Summary
Calorie restriction (CR) extends lifespan in diverse species. Mitochondria play a key role in CR adaptation, however, the molecular details remain elusive. We developed and applied a quantitative mass spectrometry method to probe the liver mitochondrial acetyl-proteome during CR vs. control diet in mice that were wild-type or lacked the protein deacetylase SIRT3. Quantification of 3,285 acetylation sites −2,193 from mitochondrial proteins rendered a comprehensive atlas of the acetyl-proteome and enabled global site-specific, relative acetyl occupancy measurements between all four experimental conditions. Bioinformatic and biochemical analyses provided additional support for the effects of specific acetylation on mitochondrial protein function. Our results (1) reveal widespread reprogramming of mitochondrial protein acetylation in response to CR and SIRT3, (2) identify three biochemically distinct classes of acetylation sites, and (3) provide evidence that SIRT3 is a prominent regulator in CR adaptation by coordinately deacetylating proteins involved in diverse pathways of metabolism and mitochondrial maintenance.
Introduction
Calorie restriction (CR) is the only regimen known to extend the lifespan and healthspan in a spectrum of organisms that include yeast, mice, and non-human primates (Colman et al., 2009; Mattison et al., 2012; Weindruch et al., 1986). Reducing food consumption of mice 25%-60% without undernutrition extends the lifespan of rodents up to 50% (Weindruch et al., 1986) and delays the onset of age-related maladies like cardiovascular disease, cancer, and diabetes in different animal models (Colman et al., 2009; Koubova and Guarente, 2003). The positive effects of CR are linked to major metabolic reprogramming towards efficient fuel utilization and a reduction in oxidative damage to macromolecules (Anderson and Weindruch, 2012; Sohal and Weindruch, 1996). Mitochondrial dysfunction plays an important role in cellular aging (Wallace, 2005) and reduces fuel utilization plasticity. As metabolic centers for fuel utilization and primary producers of cellular reactive oxygen species, mitochondria are poised to act as mediators of reprogramming under CR. However, the molecular basis for global metabolic adaptation induced by CR remains unknown.
Emerging evidence suggest that reversible protein acetylation might be a dynamic protein modification that regulates metabolism through multiple mechanisms. The sirtuin family of NAD+-dependent protein deacetylases are implicated as possible regulators of the cellular adaption to CR, and directly deacetylate specific metabolic enzymes. In yeast, sirtuins are reported as critical components of the CR dependent extension of lifespan (Lin et al., 2000), and in multiple organisms sirtuins are linked with improved healthspan and metabolic regulation. A critical link between an anti-aging phenotype associated with CR and sirtuins was recently demonstrated for the mitochondrial Sirtuin3 (SIRT3), which was essential for the prevention of age-related hearing loss in mice under CR (Someya et al., 2010; Yu et al., 2012). In response to CR or prolonged fasting, the expression of SIRT3 is induced considerably (Hirschey et al., 2010; Someya et al., 2010). SIRT3 regulates the function of several mitochondrial proteins involved in oxidative phosphorylation, fatty acid oxidation, the urea cycle, and the antioxidant response system (Hallows et al., 2006; Hallows et al., 2011; Hirschey et al., 2010; Lombard et al., 2007; Qiu et al., 2010; Schlicker et al., 2008; Schwer et al., 2006; Shimazu et al., 2010; Yu et al., 2012). SIRT3 is postulated to be the main, if only, protein mitochondrial deacetylase, but the extent of its capacity to modulate mitochondrial processes remains unclear.
Protein acetylation has emerged as a major post-translational modification and accumulating evidence indicates that acetylation rivals phosphorylation as a regulatory modification (Chen et al., 2012; Choudhary et al., 2009; Henriksen et al., 2012; Kim et al., 2006; Lundby et al., 2012; Pagliarini et al., 2008; Zhao et al., 2010). With respect to CR and the role of SIRT3, a broad and deep understanding of acetylation dynamics remains elusive. In a handful of cases, mass spectrometry (MS) has been applied to study mitochondrial acetylation in response to diet changes or SIRT3 expression on a subset of potential protein substrates (Fritz et al., 2012; Hirschey et al., 2010; Kendrick et al., 2011; Law et al., 2009; Schwer et al., 2009; Someya et al., 2010); however, to understand the diverse mechanisms of reversible acetylation that modulate mitochondrial metabolism and maintenance, a robust, quantitative approach that maps physiologically relevant changes across the mitochondrial acetylome is required.
Here we describe a quantitative acetyl-proteomic method that combines isobaric tagging for multiplexed quantification, immunoenrichment, two-dimensional chromatography, and high resolution, high accuracy MS. We applied this approach to examine the mitochondrial-wide changes to the acetyl-proteome during CR in mice and concomitantly examined the function of resident protein deacetylase SIRT3 in mediating the CR response. We identified 3,285 acetylation sites, of which 2,193 were localized to mitochondrial proteins, and achieved near complete coverage of the mitochondrial proteome. This quantitative acetylation atlas provides a key resource for understanding mitochondrial regulation and reversible protein acetylation. With these data, we establish that SIRT3 is a major regulator of the mitochondrial acetylome in response to CR and discover new processes controlled by SIRT3.
Results
Metrics
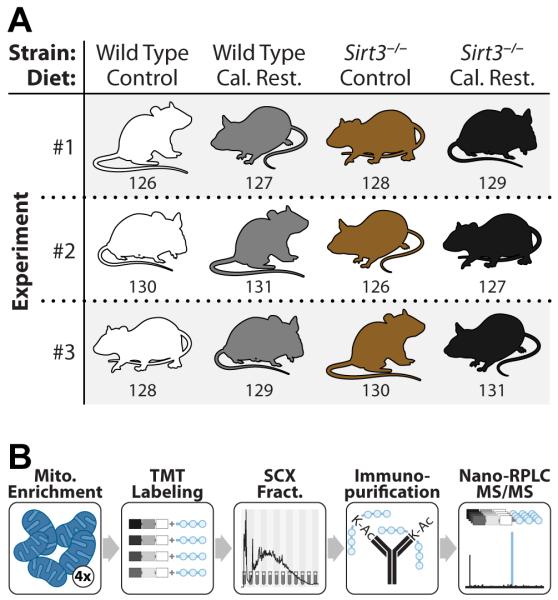
We developed and applied a quantitative acetyl-proteomic method to examine the mitochondrial-wide reprogramming of the acetylome and proteome in response to calorie restriction (CR) and the role of SIRT3 dependent regulation of the mitochondrial acetylome under CR. Our approach (Figure 1) utilized four distinct components: (1) isobaric tags for multiplexed quantification, (2) an antibody-based acetylpeptide enrichment technique, (3) two-dimensional chromatography, and (4) high resolution, high mass accuracy MS. We compared liver mitochondrial proteomes and acetyl-proteomes of 12 mice (C57BL/6J) following a CR dietary study. Specifically, 3 WT and 3 Sirt3−/− mice, at 2 months of age, had their caloric intake reduced by 25% (i.e., CR group) whilst 3 WT and 3 Sirt3−/− animals were maintained on a control diet (i.e., CD group, Figure 1). All mice were euthanized at 12 months of age. Altogether, we analyzed, in biological triplicate, the mouse liver mitochondrial proteomes and acetyl-proteomes across four conditions: WT-CD, WT-CR, Sirt3−/−-CD, and Sirt3−/−-CR. From these experiments we detected and quantified 3,285 acetylation sites – the vast majority of which (2,193) were localized to mitochondrial proteins (434). We likewise achieved very deep quantification of the mitochondrial proteome (568 of the 701 confirmed members) allowing for the most comprehensive mitochondrial acetyl-proteome study to date.
Figure 1. Experimental design and mass spectrometry workflow for quantitation of acetylated mitochondrial proteins between four biological conditions.
(A) WT and Sirt3−/− mouse liver mitochondria were compared in biological triplicate from mice fed either the control or calorie restricted (CR) diet. Numbers correspond to the isobaric mass tag used for that sample.
