Abstract
We previously reported that selenamide reagents such as ebselen and N-(phenylseleno) phthalimide (NPSP) can be used to selectively derivatize thiols for mass spectrometric analysis, and the introduced selenium tags are useful as they could survive or removed with collision-induced dissociation (CID). Described herein is the further study of the reactivity of various protein/peptide thiols toward NPSP and its application to derivatize thiol peptides in protein digests. With a modified protocol (i.e., dissolving NPSP in acetonitrile instead of aqueous solvent), we found that quantitative conversion of thiols can be obtained in seconds, using NPSP in a slight excess amount (NPSP:thiol of 1.1–2:1). Further investigation shows that the thiol reactivity toward NPSP reflects its chemical environment and accessibility in proteins/peptides. For instance, adjacent basic amino acid residues increase the thiol reactivity, probably because they could stabilize the thiolate form to facilitate the nucleophilic attack of thiol on NPSP. In the case of creatine phosphokinase, the native protein predominately has one thiol reacted with NPSP while all of four thiol groups of the denatured protein can be derivatized, in accordance with the corresponding protein conformation. In addition, thiol peptides in protein/peptide enzymatic digests can be quickly and effectively tagged by NPSP following tri-n-butylphosphine (TBP) reduction. Notably, all three thiols of the peptide QCCASVCSL in the insulin peptic digest can be modified simultaneously by NPSP. These results suggest a novel and selective method for protecting thiols in the bottom-up approach for protein structure analysis.
Keywords: N-(phenylseleno)phthalimide, Derivatization, Thiol protein/peptide, Mass spectrometry, Reactivity
Introduction
Cysteine (abbreviated as Cys or C), which contains a thiol group (-SH), is unique among 20 natural amino acids and has biological significance. For example, cysteine defines protein structure via forming covalent disulfide bond linkages. In addition, cysteine-containing peptides and proteins, such as glutathione (GSH) are critical physiological components directly or indirectly related to many important biological phenomena, including cellular antioxidant defenses and redox signaling, modulation of various cellular activities, and so forth [1, 2]. To facilitate the structure analysis of proteins, disulfide bond linkages are normally reduced [3, 4]. However, because the resulting thiols are readily oxidized back to disulfides [5], derivatization of thiol groups with a suitable chemical reagent is necessary for increasing thiol stability and improving detection selectivity.
Traditional approaches of protecting free cysteines in solution are based on thiol alkylation, Michael-addition, or thiol/disulfide exchange, but each of them has some limitations. The alkylation of protein thiols using N-ethyl-maleimide or iodoacetamide is not specific toward thiols [3, 6]. Typically, these reactions require an excess amount of derivatizing reagent (1-to 1,000-fold molar excess) and a long reaction time (15 min–8 h) [6, 7]. For the Michael-addition reaction, it refers to the addition of thiols to α, β-unsaturated carbonyl and related compounds. Although it is selective to thiols [8–10], the reaction is irreversible and takes time. In addition, reversible thiol/disulfide exchange reactions serve to oxidize one thiol while reducing one disulfide [11]. This dynamic process is random and non-directed. Therefore, the formation of undesired disulfides is often seen [12]. Our cysteine-specific tagging strategy is the selective selenylation of thiols using selenamide reagents. In our recent study, cysteines in peptides and proteins were successfully derivatized by two selenamide reagents, 2-phenyl-1,2-benzisoselenazol-3(2H)-one (ebselen) and N-(phenylseleno)phthalimides (NPSP) [13]. Among 20 amino acids, only cysteine is reactive toward the selenamide reagents. We also found that the phenylselenenyl tag (PhSe-) added to the cysteine residue via the NPSP derivatization can survive collision-induced dissociation (CID), which is useful in sequencing peptides and locating cysteine residues [14]. However, the selenium tag introduced through ebselen derivatization can be lost during CID to form the protonated ebselen, providing a quick screening method for selective detection of cysteine-containing proteins from mixtures using precursor ion scan by mass spectrometry (MS) [14].
In this work, in consideration of the instability of NPSP in aqueous solution that actually decreases its efficiency in thiol derivatization, a new protocol was adopted to acquire a high conversion yield, in which a freshly prepared NPSP in anhydrous acetonitrile was used to react with thiol substrates for the modification. We further investigated the thiol reactivity toward NPSP and explored the related analytical applications of this modification reaction. This study shows that (1) the NPSP selenylation reaction occurs to thiol peptides and proteins rapidly, selectively and quantitatively within seconds, and the reaction products are stable at room temperature in acidic solution (pH 2.5–6), suggesting that NPSP is a premium reagent for thiol derivatization; (2) the reactivities of thiols depend on the acidities of their adjacent amino acid residues as well as their accessibility, providing an approach to probe protein conformational structure; (3) thiol peptides in enzymatic digests of proteins/peptides can be rapidly and effectively modified by NPSP, indicating the potential utility of this reaction in bottom-up proteomic approaches.
Experimental
Chemicals
β-Lactoglobulin A from bovine milk (lyophilized powder), bovine serum albumin (lyophilized powder, MW ∼66 kDa), human hemoglobin (lyophilized powder, MW ∼64,500 Da), creatine phosphokinase from rabbit muscle (lyophilized powder, MW ∼81 kDa), insulin from bovine pancreas (lyophilized powder), pepsin from porcine gastric mucosa (lyophilized powder), NPSP, tri-n-butylphosphine (TBP, 97%), and N,N-dimethylformamide (anhydrous DMF, 99.8%) were all purchased from Sigma-Aldrich (St. Louis, MO, USA). Peptides Cys-Gly (CG, MW 178.2 Da), glutathione (GSH, reduced form, MW 307.3 Da), His-Cys-Lys-Phe-Trp-Trp (HCKFWW, MW 906.1 Da), and Cys-Gln-Asp-Ser-Glu-Thr-Arg-Thr-Phe-Tyr (CQRSETRTFY, MW 1249.3 Da) were all purchased from TCI America (Tokyo, Japan). Peptide [Arg8]-conopressin G (sequence CFIRNCPRC with one disulfide bond bridging the two cysteines; MW 1062.3 Da) was purchased from Bachem (King of Prussia, PA, USA).
