Abstract
The objective of this study was to detect changes in gene expression in the ventral tegmental area (VTA) following repeated excessive binge-like (‘loss-of-control’) alcohol drinking by alcohol-preferring (P) rats. Adult female P rats (n = 7) were given concurrent access to 10, 20, and 30% EtOH for 4 1-hour sessions daily for 10 weeks followed by 2 cycles of 2 weeks of abstinence and 2 weeks of EtOH access. Rats were sacrificed by decapitation 3 hours after the 4th daily EtOH-access session at the end of the second 2-week relapse period. A water-control group of female P rats (n = 8) was also sacrificed. RNA was prepared from micro-punch samples of the VTA from individual rats; analyses were conducted with Affymetrix Rat 230.2 GeneChips. Ethanol intakes were 1.2–1.7 g/kg per session, resulting in blood levels > 200 mg% at the end of the 4th session. There were 211 unique named genes that significantly differed (FDR = 0.1) between the water and EtOH groups. Bioinformatics analyses indicated alterations in a) transcription factors that reduced excitation-coupled transcription and promoted excitotoxic neuronal damage involving clusters of genes associated with Nfkbia, Fos, and Srebf1, b) genes that reduced cholesterol and fatty acid synthesis, and increased protein degradation, and c) genes involved in cell-to-cell interactions and regulation of the actin cytoskeleton. Among the named genes, there were 62 genes that showed differences between alcohol-naïve P and non-preferring (NP) rats, with 43 of the genes changing toward NP-like expression levels following excessive binge-like drinking in the P rats. These genes are involved in a pro-inflammatory response, and enhanced response to glucocorticoids and steroid hormones. Overall, the results of this study indicate that the repeated excessive binge-like alcohol drinking can change the expression of genes that may alter neuronal function in several ways, some of which may be deleterious.
Keywords: alcohol-preferring rat, binge-like alcohol drinking, ventral tegmental area, gene expression
Introduction
Examining changes in gene expression resulting from chronic ethanol drinking could provide clues to identifying genes and gene networks involved in maintaining high alcohol drinking behavior, as well as identifying genes involved in the neurotoxic consequences of chronic alcohol consumption. There have been several studies that applied genomic and proteomic analyses to examine the effects of ethanol in rodent models (Bell et al., 2006, 2009; Kerns et al., 2005; McBride et al., 2010; Mulligan et al., 2006, 2011; Rodd et al., 2008; Saito et al., 2002, 2004; Tabakoff et al., 2009; Treadwell and Singh, 2004). Studies conducted on post-mortem human tissue have examined the effects of chronic alcohol consumption (Alexander-Kaufman et al., 2006, 2007; Flatscher-Bader et al., 2005, 2010; Lewohl et al., 2000, 2004; Liu et al., 2004, 2006; Matsumoto et al., 2007; Mayfield et al., 2002). Collectively, these studies indicate that differences between alcoholics and controls could be detected in several brain regions, and these differences may represent altered neuronal function.
Alterations in gene expression produced by exposure to alcohol have been reported in several studies with rats and mice. Acute ethanol injections (6 g/kg; i.p.) produced changes in whole brain of C57BL/6J and DBA/2J mice (high- and low-alcohol drinkers, respectively) in expression of genes involved in regulating cell signaling, gene regulation, and homeostasis/stress response (Treadwell and Singh, 2004). Kerns et al. (2005) reported that acute i.p. ethanol injections altered expression of genes involved in glucocorticoid signaling, neurogenesis, myelination, neuropeptide signaling, and retinoic acid signaling in the nucleus accumbens (Acb), prefrontal cortex, and ventral tegmental area (VTA) of C57BL/6J and DBA/2J mice. Differences in expression of genes coding for oxido-reductases and ADP-ribosylation factors were found in the dorsal hippocampus of Lewis rats given 12% ethanol or water for 15 months (Saito et al., 2002). In contrast, Saito et al. (2004) found no statistically significant effects of chronic free-choice alcohol drinking on gene expression in the striatum of C57BL/6By mice. The above rodent studies were conducted using ethanol injections or 24-hour free-choice drinking. Moreover, other than the study of Kerns et al. (2005) using i.p. ethanol injections, none of the above rodent studies reported data on limbic regions that are involved in mediating alcohol drinking.
Bell et al. (2006) examined the effects of chronic alcohol drinking by alcohol-preferring (P) rats on protein levels in the Acb and amygdala, and reported a small number of changes in each region following chronic alcohol drinking. Rodd et al. (2008) examined the effects of operant ethanol self-administration on changes in gene expression in the Acb and amygdala of inbred P rats 24 hours after the last 1-hour drinking session. In the Acb, approximately 200 genes differed significantly between the ethanol and water-control groups, whereas in the amygdala, few significant differences were observed. In another study (Bell et al., 2009), gene expression changes were detected in the Acb of P rats following 2 different alcohol-drinking conditions, i.e., continuous 24-hour access and multiple 1-hour daily scheduled access periods. Significant differences in expression of genes involved in intracellular signaling pathways and transcription factors were found between the continuous-access and water-control groups, whereas no significant differences were observed in the Acb between the multiple scheduled-access and the water-control groups, when measured the day after the last drinking episode. McBride et al. (2010) reported significant changes in gene expression in the Acb-shell and central nucleus of the amygdala (CeA) of P rats after binge-like alcohol drinking (8 weeks of access to 15% and 30% ethanol in 3 1-hour daily scheduled-access sessions). Although there were some categories of biological processes in common between the two regions (e.g., synaptic transmission, neurite development), there were few genes in common between the two regions.
Few studies on changes in gene expression in the VTA following chronic alcohol drinking have been undertaken. In human studies, Flatscher-Bader et al. (2008, 2010) reported that changes in genes involved with neurotransmission and signal transduction in the VTA were associated with alcohol abuse.
The VTA is a critical part of the brain reward system and has been implicated in mediating the rewarding actions of ethanol (Gatto et al, 1994; McBride et al., 1999; Rodd et al., 2004; Rodd-Henricks et al., 2000) and in regulating alcohol drinking (Hodge et al., 1993; Hwa et al., 2012; Rodd et al., 2010) and seeking (Hauser et al., 2011). Other studies indicated that chronic alcohol drinking or repeated local administration of ethanol increased the sensitivity of the VTA to the rewarding (Rodd et al. 2005a, b) and dopamine stimulating (Ding et al., 2009) effects of ethanol.
Thus far, studies on the effects of alcohol drinking on changes in gene expression in the VTA have not been examined in an accepted animal model of alcoholism. The present study uses a unique binge-like ethanol drinking protocol that produces daily blood ethanol concentrations (BECs) in excess of 200 mg%, and examines the effects of this dangerous level of alcohol drinking on gene expression in the VTA 3 hours after the last drinking episode.
Materials and methods
Subjects were adult (approximately 90 days old at the start of the experiment) female selectively bred P rats from the 61st generation. The rats were single-housed on a reverse 12-h/12-h dark-light cycle (lights off at 9:00 AM). Animals had ad libitum access to food and water. The animals used in these experiments were maintained in facilities fully accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care (AAALAC). All research protocols were approved by the institutional animal care and use committee and are in accordance with the guidelines of the Institutional Care and Use Committee of the National Institute on Drug Abuse, National Institutes of Health, and the Guide for the Care and Use of Laboratory Animals (Institute of Laboratory Animal Resources, Commission on Life Sciences, National Research Council 1996).
The rats were self-trained in 3-lever operant chambers using essentially the same procedure previously described for a 2-lever operant paradigm (Rodd et al, 2008; Rodd-Henricks et al., 2002a, b). The P rats (n = 7) were given 4 1-hour sessions in the 3-lever operant chambers to respond for 10, 20, and 30% ethanol on an FR5 schedule of reinforcement. Water was freely available during these sessions. Water and food were freely available in the home cage. The 1-hour operant sessions were conducted during the dark cycle; there was a 1-hour interval between each operant session. Sessions were conducted 5 consecutive days each week. Following 10 weeks of ethanol access, rats were taken through 2 cycles of 2 weeks of alcohol deprivation and 2 weeks of ethanol access. Ethanol intakes were similar during the relapse periods as were observed during weeks 9 and 10. Rats were killed by decapitation 3 hours after the 4th session at the end of the second 2-week relapse period. This 3-hour time point was selected in an attempt to maximize the response to alcohol on the expression of genes in tissue from rats that have had a history of repeated excessive binge drinking. The brains were quickly removed and frozen in isopentane on solid frozen CO2. Age-matched alcohol-naïve water-control female P rats (n = 8) were killed and brains were removed in a similar manner. Brains were stored at −70°C until sectioned.
Tail-blood samples were taken following the 1st, 2nd and 4th daily-access sessions, and prior to the 3rd session to provide BECs across the drinking period. Blood samples were taken from multiple rats on different occasions to minimize the impact on overall drinking. Ethanol concentrations were determined with an Analox Analyzer.
Sample collection and microarray procedure
On the day of preparation of micro-punch samples, brains were transferred to a cryostat set at −6 to −10°C at least 2 hours prior to sectioning. Sections (300 μm) were obtained and transferred to glass slides that had been pre-cooled in the cryostat. Micro-punch sampling was done on a frozen stage (−25 to −35°C) with an anatomic microscope equipped with a cool microscope lamp. The stereotaxic atlas of Paxinos and Watson (1998) was used to identify the VTA. Micro-dissection needles (Fisher Scientific) with an inner diameter of 0.77 mm were used to obtain the VTA. This inner diameter fits within the entire region and minimizes contamination from adjacent tissue. Punches were taken bilaterally from 2–3 sections. A different fresh sterile micro-punch needle was used for each animal. After withdrawing the micro-punch sample, a distinct demarcated hole remained; this hole was used to validate the micro-dissection method. All equipment used to obtain tissue was treated with RNAse Zap (Ambion, Inc. Austin, TX) to prevent RNA degradation. A second trained individual independently verified the quality of the micro-punch dissections.
The micro-punched samples were immediately homogenized in Trizol reagent (Invitrogen, Carlsbad, CA) and processed according to the manufacturer’s protocol, but with twice the suggested ratio of Trizol to tissue (Edenberg et al., 2005). Ethanol-precipitated RNA was further purified through RNeasy columns (Qiagen, Valencia, CA) according to the manufacturer’s protocol. The yield, concentration, and purity of the RNA were determined by running a spectrum from 210 to 350 nm, and analyzing the ratio of large and small ribosomal RNA bands using an Agilent Bioanalyzer. Yields, purity, and quality of the RNA were excellent; RNA integrity numbers (RIN) averaged 8.5 for the samples, showing little or no degradation.
Separate preparations of total RNA were made for the VTA from each animal. Samples were not pooled. Standard Affymetrix protocols (GeneChip Expression Analysis Technical Manual, Rev. 5 and updates) were used to synthesize biotinylated cRNA, using the Affymetrix kits for cDNA synthesis, in vitro transcription, and sample cleanup. The fragmented, biotinylated cRNA from each independent sample was mixed into 300 μl of hybridization cocktail, of which 200 μl were used for each sample. Hybridization was carried out for 17 hours at 42°C. Samples were hybridized to the Affymetrix Rat Genome 230 2.0 GeneChips. Washing and scanning of the GeneChips were carried out according to standard protocols, as previously described (Edenberg et al., 2005; McClintick et al., 2003).