(B) Extracted protein from purified liver mitochondria was trypsin digested followed by labeling with TMT reagents. Labeled peptides were combined and fractionated by strong cation exchange chromatography. Each fraction was enriched for acetyl peptides with a pan anti-acetyl lysine antibody and analyzed via nano-RPLC-MS/MS. See also Figure S3.
To profile acetylome dynamics, as regulated by CR and/or SIRT3, intact mitochondria were purified from mouse livers (vide supra) via differential centrifugation. The purified mitochondrial proteins were subjected to trypsin digestion followed by isotopic labeling (isobaric tag, TMT). Labeled peptides were then combined (Figure 1), fractionated by strong cation exchange (SCX) chromatography, and immunopurified to isolate acetylated peptides. A small portion (~5%) was excluded from acetyl-enrichment to obtain protein abundance (Table S1). All fractions were analyzed using a nanoflow liquid chromatography-mass spectrometry method (nHPLC-MS) on a high resolution and high mass accuracy orbitrap mass spectrometer. Eluting peptide cations were isolated and subjected to tandem mass spectrometry (MS/MS) to obtain both sequence identity and quantitative information in a single scan (Figure 2). A pervasive problem that occurs during isobaric tagging is a phenomenon known as precursor interference. Briefly, when a precursor m/z is selected for dissociation it is isolated from all the other peptide m/z values present in the MS1 spectrum with approximately a 2-3 m/z window. Very often, (>60%) other peptide species are co-isolated with the intended target. And though these contaminants are usually of lower abundance and do not affect identification of the target sequence, all isolated precursors generate reporter tags and are measured collectively. The presence of these contaminants distorts the measurement and reduces both quantitative accuracy and dynamic range. To eliminate this problem we employed a recently developed gas-phase ion/ion reaction scheme, QuantMode (Wenger et al., 2011a). QuantMode subjects each isolated precursor to an extremely fast (~50 ms) gas-phase ion/ion reaction that purifies the intended target prior to MS/MS. With this strategy we collected ~1.5 million MS/MS scans, each of which was processed using a custom in-house developed software suite (COMPASS) (Phanstiel et al., 2011; Wenger et al., 2011b). Only peptide and protein identifications below 1% false discovery rate (FDR) are reported here; acetylation site assignments were subsequently localized to specific lysines using a modified A-score algorithm (Figure S1) (Wu et al., 2011).
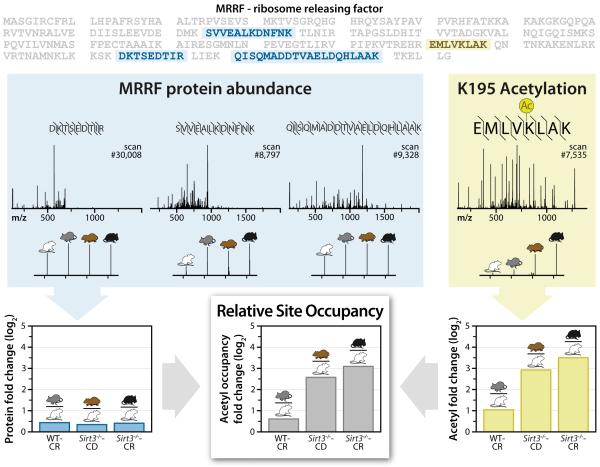
Figure 2. Example acetyl occupancy quantitation (relative), for site K195 on the MRRF protein. The peptides used for calculations are highlighted within the MRRF protein sequence.
Blue: Un-modified peptide sequences, spectra with reporter ion magnification and protein fold change for each condition compared to WT-CD condition. Note, the three unique peptides from five total identifications are shown here. Yellow: The MRRF K195 acetylated peptide sequence, spectra, and acetyl fold change for each condition compared to WT-CD. Bottom center: Results for acetyl fold change normalized to protein fold change for each condition compared to WT-CD. See also Figure S1.
Alterations of protein abundance, either in vivo or during preparation, can cause apparent acetylation changes, even when the acetylation site occupancy of a protein remains unaffected. Measuring absolute occupancy of global acetylation sites would require the synthesis of thousands of isotopically labeled internal standards and is, thus, currently impractical. Instead we implemented an experimental design to generate relative acetylation site occupancy information by quantifying both acetylation and protein abundance. The spectrum shown in Figure 2A depicts an MS/MS scan mapped to the peptide EMLVK(ac)LAK, a product of digested ribosome releasing factor (MRRF). Note the inset displays the reporter tag mass-to-charge (m/z) region, which indicates strong up-regulation of this site in WT-CR (2-fold), Sirt3−/−-CD (7.5-fold), and Sirt3−/−-CR (11-fold) animals. To confirm this difference is a result of increased acetylation, rather than an artifact of MRRF abundance, we examined all detected un-modified MRRF peptides. In total, five MS/MS scans were mapped to 3 unique un-modified peptides from MRRF. Summation of these 5 reporter ion regions provides a protein abundance measurement; combination of these data with acetylation fold-change confirms that the acetylation site occupancy is indeed dynamic (Figure 2). This process was conducted for each of the 3,285 acetylation sites documented here and is presented in whole in Table S2.
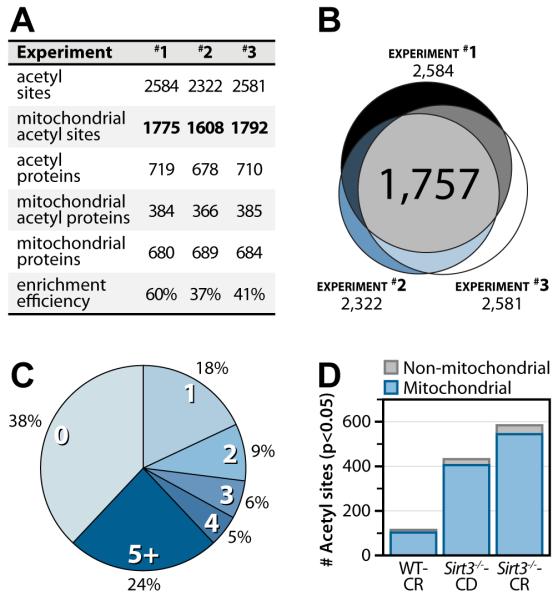
From these data we conclude our method is both specific and reproducible. In each of the 3 experimental replicates, approximately 70% of the acetylation sites were from mitochondrial proteins. The average enrichment efficiency observed in this study (46%), i.e., number of acetyl peptides identified divided by the total number of identified peptides, was considerably higher than values typically reported (Figure 3A) (Guan et al., 2010; Lundby et al., 2012). Improved enrichment efficiency may have resulted from reducing sample complexity through fractionation prior to immunoenrichment or the increased abundance of the acetylation modification in the Sirt3−/− mitochondria. Figure 3B presents the overlap of acetylation sites detected across the replicates – over half of all acetyl sites were found in every experiment. The inter-experiment overlap of our acetylation identifications is higher than what is commonly observed for phosphorylation, where only 1/3 of all phosphorylation sites would likely be observed in all 3 experiments (Wolf-Yadlin et al., 2007). The high overlap provides both a basis for strong statistical analysis, as most sites are detected in all 3 biological replicates, and strong confidence in our identifications. From this comprehensive atlas we report that 65% of mitochondrial proteins have at least one identifiable acetylation site (Figure 3C); the majority of these proteins have multiple sites of acetylation. This observation was unexpected and goes well beyond the recent estimates that ~20% of the mitochondrial proteome is acetylated (Kim et al., 2006). Site occupancy measurements indicate that of the 1,275 mitochondrial acetylation sites identified in all of the experiments 8%, 32%, and 43% exhibit ≥ 2-fold statistically significant changes (p < 0.05, Welch’s t-test with Storey correction) in the CR, Sirt3−/−, and Sirt3−/−-CR conditions, respectively (Figure 3D. Altogether, our data indicate that the mitochondrial proteome is (1) extensively acetylated and (2) pervasively altered by CR and genetic loss of SIRT3.
Figure 3. Acetylomic experimental results and metrics.