HPLC-grade methanol and acetonitrile (ACN) from GFS Chemicals (Columbus, OH) and Sigma-Aldrich (St. Louis, MO) were used and acetic acid (HOAc) was purchased from Fisher Scientific (Pittsburgh, PA, USA). The de-ionized water used for sample preparation was obtained using a Nanopure Diamond Barnstead purification system (Barnstead International, Dubuque, IA, USA).
NPSP Derivatization
Unless specified otherwise, the protein/peptide samples were typically dissolved in ACN:H2O:HOAc (49.5:49.5:1, v:v:v; denoted as solvent A). The NPSP solution was freshly prepared by adding anhydrous ACN to NPSP powder, and then added to the peptide/protein samples to effect derivatization. After vortexing the reaction mixture for 15 s, the resulting solution was analyzed by MS.
In the case of creatine phosphokinase, for the reaction with the protein under native conditions, a 40 μM creatine phosphokinase in 20 mM NH4OAc solution was mixed with 1 μL of 24 mM NPSP in ACN. For the reaction with the protein under denaturated conditions, a 30 μL of 40 μM creatine phosphokinase in MeOH:H2O:HOAc (49.5:49.5:1, v:v:v) was reacted with 1 μL of 24 mM NPSP in ACN. The reacted proteins were desalted using Vivaspin 5000 MWCO PES prior to MS analysis.
Protein/Peptide Digestion
Digestion of insulin was carried out using pepsin. A 558 μL aliquot of 0.5 mM insulin in 0.5% (v:v) formic acid (FA)/H2O was mixed with 19.5 μL of 0.29 mM pepsin in H2O, to reach a molar ratio of 50:1 (insulin:pepsin). The solution was incubated at 37 °C for 6.5 h. Digestion of [Arg8]-conopressin G was carried out using trypsin. A 1000 μL of 0.1 mM [Arg8]-conopressin G in 25 mM NH4HCO3 aqueous solution was mixed with 50 μL of 0.1 mM trypsin in H2O. The mixture was incubated at 37 °C for 16 h. The resulting digest solutions were diluted 10 times using solvent A before MS analysis.
Reduction of Enzymatic Digests
To reduce the disulfide bonds of peptides in the insulin digest, 5 μL of 0.15 % TBP in DMF was added to 100 μL of 10 μM protein digest, according to the literature [15]. The mixture was vortexed for 1 h at room temperature. Then, the resulting mixture was dried at 60 °C using a vacuum concentrator to remove the excess volatile TBP. The vacuum was released after the temperature cooled down to room temperature and the protein was redissolved in 100 μL of solvent A. Finally, a 100 μL of 10 μM reduced digested insulin solution was mixed with 100 μL of 100 μM NPSP in anhydrous ACN for derivatization.
Mass Spectrometric Analysis
Electrospray ionization (ESI) or electrosonic spray ionization (ESSI [16], a variant form of ESI) was used for analysis of the reacted protein/peptide products. ESSI-MS spectra were recorded in the positive ion mode on a Thermo Finnigan LCQ DECA Mass Spectrometer (San Jose, CA, USA) equipped with a home-built ESSI source. A +5 kV high voltage was used for ESSI and the nebulizing gas (N2) was kept at 180 psi. The sample injection flow rate was 3– 5 μL/min. The heated capillary temperature was 150 °C. CID was used for structural confirmation of the assigned product ions. Data acquisition was performed using Xcalibur (rev. 2.0.7, Thermo Scientific, San Jose, CA, USA). Deconvolution of mass spectra was carried out using Mag-Tran 1.03b2 software based on the ZScore algorithm [17].
The ESI-MS spectra of creatine phosphokinase samples were collected using a Bruker MaXis Q-TOF mass spectrometer (Bremen, Germany) in the positive ion mode. The capillary voltage was 4.5 kV, nebulizer gas was 0.4 bar, dry gas was 4.0 L/min, and the source temperature (dry temperature) was 180 °C.
Safety Precaution
As TBP and NPSP are harmful in contact with skin, swallowed, or inhaled, a cold trap was used to condense the TBP vapor during the vacuum evaporation of the reduced protein digests.
Results and Discussion
Derivatization of Various Protein/Peptide Thiols
In our previous experiment [13], NPSP was first prepared in ACN/H2O/HOAc to reach the desired concentration, and then mixed with protein/peptide thiols for reaction. In the case of glutathione (GSH, sequence r-ECG), GSH was not completely derivatized even when NPSP used was in 5-fold excess.[13] It is very likely that NPSP degraded in the original reagent solution containing water. Indeed, we noticed that the NPSP gradually develops a yellow color in the aqueous solution with time. To circumvent this issue, in this study, NPSP was freshly prepared in anhydrous ACN and then used for derivatizing thiols. Various thiol peptides and proteins were chosen to evaluate the efficiency of the derivatization reaction by NPSP using this new protocol. In the experiment, MS spectra were acquired 15 s after mixing NPSP solution with protein/peptide samples.