To minimize potential systematic errors, all stages of the experiment were balanced across experimental groups. That is, equal numbers of animals in each group were sacrificed within the same 2-hour time frame each day, and equal numbers of RNA preparations from the 2 groups were processed through the labeling, hybridization, washing, and scanning protocols on a given day, in a counterbalanced order, using pre-mixed reagents.
Statistical and neuroinformatics analysis of microarray data
Each GeneChip was scanned using an Affymetrix Model 3000 scanner and underwent image analysis using Affymetrix GCOS software. Microarray data are available from the National Center for Biotechnology Information’s Gene Expression Omnibus under accession GSE42578.
Raw cel files were imported into the statistical programming environment R (R: A language and environment for statistical computing Ver 2.13.0; R Foundation for Statistical Computing, 2005) for further analysis with tools available from the Bioconductor Project (Gentleman et al., 2004). Expression data of the 15 arrays from the VTA were normalized and converted to log(2) using the Robust Multi-chip Average (RMA) method (Irizarry et al., 2003) implemented in the Bioconductor package RMA. As a standardization step to facilitate later comparisons with other experiments, expression levels were scaled such that the mean expression of all arrays was log2(1000). As we were primarily concerned with identifying genes that could be subjected to further bioinformatic analysis, all probe sets currently annotated by Affymetrix as “expressed sequence tags” or whose gene names contain the words “riken,” “predicted,” or “similar to” were filtered out. We next filtered out probe sets that were not detectable above background in our samples; this has been shown to reduce noise in microarray experiments (McClintick and Edenberg, 2006). Probe sets that did not have at least 25% of samples with normalized scaled expression greater than 64 were not analyzed. Linear modeling to calculate gene-wise p values for the contrasts of the ethanol group versus water group was performed using the package Limma (Smyth, 2004). Probe sets were considered to be statistically significant at FDR = 0.1 (calculated according to Storey et al., 2004).
Testing for over-representation of Gene Ontology (Ashburner et al., 2000; Harris et al., 2004; GO) biological processes (BP) and Kyoto Encyclopedia of Genes and Genomes (KEGG) categories was performed using the Bioconductor package GOstats (Gentleman, 2004). Briefly, for each gene set tested, a list of unique Entrez-Gene identifiers was constructed. This list was then compared to the list of all known Entrez-Gene identifiers that are represented on the Affymetrix chipset Rat Genome 230 2.0. Identification of over-represented GO categories was then accomplished within GOstats using the hypergeometric distribution. GO and KEGG categories were called significant at p < .05.
Genes differentially expressed at FDR = 0.1 were uploaded into Ingenuity Pathway Analysis (Ingenuity Systems, www.ingenuity.com). Genes were identified by their Affymetrix probe set ID and then mapped to their corresponding objects in the Ingenuity Knowledge Base. These molecules, called Network Eligible molecules, were overlaid onto a global molecular network developed from information contained in the Ingenuity Knowledge Base. Networks of Network Eligible Molecules were then algorithmically generated based on their connectivity.
To provide a more global network analysis, a weighted gene co-expression network analysis (WGCNA) was also conducted (Zhang and Horvath, 2005), using the Bioconductor (Gentleman et al., 2004) package WGCNA (Langfelder and Horvath, 2008, 2012) within R (R: A language and environment for statistical computing Ver 2.15.0; R Foundation for Statistical Computing, 2013). Briefly, gene expression data of named genes were rank-ordered according to their ascending p values obtained from traditional t testing of the two experimental groups. For WGCNA, default values, including the use of the power function with power β, were used for all functions with the exception that signed correlation coefficients were used. Various p- and FDR-value cutoffs between FDR ≤ 0.10 and p ≤ .10 were tried in an attempt to select a set of genes whose resultant networks met the criteria of legitimacy for scale free topology (Zhang and Horvath, 2005). Resultant modules were tested for enrichment of various categories of genes using Fisher’s Exact Test. Categories tested included GO biologic process (Ashburner et al., 2000; Harris et al., 2004) and location by cell type (Cahoy et al., 2008).
Results
Ethanol intakes and BECs
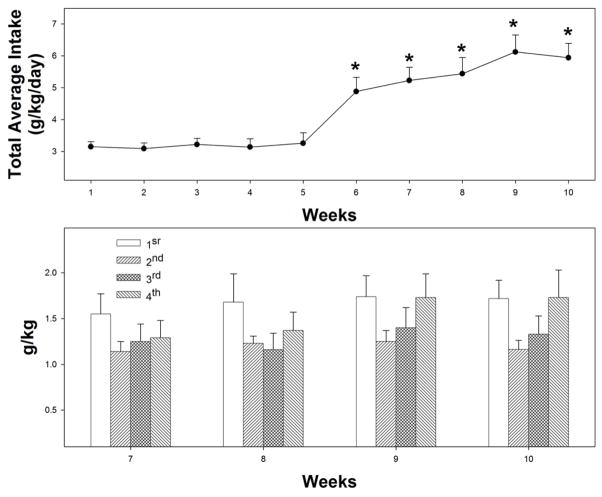
Figure 1 shows the average daily ethanol intakes during the first 10 weeks of concurrent access to 10, 20, and 30% ethanol and ethanol intakes during each of the 4 sessions during weeks 7–10. P rats readily acquired ethanol self-administration during the 2nd day of concurrent access to the 3 ethanol solutions (similar to Rodd-Henricks et al., 2002a, b). During the initial 5 weeks of responding, overall ethanol intakes were consistent (3.2 ± 0.2 g/kg for the 4 1-hour sessions; Fig. 1, top panel). Elevated ethanol intakes were observed during week 6, when total daily amount increased 48% (two-tailed t test indicated significant increase from average intake of weeks 3–5; p < .001). Ethanol intakes gradually increased until week 9 (92% increase compared to average intake of weeks 3–5; p < .001) and stabilized between weeks 9–10.
Fig. 1.
Estimated total ethanol intakes (g/kg/day; top panel) by P rats given concurrent access to 10, 20, and 30% ethanol (with water freely available) for 4 1-hour sessions each day during the dark cycle. Estimated intakes (g/kg) for each of the 4 1-hour access sessions during weeks 7–10 are given in the bottom panel. Ethanol was given 5 consecutive days each week (no ethanol on weekends) over the 10 weeks of access. *Indicates significantly (p < .001) higher ethanol intakes compared to sessions 3–5. BECs sampled across multiple sessions during weeks 8–10 indicated: 1) 124 ± 14 mg% following 1st session; 2) 177 ± 19 mg% following the 2nd session; 3) 128 ± 16 mg% immediately prior to the 3rd session; and 4) 237 ± 20 mg% following the 4th session. Data are the means ± SEM (n = 7).
During the individual sessions, ethanol intakes were slightly higher during the 1st and 4th session compared to sessions 2 and 3 (Fig. 1, bottom panel); the 1-hour session average was approximately 1.2 to 1.7 g/kg during the last two weeks of access.
BECs were sampled across multiple sessions during weeks 8–10. Samples were obtained following the 1st and 2nd session, immediately prior to the 3rd session (60 minutes post-2nd session), and following the 4th session (see legend of Fig. 1 for values). BECs were approximately 120 mg% following the 1st session, and exceeded 150 mg% following the 2nd session. Prior to the 3rd session, BECs were still higher than 100 mg% even after 60 minutes without access to ethanol. BECs following the 4th session exceeded 200 mg%. The estimated total ethanol intakes were significantly correlated with BECs obtained following the 4th session (r = 0.89).
Changes in gene expression in the VTA
There were a total of 335 probe sets that significantly differed between the ethanol and water groups in the VTA: 102 ESTs and 233 named genes (Table 1 shows a list of named genes comprised of 211 unique genes). Over 75% of the fold changes were 1.2 or higher (Table 1).
Table 1.
List of named genes in the VTA of P rats that were significantly different (FDR = 0.1) between the ethanol and water groups
| Symbol | Gene Description | F- C | QTL |
|---|---|---|---|
| Aadat | aminoadipate aminotransferase | 1.29 | Alc11 |
| Abca1 | ATP-binding cassette, sub-family A (ABC1), member 1 | 1.28 | |
| Acat2 | acetyl-Coenzyme A acetyltransferase 2 | −1.29 | |
| Aco1 | aconitase 1, soluble | −1.13 | |
| Acot4 | acyl-CoA thioesterase 4 | 1.31 | |
| Acss2 | acyl-CoA synthetase short-chain family member 2 | −1.40 | |
| Aif1 | allograft inflammatory factor 1 | 1.32 | |
| Aifm3 | apoptosis-inducing factor, mitochondrion-associated 3 | −1.20 | |
| Anxa3 | annexin A3 | 1.27 | |
| Anxa3 | annexin A3 | 1.34 | |
| Anxa3 | Annexin A3 | 1.32 | |
| Apln | apelin | −1.34 | |
| Apln | apelin | −1.42 | |
| Arhgap25 | Rho GTPase activating protein 25 | 1.28 | Alc18 |
| Arl11 | ADP-ribosylation factor-like 11 | 1.29 | |
| Azin1 | antizyme inhibitor 1 | −1.20 | |
| Azin1 | antizyme inhibitor 1 | −1.42 | |
| B2m | beta-2 microglobulin | 1.14 | |
| B2m | Beta-2 microglobulin | 1.17 | |
| Baiap2 | BAI1-associated protein 2 | −1.17 | |
| Baiap2 | BAI1-associated protein 2 | −1.21 | |
| Bin2 | bridging integrator 2 | 1.20 | |
| Birc7 | baculoviral IAP repeat-containing 7 | 1.39 | |
| C1qa | complement component 1, q subcomponent, A chain | 1.29 | |
| C1qtnf5 | C1q and tumor necrosis factor related protein 5 | −1.20 | |