(A) Experimental metrics for each experiment. Enrichment efficiency is calculated as the ratio of acetyl peptide spectral matches to the total number of peptide spectral matches for enriched samples.
(B) Venn diagram indicating overlap of the acetyl sites quantified in each experiment. Experimental overlap of acetyl sites identified in three biological replicates yields 1,757 quantifiable acetylation sites.
(C) Pie chart of the number of acetylation sites detected per mitochondrial protein.
(D) Number of acetyl sites significantly changing ≥ 2-fold for each condition compared to WT control diet (p < 0.05, Welch’s t-test with Storey Correction). See also Tables S1 and S2.
Calorie restriction dramatically alters the mitochondrial protein acetylation landscape
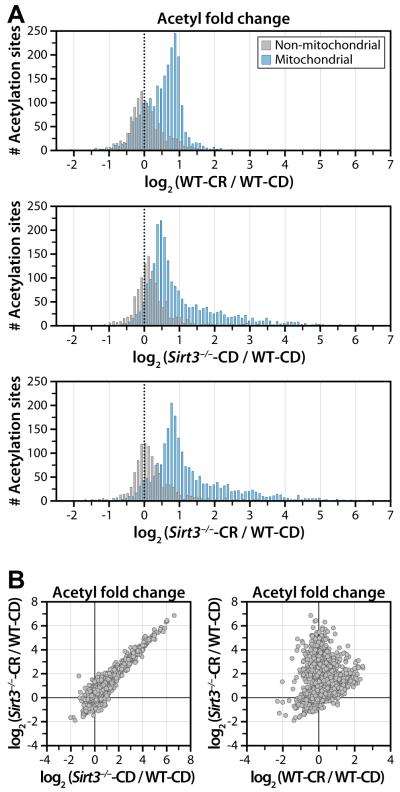
CR dynamically alters mitochondrial protein acetylation, with two notable trends. CR increases acetylation site occupancy of 135 acetyl sites (p < 0.05) by >2-fold when compared to CD (Figure 3D), while a subset of mitochondrial acetylation sites (>100) display decreased acetylation under CR. The two modes of the distribution in Figure 4A reflect both the acetylation sites that decrease under CR and a segregation based upon mitochondrial localization, with acetyl sites localized to the mitochondria exhibiting increased acetylation and the nearly 500 non-mitochondrial acetyl sites showing minimal change. These results indicate CR is a major regulator of protein acetylation, but these changes are predominantly mitochondrial. A major benefit of our approach is the quantitation of total protein in addition to acetyl sites. Compared with acetylation, there is relatively little change in the overall distribution of protein-fold changes across conditions, as only 33 proteins change more than 1.5 fold (Figure S2A), supporting a hypothesis that changes in acetylation, but not overall protein levels, drive metabolic reprogramming under CR.
Figure 4. Quantitative distribution of acetyl site data and scatterplot analysis reveals distinct populations.
(A) Bimodal distribution of acetyl sites quantified (n=3) segregates between mitochondrial (blue) and non-mitochondrial (gray) proteins. Mitochondrial acetyl sites exhibit increased acetylation, while non-mitochondrial acetyl sites show minimal change. (Top plot) Analysis of acetyl sites for WT-CR vs. WT-CD indicates CR alters mitochondrial protein acetylation. (Middle and lower plots) Analysis of acetyl sites for Sirt3−/−-CD/CR vs WT-CD indicates that loss of SIRT3 dominates the acetylation content of mitochondrial proteins with increased acetylation apparent in Sirt3−/− under both CD and CR.
(B) Scatter plot (Top) of acetyl fold change between Sirt3−/−-CR and Sirt3−/−-CD, displays near-linear correlation and indicates SIRT3 expression dominates global acetylation status of the mitochondrial proteome.
(Lower plot) Scatter plot comparison of the acetyl fold change of Sirt3−/−-CR to WT-CR exposes unique populations of acetyl sites. See also Figure S2.
CR induced significant deacetylation of a 108 acetyl sites (p < 0.05) by 20% or greater. SIRT3 expression is stimulated under chronic CR (Hallows et al., 2011; Schwer et al., 2009; Someya et al., 2010) (Figure S3) and both deacetylates and regulates the function of several mitochondrial proteins in response to fasting and/or CR, including OTC, LCAD, HMGCS2, and IDH2 (Hallows et al., 2011; Hirschey et al., 2010; Shimazu et al., 2010; Someya et al., 2010). As the only validated deacetylase in mitochondria and because SIRT3 expression is stimulated under CR, we attribute acetyl sites displaying decreased acetylation under CR as SIRT3 substrates. Further, the majority of acetyl sites with decreased acetylation in CR show dramatically increased acetylation (8- to 100-fold, p < 0.05) in Sirt3−/− (Figure 4B). We conclude that SIRT3 is responsible for the striking loss of acetylation in a number of proteins in diverse mitochondrial pathways.
SIRT3 expression is a dominant regulator of the mitochondrial acetylome
We detected a vast number (~400) of mitochondrial acetyl sites increasing (i.e., >2-fold) in the absence of SIRT3 – 120 of these exhibited >8-fold increased acetylation (Figure 3D). CR treatment of Sirt3−/− mice induces an additional 34% increase in the number of acetyl sites significantly increasing ≥ 2-fold (Figure 3D). Regardless of dietary treatment, the overall mitochondrial acetylation landscape is dominated by loss of SIRT3 (Sirt3−/−) as evinced by the near linear correlation between the acetyl fold change of Sirt3−/−-CR and Sirt3−/−-CD (Figure 4B).
Importantly, a number of acetyl sites previously identified as SIRT3 substrates were identified, including those in HMGCS2, IDH2, and SDHA. The trends we observed for HMGCS2 (Shimazu et al., 2010) are consistent with published results as regulated lysine residues (K327, K333, K473) display low acetylation in CR and considerably increased acetylation in Sirt3−/− (Figure S4). K413 of IDH2 is a SIRT3 target (Yu et al., 2012), and here we observed 50% increased acetylation in Sirt3−/−. Comparing Sirt3−/−-CR to WT-CD results, a similar analysis revealed numerous proteins exhibiting 10- to 100-fold increases in specific acetylation, while many lysine residues displayed minimal perturbation.
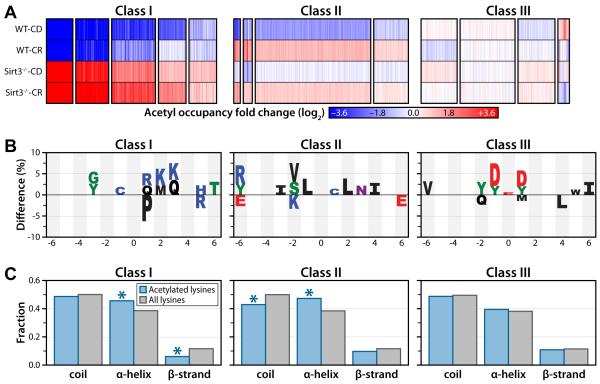
Cluster analysis reveals distinct classes of acetyl isoforms
To determine whether the quantifiable mitochondrial acetyl sites could be categorized into subpopulations based on their specific patterns across all four conditions (WT-CD, WT-CR, Sirt3−/−-CD, Sirt3−/−-CR), we used a probabilistic clustering algorithm based on a Gaussian mixture model (Hastie et al., 2009). The clustering algorithm generated 15 clusters (Figure 5A) that segregate into three distinct classes: Class 1) acetyl sites that are controlled by SIRT3 protein expression, exhibiting low acetylation under CR when SIRT3 is maximally expressed and hyperacetylation when SIRT3 expression is lost: Class 2) acetyl sites that increase in response to CR: Class 3) sites that display minimal change in response to either dietary regimen or SIRT3 expression.
Figure 5. Cluster analysis resolves acetyl sites into three distinct classes.
