seconds, even though the probability of creating state B per photon absorbed is small (∼10−7 to 10−8). Furthermore, the quenching functionality is notable for two additional reasons: (i) Identification of this quenching process illustrates that PPCs can withstand fluences much greater than that of sunlight, which shows these systems are robust to variations in their surroundings that exceed natural fluctuations. (ii) The quenching dynamics observed here are particularly relevant to the ongoing incorporation of LH2 into light harvesting devices (33), which may be installed under solar concentrators that produce intensities orders of magnitude above natural levels (34). These results show that LH2, like inorganic systems, can maintain its functionality under these high fluences.
This work demonstrates that solution-phase, single-molecule measurements reveal previously unknown states of a well-studied photosynthetic PPC, LH2. The quenching process observed here competes with fluorescence; thus, the dissipation occurs on timescales similar to those of triplet formation (3), therefore providing a photoactivated, reversible alternative to triplet formation. In the case of high light intensities, this offers a mechanism in addition to carotenoid protection to prevent the generation of deleterious singlet oxygen. The existence of multiple photoprotective mechanisms is particularly important in situations such as artificial light harvesting systems installed under concentrated sunlight that far exceeds natural levels (34). The photostability and photoprotective aspects of LH2 characterized here for wavelengths across the solar spectrum are particularly valuable for the design of solar energy devices incorporating LH2 (33). Fundamentally, the conformationally driven photoprotective mechanism found here provides an avenue by which light harvesting systems exploit interactions with their environment as an adjustable parameter to adapt to changes in the intensity of incident light.
Methods
Sample Preparation.
Purified LH2 complexes of Rps. acidophila were prepared as described previously (35). The stock solution of LH2 suspended in 20 mM Tris⋅HCl (pH 8.0) and 0.1% lauryldimethylamine oxide was kept at −80 °C and thawed immediately before the experiments. The sample was diluted to a concentration of ∼5 pM in a 2:1 (vol/vol) mixture of 50 mM PBS (pH 7.4) with 0.1% n-dodecyl-β-d-maltoside and glycerol. An enzymatic oxygen-scavenging system was added to the buffer for a final concentration of 2.5 mM of protocatechuic acid and 25 nM protocatechuate-3,4-dioxygenase. The details of sample characterization are given in SI Text and shown in Fig. S3. ABEL trap microfluidic cells made of fused silica were constructed as described previously (5) with 700-nm cell depth, which achieves confinement in z, the direction of beam propagation. Cell interiors were coated with two pairs of layers of polyethylene-imine and polyacrylic acid (PAA) ending in PAA to prevent nonspecific adsorption (36).
Excitation and Detection Optics.
Excitation was provided by doubling a mode-locked fiber laser (Mercury; PolarOnyx; 1,030 nm doubled to 515 nm, 200-fs pulse length, 43.3-MHz repetition rate) and by a mode-locked Ti:sapphire laser (Mira 900-D; Coherent; tuned to 805 nm or 840 nm, 200-fs pulse length, 76-MHz repetition rate). The Ti:sapphire pulse length was stretched to ∼10 ps because of propagation through an optical fiber. The ABEL trap apparatus was implemented as described (21). In brief, the excitation laser was sent through an orthogonal pair of acousto-optic modulators (46080-3-LTD; NEOS) to deflect the beam to produce a knight’s tour scan pattern on a 32-point square grid with a frequency of 9.8 kHz. The excitation laser then was sent into an Olympus IX71 inverted microscope equipped with a high-N.A. oil immersion objective (1.35 N.A., 60×; Olympus). The excitation spot is 0.8 µm in diameter in the sample plane, as measured by scanning a fluorescent bead through the focal volume, and the beam scans with a grid spacing of 0.4 µm in the sample plane. Sample fluorescence is collected back through the same objective and passed through a dichroic (514rdc for the 515-nm excitation, T825dcxrxt for the 805-nm excitation, ZTt860LPXR for the 840-nm excitation; Chroma) and two long-pass filters [HQ865LP (Chroma) for the 840-nm excitation, FF01-834/LP (Semrock) for the 800- and 515-nm excitation], a 400-µm pinhole to reject out-of-focus background fluorescence, and a 70:30 near infrared beamsplitter, and the transmitted component is focused onto an avalanche photodiode (APD; SPCM-AQR-15; Perkin-Elmer). Single LH2 complexes enter the trap and are maintained in an observation region until they either photobleach or reversibly enter a dark state for tens of milliseconds or more, at which point they diffuse out of the field of view.