| C3 | complement component 3 | 2.56 | |
| Casp1 | caspase 1 | 1.20 | |
| Ccdc125 | coiled-coil domain containing 125 | 1.19 | |
| Ccdc28b | coiled coil domain containing 28B | −1.18 | |
| Ccl6 | chemokine (C-C motif) ligand 6 | −1.71 | |
| Ccne2 | cyclin E2 | −1.29 | |
| Cd53 | Cd53 molecule | 1.28 | Alc15 |
| Cd9 | CD9 molecule | −1.20 | |
| Cdh23 | cadherin 23 (otocadherin) | 1.24 | |
| Cdk2ap1 | CDK2-associated protein 1 | −1.15 | Alc6 |
| Celsr2 | cadherin, EGF LAG seven-pass G-type receptor 2 (flamingo homolog, Drosophila) | 1.21 | Alc15 |
| Chfr | checkpoint with forkhead and ring finger domains | −1.14 | |
| Chmp1b | chromatin modifying protein 1B | −1.12 | |
| Chordc1 | cysteine and histidine-rich domain (CHORD)-containing 1 | −1.23 | |
| Cldn11 | claudin 11 | −1.37 | |
| Cldn11 | claudin 11 | −1.21 | |
| Cmip | c-Maf-inducing protein | 1.13 | |
| Col14a1 | collagen, type XIV, alpha 1 | 1.25 | |
| Col1a1 | collagen, type I, alpha 1 | −1.40 | |
| Col3a1 | collagen, type III, alpha 1 | −1.60 | |
| Col4a5 | collagen, type IV, alpha 5 | −1.30 | |
| Crcp | CGRP receptor component | 1.20 | Alc10 |
| Csrp2 | cysteine and glycine-rich protein 2 | −1.37 | |
| Ctse | cathepsin E | 1.22 | |
| Ctsl1 | cathepsin L1 | −1.17 | |
| Ctss | cathepsin S | 1.28 | |
| Ctsz | cathepsin Z | 1.27 | |
| Cxcr4 | chemokine (C-X-C motif) receptor 4 | 1.21 | |
| Cyp51 | cytochrome P450, family 51 | −1.40 | |
| Cyp51 | cytochrome P450, family 51 | −1.35 | |
| Dctd | dCMP deaminase | 1.30 | Alc11 |
| Dcxr | dicarbonyl L-xylulose reductase | 1.15 | |
| Ddit4 | DNA-damage-inducible transcript 4 | −1.56 | |
| Decr1 | 2,4-dienoyl CoA reductase 1, mitochondrial | 1.14 | |
| Dhcr7 | 7-dehydrocholesterol reductase | −1.23 | |
| Dock6 | dedicator of cytokinesis 6 | −1.23 | |
| Dock8 | dedicator of cytokinesis 8 | 1.20 | |
| Dusp1 | dual specificity phosphatase 1 | −1.40 | Alc5 |
| Ece2 | endothelin-converting enzyme 2 | 1.16 | |
| Egr1 | early growth response 1 | −1.54 | |
| Eif4b | eukaryotic translation initiation factor 4B | 1.15 | |
| Emr1 | EGF-like module containing, mucin-like, hormone receptor-like 1 | 1.57 | |
| Errfi1 | ERBB receptor feedback inhibitor 1 | −1.35 | |
| Ezh2 | enhancer of zeste homolog 2 (Drosophila) | −1.33 | Alc18 |
| Ezh2 | enhancer of zeste homolog 2 (Drosophila) | −1.27 | Alc18 |
| Fa2h | fatty acid 2-hydroxylase | −1.36 | |
| Fads1 | fatty acid desaturase 1 | −1.15 | |
| Fam181b | family with sequence similarity 181, member B | −1.16 | |
| Fcer1g | Fc fragment of IgE, high affinity I, receptor for; gamma polypeptide | 1.33 | |
| Fcgr2a | Fc fragment of IgG, low affinity IIa, receptor (CD32) | 1.32 | |
| Fcgr2a | Fc fragment of IgG, low affinity IIa, receptor (CD32) | 1.69 | |
| Fcgr2a | Fc fragment of IgG, low affinity IIa, receptor (CD32) | 1.33 | |
| Fcgr3a | Fc fragment of IgG, low affinity IIIa, receptor | 1.28 | |
| Fgfr3 | fibroblast growth factor receptor 3 | −1.17 | |
| Fgfr3 | Fibroblast growth factor receptor 3 | −1.13 | |
| Fos | FBJ osteosarcoma oncogene | −1.54 | |
| Gadd45b | growth arrest and DNA-damage-inducible, beta | 1.24 | |
| Gamt | guanidinoacetate N-methyltransferase | −1.24 | |
| Gng12 | guanine nucleotide binding protein (G protein), gamma 12 | −1.34 | Alc18 |
| Gpatch4 | G patch domain containing 4 | −1.31 | |
| Gpatch4 | G patch domain containing 4 | −1.35 | |
| Gpd1 | glycerol-3-phosphate dehydrogenase 1 (soluble) | −1.42 | |
| Gpd1 | glycerol-3-phosphate dehydrogenase 1 (soluble) | −1.76 | |
| Gpr34 | G protein-coupled receptor 34 | 1.81 | |
| Gpr84 | G protein-coupled receptor 84 | 1.30 | |
| Gramd3 | GRAM domain containing 3 | −1.22 | |
| Gramd3 | GRAM domain containing 3 | −1.29 | |
| Grxcr1 | glutaredoxin, cysteine rich 1 | 2.64 | |
| Gstk1 | glutathione S-transferase kappa 1 | 1.13 | Alc18 |
| Hhip | Hedgehog-interacting protein | −1.27 | |
| Hip1 | huntingtin interacting protein 1 | −1.25 | Alc10 |
| Hip1 | huntingtin interacting protein 1 | −1.29 | Alc10 |
| Hist1h2bh | histone cluster 1, H2bh | −1.28 | |
| Hmgcr | 3-hydroxy-3-methylglutaryl-Coenzyme A reductase | −1.20 | |
| Hmgcs1 | 3-hydroxy-3-methylglutaryl-Coenzyme A synthase 1 (soluble) | −1.34 | |
| Hsd17b7 | hydroxysteroid (17-beta) dehydrogenase 7 | −1.28 | |
| Idi1 | isopentenyl-diphosphate delta isomerase 1 | −1.31 | |
| Ifrd1 | interferon-related developmental regulator 1 | −1.16 | |
| Igf2bp2 | insulin-like growth factor 2 mRNA binding protein 2 | −1.16 | |
| Il10rb | interleukin 10 receptor, beta | 1.18 | |
| Irf1 | interferon regulatory factor 1 | 1.18 | |
| Irf8 | Interferon regulatory factor 8 | 1.33 | |
| Itgb1 | integrin, beta 1 | −1.15 | |
| Itgb2 | integrin, beta 2 | 1.26 | |
| Kcnj8 | potassium inwardly-rectifying channel, subfamily J, member 8 | 1.31 | |
| Klf9 | Kruppel-like factor 9 | −1.24 | |
| Laptm5 | lysosomal protein transmembrane 5 | 1.25 | |
| Laptm5 | lysosomal protein transmembrane 5 | 1.24 | |
| Lgals3bp | lectin, galactoside-binding, soluble, 3 binding protein | 1.29 | |
| Lims2 | LIM and senescent cell antigen like domains 2 | −1.29 | |
| LOC100362458 | rCG23949-like | 1.21 | |
| LOC290595 | hypothetical gene supported by AF152002 | 1.48 | Alc13 |
| LOC64038 | Sertolin | 1.11 | |
| LOC691777 | hypothetical protein LOC691777 | 1.16 | |
| Lppr5 | lipid phosphate phosphatase-related protein type 5 | −1.22 | Alc15 |
| Lrpap1 | low density lipoprotein receptor-related protein associated protein 1 | 1.17 | |
| Lrrc33 | leucine rich repeat containing 33 | 1.23 | |
| Ly86 | lymphocyte antigen 86 | 1.24 | |
| Mal | mal, T-cell differentiation protein | −1.33 | |
| Mfsd2 | major facilitator superfamily domain containing 2 | −1.29 | |
| Mog | myelin oligodendrocyte glycoprotein | −1.23 | |
| Mpg | N-methylpurine-DNA glycosylase | 1.15 | Alc5 |
| Mthfd1l | methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like | −1.26 | |
| Mtmr2 | myotubularin related protein 2 | −1.27 | |
| Mysm1 | myb-like, SWIRM and MPN domains 1 | −1.18 | |
| Naaa | N-acylethanolamine acid amidase | 1.15 | |
| Ndfip2 | Nedd4 family interacting protein 2 | −1.12 | |
| Nfasc | neurofascin | −1.23 | |
| Nfasc | neurofascin | −1.25 | |
| Nfil3 | nuclear factor, interleukin 3 regulated | −1.40 | |
| Nfkbia | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha | −1.32 | |
| Nol3 | nucleolar protein 3 (apoptosis repressor with CARD domain) | 1.11 | |
| Npc2 | Niemann-Pick disease, type C2 | 1.20 | |
| Nr1d1 | nuclear receptor subfamily 1, group D, member 1 | 1.25 | |
| Nsdhl | NAD(P) dependent steroid dehydrogenase-like | −1.24 | |
| Olig1 | oligodendrocyte transcription factor 1 | −1.22 | |
| Pald | paladin | 1.18 | |
| Pcdh20 | protocadherin 20 | −1.46 | |
| Pcdh20 | protocadherin 20 | −1.47 | |
| Pcdhb9 | protocadherin beta 9 | 1.33 | |
| Pck2 | phosphoenolpyruvate carboxykinase 2 (mitochondrial) | 1.16 | |
| Pex11a | peroxisomal biogenesis factor 11 alpha | −1.44 | |
| Phactr3 | phosphatase and actin regulator 3 | −1.34 | |
| Plcd4 | phospholipase C, delta 4 | −1.21 | |
| Plekha1 | pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1 | −1.16 | |
| Plekhf1 | pleckstrin homology domain containing, family F (with FYVE domain) member 1 | −1.34 | |
| Plod1 | procollagen-lysine 1, 2-oxoglutarate 5-dioxygenase 1 | −1.14 | |
| Pnlip | pancreatic lipase | −1.52 | |
| Pppde2 | PPPDE peptidase domain containing 2 | −1.18 | |
| Prickle1 | prickle homolog 1 (Drosophila) | −1.16 | |
| Prkcd | protein kinase C, delta | 1.19 | Alc13 |
| Prkd2 | protein kinase D2 | −1.26 | |
| Psmb9 | proteasome (prosome, macropain) subunit, beta type 9 (large multifunctional peptidase 2) | 1.30 | |
| Psme1 | proteasome (prosome, macropain) activator subunit 1 | 1.18 | |
| Ptpla | protein tyrosine phosphatase-like (proline instead of catalytic arginine), member a | −1.29 | |
| Ptpla | protein tyrosine phosphatase-like (proline instead of catalytic arginine), member a | −1.22 | |
| Ptprc | protein tyrosine phosphatase, receptor type, C | 1.29 | |
| Pycard | PYD and CARD domain containing | 1.23 | |
| Rab27a | RAB27A, member RAS oncogene family | 1.20 | |
| Rac2 | ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) | 1.21 | |
| Ralgds | ral guanine nucleotide dissociation stimulator | −1.22 | Alc8 |
| Rem2 | RAS (RAD and GEM) like GTP binding 2 | −1.22 | |
| RGD1566254 | RGD1566254 | 1.29 | |
| Rhobtb3 | Rho-related BTB domain containing 3 | −1.22 | |
| Ril | reversion induced LIM gene | 1.15 | |
| Rnasel | Ribonuclease L (2′,5′-oligoisoadenylate synthetase-dependent) | 1.17 | |
| Rnaset2 | ribonuclease T2 | 1.21 | |
| Rpe | ribulose-5-phosphate-3-epimerase | −1.12 | |
| Rras2 | related RAS viral (r-ras) oncogene homolog 2 | −1.21 | |
| Rras2 | related RAS viral (r-ras) oncogene homolog 2 | −1.25 | |
| Sc4mol | sterol-C4-methyl oxidase-like | −1.32 | Alc11 |
| Sc5dl | sterol-C5-desaturase (ERG3 delta-5-desaturase homolog, S. cerevisiae)-like | −1.19 | |
| Sc5dl | sterol-C5-desaturase (ERG3 delta-5-desaturase homolog, S. cerevisiae)-like | −1.17 | |
| Scarb2 | scavenger receptor class B, member 2 | −1.15 | |