(A) Cluster analysis identified 15 clusters, with 13 grouped into 3 classes based on their overall trends between the four biological conditions: Class I - Acetylation sites that respond dynamically to SIRT3 expression. Class II - Acetylation sites that are largely independent of SIRT3 expression but respond to CR. Class III - Acetylation sites that display minimal perturbation in response to either loss of SIRT3 expression or CR. Each column represents a quantified acetyl site. Blue: decreased acetyl occupancy, White: no change, Red: increased acetyl occupancy.
(B) Amino acid motifs identified for clusters displaying the largest magnitude fold change for each class (from A). The motif shows significant (p < 0.05) under or over representation of a particular amino acid at each site flanking the acetylated lysine (position 0).
(C) Predicted protein secondary structures of acetylation sites identified in each class. Probabilities for coil, α-helix and β-strands were compared with the probabilities of these secondary structure elements in all lysines identified in the study. (Class I α-helix: p < 3.0 × 10−3; Class I β-strand: p < 2.0 × 10−4; Class II coil: p < 8.0 × 10−4; Class II α-helix: p < 2.5 × 10−5, hyper-geometric analysis).
We postulated that each major acetylation class might comprise unique physical properties. To investigate this we interrogated the sequence and secondary structure of acetyl sites among each class sub-population. Initially, the frequencies of neighboring amino acids were determined for acetyl sites from each class and compared against all acetyl peptides quantified, avoiding false motifs that may arise from antibody bias by immunoenrichment. We observed distinct preferences for amino acids localized near the acetyl lysine for each class (Figure 5B). Positively charged residues (i.e., Lys and Arg) near the acetyl lysine are more prevalent in clusters (Class 1) with increased acetylation in the absence of SIRT3. A strong preference for basic peptides is consistent with previous SPOT-library and machine learning screens of SIRT3 substrate specificity (Smith et al., 2011) and with the structure of SIRT3. The peptide-binding pocket of SIRT3 contains negatively charged residues, particularly C-terminal to the acetyl lysine binding site (Jin et al., 2009). In Class 2 (acetyl sites displaying increased acetylation in response to CR), surrounding sequences are largely hydrophobic and uncharged residues. The nature of Class 2 sites and the inability of SIRT3 to induce deacetylation suggests that these acetyl lysines are within hydrophobic regions and inaccessible to SIRT3. In Class 3 (acetyl sites displaying little change in response to dietary treatment or SIRT3 expression), amino acids surrounding acetyl lysine are enriched for negatively charged residues at positions −1 and +1. Given the known in vitro peptide specificity of SIRT3 (Smith et al., 2011), Class 3 acetyl lysines are expected to be poor SIRT3 substrates.
For each acetyl isoform, the conformational tendencies for α helix, β strand, or coil were determined using PSIPRED (Jones, 1999), a protein structure server used to predict protein secondary structure, against a background set containing all lysines of mitochondrial proteins identified in this study. The local secondary structures allow for interrogation of differentially enriched regions for acetyl lysine in each cluster (Figure 5C). Previous global acetyl-proteomics studies indicated that acetyl lysine sites are significantly enriched in α-helical and β-strand regions (Kim et al., 2006). Our data reveal distinct secondary structure preferences among different subpopulations of acetyl sites. Class 1 acetyl sites are significantly enriched in α-helical regions (p < 3.0 × 10−3, hyper-geometric test) and depleted in β-strand regions (p < 2.0 × 10−4) (Figure 5C). Acetyl sites increasing in response to CR (Class 2) are enriched in α-helical regions (p < 2.5 × 10−5) and depleted in coils (p < 8.0 × 10−4). Interestingly, Class 3 acetyl sites, which are unchanging across conditions, display no significant enrichment in any secondary structure element.
SIRT3 coordinately deacetylates multiple targets in mitochondrial metabolic pathways
Dysregulation of mitochondrial energy metabolism is a hallmark of aging and disease; however, CR apposes these trends and leads to increased metabolic gene expression and changes in metabolism that protect cells and tissues from aging. The reprogramming of metabolism towards more efficient fuel utilization and energy production requires coordinate regulation of a number of mitochondrial metabolic pathways. Cluster (Figure 5) and scatterplot (Figure 4B) analysis identified a large pool of regulated sites dominated by SIRT3. To identify affected pathways and understand the biological functions of these acetyl sites responding to CR and Sirt3−/−, we performed a network analysis using functional association data (Warde-Farley et al., 2010). Proteins were grouped according to their involvement in biological processes using Gene Ontology (GO) annotations and networks were visualized in Cytoscape (Smoot et al., 2011). Significant enrichment of major mitochondrial processes was apparent, including fatty acid metabolism, electron transport / ATP production, acetyl-CoA metabolism, amino acid catabolism, and mitochondrial integrity (Figure S5). Strikingly, these results reveal two major features: (1) SIRT3 regulates the acetylation status of multiple proteins within a given pathway, and (2) SIRT3 regulates pathways not previously linked to SIRT3 function. These include branched-chain amino-acid (BCAA) catabolism, one-carbon metabolism, mitochondrial genome maintenance and transcription, and iron homeostasis.
Metabolite analysis of Sirt3−/− MEFs verifies pathways regulated by acetylation
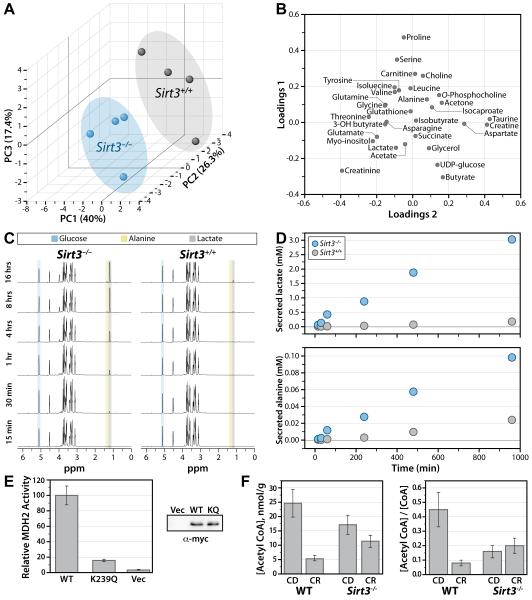
To provide complementary evidence of pathways affected by SIRT3, we profiled metabolite changes resulting from genetic loss of SIRT3. This was accomplished by comparing, over a time course, the metabolic differences in Sirt3−/− and Sirt3+/+ mouse embryonic fibroblasts (MEFs). Although a number metabolites in liver tissue and plasma were determined in previous analyses of Sirt3−/− mice (Hallows et al., 2011; Hirschey et al., 2010), here the utilization of homogenous MEFs provided a number of benefits: (1) eliminated complexity of entire organism analysis and multi-tissue compensation; (2) allowed for controlled growth conditions; and (3) provided the opportunity to utilize isotopic metabolites and quantitatively monitor flux; ultimately resulting in clear identification of pathways affected by loss of SIRT3 and subsequent increased protein acetylation. Briefly, MEFs were cultured to 80% confluence and media was replaced with modified minimal medium supplemented with 2 mM [U-13C]-glucose and 10 mM glutamine. Cells and their corresponding conditioned media were collected at 0.25, 0.5, 1, 4, 8 and 16 hours. Lysed cells were subjected to NMR-based metabolomics analysis. A total of 44 cellular metabolites were identified in 1H spectra (Table S3). To determine the distribution of experimental groups, an unsupervised principal component analysis (PCA) (Bathe et al., 2011; Janes and Yaffe, 2006) was performed with 31 metabolites that were quantifiable in all samples (Figure 6). Three principal components (PC) were used in the analysis, capturing 84% of the total variance in the data (Figure S6). PCA analysis resulted in a clear segregation between Sirt3−/− and Sirt3+/+ MEFs (Figure 6A), demonstrating these populations are metabolically distinct. The loading plot (Figure 6B) identified metabolites that significantly contribute to the biochemical segregation of Sirt3−/− and Sirt3+/+ MEFs. This metabolomic analysis identified perturbations in many metabolic pathways, which likewise were revealed from our SIRT3-dependent acetylome investigation, providing corroborating evidence that SIRT3 regulates the acetylation and activity of diverse proteins in many mitochondrial processes. Below, we highlight these pathways and briefly discuss supporting biochemical, structural and biological evidence.