Time-correlated single-photon counting is achieved using a timing module (PicoHarp 300; PicoQuant). An instrument response function of 0.2–0.4 ns was measured from scatter off a glass coverslip. Continuous wave experiments were performed with a 514-nm line from argon ion laser (Innova 90; Coherent) and 805-nm diode laser (Axcel) excitation sources, and showed longer survival times. Details of the fitting procedure are given in SI Text, with representative fits of each state in Fig. S4.
The fluorescence emission spectra were recorded with the reflected component (30%) of the fluorescence emission. The emission was spectrally dispersed using a volume-phase holographic grating (600 lines per millimeter, λc = 900 nm; Wasatch). The dispersed beam was focused onto an electron multiplying charge coupled device camera (Ixon Ultra 897; Andor) and recorded at a 10-Hz frame rate in sync with the APD photon record. The spectrometer was calibrated using an argon discharge lamp. The resolution of the spectrometer was determined to be 1 nm by finding the SD of the spectral width of the Ti:sapphire laser beam in continuous wave mode.
ABEL Trap Position Algorithm and Implementation.
The initial estimate of the position of the protein is found in real time by taking the beam position in the knight’s tour scanning pattern at the moment of each photon detection event. The position estimate is refined using a Kalman filter algorithm (37) given previous estimates, applied voltages, and the protein’s estimated dynamical parameters (diffusion coefficient and mobility). The algorithm is implemented on a field-programmable gate array (FPGA; PCI-7833R; National Instruments) through a Labview interface. Two-dimensional feedback voltages are calculated to cancel Brownian motion displacements through electrokinetic flow, and are produced by amplifying the FPGA analog output with two home-built high-voltage amplifiers (OPA453; Texas Instruments). Four platinum electrodes are inserted into the sample cell. The resulting electric field falls primarily across the ∼20-µm transverse width of the shallow region of the sample cell and is approximately uniform. The relatively small electric fields have been shown to have no effect on the photophysics (15).
Supplementary Material
Acknowledgments
The authors thank Samuel Bockenhauer, Dr. Yan Jiang, and Prof. Randall Goldsmith for helpful discussions. The authors acknowledge the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the US Department of Energy for funding the single-molecule studies through Grant DE-FG02-07ER15892 (to W.E.M) and the Biotechnology and Biological Sciences Research Council (R.J.C.) for funding the protein preparation work.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1310222110/-/DCSupplemental.
References
- 1.Blankenship RE. Molecular Mechanisms of Photosynthesis. Oxford: Blackwell Scientific; 2002. [Google Scholar]
- 2.Cogdell RJ, Gall A, Köhler J. The architecture and function of the light-harvesting apparatus of purple bacteria: From single molecules to in vivo membranes. Q Rev Biophys. 2006;39(3):227–324. doi: 10.1017/S0033583506004434. [DOI] [PubMed] [Google Scholar]
- 3.Cogdell RJ, Frank HA. How carotenoids function in photosynthetic bacteria. Biochim Biophys Acta. 1987;895(2):63–79. doi: 10.1016/s0304-4173(87)80008-3. [DOI] [PubMed] [Google Scholar]
- 4.Ishizaki A, Calhoun TR, Schlau-Cohen GS, Fleming GR. Quantum coherence and its interplay with protein environments in photosynthetic electronic energy transfer. Phys Chem Chem Phys. 2010;12(27):7319–7337. doi: 10.1039/c003389h. [DOI] [PubMed] [Google Scholar]