| Scd1 | stearoyl-Coenzyme A desaturase 1 | −1.48 | |
| Scrg1 | stimulator of chondrogenesis 1 | −1.26 | Alc11 |
| Sephs2 | selenophosphate synthetase 2 | −1.15 | |
| Serpinb9 | serine (or cysteine) peptidase inhibitor, clade B, member 9 | 1.22 | |
| Serpinb9 | Serine (or cysteine) peptidase inhibitor, clade B, member 9 | 1.17 | |
| Serpinh1 | serine (or cysteine) peptidase inhibitor, clade H, member 1 | −1.28 | |
| Sfrs7 | splicing factor, arginine/serine-rich 7 | 1.12 | Alc17 |
| Sgk1 | serum/glucocorticoid regulated kinase 1 | −1.60 | |
| Sh3tc2 | SH3 domain and tetratricopeptide repeats 2 | 1.19 | |
| Slain2 | SLAIN motif family, member 2 | −1.18 | |
| Slc15a3 | solute carrier family 15, member 3 | 1.38 | |
| Slc22a23 | solute carrier family 22, member 23 | −1.12 | |
| Slc22a4 | solute carrier family 22 (organic cation transporter), member 4 | 1.59 | |
| Slc25a1 | solute carrier family 25 (mitochondrial carrier, citrate transporter), member 1 | −1.21 | |
| Slc29a2 | solute carrier family 29 (nucleoside transporters), member 2 | −1.12 | |
| Slc38a2 | solute carrier family 38, member 2 | −1.16 | |
| Slc6a8 | solute carrier family 6 (neurotransmitter transporter, creatine), member 8 | −1.17 | |
| Slc9a3 | solute carrier family 9 (sodium/hydrogen exchanger), member 3 | 1.31 | |
| Slc9a3r2 | solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2 | −1.21 | Alc5 |
| Smagp | small trans-membrane and glycosylated protein | 1.31 | |
| Smarcd2 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 2 | −1.22 | |
| Smc4 | structural maintenance of chromosomes 4 | −1.18 | |
| Spint2 | serine peptidase inhibitor, Kunitz type, 2 | 1.29 | |
| Sqle | squalene epoxidase | −1.35 | |
| Srebf1 | sterol regulatory element binding transcription factor 1 | 1.30 | |
| Svep1 | Sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1 | −1.29 | |
| Tc2n | tandem C2 domains, nuclear | 1.25 | |
| Tekt4 | tektin 4 | −1.32 | Alc5 |
| Timm8a1 | translocase of inner mitochondrial membrane 8 homolog a1 (yeast) | −1.31 | |
| Timm8a1 | translocase of inner mitochondrial membrane 8 homolog a1 (yeast) | −1.26 | |
| Timp2 | TIMP metallopeptidase inhibitor 2 | 1.23 | |
| Tinagl1 | tubulointerstitial nephritis antigen-like 1 | −1.30 | |
| Tjap1 | tight junction associated protein 1 | −1.19 | |
| Tm7sf2 | transmembrane 7 superfamily member 2 | −1.47 | |
| Tmem176a | transmembrane protein 176A | 1.24 | Alc18 |
| Tmem176b | transmembrane protein 176B | 1.27 | Alc18 |
| Tmem81 | transmembrane protein 81 | 1.14 | |
| Tnfaip6 | tumor necrosis factor alpha induced protein 6 | −1.35 | |
| Trem2 | triggering receptor expressed on myeloid cells 2 | 1.43 | |
| Tsc22d1 | TSC22 domain family, member 1 | −1.10 | |
| Tsc22d3 | TSC22 domain family, member 3 | −1.27 | |
| Ttk | Ttk protein kinase | 1.21 | |
| Tyrobp | Tyro protein tyrosine kinase binding protein | 1.34 | |
| Ube2d2 | ubiquitin-conjugating enzyme E2D 2 | −1.14 | |
| Ube2g1 | ubiquitin-conjugating enzyme E2G 1 | −1.12 | |
| Ugt8 | UDP glycosyltransferase 8 | −1.64 | Alc15 |
| Usp2 | ubiquitin specific peptidase 2 | −1.41 | |
| Vav1 | vav 1 guanine nucleotide exchange factor | 1.14 | |
| Vcl | vinculin | 1.20 | |
| Wfdc1 | WAP four-disulfide core domain 1 | −1.22 | |
| Xdh | xanthine dehydrogenase | −1.24 | Alc17 |
| Xylt2 | xylosyltransferase II | 1.14 | |
| Zfp189 | zinc finger protein 189 | −1.20 | |
| Zfp90 | zinc finger protein 90 | 1.12 |
F-C = fold change ethanol/water; positive sign – ethanol > water; negative sign – ethanol < water QTLs are for rat.
GO analysis indicated there were 8 biological processes categories with 10 or more genes that differed significantly between the water and ethanol groups (Table 2). Three categories involved metabolic processes, the first for steroids, the second for alcohol and the third for proteins. Two other categories (cellular adhesion and transcription) contain genes that could influence neuronal function.
Table 2.
List of significant GO biological processes categories containing 10 or more genes in the VTA of the ethanol vs. water group
| Category_ID | Term | P-value | OddsRatio | ExpCount | Count | Size |
|---|---|---|---|---|---|---|
| GO:0008202 | steroid metabolic process | 0 | 7.09 | 1.9 | 11 | 73 |
| GO:0045321 | leukocyte activation | 0 | 4.66 | 3.7 | 15 | 144 |
| GO:0006066 | alcohol metabolic process | 0.00012 | 4.05 | 3.3 | 12 | 128 |
| GO:0007155 | cell adhesion | 0.00087 | 2.67 | 6.6 | 16 | 258 |
| GO:0032268 | regulation of cellular protein metabolic process | 0.01403 | 2.05 | 7.3 | 14 | 287 |
| GO:0003008 | system process | 0.01659 | 2.32 | 4.6 | 10 | 180 |
| GO:0000122 | negative regulation of transcription from RNA polymerase II promoter | 0.0182 | 2.1 | 6.1 | 12 | 233 |
| GO:0001568 | blood vessel development | 0.02795 | 1.97 | 6.4 | 12 | 249 |
KEGG analysis of the VTA gene data indicated 11 significant categories containing 6 or more genes each (Table 3), some of which could alter neuronal function. Two categories, steroid biosynthesis and extracellular matrix- (ECM) receptor interaction, are consistent with the results of the GO categories of ‘steroid metabolic process’ and ‘cell adhesion.’ Genes in the category of ‘regulation of actin cytoskeleton’ could influence dendritic structure and function, including synaptic plasticity.
Table 3.
List of significant KEGG categories with 6 or more genes in the VTA of the ethanol vs. water group
| Category_ID | Term | Pvalue | OddsRatio | ExpCount | Count | Size |
|---|---|---|---|---|---|---|
| 00100 | Steroid biosynthesis | 0 | 24.43 | 0.5 | 7 | 16 |
| 05140 | Leishmaniasis | 0.00214 | 5.15 | 1.4 | 6 | 42 |
| 04512 | ECM-receptor interaction | 0.00473 | 4.3 | 1.6 | 6 | 49 |
| 04510 | Focal adhesion | 0.00514 | 2.71 | 4.5 | 11 | 139 |
| 05146 | Amoebiasis | 0.00523 | 3.67 | 2.2 | 7 | 66 |
| 04810 | Regulation of actin cytoskeleton | 0.00603 | 2.64 | 4.6 | 11 | 142 |
| 04142 | Lysosome | 0.01022 | 2.92 | 3 | 8 | 93 |
| 04380 | Osteoclast differentiation | 0.01122 | 3.13 | 2.5 | 7 | 76 |
| 04670 | Leukocyte transendothelial migration | 0.01561 | 2.92 | 2.6 | 7 | 81 |
| 04650 | Natural killer cell mediated cytotoxicity | 0.01589 | 3.23 | 2.1 | 6 | 63 |
| 04145 | Phagosome | 0.03419 | 2.44 | 3.1 | 7 | 95 |
Ingenuity pathways analysis indicated different networks were altered by ethanol drinking, one of which had clusters around Fos and Srebf1 and a second which had clusters of genes around Nfkb1a (Table 4). There were 16 genes clustered around Fos and 15 genes clustered around Srebf1; 11 of the 16 genes clustered around Fos and 13 of the 15 genes clustered around Srebf1 had lower expression levels in the ethanol group than in the water group. In contrast, 10 of the 14 genes clustered around Nfkb1a had higher expression levels in the ethanol drinking group vs. water controls.
Table 4.
Summary of Ingenuity Pathway Analysis on effects of excessive binge-like drinking in the VTA of P rats with genes clustering around transcription factors involved with steroid metabolism, excitation-coupled transcription and excitotoxicity
| Transcription factor | Gene Cluster |
|---|---|
| Sterol regulatory element binding transcription factor 1 (Srebf1) | UP: Abca1, Decr1 |
| DOWN: Acss2, Cyp51a1, Dhcr7, Fads1, Hmgcr, Hmgcs1, Idi1, Msmo1, Nsdhl, Sc5dl, Scd, Sqle, Tm7sf2 | |
| FBJ osteosarcoma oncogene (Fos) | UP: Git2, Golm1, Rac2, Serpinb9, Vav1 |
| DOWN: Apln, Cry1, Ctsl, Fstl1, Gamt, Gjb6, Mfil3, Nsdhl, Scd, Smarcd2, Xdh | |
| Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha (Nfkb1a) | UP: B2m, C3, Cxcr4, Fcer1g, Fcgr2a, Irf1, Itgb2, Psmb9, Pycard, Tmem76b |
| DOWN: Ccne2, Col3a1, Errf1, Tnfrsf11a |
UP: Indicates higher gene expression in the ethanol than water group; DOWN: Indicates lower gene expression in the ethanol than water group.
See Table 1 for descriptions of gene symbols.
For the WGCNA, only by using a very liberal p value of .10 as the cutoff for gene inclusion (2137 genes) did the resultant network meet the criteria for scale free topology (Zhang and Horvath, 2005). With β = 12, the soft-threshold R2 value was 0.85 with a slope of −1.79. Median connectivity was 3.18 and maximum connectivity was 38.63. Not including the grey module of uncorrelated genes, there were 4 modules with 100 genes or more (Yellow, Blue, Turquoise, and Brown). The Yellow module is a neuron-enriched module and contained 350 genes. The Blue module is an astrocyte- and oligodendrocyte-enriched module and contained 597 genes. The Yellow module had several GO categories (Table 5) that did not appear in the other GO analysis, e.g., G-protein coupled receptor signaling, transmission of nerve impulses, and regulation of membrane potential. The Blue module had some significant categories that were consistent with the results of the GO, KEGG, and Ingenuity analyses of significant (FDR = 0.10) genes (Tables 2–4), e.g., steroid biosynthetic process, sterol metabolic process. In addition, the Blue module had significant GO categories associated with lipid, fatty acid, and phospholipid metabolism (Table 5).
Table 5.