Figure 6. Functional validation of SIRT3 in mitochondrial metabolism.
(A-D) NMR-metabolomics analysis of Sirt3+/+ and Sirt3−/− MEFs. (A) PCA analysis 3-D score plot for the first 3 principal components (PC) with corresponding contribution percentage for each PC. PCA segregates Sirt3+/+ and Sirt3−/− MEFs. (B) Metabolites contributing to the segregation of Sirt3+/+ and Sirt3−/− MEFs are identified in the loading plot for PC1 and PC2. Metabolites localized further from the origin (0,0) have a greater contribution to the segregation of Sirt3+/+ and Sirt3−/−. (C) 13C-edited 1H-NMR spectra of metabolites secreted into the media. 13C-Glucose, 13C-lactate and 13C-alanine peaks are identified. (D) Time-dependent increases in 13C-lactate (top plot) and 13C-alanine (bottom plot) concentrations relative to glucose concentration. (E) Acetyl CoA concentration (left) from mouse liver tissue (one per biological condition) displayed as nmol/g wet weight of tissue and acetyl CoA / CoA ratio (right). Error bars represent percent error calculated from isotopic acetyl CoA standard. (F) MDH2 activity is reduced by K239 acetyl mimic. Rates are calculated relative to WT, error bars represent standard deviation (n = 3, n=2 for Vector control). See also Figure S6, Table S3.
Amino acid catabolism
CR requires metabolic adaption to lowered energy input, commonly resulting in enhanced gluconeogenesis fueled by precursors generated from protein breakdown. In CR mice, the transcription and enzymatic activity of transamination and nitrogen disposal enzymes are increased, supporting the importance of amino acid catabolism in the adaption to CR (Hagopian et al., 2003). Our analysis identified 8 highly regulated acetyl sites on proteins involved in transamination and nitrogen detoxification, including carbamoyl phosphate synthase 1 (CPS1, K1356), glutaminase (GLS2, K329), glutamate dehydrogenase (GLUD1, K480), and ornithine carbamoyltransferase (OTC, K275), demonstrating the extent to which SIRT3 regulates amino acid catabolism and the urea cycle in response to CR. Metabolite analysis revealed an accumulation of many amino acids in Sirt3−/− MEFs, including glutamate, glutamine, serine, threonine and tyrosine (Table 1, S3). These data are supportive of significant defects in amino acid catabolism resulting from the loss of SIRT3. The accumulation of glutamine over time in Sirt3−/− MEFs provides evidence that acetylation down-regulates GLS2 activity, a primary enzyme responsible for glutamine catabolism. Here, we identified K329 within GLS2 with a 4.7-fold increase in acetyl occupancy in Sirt3−/−-CR. Based on the homologous site in the cytoplasmic isoform of GLS2, K329 lies in the homodimeric interface and would be predicted to affect activity (DeLaBarre et al., 2011).
Table 1.
Examples of new SIRT3 targets and metabolite perturbations in Sirt3−/−
| Pathways | Protein | Site | Acetyl fold ↑ Sirt3−/−-CR |
Proposed function | Altered metabolites: WT vs Sirt3−/− MEFs |
Altered metabolites: WT vs Sirt3−/− |
|---|---|---|---|---|---|---|
|
Amino acid catabolism Branched chain amino acid catabolism |
||||||
| ACAT1 | K265 [4] | 26 | Altered substrate binding (CoA) | Isoleucine, leucine, valine, 2-oxocaproate*, 3-hydroxyisovalerate* |
(Isoleucinea, Leucinea, Valinea) Methylsuccinateb, Isobutyrylglycineb |
|
| AUH | K304 [1] | 25 | Stability/protein interactions/localization | |||
| DBT | K261 [1] | 7.3 | BCKDH complex formation | |||
| MCCC2 | K495 [1] | 21 | No structural data | |||
| BCKDHA | K199 | 7.1 | BCKDH complex formation | |||
| HSD17B10 | K104|105 | 51 | Tetramer interface | |||
| HIBCH | K100 [2] | 7.7 | Stability/protein interactions/localization | |||
| Transamination and ammonia detoxification |
CPS1 | K1356 [3] | 7.6 | Allosteric regulator binding | Aspartate, Glutamate Glutamine, 3-Phenylacetate* |
Aspartatea, Ornithinea, Uracila, Uridinea, Glutaminea, Citrullinea, Arginosuccinatea, |
| GLS2 | K329 | 4.7 | Dimer formation | |||
| GLUD1 | K480 [1] | 9.6 | Stability/protein interactions/localization | |||
| OTC | K275 | 44 | Stability/protein interactions/localization | |||
|
Mitochondrial integrity mtDNA transcription and translation |
||||||
| CLPX | K127 | 6.4 | Protein-protein interaction | |||
| HSPA9 | K653 [5] | 16 | Protein-protein interaction | |||
| HSPE1 | K36 | 29 | Protein-protein interaction | |||
| LETM1 | K458 | 6.8 | No structural data | |||
| LONP1 | K378 | 15 | No structural data | |||
| MRPL12 | K145 [1] | 18 | mtDNA interaction | |||
| MRPS9 | K174 | 5.8 | RNA binding | |||
| PNPT1 | K421 | 6 | No structural data | |||
| TFAM | K70 [1] | 8.2 | mtDNA binding | |||
| TACO1 | K162 | 8.1 | No structural data | |||
| Iron homeostasis | ACO2 | K144 | 7.3 | Substrate binding | ||
| CISD3 | K96 | 6.9 | Iron binding; Zn finger region | |||
| FXN | K194 | 11 | Ubiquitin/acetylation cross talk | |||
| GLRX5 | K147 | 14 | Stability/protein interactions/localization | |||
| TST | K175 | 10 | No structural data | |||
| Antioxidant response | PRDX3 | K254 | 11 | Ubiquitin/acetylation cross talk | ROSd, Glutathioned, NADPHd |
|
| TXN2 | K110 | 8.7 | Protein-protein interaction | |||
| Citric acid cycle | ACO2 | K144 [2] | 7.3 | Substrate binding | Glutamate, Lactate, Succinate, Fumarate* |
|
| CS | K49|52 [2] | 32 | Stability/protein interactions/localization | |||
| FH | K424 | 9.7 | Protein-protein interaction | |||
| MDH2 | K239 [1] | 83 | Dimer formation | |||
| SDHA | K179 | 22 | FAD binding | |||
|
Acetyl CoA metabolism Ketone body synthesis |
||||||
| ACAA2 | K209 | 58 | No structural data | 3-hydroxybutyrate | Malonyl carnitinea, Hydroxybutyryl carnitinea, 3-hydroxybutyratec |
|
| ACAT1 | K265 [4] | 26 | Altered substrate binding (CoA) | |||
| ECH1 | K316 | 10 | No structural data | |||
| HMGCS2 | K333 [6] | 11 | Protein conformation | |||
| Fatty acid oxidation | ACADVL | K299 [3] | 7.7 | Stability/protein interactions/localization | Isobutyrate, Taurine | Carnitinea, Malonyl carnitinea, Hydroxybutyryl carnitinea, C6a, C14a, C14:1a, C16a, C16:1b, C16:2b, C18-OHb, C18:2-OHb, C20:4b, Ethylmalonateb |
| ECHS1 | K288 [1] | 23 | Stability/protein interactions/localization | |||
| HADH | K312 [2] | 29 | No structural data | |||
| HADHA | K406|411 [14] | 45 | No structural data | |||
| HADHB | K189|191 [1] | 4.4 | No structural data | |||
| One-carbon metabolism | DMGDH | K852 [1] | 37 | No structural data | Phosphocholine, Serine |
Cholinea, Betainea, Homocysteinea, Methioninea |
| CHDH | K582 | 6.8 | No structural data | |||
| SHMT2 | K464 [1] | 4.7 | Stability/protein interactions/localization |
Hallows, W.C. et al. (2011). Mol Cell 41, 139-149.