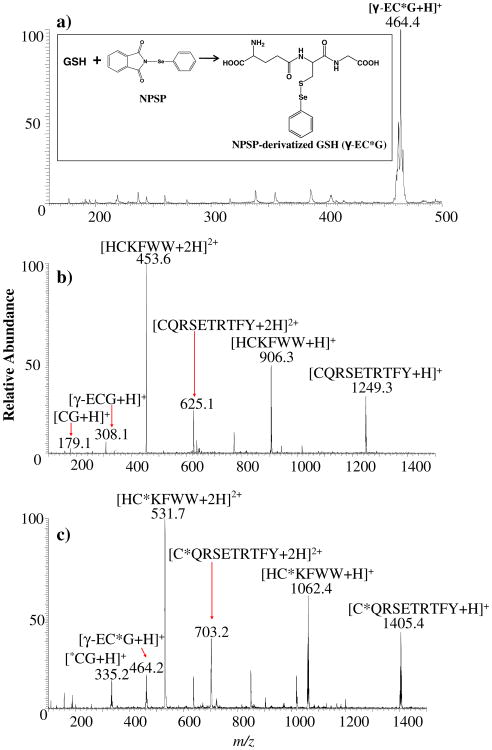
Figure 1a is the ESSI-MS spectrum of the reaction products after mixing NPSP (11 μM in ACN) with GSH (10 μM in solvent A). We found that GSH was completely derivatized and there was no longer a peak at m/z 308 in the spectrum corresponding to the protonated GSH. Instead, a dominant peak at m/z 464 corresponding to the protonated NPSP-derivatized GSH ion [r-EC*G+H]+ appears (the labeling asterisk * indicates a derivatized cysteine with a PhSe tag; see the reaction equation in the Figure 1a inset). A mass shift of 156 Da occurring for this peptide ion after derivatization agrees with the addition of one phenyl-selenenyl tag PhSe- to GSH containing only one free thiol. The characteristic selenium isotope distribution of the derivatized GSH ion helps verify its identification. Also, upon CID of m/z 464, backbone cleavages (the formation of m/z 318, 335, and 389) as well as water loss (the formation of m/z 446) were observed, confirming the product ion structure and the covalent nature of the newly formed Se–S bond in the derivatized peptide. Therefore, one can see that the new protocol of NPSP derivatization does enhance the reaction efficiency, in which quantitative conversion of thiols with a slightly excess amount of NPSP is possible.
Figure 1.
(a) ESSI-MS spectrum showing the products from the NPSP derivatization of GSH via mixing 100 μL of 11 μM NPSP in anhydrous ACN with 100 μL of 10 μM γ-ECG prepared in solvent A for 15 s. (b) ESSI-MS spectrum of 100 μL of HCKFWW/CQRSETRTFY/CG/γ-ECG (10 μM each) in solvent A. (c) ESSI-MS spectrum showing the products from the NPSP derivatization of a peptide mixture via mixing 100 μL of 44 μM NPSP in anhydrous ACN with 100 μL of HCKFWW/CQRSETRTFY/CG/γ-ECG (10 μM each) in solvent A for 15 s. Note that the asterisk * labeling indicates a PhSe- tag
The stability of the derivatized peptide products was also examined. It was found that the derivatized GSH was stable (as tested by ESI-MS) after 2-d storage at room temperature at a pH of 2.5–6.0. However, when the pH of the product solution was adjusted to a pH of 10, the derivatized peptide was not stable and the ion at m/z 464 was no longer apparent in the acquired mass spectrum.
Using the new protocol, we further reacted NPSP with a thiol peptide mixture composed of HCKFWW, CQRSETRTFY, CG, and GSH. Figure 1b is the ESSI-MS spectrum of the original peptide mixture, in which +1 or +2 peptide ions are present. After the modification using NPSP (Figure 1c), the ions of all derivatized peptides [HC*KFWW+H]2+ (m/z 532), [HC*KFWW+H]+ (m/z 1062), [C*QRSETRTFY+H]2+ (m/z 703), [C*QRSETRTFY+H]+ (m/z 1406), [C*G+H]+(m/z 335), and [r-EC*G+H]+ (m/z 464) were observed. The identities of these ions were confirmed by CID (data not shown). In addition, peaks corresponding to the intact peptide ions such as [HCKFWW+H]+ (m/z 906), [CQRSETRTFY+H]+ (m/z 1250), [CG+H]+ (m/z 179), and [r-ECG+H]+ (m/z 308) were not observed, indicating that all of these thiol peptides had been reacted with NPSP. These results further demonstrate the complete tagging of all free cysteines of peptides in mixtures, even with nearly stoichiometric amount of NPSP (NPSP:thiol of 1.1:1). Note that there are some unlabeled peaks in Figure 1b and c. These ions arose from background contamination as determined by the analysis of blank samples.
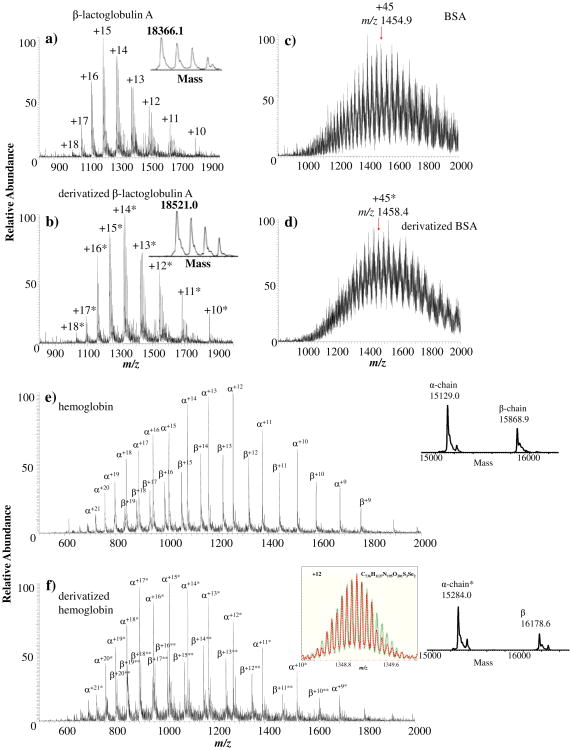
To go a step further, three representative thiol proteins were used as substrates to react with NPSP. The first case was β-lactoglobulin A from bovine milk. Being a common variant of β-lactoglobulin, β-lactoglobulin A contains one free thiol group at Cys121 and two disulfide bonds at Cys66-Cys160 and Cys106-Cys119. Figure 2a displays the ESSI-MS spectrum of 10 μM β-lactoglobulin A in solvent A and Figure 2b is the ESSI-MS spectrum of β-lactoglobulin A (10 μM in solvent A) reacted with NPSP (20 μM in ACN). In the deconvoluted spectra (insets of Figure 1a and b), the mass difference between untagged and tagged protein ions is 155 Da, indicating the addition of a PhSe- tag to the sole free cysteine site of the protein (the theoretical shift is 156 Da and the small mass discrepancy of 1 Da may be caused by a mass error in measuring multiply charged protein ions by our LCQ instrument with low mass resolution). For the second case, a much larger protein, bovine serum albumin (BSA, MW ∼66 kDa) containing one free thiol group, was reacted with NPSP. Figure 2c is the ESSI-MS spectrum of 10μM BSA in solvent A while Figure 2d is the ESSI-MS spectrum of its derivatization product from reaction with NPSP. Again, a mass shift of 157.5 Da appears after derivatization in the acquired spectra that indicates the tagging of the free thiol in BSA. Likewise, the deviation from the theoretical mass shift of 156 Da is likely to be caused by the low resolution of our LCQ instrument.