WGCNA list of significant GO biological categories containing 10 or more genes in the Yellow and Blue modules for the VTA of the ethanol vs. water group
| Category_ID | Term | P-value | OddsRatio | ExpCount | Count | Size |
|---|---|---|---|---|---|---|
| YELLOW MODULE (neuron enriched) | ||||||
| GO:0007186 | G-protein coupled receptor signaling pathway | 0.0012 | 2.31 | 9.4 | 20 | 268 |
| GO:0008284 | positive regulation of cell proliferation | 0.0065 | 1.86 | 13.2 | 23 | 375 |
| GO:0019226 | transmission of nerve impulse | 0.0062 | 1.82 | 14.7 | 25 | 417 |
| GO:0019725 | cellular homeostasis | 0.0087 | 1.73 | 16.7 | 27 | 474 |
| GO:0042391 | regulation of membrane potential | 0.0084 | 2.19 | 6.8 | 14 | 194 |
| GO:0050801 | ion homeostasis | 0.0027 | 1.93 | 14.5 | 26 | 413 |
| GO:0071702 | organic substance transport | 0.0094 | 1.92 | 10.6 | 19 | 300 |
| BLUE MODULE (glia enriched) | ||||||
| GO:0001508 | regulation of action potential | 0.0085 | 2.21 | 6.9 | 14 | 113 |
| GO:0006629 | lipid metabolic process | 0.0000 | 3.07 | 8.7 | 23 | 147 |
| GO:0006633 | fatty acid biosynthetic process | 0.0016 | 2.87 | 5.1 | 13 | 84 |
| GO:0006694 | steroid biosynthetic process | 0.0000 | 4.76 | 4.8 | 18 | 78 |
| GO:0006695 | cholesterol biosynthetic process | 0.0000 | 8.59 | 2.1 | 12 | 34 |
| GO:0007272 | ensheathment of neurons | 0.0005 | 3.35 | 4.5 | 13 | 74 |
| GO:0008285 | negative regulation of cell proliferation | 0.0034 | 1.83 | 17.1 | 29 | 280 |
| GO:0008654 | phospholipid biosynthetic process | 0.0075 | 2.44 | 5.4 | 12 | 89 |
| GO:0016125 | sterol metabolic process | 0.0000 | 4.01 | 4.5 | 15 | 74 |
| GO:0030099 | myeloid cell differentiation | 0.0004 | 2.57 | 8.7 | 20 | 143 |
| GO:0042552 | myelination | 0.0012 | 3.13 | 4.4 | 12 | 72 |
| GO:0044255 | cellular lipid metabolic process | 0.0033 | 1.65 | 27.4 | 42 | 449 |
| GO:0044283 | small molecule biosynthetic process | 0.0041 | 1.92 | 13.5 | 24 | 221 |
| GO:0048634 | regulation of muscle organ development | 0.0045 | 2.95 | 3.8 | 10 | 63 |
| GO:0048741 | skeletal muscle fiber development | 0.0018 | 3.4 | 3.4 | 10 | 56 |
| GO:0055002 | striated muscle cell development | 0.0031 | 2.76 | 4.9 | 12 | 80 |
| GO:0061061 | muscle structure development | 0.0015 | 1.95 | 16.2 | 29 | 265 |
| GO:0070507 | regulation microtubule cytoskeleton organization | 0.0012 | 3.64 | 3.2 | 10 | 53 |
Among the 211 unique named genes that differed (FDR = 0.1) between the ethanol and water groups in the VTA, there were 26 genes located within rat ethanol-preference QTLs (Table 1). There were 4 rat ethanol-preference QTLs that contained 4 or more genes: Alc18 (Ezh2, Gstk1. Gng12, Arhgap25, Tmem176a, Tmem176b); Alc11 (Aadat, Dctd, Sc4mol, Scrg1); Alc15 (Celsr2, Cd53, Lppr5, Ugt8); and Alc5 (Dusp1, Mpg, Slc9a3r2, Tekt4).
Changes in expression of genes that were altered by alcohol drinking and also significantly differed between alcohol-naïve P and NP rats
There were 62 genes that significantly differed between the ethanol and water groups and also differed between naïve P and NP rats (McBride et al., 2012) in the VTA (Table 6). Among these 62 genes, 7 were located in 4 different rat QTLs: a) Dctd, Sc4mol, Scrg1 in Alc11, b) Tmem176a, Tmem176b in Alc18, c) Cd53 in Alc15, and (d) Ralgds in Alc8.
Table 6.
List of genes that were altered in the VTA by alcohol drinking and were also significantly different between naïve alcohol-preferring (P) and –non-preferring (NP) rats
| Symbol | Gene Description | Lower/upa | Higher/down | Higher/up | Lower/down |
|---|---|---|---|---|---|
| Abca1 | ATP-binding cassette, sub-family A (ABC1), member 1 | X | |||
| Aif1 | allograft inflammatory factor 1 | X | |||
| B2m | beta-2 microglobulin | X | |||
| C1qa | complement component 1, q subcomponent, A chain | X | |||
| C3 | complement component 3 | X | |||
| Cd53 | Cd53 molecule | X | |||
| Celsr2 | cadherin, EGF LAG seven-pass G-type receptor 2 (flamingo homolog, Drosophila) | X | |||
| Chmp1b | chromatin modifying protein 1B | X | |||
| Cldn11 | claudin 11 | X | |||
| Coq6 | Coenzyme Q6 homolog (yeast) | X | |||
| Cry1 | cryptochrome 1 (photolyase-like) | X | |||
| Ctsl1 | cathepsin L1 | X | |||
| Ctss | cathepsin S | X | |||
| Dctd | dCMP deaminase | X | |||
| Dcxr | dicarbonyl L-xylulose reductase | X | |||
| Eif4ebp1 | eukaryotic translation initiation factor 4E binding protein 1 | X | |||
| Emr1 | EGF-like module containing, mucin-like, hormone receptor-like 1 | X | |||
| Errfi1 | ERBB receptor feedback inhibitor 1 | X | |||
| Fcer1g | Fc fragment of IgE, high affinity I, receptor for; gamma polypeptide | X | |||
| Fcgr3a | Fc fragment of IgG, low affinity IIIa, receptor | X | |||
| Gamt | guanidinoacetate N-methyltransferase | X | |||
| Gpd1 | glycerol-3-phosphate dehydrogenase 1 (soluble) | X | |||
| Gpr84 | G protein-coupled receptor 84 | X | |||
| Grxcr1 | glutaredoxin, cysteine rich 1 | X | |||
| Hhip | Hedgehog-interacting protein | X | |||
| Hist1h2bh | histone cluster 1, H2bh | X | |||
| Hist2h2aa3 | histone cluster 2, H2aa3 | X | |||
| Idi1 | isopentenyl-diphosphate delta isomerase 1 | X | |||
| Irf8 | Interferon regulatory factor 8 | X | |||
| Itgb2 | integrin, beta 2 | X | |||
| Klf9 | Kruppel-like factor 9 | X | |||
| Laptm5 | lysosomal protein transmembrane 5 | X | |||
| Mal | mal, T-cell differentiation protein | X | |||
| Mtmr2 | myotubularin related protein 2 | X | |||
| Mysm1 | myb-like, SWIRM and MPN domains 1 | X | |||
| Npc2 | Niemann-Pick disease, type C2 | X | |||
| Pald | paladin | X | |||
| Pcdh20 | protocadherin 20 | X | |||
| Pex11a | peroxisomal biogenesis factor 11 alpha | X | |||
| Plekhf1 | pleckstrin homology domain containing, family F (with FYVE domain) member 1 | X | |||
| Psme1 | proteasome (prosome, macropain) activator subunit 1 | X | |||
| Ptpla | protein tyrosine phosphatase-like (proline instead of catalytic arginine), member a | X | |||
| Pycard | PYD and CARD domain containing | X | |||
| Qdpr | quinoid dihydropteridine reductase | X | |||
| Ralgds | ral guanine nucleotide dissociation stimulator | X | |||
| Rcn3 | reticulocalbin 3, EF-hand calcium binding domain | X | |||
| Sc4mol | sterol-C4-methyl oxidase-like | X | |||
| Sc65 | synaptonemal complex protein SC65 | X | |||
| Scarb2 | scavenger receptor class B, member 2 | X | |||
| Scrg1 | stimulator of chondrogenesis 1 | X | |||
| Serpinb9 | Serine (or cysteine) peptidase inhibitor, clade B, member 9 | X | |||
| Sgk1 | serum/glucocorticoid regulated kinase 1 | X | |||
| Slc22a4 | solute carrier family 22 (organic cation transporter), member 4 | X | |||
| Slc29a2 | solute carrier family 29 (nucleoside transporters), member 2 | X | |||
| Slc38a2 | solute carrier family 38, member 2 | X | |||
| Tjap1 | tight junction associated protein 1 | X | |||
| Tmem176a | transmembrane protein 176A | X | |||
| Tmem176b | transmembrane protein 176B | X | |||
| Tsc22d3 | TSC22 domain family, member 3 | X | |||
| Ube2g1 | ubiquitin-conjugating enzyme E2G 1 | X | |||
| Unc93b1 | unc-93 homolog B1 (C. elegans) | X | |||
| Zfp189 | zinc finger protein 189 | X |
Lower/up = lower expression levels in P vs. NP and up-regulation of expression in the ethanol vs. water group; Higher/down = higher expression levels in P vs. NP and down-regulation of expression in the ethanol vs. water group; Higher/up = higher expression levels in P vs. NP and up-regulation of expression in the ethanol vs. water group; Lower/down = lower expression levels in P vs. NP and down-regulation of expression in the ethanol vs. water group
KEGG analysis of the 62 genes indicated only an antigen-presentation category and an immune category, whereas the GO biological processes gene enrichment analysis revealed several significant categories with 5 or more genes (Table 7). Prominent among these GO categories were those involved in immune and inflammatory responses, as well as responses to glucocorticoid and steroid hormone stimuli.
Table 7.
List of GO biological process categories for common genes that were significantly different between the P vs. NP line and also changed significantly with alcohol drinking
| GO Biological Processes | Genes |
|---|---|
| Immune effector process | Up: C3, Serpinb9, Anax3a, B2m, C1qa, Fcer1g |
| Inflammatory response | Up: C3, Serpinb9, Fcer1g, Itgb2, Pycard |
| Induction programmed cell death | Up: Serpinb9, B2m, Mal, Pycard |
| Down: Pleckhf1 | |
| Cellular response to hormone stimulus | Up: Serpinb9, Aif1, Ctss |
| Down: Klf9, Sgk1, Slc29a2, Ctsl1 | |
| Leukocyte activation | Up: Cxcr4, Aif1, Anxa3, B2m, Fcer1g, Fcgr3a, Itgb2 |
| Response to steroid hormone stimulus | Up: C3, Serpinb9, Aif1, Anxa3 |
| Down: Sgk1, Ctsl1 | |
| Response to glucocorticoid stimulus | Up: C3, Aif1, Anxa3 |
| Down: Sgk1, Ctsl1 | |
| Response to other organisms | Up: Abca1, Serpinb9, Anxa3, B2m, Fcer1g, Fcgr3a, Npc2 |
| Organic substance transport | Up: Abca1, Slc38a2, Fcer1g, Npc2, Slc22a4 |
Up = genes with higher expression levels in ethanol compared to water group. Down = genes with lower expression levels in ethanol compared to water group
Among the 62 genes, 43 differed between the alcohol-naïve P and NP rats and changed toward NP expression levels as a result of binge drinking in the P rats (Table 6). There were 28 genes lower in the P than NP rats that were up-regulated in the ethanol group compared to the water group, and 15 genes higher in the VTA of the P than NP rats that were down-regulated with alcohol drinking.
There were 16 genes down-regulated with alcohol drinking that were also lower in the VTA of the P compared to the NP rats (Table 6). Only 3 genes were up-regulated with alcohol drinking and were also higher in the P than NP rats.
Although Ingenuity pathway analysis did not reveal significant networks when all 62 genes were used in the analysis, one network emerged using only genes that changed in the opposite direction in the alcohol drinking group compared to the difference between the alcohol-naïve P vs. NP rats. There were several genes associated with Ubc (ubiquitin conjugating gene) that were lower in the P vs. NP rats and were higher in the ethanol vs. water group (listed in protein metabolic processes, cell adhesion, and inflammatory response categories; Table 8). Within this same network, there were several genes associated with Il6 (Interleuken 6) that were also significantly changed in the opposite direction with alcohol drinking relative to the naïve group (listed in inflammatory response category; Table 8).
Table 8.