Hirschey, M.D. et al. (2010). Nature 464, 121-125.
Shimazu, T. et al. (2010). Cell Metab 12, 654-661.
Someya, S. et al. (2010). Cell 143, 802-812.
Number of additional acetyl sites increasing 4-fold
Metabolite identified in MEFs but not quantified
( ) Metabolite exhibiting change but not statistically significant ( p < 0.05)
Amino acid catabolism is an important anapleurotic pathway for other mitochondrial processes. The breakdown of BCAA (isoleucine, leucine, valine) results in the production of acetyl-CoA and succinyl CoA, metabolites used in the citric acid cycle (TCA) or ketone body synthesis, pathways important in CR responses. Acetylome data reveal BCAA catabolism as a highly enriched KEGG pathway among mitochondrial proteins regulated by SIRT3 in response to CR (Figure S7). Nearly half of the proteins involved in this pathway contain significantly increasing acetyl sites in Sirt3−/−-CR (Figure S5 and Table 1). These include two subunits of branched-chain alpha-keto acid dehydrogenase (BCKDHA and AUH), the enzyme catalyzing the first, shared step in breakdown of all BCAA in the liver. Supporting the conclusion that acetylation decreases BCKDHA activity, isoleucine, leucine, and valine are elevated in Sirt3−/− MEFs (Table S3) and in Sirt3−/− liver tissue (Table 1). Deficiency in acetyl-CoA acetyltransferase (ACAT1) in humans has been associated with a buildup of branched chain amino-acid metabolites (Fontaine et al., 1996). ACAT1 plays a critical role in BCAA catabolism as well as acetyl-CoA metabolism and ketone body synthesis (Haapalainen et al., 2007). Our acetylome analysis provided strong evidence that ACAT1 is a major target of SIRT3 (Figure S4). Of the 22 acetyl lysines quantified in ACAT1, three (K260, K263, K265) display a 16- to 32-fold acetylation increase in the absence of SIRT3. These residues are located within the CoA binding pocket of ACAT1, within 3-5 Å from the ribosyl-phosphate group of CoA (Dombrovski et al., 2011), thus we predict acetylation of these residues will alter CoA binding and regulate ACAT1 activity.
Additional evidence for SIRT3-dependent regulation of BCAA catabolism comes from the elevated levels of 3-hydroxyisovalerate in Sirt3−/− MEFs at early time points (Table S3). Increased excretion of 3-hydroxyisovalerate is an indicator of BCAA catabolic deficiency, organic acidemia, and is associated with a rare human recessive autosomal disorder caused by isovaleryl-CoA dehydrogenease (IVD) deficiency (Lin et al., 2007). Our acetylome analysis revealed that the acetylation state of IVD at K316 is controlled by SIRT3 in a CR-dependent manner. K316 undergoes deacetylation during CR, but a 3.27-fold increase in acetylation in the absence of SIRT3. K316 is found near the subunit interface of IVD and may affect tetrameric assembly (Tiffany et al., 1997).
Citric acid cycle
An increase in respiration efficiency is a hallmark of the mitochondrial response to CR (Civitarese et al., 2007). This relies on precisely controlled production and consumption of redox coenzymes NADH and FADH2, produced in the TCA cycle. We identified nine acetyl sites on four enzymes of the TCA cycle that display increased acetylation of >4-fold, including citrate synthase (CS, K49|K52), aconitase (ACO2, K144), malate dehydrogenase (MDH2, K239) and succinate dehydrogenase (SDHA, K179). Metabolite analysis identified an increase in succinate in Sirt3−/− MEFs (Table S3) and provides further evidence for acetylation decreasing succinate dehydrogenase activity. Consistent with a role for dynamic acetylation in the TCA cycle, alterations in glutamate, lactate, and aspartate were also apparent in Sirt3−/− MEFs.
One of the most striking examples of hyperacetylation upon loss of SIRT3 is malate dehydrogenase (MDH2). Among seven quantified acetyl sites on MDH2, K239 displayed a 66-fold increase in acetylation in Sirt3−/− mice. This same site exhibited decreased acetylation under CR with WT animals. Within the MDH2 structure, K239 lies within the dimeric interface and near to the substrate-binding channel (PDB 2DFD). Thus K239 is a likely target of SIRT3 in response to CR (Figure S4). Within the MDH2 structure, K239 lies within the dimeric interface and near to the substrate-binding channel (PDB 2DFD). To provide evidence that acetylation of K239 would affect MDH2 activity, we generated the acetyl mimic (K239Q) and compared activity against WT protein. Indeed, the K239Q mutant exhibited a dramatic loss in activity (>5-fold) when compared to WT, suggesting acetylation of this site dynamically regulates MDH2 activity (Figure 6E). This example provides additional validation of our acetylome atlas, and demonstrates the utility of mining the data for functional importance and defining specific roles for SIRT3.
One-carbon metabolism
One-carbon (1C) transfer reactions are essential for amino acid catabolism, transfer of methyl groups to DNA, RNA, and proteins, and in formylation of the initiator tRNA required for mitochondrial protein synthesis (Tibbetts and Appling, 2010). Serine, glycine, dimethylglycine, sarcosine, and choline serve as mitochondrial 1C donors. Four key proteins involved in 1C transfer reactions contain acetyl sites responsive to CR and Sirt3−/− (Figure S5 and Table 1). Serine hydroxymethyltransferase (SHMT2, K464) is involved in breakdown of serine to glycine, with concomitant conversion of tetrahydrofolate to 5,10-methylenetetrahydrofolate. Lysine 464 displays 4-fold increased acetylation in response to Sirt3−/−. Serine contributes significantly to the segregation between Sirt3−/− and Sirt3+/+ MEFs by PCA and is elevated in Sirt3−/− MEFs (Table S3), supporting the role of SIRT3 in regulating SHMT2 activity and a defect in serine utilization. Mitochondrial dimethylglycine dehydrogenase (DMGDH) converts dimethylglycine to sarcosine and contains an acetyl site (K852) whose relative acetyl occupancy increases a dramatic 36-fold in Sirt3−/−-CR.
Choline is a major source of dietary methyl groups in mammals, and oxidation of choline to betaine, which ultimately supplies 1C units to the mitochondria as dimethylgylcine, takes place primarily in the liver. Choline dehydrogenase (CHDH, K582) contains an acetyl site displaying increased acetylation of 6.8 fold. Betaine, choline, and homocysteine, a metabolite necessary for the conversion of betaine to dimethylglycine, are all altered in liver tissue in response to CR and Sirt3−/− (Figure S5 and Table 1). Together these results provide compelling evidence that SIRT3 controls donation of 1C units to carbon carriers in the mitochondria.
Mitochondrial integrity
CR is reported to enhance mitochondrial proliferation (Civitarese et al., 2007; Lopez-Lluch et al., 2006). Consistent with new protein synthesis, over 40% of gene transcripts overexpressed in response to CR (Plank et al., 2012) were identified in the current study with increased (≥40%) relative protein level in CR (Table S1). However, overall proteome quantitation argues against widespread mitochondrial biogenesis during CR (Figure S2), as discussed previously (Miller et al., 2012). We observed a slight overall enrichment of mitochondrial proteins relative to non-mitochondrial proteins during CR. Our data support reversible acetylation as a major mechanism that drives CR-dependent metabolic reprogramming in mitochondria.
It is known that CR-adapted mitochondria use less oxygen and produce less ROS, while maintaining ATP production. One molecular mechanism to enhance mitochondrial performance while minimizing new protein synthesis might be the stimulation of effective quality control pathways for mitochondrial proteins. Our study supports this hypothesis. We identified significant SIRT3-dependent changes in acetylation (>6-fold, p<0.05) on major enzymatic regulators of mitochondrial proteome homeostasis, including the ATP-dependent Lon-like protease (LONP1, K378), and the chaperone proteins, 100 kDa chaperone (CLPX, K127), heat shock protein 70 (HSPA9, K653), 10 kDa heat shock protein (HSPE1, K36).