Figure 2.
ESSI-MS spectra showing free thiol proteins of (a) 10 μM β-lactoglobulin A in solvent A, (c) 10 μM BSA in solvent A, and (e) 10 μM human hemoglobin in solvent A. ESSI-MS spectra containing the products of the NPSP derivatization of thiol proteins via (b) mixing 100 μL of 20 μM NPSP in anhydrous ACN with 100 μL of 10 μM β-lactoglobulin A in solvent A, (d) mixing 100 μL of 20 μM NPSP in anhydrous ACN with 100 μL of 10 μM BSA in solvent A, and (f) mixing 100 μL of 65 μM NPSP in anhydrous ACN with 100 μL of 10 μM human hemoglobin in solvent A. Note that two asterisks ** refer to two PhSe- tags. The left inset of Figure 2f gives the comparison between the measured isotope peak distribution of +12 NPSP-modified hemoglobin β-chain ion (red dashed curve) and the simulated isotope peak distribution of the corresponding ion (green solid curve). The two distributions overlap well
Furthermore, human hemoglobin containing multiple free thiol groups was chosen to be reacted with NPSP. Human hemoglobin is a tetramer, consisting of two α-chains and two β-chains; each α-chain contains a single cysteine residue while each β-chain contains two [18]. Figure 2e is the ESSI-MS spectrum of 10 μM human hemoglobin (in solvent A), in which multiply charged α-chain and β-chain ions were observed under the denaturing condition. Figure 2f displays the ESSI-MS spectrum of derivatized human hemoglobin generated from the selenylation reaction of 100 μL of human hemoglobin 10 μM in solvent A and 100 μL of NPSP (65 μM in ACN). By comparison of the deconvoluted spectra before and after derivatization (insets of Figure 2e and f), the α-chain obtained one PhSe- tag and the β-chain had acquired two tags. This result is expected for the reason that there is only one free thiol group in the α-chain but two free thiol groups in the β-chain. A high resolution ESI-MS profile was collected on a Bruker 12 Tesla FT-ICR mass spectrometer (illustrated in the inset of Figure 2f), the measured isotope peak distribution of +12 ion of the NPSP-derivatized β-chain (red dashed curve) matches the calculated isotope peak distribution of the corresponding ion (green solid curve), confirming the addition of two PhSe-tags onto the β-chain.
In a recent literature about selective modification of cysteine residues in hemoglobin using 5.8-fold excess amount of 1,4-naphthoquinone, the reaction mixture was allowed to react for 15 min and partially modified β chain ions were observed [18]. In comparison, NPSP seems to be a more potent reagent than the Michael-addition reagent in modifying protein thiols. These examples of derivatized thiol proteins and peptides demonstrated that a slight excessive amount of NPSP (NPSP:thiol= 1.1−2:1) could completely convert protein/peptide thiols, showing the high efficiency of the derivatization and the high reaction yield. The number of added tags agrees with that of the free thiols available in the proteins/peptides, indicating the high reaction specificity. The high selectivity is important and is consistent with our previous observation that among 20 amino acids, only cysteine is reactive to NPSP. Equally important, the time needed for derivatization is seconds, representing another advantage in comparison to other traditional thiol derivatization reagents such as alkylation or Michael-addition reagents.
NPSP-derivatized hemoglobin was further digested with pepsin, and MS/MS was used to identify the modification sites. The ESSI spectrum of the digest shows two derivatized peptide ions [KLLSHC*LL+H]+ (m/z 1082) and [KLLSHC*LL+2H]2+ (m/z 542; Figure 1S-a, Supporting Information). CID of these ions indicates that the tags are located in the cysteine residues (Figures 1S-b and c, Supporting Information). This result confirms the high selectivity of NPSP toward cysteines, even in protein substrates.
Probing Thiol Reactivity
Thiol reactivity has attracted increasing attention [19]. Cysteine-specific chemical modification in conjunction with mass spectrometry can be a valuable way to obtain useful protein structural information [6]. It has been reported that solvent accessibility is not the only factor that controls cysteine reactivity, and certain relationships between the chemical environment of cysteine in a protein and its reactivity have been noted [6, 20]. Recently, globally profiling of cysteine reactivity in proteomes even showed that the most hyper-reactive cysteines were remarkably enriched in functional residues of proteins [19].