Overall summary of bioinformatics analyses of changes in gene expression in the VTA resulting from loss-of-control drinking by P rats
| Biological Categories | Genes | Function |
|---|---|---|
| Transcription factors | Multiple genes centered around Nfkbia, Fos, Srebf1 | Reduced excitation-coupled transcription; promotion of excitotoxic neuronal damage |
| Steroid & protein metabolic processes | Up (protein): Rcn3, Anxa3, Pycard, Serpinb9, Dcxr, Psme1, Laptm5, Eif4ebp1, Unc93b1, Celsr2 | Reduced cholesterol & fatty acid synthesis; increased protein degradation |
| Down (steroid): Acss2, Sc5dl, Sqle, Cyp51a1, Hmgcr, Dhcr7, Msmo1, Scd, Tm7sf2, Hmgcs1, Idl1 | ||
| Cell adhesion; regulation of actin cytoskeleton | Up: Emr1, C3, C1qa, Leprel4, Cd53, Serpinb9, Cxcr4, Laptm5, Ctss, Tmem176b | Altered cell-to-cell interactions & dendrite development |
| Down: Sgk1, Gpd, Pcdh20, Gpatch4, Klf9, Plekhf1, Errfi1, Ube2g1, Slc38a2, Tsc22d3 | ||
| Inflammatory response & immune process | Up: Emr1, Ctss, Npc2, Eif4ebp1, Psme1, Pycard, Cd53, C3, Serpinb9, Anax3a, B2m, C1qa, Fcer1g, Itgb2 | Pro-inflammatory |
| Response to glucocorticoid or steroid hormone stimulus | Up: C3, Serpinb9, Aif1, Anxa3 | Altered response to stimulus |
| Down: Sgk1, Ctsl1 |
Up = genes with higher expression levels in ethanol compared to water group. Down = genes with lower expression levels in ethanol compared to water group
Discussion
This study examined changes in gene expression in response to alcohol in the VTA of P rats with a history of excessive binge drinking. The changes reflect the effects of the combination of a high brain ethanol concentration plus a history of binge drinking on gene expression in the VTA examined 3 hours after the last drinking episode. Samples taken at later time points after the last binge drinking episode will likely show different patterns of genes being expressed between the ethanol and water groups.
The major findings of this study are that multiple scheduled (4 × 1 hour sessions during the dark cycle) sessions of concurrent access to 10, 20 and 30% ethanol a) resulted in ‘loss-of-control’ alcohol drinking (Fig. 1), defined as drinking that routinely produces BECs exceeding 200 mg%, and b) produced major changes (mainly 1.2-fold and higher in 211 unique genes) in expression of genes in the VTA (Table 1). Genes that are involved in transcription, metabolic processes, cell-to-cell interactions, inflammatory response, and response to hormone stimuli were particularly affected (Tables 7 and 8). The overall results support the idea that this level of drinking is producing major changes in gene expression within the VTA that could promote neuroadaptations mediating excessive alcohol drinking and alcohol-induced neuronal damage.
The reduction in Fos (FBJ osteosarcoma oncogene) expression with alcohol drinking along with the reduction in gene expression of 11 of the 16 genes associated with the Fos (Table 4) supports the idea that there is reduced excitation-coupled transcription in the alcohol group compared to controls (George et al., 2012; Schiavone et al., 2011). The findings that 10 of 14 genes are up-regulated in the Nfkbia (nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha) cluster (Table 4) of the alcohol group suggest promotion of excitotoxic neuronal damage (Himadri et al., 2010; Koltsova et al., 2012). It is noteworthy that glucocorticoid receptor activity, implicated in the present study, modulates transcription of Nfkbia and Tsc22d3 (e.g., D’Adamio et al., 1997; Scheinman et al., 1995). Moreover, there is some evidence that morphine may co-regulate Tsc22d3, Sgk, Klf15, and Nfkbia (Korostynski et al., 2007); paralleling a similar pattern of Tsc22d3, Sgk1, Klf9, and Nfkbia all being down-regulated by ethanol in the present study.
The reduced expression of 13 of 15 genes (Table 4) associated with Srebf1 (sterol regulatory element binding transcription factor 1) suggests reduced cholesterol and fatty acid synthesis (Ong et al., 2000; Tabernero et al., 2002), which is also consistent with excessive binge drinking having a negative impact on neuronal function. In addition, Srebf1, Hmgcr, and Hmgcs1 are all implicated in white matter integrity (e.g., Carter, 2007; Xiang et al., 2011), which is disrupted after chronic alcohol abuse (Harris et al., 2008). Neither the GO (Table 2) nor the KEGG (Table 3) analysis provided a clear answer on the effects of repeated excessive binge drinking on neuronal function in the VTA, since both positive (e.g., regulation of actin cytoskeleton and cell adhesion) and negative (e.g., killer cell mediated cytotoxicity, negative regulation of transcription, and steroid biosynthesis with all genes down-regulated) effects on cellular function are evident.
Many of the genes, which differed between the alcohol-naive P vs. NP rats and changed in the opposite direction with alcohol drinking (Table 6), promoted pro-inflammatory responses and induced programmed cell death (Table 7). The innate differences in the lower expression of genes involved in the immune and inflammatory responses in the VTA of the P compared to the NP line (e.g., C3, Serpinb9, B2m, C1qa, Fcer1g, Itgb2, Pycard, Aif1; Tables 6 and 7) could be factors contributing to the vulnerability of the P line to high alcohol drinking behavior. Recent findings (Crews et al., 2013) are compatible with the present results of increased brain neuroimmune activation in alcohol dependence. These results (Tables 6 and 7) are in agreement with the overall analysis of the data indicating a negative impact of this excessive binge-like drinking protocol by P rats (Table 8). On the other hand, the complement system has been implicated in synaptic plasticity (Stephan et al., 2012), and some of the changes observed in the immune process and inflammatory response may result in positive alterations in synaptic function. The results with WGCNA (Table 5, Yellow module) support the contention that positive alterations in synaptic function may have occurred under the excessive binge-like drinking conditions utilized in the present study. However, with few exceptions, the individual genes in this module were not significantly different at a modest p value of .01.
Previous studies indicated that chronic 24-hour free-choice alcohol drinking increased the sensitivity and response of the posterior VTA to the rewarding effects of ethanol (Rodd et al., 2005a, b). These results suggest that ethanol drinking is producing positive effects on neuronal function within the VTA. In addition, repeated binge-like ethanol intakes of 1.5–2 g/kg/session (3 × 1-hour daily sessions) produced changes in expression of genes that could alter transcription, synaptic function, and neuronal plasticity in a generally positive manner in the Acb-shell and central nucleus of the amygdala (McBride et al., 2010). In contrast to the above results, the findings of the present study suggest a more negative effect of alcohol drinking on cellular function. Main differences between the current study and the previous 3 reports are that BECs in excess of 200 mg% were attained on a daily basis over several weeks in the present study, that different time points after drinking were measured, and that different tissues were assayed. Such chronically high brain levels of alcohol may produce excessive cellular oxidative stress and eventually cause neuronal damage within the VTA. The WGCNA provided additional evidence that excessive binge drinking produces alterations in glia cells within the VTA (Table 5; Blue module GO categories). Alterations in fatty acid, steroid, cholesterol, and phospholipid biosynthetic processes could be indicators of the damaging effects of the very high daily concentrations of ethanol on brain function.
The effects of binge-like alcohol drinking on gene expression have been studied in the Acb of inbred P rats (Rodd et al., 2008), the extended amygdala of P rats (McBride et al., 2010), and several brain regions (olfactory bulb, frontal cortex, striatum, cerebellum, ventral midbrain, and hippocampus) of C57 mice using a single-day procedure (Mulligan et al., 2011). Comparison of the present results with those obtained with the C57 mice or the 2 rat binge-drinking studies did not indicate any overlap in the top annotated genes, with one exception: B2m; beta-2 microglobin was common in the present study and that of Rodd et al., 2008. The lack of overlap likely reflects a combination of factors, such as different regions analyzed, duration and level of binge-like alcohol drinking, time of sampling after ethanol drinking, and, in the case of mice, species differences.
There were no genes in common between those identified as possible candidate genes in the expression profiling of congenic rat strains (Carr et al., 2007) and the present study (Table 1). In addition, there were no genes in common between the present study (Table 1) and the candidate genes within the chromosome 10 QTL (Bice et al., 2010).
Flatscher-Bader et al. (2010) reported on differences in gene expression in the VTA of alcoholics vs. control subjects. There was a general overlap (see Table 7) in a canonical pathway related to “regulation of the actin skeleton”. In both studies, the changes in gene expression may reflect altered cellular organization and structure.
QTL Alc18 contained 6 significant genes (Ezh2, Gstk1, Gng12, Arhgap25, Tmem176a, and Tmem176b) altered by excessive binge drinking (Table 2), 2 of which (Tmem176a, Tmem176b) were also common in the differences between the alcohol-naïve P vs. NP rats (Table 5). Tmem176a and Tmem176b are trans-membrane proteins involved in the immune system (Cuajungco et al., 2012). Ezh2 is the catalytic subunit of Polycomb repressive complex, which is a highly conserved histone methyltransferase that targets lysine-27 of histone H3 (Simon and Lange, 2008). Gstk1 (glutathione S-transferase kappa 1), located in mitochondria and peroxisomes, is involved in energy and lipid metabolism (Morel and Aninat, 2011). Gng12 (guanine nucleotide binding protein, gamma 12) is a negative regulator of the inflammatory response (Larson et al., 2010). Arhgap25 (Rho GTPase activating protein 25) is a member of the RhoGAP family, which are negative regulators of Rho family GTPases implicated in actin remodeling (Katoh and Katoh, 2004). Further research is required to determine how these genes could impact a predisposition for high alcohol drinking or be involved in maintaining high alcohol consumption.
All 3 genes within Alc 11 (Dctd, Sc4mol, Scrg1; Table 1) were also common in the differences between alcohol- naïve P vs. NP rats (Table 5). Dctd (cCMP [deoxycytidine-5′-monophosphate] deaminase) provides the main nucleotide substrate for thymidylate synthase, which is important for DNA synthesis (Hou et al., 2008). Sc4mol (sterol-C4-methyl oxidase-like) is involved in cholesterol synthesis (He et al., 2011) and the growth and guidance of axons (Yu et al., 2008). Scrg1 (stimulator of chondrogenesis 1) encodes a highly conserved cysteine-rich protein, is principally expressed in the CNS and is associated with large dense-core vesicles in neurons (Dandoy-Dron et al., 2003). Overall, the findings may point to segments of QTLs Alc11 and Alc18 being important for containing genes involved in vulnerability to alcohol abuse and contributing to the maintenance of high alcohol intake.
The study of Rodd et al. (2008) examined the effects of operant responding for a 0.0125% saccharin solution (approximately 500 lever presses/session for 10 weeks) and did not observe any significant effects on gene expression in the Acb of inbred P rats, suggesting that instrument responding per se is not producing alterations in gene expression. The results of the current study are consistent with the observed effects being due to repeated excessive binge-like alcohol drinking, and not mainly a result of instrument responding.
Validation studies on key genes, using qRT-PCR, could not be conducted because there was not sufficient sample remaining after the microarray procedure. Previous studies from our laboratory indicated good agreement between the data obtained with microarrays and the results found with qRT-PCR (Bell et al., 2009; Kimpel et al., 2007; Rodd et al., 2008).