Our analysis reveals a potential role for SIRT3 in coordinating the regulatory network required for maintaining mitochondrial integrity, including protein quality control, mtDNA transcription and translation, and iron homeostasis. Mitochondria play a central role in iron metabolism and serve as a key location for iron sulfur cluster (Fe-S) and heme biogenesis (Richardson et al., 2010). Several proteins involve in homeostasis are well-represented as major targets of SIRT3 deacetylation (Figure S5 and Table 1). These include frataxin (FXN), ACO2 and GLRX5.
Mitochondrial genome maintenance, transcription and translation
A number of components from mitochondrial transcription and translational machinery are highlighted in the network analysis of SIRT3-regulated acetylation sites (Figure S5). Mitochondrial transcription factor A (TFAM, K70) plays an essential role in maintenance, expression, and organization of mitochondrial DNA (Ngo et al., 2011). TFAM is required for efficient promoter recognition by mitochondrial RNA polymerase and binds and induces significant conformational changes in mtDNA (Campbell et al., 2012). We identified a previously unreported acetyl lysine in TFAM, K70 that exhibits a 8-fold increase in acetylation in Sirt3−/−, and a slight decrease in acetylation under CR when SIRT3 expression is induced. Reversible acetylation of TFAM could control mtDNA binding and subsequent transcription, akin to the role of histone acetylation regulating nuclear gene expression. Mitochondrial ribosomal proteins (MRPS9, K174 and MRPL12, K145) play an important role in coordinating mitochondrial transcription and translation and contain acetyl sites that increase 5- and 18-fold, respectively in the Sirt3−/−-CR condition. MRPL12 interacts with mitochondrial RNA polymerase and is thought to enhance mitochondrial transcription (Wang et al., 2007). The essential mitochondrial elongation factor Ts (TSFM, K83) is also a target of SIRT3-dependent acetylation. Mutations in TSFM result in severe infantile liver failure (Vedrenne et al., 2012). TSFM functions as an activator of elongation factor Tu, which delivers the aminoacyl-tRNA to the ribosome during elongation of protein synthesis. K83 exists at a kink in alpha helix 3 of TSFM, connecting two major contact regions with elongation factor Tu, and would be predicted to alter protein:protein interaction (Jeppesen et al., 2005).
13C-glucose flux analysis confirms major defects in oxidative glucose metabolism in Sirt3−/− MEFs
Our acetylome and metabolite analyses suggest a profound up-regulation of oxidative metabolism by the dynamic regulation of protein acetylation induced by CR and mediated by SIRT3. To provide support that wholesale alteration to the acetylome results in a major shift in glucose metabolism, we measured the metabolic flux of [U-13C]-glucose in Sirt3−/− and Sirt3+/+ MEFs. Conditioned-media from cells treated with [U-13C]-glucose were analyzed by NMR spectroscopy and the consumption of glucose and production of 13C-labeled metabolites was monitored over 16 hours (Figure 6D). Strikingly, the rate of 13C-lactate and 13C-alanine production and excretion to media was increased dramatically in Sirt3−/− MEFs 17- and 4-fold, respectively, compared to Sirt3+/+ (Figure 6D). These flux studies indicate a major defect in oxidative glucose metabolism in the absence of SIRT3. Glucose uptake is slightly increased in Sirt3−/− MEFs and a buildup of glucose derived metabolites such as myo-inositol and UDP-glucose, was apparent in 1H-spectral analysis (Figure 6B, Table S3). These results reinforce the hypothesis that SIRT3 plays a critical role in mitochondrial metabolic reprogramming towards oxidative metabolism and are consistent with previous reports (Finley et al., 2011).
Sirt3−/− mice display inability to lower liver acetyl-CoA levels during CR
The metabolic flux results suggest decreased ability of Sirt3−/− MEFs cells to perform full oxidation of glucose. The consequences on acetyl-CoA and CoA levels were assessed in liver from Sirt3−/− and WT mice, under either CD or CR diet. In WT animals, we observed a considerable (5x) decrease in liver acetyl-CoA under CR compared with CD (Figure 6F). In contrast, Sirt3−/− mice were unable to lower their acetyl-CoA levels in response to CR (Figure 6F). The ratios of acetyl-CoA/CoA displayed similar trends, indicating that the difference in these ratios reflect differences primarily in acetyl-CoA. Lower levels of acetyl-CoA during CR are consistent with increased acetyl-CoA utilization in energy requiring processes, and validate the role of SIRT3 in mitochondrial reprogramming towards efficient oxidative metabolism. Sirt3−/− mice display lower acetyl CoA under either dietary condition and were unable to lower their acetyl CoA levels in response to CR. These findings are consistent with a deficiency in mitochondrial oxidative metabolism due to misregulation of protein acetylation in the absence of SIRT3.
Concluding Remarks
We developed and applied a method for multiplexed, global acetyl-proteome quantification directly from mammalian tissues. Our approach uses tandem mass tags for isotope-based isobaric tagging, fractionation using strong cation exchange chromatography, highly efficient immunopurification of acetyl-lysine residues, and finally, reversed-phase nanoflow liquid chromatography coupled with high resolution and high mass accuracy MS. We used this approach to provide a comprehensive and quantitative analysis of 3,285 acetylation sites altered by CR and/or the genetic loss of SIRT3. Biochemical and computational analyses revealed that the 2,193 acetylation sites from mitochondrial proteins fell into three general classes, each with distinct functional and physical features. Given their dynamic nature, Class 1 and 2 acetylation sites provide new insight into regulation of mitochondrial processes and the mechanisms of protein (de)acetylation, while Class 3 acetylation sites appear to represent low-level, spurious acetylation of surface exposed lysine residues. Importantly, these data provide the foundation for understanding the breadth and depth of the mitochondrial proteome and acetylome in response to such profound reprogramming input as CR and SIRT3 expression.
Experimental procedures
See supplemental information for detailed experimental procedures.
Sample preparation, Mass Spectrometry, Data analysis
Mitochondria were isolated from homogenized mouse liver tissue by differential centrifugation. Protein was extracted by sonication and quantified by a BCA assay. Proteins were reduced, alkylated, and digested with LysC followed by trypsin. The resulting peptides were labeled with TMT isobaric labels, mixed and fractionated by strong cation exchange chromatography. Acetylated peptides were enriched with pan-acetyl lysine antibody-agarose conjugate. Enriched and non-enriched fractions were analyzed by nano reverse phase liquid chromatography coupled to an Orbitrap Elite (Thermo). Spectra were searched using OMSSA and results were filtered to 1% FDR at the unique peptide level using the COMPASS software suite. TMT quantitation and protein grouping were performed according to previously reported rules (Phanstiel et al., 2011). Proteins were identified as mitochondrial or non-mitochondrial based on inclusion or exclusion from the mitoCarta compendium of mitochondrial mouse proteins (Pagliarini et al., 2008).
Bioinformatics
To cluster the acetyl sites we used a Gaussian mixture model clustering algorithm (Hastie et al., 2009). The frequencies of amino acids surrounding acetylated lysines were compared for acetyl sites from each class with all acetyl sites quantified in the study using iceLogo (Colaert et al., 2009). Mouse protein FASTA sequences were downloaded from EMBL and the PSIPRED program was used to predict three types of secondary structures (Jones, 1999).
Metabolite analysis
Sirt3+/+ or Sirt3−/− MEFs were cultured to 80% confluence. Cells were treated with 2 mM [U-13C]-glucose and 10 mM glutamine. Cells were harvested and conditioned media were collected at 15 minutes, 30 minutes, 1, 4, 8 and 16 hours. NMR spectra were collected using a Varian 600 NMR spectrometer and data was processed using software package vnmrJ 1.1D (Varian, Inc).
Acetyl CoA and CoA Measurement
Metabolites were extracted from liver tissue in a cold solution of methanol and 1% TFA in the presence of acetyl-1,2-13C2 coenzyme A. Samples were prepared using solid phase chromatography and analyzed by HPLC coupled to a Qtrap MS (Applied Biosystems).
Supplementary Material
Highlights.