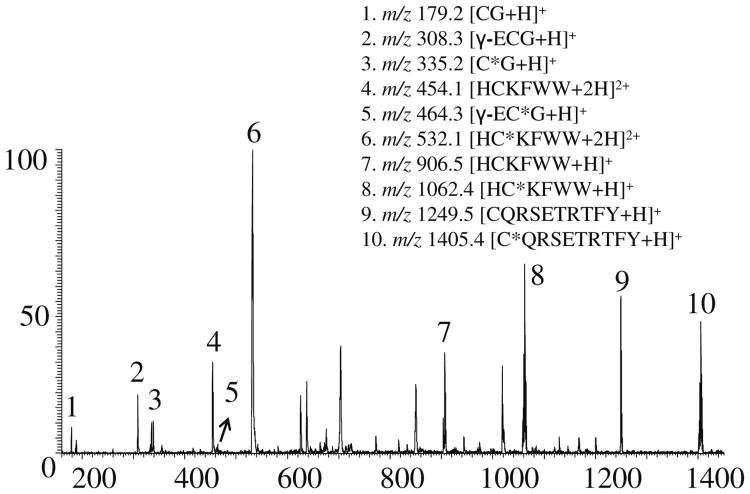
In this study, reactivities of different peptide thiols toward NPSP were investigated. In the experiment, an insufficient amount of NPSP was added to a thiol peptide mixture to generate a competition among different thiol peptides for NPSP (i.e., 100 μL of 20 μM NPSP in anhydrous ACN was added to 100 μL of a mixture containing HCKFWW, CQRSETRTFY, CG, and GSH, 10 μM each) in solvent A. As illustrated in Figure 3a, all the thiol peptides were partially derivatized because of insufficient NPSP in the solution (in contrast with the complete derivatization in Figure 1c). A reaction yield used to evaluate the thiol reactivity of thiol is defined as
Figure 3. ESSI-MS spectrum of 100 μL of 20 μM NPSP in anhydrous ACN mixed with 100 μL of a peptide mixture containing HCKFWW, CQRSETRTFY, CG, and GSH (10 μM each) in solvent A.
for which RAunreacted-peptide is the relative abundance of the underivatized peptide ion and RAreacted- peptide is the relative abundance of the derivatized peptide ion. Under the same condition, the more derivatized peptide there is, the higher the ratio is, and the more reactive this peptide is. In this case, we did not take into account the change in the peptide ionization efficiency before and after derivatization, as the tag PhSe- is a relatively small neutral functional group in comparison to the peptides themselves. From this point of view, the thiol reactivity sequence (from high to low) is HCKFWW, CG, CQDSETRTFY, and GSH, as reported in Table 1.
Table 1. Thiol Peptide Reactivity Toward NPSP.
| Peptides | Underivatized | Derivatized |
|
|||
|---|---|---|---|---|---|---|
|
|
|
|||||
| m/z | RA* (104) | m/z | RA (104) | |||
| HCKFWW | 906.5 | 4.78 | 1062.4 | 16.00 | 0.77 | |
| CG | 179.2 | 1.96 | 335.2 | 1.99 | 0.51 | |
| CQRSETRTFY | 1249.5 | 13.80 | 1405.4 | 10.70 | 0.44 | |
| GSH (γ-ECG) | 308.3 | 4.15 | 464.3 | 0.87 | 0.17 | |
RA=the relative ion abundance
We found that less reactive cysteines occur nearby acidic residues, which tend to suppress thiol dissociation and raise the pKa of the cysteine residues; on the contrary, if cysteine was adjacent to a basic amino acid, its reactivity would be enhanced. This phenomenon is in accordance with the previous report [6]. In our experiment, HCKFWW is more reactive than other thiol peptides toward NPSP, probably because of the factor that its cysteine residue is adjacent to a basic amino acid on both sides (His and Lys). GSH has the lowest reactivity toward NPSP because its cysteine residue is next to an acidic amino acid (r-ECG). In the case of CG and CQDSETRTFY, both of their cysteine residues are adjacent to a neutral amino acid. CG is more reactive with NPSP than CQDSETRTFY probably because it has a much smaller size and, therefore, a much lower steric hindrance.
Probing Protein Conformational Change
Site-specific labeling with molecular probes in conjunction with MS detection can facilitate the visualization of protein conformational changes [15, 21, 22]. In this study, we present another experiment to study the reactivity of different cysteine residues in the same protein, which can provide clues to the protein conformational alteration.
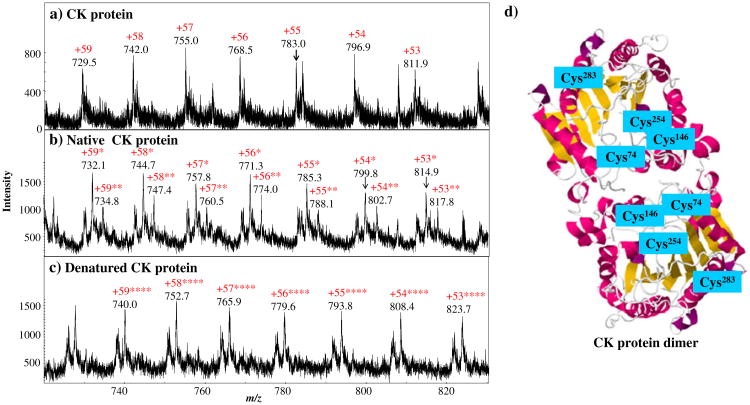
Creatine phosphokinase (CK) was used in this experiment. CK is a dimer consisting of two identical subunits (see Figure 4d). In each subunit, there are four free cysteine residues, three of which are embedded inside the protein (Cys74, Cys146, and Cys254) [23, 24]. According to the crystal structure of CK provided in Protein Data Bank (RCSB PDB), indeed, the total solvent accessibility areas calculated by the GETAREA software [25], for Cys74, Cys146, Cys254, and Cys283 are 0.00, 2.21, 4.10, and 35.76 Å2, respectively. From these data, Cys283 is the most accessible for derivatization. Figure 4a is the ESI-MS spectrum of intact CK protein in which the multiply charged ions +53 to +59 were detected. Figure 4b is the ESI-MS spectrum of the derivatized CK protein from the reaction with NPSP under native conditions (in 20 mM ammonium acetate). In Figure 4b, the ions of major products +53* to +59*, arose, corresponding to the CK ions with one PhSe-tag. This result is in agreement with the previously reported result using the derivatizing reagent of methoxycarbonyl-1,4-hydroquinone.[15] Also, minor product ions, the CK ions with two tags, also appear probably due to the modification of the Cys254, the second most accessible residue.