- 5.Cohen AE, Moerner WE. Controlling Brownian motion of single protein molecules and single fluorophores in aqueous buffer. Opt Express. 2008;16(10):6941–6956. doi: 10.1364/oe.16.006941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Q, Goldsmith RH, Jiang Y, Bockenhauer SD, Moerner WE. Probing single biomolecules in solution using the anti-Brownian electrokinetic (ABEL) trap. Acc Chem Res. 2012;45(11):1955–1964. doi: 10.1021/ar200304t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van Amerongen H, Valkunas L, Van Grondelle R. Photosynthetic Excitons. Singapore: World Scientific; 2000. [Google Scholar]
- 8.van Oijen AM, Ketelaars M, Kohler J, Aartsma TJ, Schmidt J. Unraveling the electronic structure of individual photosynthetic pigment-protein complexes. Science. 1999;285(5426):400–402. doi: 10.1126/science.285.5426.400. [DOI] [PubMed] [Google Scholar]
- 9.Rutkauskas D, Novoderezkhin V, Cogdell RJ, van Grondelle R. Fluorescence spectral fluctuations of single LH2 complexes from Rhodopseudomonas acidophila strain 10050. Biochemistry. 2004;43(15):4431–4438. doi: 10.1021/bi0497648. [DOI] [PubMed] [Google Scholar]
- 10.Bopp MA, Jia Y, Li L, Cogdell RJ, Hochstrasser RM. Fluorescence and photobleaching dynamics of single light-harvesting complexes. Proc Natl Acad Sci USA. 1997;94(20):10630–10635. doi: 10.1073/pnas.94.20.10630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friedel M, Baumketner A, Shea JE. Effects of surface tethering on protein folding mechanisms. Proc Natl Acad Sci USA. 2006;103(22):8396–8401. doi: 10.1073/pnas.0601210103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bai H, et al. Remote control of DNA-acting enzymes by varying the Brownian dynamics of a distant DNA end. Proc Natl Acad Sci USA. 2012;109(41):16546–16551. doi: 10.1073/pnas.1203118109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rasnik I, McKinney SA, Ha T. Surfaces and orientations: Much to FRET about? Acc Chem Res. 2005;38(7):542–548. doi: 10.1021/ar040138c. [DOI] [PubMed] [Google Scholar]
- 14.Novoderezhkin VI, Rutkauskas D, van Grondelle R. Dynamics of the emission spectrum of a single LH2 complex: Interplay of slow and fast nuclear motions. Biophys J. 2006;90(8):2890–2902. doi: 10.1529/biophysj.105.072652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldsmith RH, Moerner WE. Watching conformational- and photo-dynamics of single fluorescent proteins in solution. Nat Chem. 2010;2(3):179–186. doi: 10.1038/nchem.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McDermott G, et al. Crystal structure of an integral membrane light-harvesting complex from photosynthetic bacteria. Nature. 1995;374(6522):517–521. [Google Scholar]
- 17.Ostroumov EE, Mulvaney RM, Cogdell RJ, Scholes GD. Broadband 2D electronic spectroscopy reveals a carotenoid dark state in purple bacteria. Science. 2013;340(6128):52–56. doi: 10.1126/science.1230106. [DOI] [PubMed] [Google Scholar]
- 18.Ma YZ, Cogdell RJ, Gillbro T. Energy transfer and exciton annihilation in the B800-850 antenna complex of the photosynthetic purple bacterium Rhodopseudomonas acidophila (strain 10050). A femtosecond transient absorption study. J Phys Chem B. 1997;101(6):1087–1095. [Google Scholar]
- 19.Jimenez R, Dikshit SN, Bradforth SE, Fleming GR. Electronic excitation transfer in the LH2 complex of rhodobacter sphaeroides. J Phys Chem. 1996;100(16):6825–6834. [Google Scholar]
- 20.Harel E, Long PD, Engel GS. Single-shot ultrabroadband two-dimensional electronic spectroscopy of the light-harvesting complex LH2. Opt Lett. 2011;36(9):1665–1667. doi: 10.1364/OL.36.001665. [DOI] [PubMed] [Google Scholar]
- 21.Wang Q, Moerner WE. An Adaptive Anti-Brownian ELectrokinetic trap with real-time information on single-molecule diffusivity and mobility. ACS Nano. 2011;5(7):5792–5799. doi: 10.1021/nn2014968. [DOI] [PubMed] [Google Scholar]
- 22.Wang Q, Moerner WE. Lifetime and spectrally resolved characterization of the photodynamics of single fluorophores in solution using the anti-brownian electrokinetic trap. J Phys Chem B. 2013;117(16):4641–4648. doi: 10.1021/jp308949d. [DOI] [PubMed] [Google Scholar]