In summary, the overall results of the bioinformatic analyses (Table 8) indicated the up-regulation of a number of genes that could produce a pro-inflammatory response, promote excitotoxic neuronal damage, and increase protein degradation. These results suggest that the high BECs repeatedly attained in the present study could be causing neuronal damage in the VTA. Consistent with this interpretation are the findings that several genes involved in cholesterol and fatty acid synthesis were down-regulated (Table 8). Also, alterations in expression of genes around 3 transcription factors suggested reduced excitation-coupled transcription. Thus, the combination of alterations in transcription factors, metabolism of proteins and steroid, and pro-inflammatory response are indicators that this level of alcohol drinking may be repeatedly producing brain ethanol levels that could cause cellular damage.
Acknowledgments
This study was supported by AA07611, and INIA projects AA013521, AA013522, AA016652, AA016660 and AA020892, and INGEN® (which is partially funded by Lilly Endowment Inc.). The content of this manuscript is solely the responsibility of the authors and does not necessarily represent the official views of the NIAAA or NIH.
Footnotes
None of the authors has a conflict of interest associated with this research.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alexander-Kaufman K, Harper C, Wilce P, Matsumoto I. Cerebellar vermis proteome of chronic alcoholic individuals. Alcohol Clin Exp Res. 2007;31:1286–1296. doi: 10.1111/j.1530-0277.2007.00437.x. [DOI] [PubMed] [Google Scholar]
- Alexander-Kaufman K, James G, Sheedy D, Harper C, Matsumoto I. Differential protein expression in the prefrontal white matter of human alcoholics: a proteomics study. Molecular Psych. 2006;11:56–65. doi: 10.1038/sj.mp.4001741. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell RL, Kimpel MW, McClintick JN, Strother WN, Carr LG, Liang T, Rodd ZA, Mayfield RD, Edenberg HJ, McBride WJ. Gene expression changes in the nucleus accumbens of alcohol-preferring rats following chronic ethanol consumption. Pharmacol Biochem Behav. 2009;94:131–147. doi: 10.1016/j.pbb.2009.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell RL, Kimpel MW, Rodd ZA, Strother WN, Bai F, Peper CL, Mayfield RD, Lumeng L, Crabb DW, McBride WJ, et al. Protein expression changes in the nucleus accumbens and amygdala of inbred alcohol-preferring rats given either continuous or scheduled access to ethanol. Alcohol. 2006;40:3–17. doi: 10.1016/j.alcohol.2006.10.001. [DOI] [PubMed] [Google Scholar]
- Bice PJ, Liang T, Zhang L, Graves TJ, Carr LG, Lai D, Kimpel MW, Foroud T. Fine mapping and expression of candidate genes within the chromosome 10 QTL region of the high and low alcohol-drinking rats. Alcohol. 2010;44:477–485. doi: 10.1016/j.alcohol.2010.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, Barres BA. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr LG, Kimpel MW, Liang T, McClintick JN, McCall K, Morse M, Edenberg HJ. Identification of candidate genes for alcohol preference by expression profiling of congenic rat strains. Alcohol Clin Exp Res. 2007;31:1089–1098. doi: 10.1111/j.1530-0277.2007.00397.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter CJ. Convergence of genes implicated in Alzheimer’s disease on the cerebral cholesterol shuttle: APP, cholesterol, lipoproteins, and atherosclerosis. Neurochem Intl. 2007;50:12–38. doi: 10.1016/j.neuint.2006.07.007. [DOI] [PubMed] [Google Scholar]
- Crews FT, Qin L, Sheedy D, Vetreno RP, Zou J. High mobility group box 1/toll-like receptor danger signaling increases brain neuroimmune activation in alcohol dependence. Biol Psychiatry. 2013;73:602–612. doi: 10.1016/j.biopsych.2012.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuajungco MP, Podevin W, Valluri VK, Bui Q, Nguyen VH, Taylor K. Abnormal accumulation of human transmembrane (TMEM)-176A and 176B proteins is associated with cancer pathology. Acta Histochem. 2012;114:705–712. doi: 10.1016/j.acthis.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Adamio F, Zollo O, Moraca R, Ayroldi E, Bruscoli S, Bartoli A, Cannarile L, Migliorati G, Riccardi C. A new dexamethasone-induced gene of the leucine zipper family protects T lymphocytes from TCR/CD3-activated cell death. Immunity. 1997;7:803–812. doi: 10.1016/s1074-7613(00)80398-2. [DOI] [PubMed] [Google Scholar]
- Dandoy-Dron F, Griffond B, Mishal Z, Tovey MG, Dron M. Scrg1, a novel protein of the CNS, is targeted to the large dense-core vesicles in neuronal cells. Eur J Neurosci. 2003;18:2449–2459. doi: 10.1046/j.1460-9568.2003.03009.x. [DOI] [PubMed] [Google Scholar]
- Ding ZM, Rodd ZA, Engleman EA, McBride WJ. Sensitization of ventral tegmental area dopamine neurons to the stimulating effects of ethanol. Alcohol Clin Exp Res. 2009;33:1571–1581. doi: 10.1111/j.1530-0277.2009.00985.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Strother WN, McClintick JN, Tian H, Stephans M, Jerome RE, Lumeng L, Li TK, McBride WJ. Gene expression in the hippocampus of inbred alcohol-preferring and -nonpreferring rats. Genes Brain Behav. 2005;4:20–30. doi: 10.1111/j.1601-183X.2004.00091.x. [DOI] [PubMed] [Google Scholar]
- Flatscher-Bader T, van der Brug M, Hwang JW, Gochee PA, Matsumoto I, Niwa S, Wilce PA. Alcohol-responsive genes in the frontal cortex and nucleus accumbens of human alcoholics. J Neurochem. 2005;93:359–370. doi: 10.1111/j.1471-4159.2004.03021.x. [DOI] [PubMed] [Google Scholar]
- Flatscher-Bader T, Zuvela N, Landis N, Wilce PA. Smoking and alcoholism target genes associated with plasticity and glutamate transmission in the human ventral tegmental area. Human Mol Genetics. 2008;17:38–51. doi: 10.1093/hmg/ddm283. [DOI] [PubMed] [Google Scholar]
- Flatscher-Bader T, Harrison E, Matsumoto I, Wilce PA. Genes associated with alcohol abuse and tobacco smoking in the human nucleus accumbens and ventral tegmental area. Alcohol Clin Exp Res. 2010;34:1291–1302. doi: 10.1111/j.1530-0277.2010.01207.x. [DOI] [PubMed] [Google Scholar]
- Gatto GJ, McBride WJ, Murphy JM, Lumeng L, Li TK. Ethanol self-infusion into the ventral tegmental area by alcohol-preferring rats. Alcohol. 1994;11:557–564. doi: 10.1016/0741-8329(94)90083-3. [DOI] [PubMed] [Google Scholar]
- Gentleman RC. Using GO for statistical analysis. Proc COMPSTAT. 2004;2004:171–180. [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George O, Sanders C, Freiling J, Grigoryan E, Vu S, Allen CD, Crawford E, Mandyam CD, Koob GF. Recruitment of medial prefrontal cortex neurons during alcohol withdrawal predicts cognitive impairment and excessive alcohol drinking. Proc Natl Acad Sci USA. 2012 doi: 10.1073/pnas.1116523109. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris GJ, Jaffin SK, Hodge SM, Kennedy D, Caviness VS, Marinkovic K, Papadimitriou GM, Makris N, Oscar-Berman M. Frontal white matter and cingulum diffusion tensor imaging deficits in alcoholism. Alcohol Clin Exp Res. 2008;32:1001–1013. doi: 10.1111/j.1530-0277.2008.00661.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, Mungall C, et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32(Database issue):D258–261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser SR, Ding ZM, Getachew B, Toalston JE, Oster SM, McBride WJ, Rodd ZA. The posterior ventral tegmental area mediates alcohol-seeking behavior in alcohol-preferring rats. J Pharmacol Exp Ther. 2011;336:857–865. doi: 10.1124/jpet.110.168260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M, Kratz LE, Michel JJ, Vallejo AN, Ferris L, Kelley RI, Hoover JJ, Jukic D, Gibson KM, Wolfe LA, et al. Mutations in the human SC4MOL gene encoding a methyl sterol oxidase cause psoriasiform dermatitis, microcephaly, and developmental delay. J Clin Invest. 2011;121:976–984. doi: 10.1172/JCI42650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himadri P, Kumari SS, Chitharanjan S, Dhananjay S. Role of oxidative stress and inflammation in hypoxia-induced cerebral edema: a molecular approach. High Alt Med Biol. 2010;11:231–244. doi: 10.1089/ham.2009.1057. [DOI] [PubMed] [Google Scholar]
- Hodge CW, Haraguchi M, Erickson H, Samson HH. Ventral tegmental microinjections of quinpirole decrease ethanol and sucrose-reinforced responding. Alcohol Clin Exp Res. 1993;17:370–375. doi: 10.1111/j.1530-0277.1993.tb00778.x. [DOI] [PubMed] [Google Scholar]
- Hou HF, Liang YH, Li LF, Su XD, Dong YH. Crystal structures of Sterptococcus mutans 2′-desoxycytidylate deaminase and its complex with substrate analog and allosteric regulator dCTP x Mg2+ J Mol Biol. 2008;377:220–231. doi: 10.1016/j.jmb.2007.12.064. [DOI] [PubMed] [Google Scholar]
- Hwa LS, Debold JF, Miczek KA. Alcohol in excess: CRF (1) receptors in the rat and mouse VTA and DRN. Psychopharmacology. 2012 doi: 10.1007/s00213-012-2820-z. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Katoh M, Katoh M. Identification and characterization of ARHGAP24 and ARHGAP25 genes in silico. Int J Mol Med. 2004;14:333–338. [PubMed] [Google Scholar]
- Kerns RT, Ravindranathan A, Hassan S, Cage MP, York T, Sikela JM, Williams RW, Miles MF. Ethanol-responsive brain region expression networks: Implications for behavioral responses to acute ethanol in DBA/2J versus C57BL/6J mice. J Neurosci. 2005;25:2255–2266. doi: 10.1523/JNEUROSCI.4372-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimpel MW, Strother WN, McClintick JN, Carr LG, Edenberg HJ, McBride WJ. Functional gene expression differences between inbred alcohol-preferring (iP) and –non-preferring (iNP) rats in five brain regions. Alcohol. 2007;41:95–132. doi: 10.1016/j.alcohol.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koltsova SV, Truchina Y, Haloui M, Akimova OA, Tremblay J, Harnet P, Orlov SN. Unbiquitous [Na+]i/[K+]i-sensitive transcriptome in mammalian cells: evidence for Ca(2+)i-independent excitation-transcription coupling. PloS One. 2012;7:e38032. doi: 10.1371/journal.pone.0038032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korostynski M, Piechota M, Kaminska D, Solecki W, Przewlocki R. Morphine effects on striatal transcriptome in mice. Genome Biol. 2007;8:R128. doi: 10.1186/gb-2007-8-6-r128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. Fast R functions for robust correlations and hierarchical clustering. J Stat Softw. 2012:46. [PMC free article] [PubMed] [Google Scholar]