Figure 4.
ESI-MS spectra of (a) intact creatine phosphokinase, (b) the creatine phosphokinase derivatized by NPSP under native conditions, and (c) the creatine phosphokinase derivatized by NPSP under denaturated conditions. (d) The crystal structure of the rabbit muscle creatine kinase [24] showing the four free cysteine residues in each monomer of the creatine phosphokinase was obtained from the RCSB Protein Data Bank and generated by PQS software (Jmol ver. 12.0.41)
By contrast, upon denaturation in an acidic solvent containing methanol, the CK displayed different reaction behavior with NPSP. In the latter case, the CK protein ions with four PhSe- tags were observed and no underivatized or partially labeled CK ions were observed (Figure 4c). These results indicate that only one free thiol is most accessible in a native folded structure while all of the four thiols become accessible upon protein denaturation. The complete conversion of CK protein by NPSP after denaturation further shows the potency of this selenamide reagent in modifying thiols. Evidently, the thiol reactivities depend on protein conformation. It is therefore possible to monitor the protein conformational alteration by examining the number of thiols that can be derivatized by NPSP. It is advantageous to use a fast derivatizing reagent like NPSP for such a purpose, as undesirable conformational changes resulting from a long derivatization process needs to be avoided [26].
Derivatization of Protein/Peptide Digests by NPSP Following Reduction
Bottom-up approach is a common method to identify and sequence proteins by proteolytic digestion of proteins followed with MS analysis. In traditional protocols, disulfide bridges of peptides in the digest are first reduced using reducing agents such as dithiothreitol (DTT) and the resulting free thiols are protected via iodoacetamide derivatization. However, such a derivatization step usually takes half an hour. In consideration that NPSP tagging takes place in seconds with high selectivity and yield, it would be necessary to explore an alternative derivatization method using NPSP in the bottom-up approach. However, it is challenging to derivatize thiols by NPSP in the presence of excess amount of reductants because chemical reductants can also reduce Se– S bond[13] (i.e., the NPSP-derivatized peptide products). One has to remove the excess reductant prior to the addition of NPSP into the digest samples for derivatization and removal of reductant such as DTT from the digests could be troublesome. To overcome this problem, we used TBP, a volatile reductant, to reduce disulfide bonds of peptides in the digests so that the excess TBP can simply removed by vacuum evaporation [15, 27].
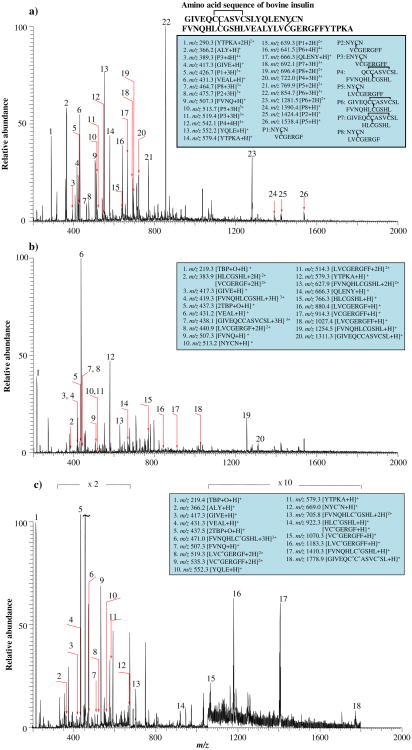
In this experiment, insulin was chosen as an example to test the methodology feasibility. After digestion by pepsin and reduced by TBP, peptides containing free cysteine residues were generated. Then, the sample was evaporated in vacuum to remove excess TBP. Subsequently, the protein digest sample was redissolved in solvent A and added with NPSP. Figure 5a is the ESSI-MS spectrum of digested insulin. Eight disulfide bond-containing peptides, NYCN/VCGERGF, NYCN/VCGERGFF, ENYCN/VCGERGFF, QCCASVCSL/FVNQHLCGSHL, NYCN/LVCGERGFF, GIVE-QCCASVCSL/FVNQHLCGSHL, GIVEQCCASVCSL/HLCGSHL, and NYCN/LVCGERGF (denoted as P1–P8, respectively, in the figure inset; the sequences before and after the slash indicate the two chains of the peptides) were present in the insulin peptic digest. In addition, peptides without disulfide bonds, such as YTPKA, ALY, GIVE, VEAL, FVNQ, YQLE, and QLENY, were also observed (see the list of detected peptide ions in the figure inset). After the reduction of disulfide bonds by TBP, all these peptides P1–P8 were reduced successfully and reduced peptides NYCN, VCGERGF, VCGERGFF, FVNQHLCGSHL, LVCGERGFF, GIVE-QCCASVSCL, HLCGSHL, and LVCGERGF were observed, as expected (shown in Figure 5b). Two reduced peptides, ENYCN and QCCASVCSL, were not detected in the positive ion mode, probably due to either high acidities or the lack of basic residues. On the other hand, peptides without disulfide bonds (e.g., GIVE, VEAL, FVNQ, YTPKA, and QLENY) were insensitive to TBP reduction and remained intact. Two major peaks at m/z 219 ([TBP+O+H]+) and 437 ([2TBP+O+H]+) in Figure 5b arose from the ionization of the oxidized TBP.