- 23.Pflock TJ, et al. The electronically excited states of LH2 complexes from Rhodopseudomonas acidophila strain 10050 studied by time-resolved spectroscopy and dynamic Monte Carlo simulations. I. Isolated, non-interacting LH2 complexes. J Phys Chem B. 2011;115(28):8813–8820. doi: 10.1021/jp202353c. [DOI] [PubMed] [Google Scholar]
- 24.Bopp MA, Sytnik A, Howard TD, Cogdell RJ, Hochstrasser RM. The dynamics of structural deformations of immobilized single light-harvesting complexes. Proc Natl Acad Sci USA. 1999;96(20):11271–11276. doi: 10.1073/pnas.96.20.11271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen XH, et al. Protein structural deformation induced lifetime shortening of photosynthetic bacteria light-harvesting complex LH2 excited state. Biophys J. 2005;88(6):4262–4273. doi: 10.1529/biophysj.104.053868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Polívka T, Pullerits T, Frank HA, Cogdell RJ, Sundström V. Ultrafast formation of a carotenoid radical in LH2 antenna complexes of purple bacteria. J Phys Chem B. 2004;108(39):15398–15407. [Google Scholar]
- 27.Liu LN, Duquesne K, Oesterhelt F, Sturgis JN, Scheuring S. Forces guiding assembly of light-harvesting complex 2 in native membranes. Proc Natl Acad Sci USA. 2011;108(23):9455–9459. doi: 10.1073/pnas.1004205108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Monshouwer R, Abrahamsson M, van Mourik F, van Grondelle R. Superradiance and exciton delocalization in bacterial photosynthetic light-harvesting systems. J Phys Chem B. 1997;101(37):7241–7248. [Google Scholar]
- 29.Novoderezhkin V, Monshouwer R, van Grondelle R. Exciton (de) localization in the LH2 antenna of rhodobacter sphaeroides as revealed by relative difference absorption measurements of the LH2 antenna and the B820 subunit. J Phys Chem B. 1999;103(47):10540–10548. [Google Scholar]
- 30.Richter MF, et al. Symmetry matters for the electronic structure of core complexes from Rhodopseudomonas palustris and Rhodobacter sphaeroides PufX- Proc Natl Acad Sci USA. 2007;104(16):6661–6665. doi: 10.1073/pnas.0611115104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Palacios MA, de Weerd FL, Ihalainen JA, van Grondelle R, van Amerongen H. Superradiance and exciton (de) localization in light-harvesting complex II from green plants? J Phys Chem B. 2002;106(22):5782–5787. [Google Scholar]
- 32.Blankenship RE, et al. Comparing photosynthetic and photovoltaic efficiencies and recognizing the potential for improvement. Science. 2011;332(6031):805–809. doi: 10.1126/science.1200165. [DOI] [PubMed] [Google Scholar]
- 33.Reynolds NP, et al. Directed formation of micro- and nanoscale patterns of functional light-harvesting LH2 complexes. J Am Chem Soc. 2007;129(47):14625–14631. doi: 10.1021/ja073658m. [DOI] [PubMed] [Google Scholar]
- 34.Luque A, Hegedus S. Handbook of Photovoltaic Science and Engineering. New York: Wiley; 2003. [Google Scholar]
- 35.Cogdell RJ, Hawthornthwaite AM. The preparation, purification and crystallisation of purple bacterial antenna complexes. In: Deisenhofer J, Norris JR, editors. Photosynthetic Reaction Center. New York: Academic; 1993. pp. 23–42. [Google Scholar]
- 36.Kartalov EP, Unger MA, Quake SR. Polyelectrolyte surface interface for single-molecule fluorescence studies of DNA polymerase. Biotechniques. 2003;34(3):505–510. doi: 10.2144/03343st02. [DOI] [PubMed] [Google Scholar]
- 37.Wang Q, Moerner WE. Optimal strategy for trapping single fluorescent molecules in solution using the ABEL trap. Appl Phys B. 2010;99(1-2):23–30. doi: 10.1007/s00340-009-3843-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Watkins LP, Yang H. Detection of intensity change points in time-resolved single-molecule measurements. J Phys Chem B. 2005;109(1):617–628. doi: 10.1021/jp0467548. [DOI] [PubMed] [Google Scholar]
- 39.Moerner WE, Fromm DP. Methods of single-molecule fluorescence spectroscopy and microscopy. Rev Sci Instrum. 2003;74(8):3597–3619. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.