- Larson KC, Lipko M, Dabrowski M, Draper MP. Gng12 is a novel negative regulator of LPS-induced inflammation in the microglial cell line BV-2. Inflamm Res. 2010;59 doi: 10.1007/s00011-009-0062-2. Epub 2009 Jul 1. [DOI] [PubMed] [Google Scholar]
- Lewohl JM, Wang L, Miles MF, Zhang L, Dodd PR, Harris RA. Gene expression in human alcoholism: microarray analysis of frontal cortex. Alcohol Clin Exp Res. 2000;24:1873–1882. [PubMed] [Google Scholar]
- Lewohl JM, Van Dyk DD, Craft GE, Innes DJ, Mayfield RD, Cobon G, Harris RA, Dodd PR. The application of proteomics to the human alcoholic brain. Ann NY Acad Sci. 2004;1025:14–26. doi: 10.1196/annals.1316.002. [DOI] [PubMed] [Google Scholar]
- Liu J, Lewohl JM, Dodd PR, Randall PK, Harris RA, Mayfield RD. Gene expression profiling of individual cases reveals consistent transcriptional changes in alcoholic human brain. J Neurochem. 2004;90:1050–1058. doi: 10.1111/j.1471-4159.2004.02570.x. [DOI] [PubMed] [Google Scholar]
- Liu J, Lewohl JM, Harris RA, Iyer VR, Dodd PR, Randall PK, Mayfield RD. Patterns of gene expression in the frontal cortex discriminate alcoholic from nonalcoholic individuals. Neuropsychopharmacology. 2006;31:1574–1582. doi: 10.1038/sj.npp.1300947. [DOI] [PubMed] [Google Scholar]
- Matsumoto I, Alexander-Kaufman K, Iwazaki T, Kashem MA, Matsuda-Matsumoto H. CNS proteomes in alcohol and drug abuse and dependence. Expert Rev Proteomics. 2007;4:539–552. doi: 10.1586/14789450.4.4.539. [DOI] [PubMed] [Google Scholar]
- Mayfield RD, Lewohl JM, Dodd PR, Herlihy A, Liu J, Harris RA. Patterns of gene expression are altered in the frontal and motor cortices of human alcoholics. J Neurochem. 2002;81:802–813. doi: 10.1046/j.1471-4159.2002.00860.x. [DOI] [PubMed] [Google Scholar]
- McBride WJ, Murphy JM, Ikemoto S. Localization of brain reinforcement mechanism: intracranial self-administration and intracranial place-conditioning studies. Behav Brain Res. 1999;101:129–152. doi: 10.1016/s0166-4328(99)00022-4. [DOI] [PubMed] [Google Scholar]
- McBride WJ, Kimpel MW, Schultz JA, McClintick JN, Edenberg HJ, Bell RL. Changes in gene expression in regions of the extended amygdala of alcohol-preferring rats after binge-like alcohol drinking. Alcohol. 2010;44:171–183. doi: 10.1016/j.alcohol.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride WJ, Kimpel MW, McClintick JN, Ding ZM, Hyytia P, Colombo G, Edenberg HJ, Lumeng L, Bell RL. Gene expression in the ventral tegmental area of 5 pairs of rat lines selectively bred for high or low ethanol consumption. Pharmacol Biochem Behav. 2012;102:275–285. doi: 10.1016/j.pbb.2012.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintick JN, Edenberg HJ. Effects of filtering by Present call on analysis of microarray experiments. BMC Bioinformatics. 2006;7:49. doi: 10.1186/1471-2105-7-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintick JN, Jerome RE, Nicholson CR, Crabb DW, Edenberg HJ. Reproducibility of oligonucleotide arrays using small samples. BMC Genomics. 2003;4:1–15. doi: 10.1186/1471-2164-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel F, Aninat C. The glutathione transferase kappa family. Drug Metab Rev. 2011;43:281–291. doi: 10.3109/03602532.2011.556122. [DOI] [PubMed] [Google Scholar]
- Mulligan MK, Ponomerav I, Hitzemann RJ, Belknap JK, Tabakoff B, Harris RA, Crabbe JC, Blednov YA, Grahame NJ, Phillips TJ, et al. Toward understanding the genetics of alcohol drinking through transcriptome meta-analysis. Proc Natl Acad Sci USA. 2006;103:6368–6373. doi: 10.1073/pnas.0510188103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan MK, Rhodes JS, Crabbe JC, Mayfield RD, Harris RA, Ponomarev I. Molecular profiles of drinking alcohol to intoxication in C57BL/6J mice. Alcohol Clin Exp Res. 2011;35:659–670. doi: 10.1111/j.1530-0277.2010.01384.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong WY, Hu CY, Soh YP, Lim TM, Pentchev PG, Patel SC. Neuronal localization of sterol regulatory element binding protein-1 in the rodent and primate brain: a light and electron microscopic immunocytochemical study. Neuroscience. 2000;97:143–53. doi: 10.1016/s0306-4522(00)00031-2. [DOI] [PubMed] [Google Scholar]
- Paxinos G, Watson C. The Rat Brain in Stereotaxic Coordinates. New York: Academic Press; 1998. [Google Scholar]
- Rodd ZA, Bell RL, Melendez RI, Kuc KA, Lumeng L, Li TK, Murphy JM, McBride WJ. Comparison of intracranial self-administration of ethanol within the posterior ventral tegmental area between alcohol-preferring and Wistar rats. Alcohol Clin Exp Res. 2004;28:1212–1219. doi: 10.1097/01.alc.0000134401.30394.7f. [DOI] [PubMed] [Google Scholar]
- Rodd ZA, Bell RL, McQueen VK, Davids MR, Hsu CC, Murphy JM, Li TK, Lumeng L, McBride WJ. Chronic alcohol drinking by alcohol-preferring rats increases the sensitivity of the posterior ventral tegmental area to the reinforcing effects of ethanol. Alcohol Clin Exp Res. 2005a;29:358–366. doi: 10.1097/01.alc.0000156127.30983.9d. [DOI] [PubMed] [Google Scholar]
- Rodd ZA, Bell RL, McQueen VK, Davids MR, Hsu CC, Murphy JM, Li TK, Lumeng L, McBride WJ. Prolonged increases in the sensitivity of the posterior ventral tegmental area to the reinforcing effects of ethanol following repeated exposure to cycles of ethanol access and deprivation. J Pharmacol Exp Ther. 2005b;315:648–657. doi: 10.1124/jpet.105.084350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodd ZA, Kimpel MW, Edenberg HJ, Bell RL, Strother WN, McClintick JN, Carr LG, Liang T, McBride WJ. Differential gene expression in the nucleus accumbens with ethanol self-administration in inbred alcohol-preferring rats. Pharmacol Biochem Behav. 2008;89:481–498. doi: 10.1016/j.pbb.2008.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodd ZA, Bell RL, Oster SM, Toalston JE, Pommer TJ, McBride WJ, Murphy JM. Serotonin-3 receptors in the posterior ventral tegmental area regulate ethanol self-administration of alcohol-preferring (P) rats. Alcohol. 2010;44:245–255. doi: 10.1016/j.alcohol.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodd-Henricks ZA, McKinzie DL, Crile RS, Murphy JM, McBride WJ. Regional heterogeneity for the intracranial self-administration of ethanol within the ventral tegmental area of female Wistar rats. Psychopharmacology. 2000;149:217–224. doi: 10.1007/s002139900347. [DOI] [PubMed] [Google Scholar]
- Rodd-Henricks ZA, Bell RL, Kuc KA, Murphy JM, McBride WJ, Lumeng L, Li TK. Effects of ethanol exposure on subsequent acquisition and extinction of ethanol self-administration and expression of alcohol-seeking behavior in adult alcohol-preferring (P) rats: I. Peri-adolescent exposure. Alcohol Clin Exp Res. 2002a;26:1632–1641. doi: 10.1097/01.ALC.0000036301.36192.BC. [DOI] [PubMed] [Google Scholar]
- Rodd-Henricks ZA, Bell RL, Kuc KA, Murphy JM, McBride WJ, Lumeng L, Li TK. Effects of ethanol exposure on subsequent acquisition and extinction of ethanol self-administration and expression of alcohol-seeking behavior in adult alcohol-preferring (P) rats: I. Adult exposure. Alcohol Clin Exp Res. 2002b;26:1642–1652. doi: 10.1097/01.ALC.0000036302.73712.9D. [DOI] [PubMed] [Google Scholar]
- Saito M, Smiley J, Toth R, Vadasz C. Microarray analysis of gene expression in rat hippocampus after chronic ethanol treatment. Neurochem Res. 2002;27:1221–1229. doi: 10.1023/a:1020937728506. [DOI] [PubMed] [Google Scholar]
- Saito M, Szakall I, Toth R, Kovacs KM, Oros M, Prasad VV, Blumenberg M, Vadasz C. Mouse striatal transcriptome analysis: effects of oral self-administration of alcohol. Alcohol. 2004;32:223–241. doi: 10.1016/j.alcohol.2004.02.005. [DOI] [PubMed] [Google Scholar]
- Scheinman RI, Cogswell PC, Lofquist AK, Baldwin AS., Jr Role of transcriptional activation of I kappa B alpha in mediation of immunosuppression by glucocorticoids. Science. 1995;270:283–286. doi: 10.1126/science.270.5234.283. [DOI] [PubMed] [Google Scholar]
- Schiavone D, Avalle L, Dewilde S, Poli V. The immediate early genes Fos and Egr1 become STAT1 transcriptional targets in the absence of STAT3. FEBS Lett. 2011;585:2455–2460. doi: 10.1016/j.febslet.2011.06.020. [DOI] [PubMed] [Google Scholar]
- Simon JA, Lange CA. Roles of the EZH2 histone methyltransferase in cancer epigenetics. Mutat Res. 2008;647:21–29. doi: 10.1016/j.mrfmmm.2008.07.010. [DOI] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3(1) doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- Stephan AH, Barres BA, Stevens B. The complement system: an unexpected role in synaptic pruning during development and disease. Annu Rev Neurosci. 2012;35:369–389. doi: 10.1146/annurev-neuro-061010-113810. [DOI] [PubMed] [Google Scholar]
- Storey JD, Taylor JE, Siegmund D. Strong control, conservative point estimation, and simultaneous conservative consistency of false discovery rates: A unified approach. J Royal Statistical Soc, Series B. 2004;66:187–205. [Google Scholar]
- Tabakoff B, Saba L, Printz M, Flodman P, Hodgkinson C, Goldman D, Koob G, Richardson HN, Kechris K, Bell, et al. Genetical genomic determinants of alcohol consumption in rats and humans. BMC Biol. 2009;7:70–78. doi: 10.1186/1741-7007-7-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabernero A, Velasco A, Granda B, Lavado EM, Medina JM. Transcytosis of albumin in astrocytes activates the sterol regulatory element-binding protein-1, which promotes the synthesis of the neurotrophic factor oleic acid. J Biol Chem. 2002;277:4240–4246. doi: 10.1074/jbc.M108760200. [DOI] [PubMed] [Google Scholar]
- Treadwell JA, Singh SM. Microarray analysis of mouse brain gene expression following acute ethanol treatment. Neurochem Res. 2004;29:357–369. doi: 10.1023/b:nere.0000013738.06437.a6. [DOI] [PubMed] [Google Scholar]
- Xiang Z, Valenza M, Cui L, Leoni V, Jeong HK, Brilli E, Zhang J, Peng Q, Duan W, Reeves SA, et al. Peroxisome-proliferator-activated receptor gamma coactivator 1 alpha contributes to dysmyelination in experimental models of Huntington’s disease. J Neurosci. 2011;31:9544–9553. doi: 10.1523/JNEUROSCI.1291-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Cook MC, Shin DM, Silva DG, Marshall J, Toellner KM, Havran WL, Caroni P, Cooke MP, Morse, et al. Axon growth and guidance genes identify T-dependent germinal centre B cells. Immunol Cell Biol. 2008;86:3–14. doi: 10.1038/sj.icb.7100123. [DOI] [PubMed] [Google Scholar]
- Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005:4. doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]