Figure 5. ESSI-MS spectra of (a) the pepsin-digested insulin, (b) the insulin digest after TBP reduction, and (c) the reduced insulin digest after NPSP derivatization.
Figure 5c is the spectrum of the peptides detected after the NPSP derivatization. All the free thiol-containing peptides produced from the reduction of P1–P8 appear in Figure 5b (i.e., NYCN, VCGERGF, VCGERGFF, FVNQHLCGSHL, LVCGERGF, LVCGERGFF, GIVE-QCCASVCSL, and HLCGSHL), and have acquired PhSe-tags from NPSP. The peak assignment was also confirmed by CID. For example, the single charged ion [NYC*N+H]+ (m/z 669) gave rise to fragment ions b3 and y3 and the doubly charged ion [FVNQHLC*GSHL+2H]2+ (m/z 706) produced fragment ions b6, b7, b9, y4, y9, and y7. In particular, the peak at m/z 1779 corresponding to [GIVE-QC*C*ASVC*SL+H]+ was detected (Figure 5c), indicating that all the three thiols of the peptide GIVEQCCASVCSL were successfully modified simultaneously by NPSP. The m/z 1779 ion gave rise to fragment ions of b10, b11, and b12 upon CID (Figure 2S-d, Supporting Information), confirming its structure. It is also noted that the PhSe- tags survive the CID process of [GIVEQC*C*ASVC*SL+H]+, in agreement with our previous observation [14]. This phenomenon was also observed in CID spectra of other derivatized peptides (Figures 2S-a, b, and c, Supporting Information). This feature is useful in pinpointing the location of thiols in examined peptides. The successful derivatization of thiol peptides in the digest via the reaction with NPSP within 15 s emphasizes that NPSP is a very effective derivatization reagent.
Another tryptic digest of [Arg8]-conopressin G was also tested using the same approach and the reduced peptides were also modified by NPSP (Figures 3S, Supporting Information), further confirming the viability of the usage of NPSP as an alternative reagent for thiol protection in the bottom-up method. The resulting ion [C*FIR+H]+ (m/z 694) was confirmed by CID which showed the formation of fragment ions b2, b3, b4, b4-NH3, y2, and y3-NH3. Another resulting ion [NC*PR+H]+ (m/z 645) yielded fragment ions b3-NH3, b4, y2, y2-NH3, y3, and y3-NH3, upon CID.
To further expedite the process for the reduction of protein digest, we also tested an electrolytic method to reduce disulfide bonds. Besides the benefit of greatly shortened reduction time, no chemicals were introduced into the sample solution; therefore this approach could dispense with removing excess reducing reagents. Recently we reported that online electrochemistry coupled with mass spectrometry can be used for fast structural analysis of proteins/peptides [28]. In this study, as a preliminary trial, we passed the insulin digest solution into an electrochemical cell for electrolytic reduction and then derivatize the resulting products using fast NPSP derivatization. We did observe some derivatized peptides such as [FVNQHLC*GSHL+3Hz]3+ (Figure 4S, Supporting Information). As electrolysis is much faster than TBP reduction and takes only seconds, it would further expedite the whole process and eliminate a hazardous reagent from the protocol. Further investigation is underway.
Conclusions
NPSP is shown to be a premium thiol derivatization reagent. It reacts rapidly with protein/peptide thiols in seconds and allows a complete conversion of the free thiol groups with nearly stoichiometric amount of reagent. In addition, the NPSP reaction can be used to study thiol reactivity for obtaining structural and conformational information of proteins. The derivatization reaction is also applicable to protein enzymatic digests, suggesting its prospective application to the bottom-up proteomic approach. In this regard, the benefits include: (1) the derivatization step is faster than using traditional derivatizing reagents like iodoacetamide. Although the evaporation for the removal of excess amount of TBP takes time at this stage, it helps the ionization of the resulting products and it could be avoided if one replaces the chemical reduction by online fast electrochemical reduction; (2) the reaction of NPSP is highly selective and efficient; (3) the added tag via NPSP derivatization can survive the CID process, which is useful for locating thiols in peptides. Given the biological significance of thiols and the important features of selenylation reactions revealed in this study, it is expected that there will be many other applications of this thiol-selenium chemistry.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge support of this work by NSF (CHE-0911160) and ASMS Research Award. In particular, the authors are thankful to Professor Michael L. Gross for the access to the NIH/NCRR Mass Spectrometry Resources at Washington University in St. Louis (grant number 2P41RR000954) and High-End Instrument Program of the NCRR (grant No. 1S10 025101).
Footnotes
Electronic supplementary material: The online version of this article (doi:10.1007/s13361-011-0317-3) contains supplementary material, which is available to authorized users.
References
- 1.Meister A, Anderson ME. Glutathiemoone. Ann Rev Biochem. 1983;52:711–760. doi: 10.1146/annurev.bi.52.070183.003431. [DOI] [PubMed] [Google Scholar]
- 2.Baty JW, Hampton MB, Winterbourn CC. Proteomic detection of hydrogen peroxide-sensitive thiol proteins in jurkat cells. Biochem J. 2005;389:785–795. doi: 10.1042/BJ20050337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sechi S, Chait BT. Modification of cysteine residues by alkylation. A tool in peptide mapping and protein identification. Anal Chem. 1998;70:5150–5158. doi: 10.1021/ac9806005. [DOI] [PubMed] [Google Scholar]
- 4.Moritz RL, Eddes JS, Reid GE, Simpson RJ. S-pyridylethylation of intact polyacrylamide gels and in situ digestion of electrophoretically separated proteins: a rapid mass spectrometric method for identifying cysteine-containing peptides. Electrophoresis. 1996;17:907–917. doi: 10.1002/elps.1150170512. [DOI] [PubMed] [Google Scholar]
- 5.Cleland WW. Dithiothreitol, a new protective reagent for SH groups. Biochemistry. 1964;3:480–482. doi: 10.1021/bi00892a002. [DOI] [PubMed] [Google Scholar]
- 6.Mendoza VL, Vachet RW. Probing protein structure by amino acid-specific covalent labeling and mass spectrometry. Mass Spectrom Rev. 2009;28:785–815. doi: 10.1002/mas.20203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rogers LK, Leinweber BL, Smith CV. Detection of reversible protein thiol modifications in tissues. Anal Biochem. 2006;358:171–184. doi: 10.1016/j.ab.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 8.Tokoroyama T. Discovery of the Michael reaction. Eur J Org Chem. 2010;10:2009–2016. [Google Scholar]
- 9.Ishii T, Fujioka S, Sekiguchi Y, Kotsuki H. A new class of chiral pyrrolidine-pridine conjugate base catalysts for use in asymmetric Michael addition reactions. J Am Chem Soc. 2004;126:9558–9559. doi: 10.1021/ja046871g. [DOI] [PubMed] [Google Scholar]
- 10.Luo S, Mi X, Zhang L, Liu S, Xu H, Cheng J. Functionalized chiral ionic liquids as highly efficient asymmetric organocatalysts for Michael addition to nitroolefins. Angew Chem Int Ed. 2006;45:3093–3097. doi: 10.1002/anie.200600048. [DOI] [PubMed] [Google Scholar]
- 11.Gilbert HF. Thiol/disulfide exchange equilibria and disulfide bond stability. Method Enzymol. 1995;251:8–28. doi: 10.1016/0076-6879(95)51107-5. [DOI] [PubMed] [Google Scholar]
- 12.Szajewski RP, Whitesides GM. Rate constants and equilibrium constants for thiol-disulfide interchange reactions involving oxidized glutathione. J Am Chem Soc. 1980;102:2011–2026. [Google Scholar]
- 13.Xu K, Zhang Y, Tang B, Laskin J, Roach P, Chen H. Study of highly selective and efficient thiol derivatization using selenium reagents by mass spectrometry. Anal Chem. 2010;82:6926–6932. doi: 10.1021/ac1011602. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y, Zhang H, Cui W, Chen H. Tandem MS analysis of selenamide-derivatized peptide ions. J Am Soc Mass Spectrom. 2011;22:1610–1621. doi: 10.1007/s13361-011-0170-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dayon L, Roussel C, Girault H. Probing cysteine reactivity in proteins by mass spectrometric EC-tagging. J Proteome Res. 2006;5:793–800. doi: 10.1021/pr050365o. [DOI] [PubMed] [Google Scholar]
- 16.Takáts Z, Wiseman JM, Gologan B, Cooks RG. Mass spectrometry sampling under ambient conditions with desorption electrospray ionization. Anal Chem. 2004;76:4050–4058. doi: 10.1126/science.1104404. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Z, Marshall A. A universal algorithm for fast and automated charge state deconvolution of electrospray mass-to-charge ratio spectra. J Am Soc Mass Spectrom. 1998;9:225–233. doi: 10.1016/S1044-0305(97)00284-5. [DOI] [PubMed] [Google Scholar]
- 18.Diedrich JK, Julian RR. Site-selective fragmentation of peptides and proteins at quinone-modified cysteine residues investigated by ESI-MS. Anal Chem. 2010;82:4006–4014. doi: 10.1021/ac902786q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weerapana E, Wang C, Simon GM, Richter F, Khare S, Dillon MBD, Bachovchin DA, Mowen K, Baker D, Cravatt B. Quantitative reactivity profiling predicts funtional cysteines in proteomics. Nature. 2010;468:790–795. doi: 10.1038/nature09472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Britto PJ, Knipling L, Wolff J. The local electrostatic environment determines cysteine reactivity of tubulin. J Biol Chem. 2002;277:29018–29027. doi: 10.1074/jbc.M204263200. [DOI] [PubMed] [Google Scholar]
- 21.Daggett KA, Sakmar TP. Site-specific in vitro and in vivo incorporation of molecular probes to study G-protein-coupled receptors. Curr Opin Chem Biol. 2011;15:392–398. doi: 10.1016/j.cbpa.2011.03.010. [DOI] [PubMed] [Google Scholar]
- 22.Miao Z, Wu S, Chen H. The study of protein conformation in solution via direct sampling by desorption electrospray ionization mass spectrometry. J Am Soc Mass Spectrom. 2010;21:1730–1736. doi: 10.1016/j.jasms.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang HR, Bai JH, Zheng SY, Wang ZW, Zhou HM. Ascertaining the number of essential thiol groups for the folding of creatine kinase. Biochem Biophys Res Commun. 1996;221:174–180. doi: 10.1006/bbrc.1996.0565. [DOI] [PubMed] [Google Scholar]
- 24.Rao JK, Bujacz G, Wlodawer A. Crystal structure of rabbit muscle creatine kinase. FEBS Lett. 1998;439:133–137. doi: 10.1016/s0014-5793(98)01355-6. [DOI] [PubMed] [Google Scholar]
- 25.Fraczkiewicz R, Braun W. Exact and efficient analytical calculation of the accessible surface areas and their gradients for macromolecules. J Comp Chem. 1998;19:319–333. [Google Scholar]
- 26.Gau BC, Chen H, Zhang Y, Gross ML. Sulfate radical anion as a new reagent for fast photochemical oxidation of proteins. Anal Chem. 2010;82:7821–7827. doi: 10.1021/ac101760y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carru C, Deiana L, Sotgia S, Pes GM, Zinellu A. Plasma thiols redox status by laser-induced fluorescence capillary electrophoresis. Electrophoresis. 2004;25:882–889. doi: 10.1002/elps.200305768. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Y, Dewald HD, Chen H. Online mass spectrometric analysis of proteins/peptides following electrolytic cleavage of disulfide bonds. J Proteome Res. 2011;10:1293–1304. doi: 10.1021/pr101053q. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.