MS quantifies 1,578 mitochondrial acetyl sites altered during CR and loss of SIRT3
SIRT3 functions as a prominent regulator in CR adaptation
CR and SIRT3 regulate previously unrecognized processes in mitochondria
We provide an acetylation atlas for understanding mitochondrial regulation in CR
Acknowledgments
This work was funded by NIA grant AG038679 to J.M.D. and T.A.P, NIH grant GM065386 to J.M.D., NIH GM080148 to J.J.C., and Searle Scholars Award and NIH grant RC1DK086410 to D.J.P. K.E.D. was funded by a NSF Graduate Research Fellowship and NIH traineeship (5T32GM08349). We thank AJ Bureta for figure design.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson RM, Weindruch R. The caloric restriction paradigm: implications for healthy human aging. Am J Hum Biol. 2012;24:101–106. doi: 10.1002/ajhb.22243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bathe OF, Shaykhutdinov R, Kopciuk K, Weljie AM, McKay A, Sutherland FR, Dixon E, Dunse N, Sotiropoulos D, Vogel HJ. Feasibility of identifying pancreatic cancer based on serum metabolomics. Cancer Epidemiol Biomarkers Prev. 2011;20:140–147. doi: 10.1158/1055-9965.EPI-10-0712. [DOI] [PubMed] [Google Scholar]
- Campbell CT, Kolesar JE, Kaufman BA. Mitochondrial transcription factor A regulates mitochondrial transcription initiation, DNA packaging, and genome copy number. Biochim Biophys Acta. 2012 doi: 10.1016/j.bbagrm.2012.03.002. [DOI] [PubMed] [Google Scholar]
- Chen Y, Zhao W, Yang JS, Cheng Z, Luo H, Lu Z, Tan M, Gu W, Zhao Y. Quantitative acetylome analysis reveals the roles of SIRT1 in regulating diverse substrates and cellular pathways. Mol Cell Proteomics. 2012 doi: 10.1074/mcp.M112.019547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- Civitarese AE, Carling S, Heilbronn LK, Hulver MH, Ukropcova B, Deutsch WA, Smith SR, Ravussin E. Calorie restriction increases muscle mitochondrial biogenesis in healthy humans. Plos Med. 2007;4:485–494. doi: 10.1371/journal.pmed.0040076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colaert N, Helsens K, Martens L, Vandekerckhove J, Gevaert K. Improved visualization of protein consensus sequences by iceLogo. Nat Methods. 2009;6:786–787. doi: 10.1038/nmeth1109-786. [DOI] [PubMed] [Google Scholar]
- Colman RJ, Anderson RM, Johnson SC, Kastman EK, Kosmatka KJ, Beasley TM, Allison DB, Cruzen C, Simmons HA, Kemnitz JW, et al. Caloric restriction delays disease onset and mortality in rhesus monkeys. Science. 2009;325:201–204. doi: 10.1126/science.1173635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLaBarre B, Gross S, Fang C, Gao Y, Jha A, Jiang F, Song JJ, Wei W, Hurov JB. Full-length human glutaminase in complex with an allosteric inhibitor. Biochemistry-Us. 2011;50:10764–10770. doi: 10.1021/bi201613d. [DOI] [PubMed] [Google Scholar]
- Dombrovski L, Min JR, Antoshenko T, Wu H, Loppnau P, Edwards AM, Arrowsmith CH, Bochkarve A, Plotnikov AN. Human mitochondrial acetylacetyl-CoA thiolase (RCSB) 2011.
- Finley LW, Carracedo A, Lee J, Souza A, Egia A, Zhang J, Teruya-Feldstein J, Moreira PI, Cardoso SM, Clish CB, et al. SIRT3 opposes reprogramming of cancer cell metabolism through HIF1alpha destabilization. Cancer Cell. 2011;19:416–428. doi: 10.1016/j.ccr.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaine M, Briand G, Ser N, Armelin I, Rolland MO, Degand P, Vamecq J. Metabolic studies in twin brothers with 2-methylacetoacetyl-CoA thiolase deficiency. Clin Chim Acta. 1996;255:67–83. doi: 10.1016/0009-8981(96)06391-7. [DOI] [PubMed] [Google Scholar]
- Fritz KS, Galligan JJ, Hirschey MD, Verdin E, Petersen DR. Mitochondrial acetylome analysis in a mouse model of alcohol-induced liver injury utilizing SIRT3 knockout mice. J Proteome Res. 2012;11:1633–1643. doi: 10.1021/pr2008384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan KL, Yu W, Lin Y, Xiong Y, Zhao S. Generation of acetyllysine antibodies and affinity enrichment of acetylated peptides. Nat Protoc. 2010;5:1583–1595. doi: 10.1038/nprot.2010.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haapalainen AM, Merilainen G, Pirila PL, Kondo N, Fukao T, Wierenga RK. Crystallographic and kinetic studies of human mitochondrial acetoacetyl-CoA thiolase: the importance of potassium and chloride ions for its structure and function. Biochemistry-Us. 2007;46:4305–4321. doi: 10.1021/bi6026192. [DOI] [PubMed] [Google Scholar]
- Hagopian K, Ramsey JJ, Weindruch R. Caloric restriction increases gluconeogenic and transaminase enzyme activities in mouse liver. Exp Gerontol. 2003;38:267–278. doi: 10.1016/s0531-5565(02)00202-4. [DOI] [PubMed] [Google Scholar]
- Hallows WC, Lee S, Denu JM. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc Natl Acad Sci U S A. 2006;103:10230–10235. doi: 10.1073/pnas.0604392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallows WC, Yu W, Smith BC, Devries MK, Ellinger JJ, Someya S, Shortreed MR, Prolla T, Markley JL, Smith LM, et al. Sirt3 promotes the urea cycle and fatty acid oxidation during dietary restriction. Mol Cell. 2011;41:139–149. doi: 10.1016/j.molcel.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie T, Tibshirani R, Friedman J. The Elements of Statistcal Learning. Springer; New York: 2009. [Google Scholar]
- Henriksen P, Wagner SA, Weinert BT, Sharma S, Bacinskaja G, Rehman M, Juffer AH, Walther TC, Lisby M, Choudhary C. Proteome-wide analysis of lysine acetylation suggests its broad regulatory scope in Saccharomyces cerevisiae. Mol Cell Proteomics. 2012 doi: 10.1074/mcp.M112.017251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschey MD, Shimazu T, Goetzman E, Jing E, Schwer B, Lombard DB, Grueter CA, Harris C, Biddinger S, Ilkayeva OR, et al. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature. 2010;464:121–125. doi: 10.1038/nature08778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janes KA, Yaffe MB. Data-driven modelling of signal-transduction networks. Nat Rev Mol Cell Biol. 2006;7:820–828. doi: 10.1038/nrm2041. [DOI] [PubMed] [Google Scholar]
- Jeppesen MG, Navratil T, Spremulli LL, Nyborg J. Crystal structure of the bovine mitochondrial elongation factor Tu.Ts complex. J Biol Chem. 2005;280:5071–5081. doi: 10.1074/jbc.M411782200. [DOI] [PubMed] [Google Scholar]
- Jin L, Wei W, Jiang Y, Peng H, Cai J, Mao C, Dai H, Choy W, Bemis JE, Jirousek MR, et al. Crystal structures of human SIRT3 displaying substrate-induced conformational changes. J Biol Chem. 2009;284:24394–24405. doi: 10.1074/jbc.M109.014928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- Kendrick AA, Choudhury M, Rahman SM, McCurdy CE, Friederich M, Van Hove JL, Watson PA, Birdsey N, Bao J, Gius D, et al. Fatty liver is associated with reduced SIRT3 activity and mitochondrial protein hyperacetylation. Biochem J. 2011;433:505–514. doi: 10.1042/BJ20100791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- Koubova J, Guarente L. How does calorie restriction work? Genes Dev. 2003;17:313–321. doi: 10.1101/gad.1052903. [DOI] [PubMed] [Google Scholar]
- Law IK, Liu L, Xu A, Lam KS, Vanhoutte PM, Che CM, Leung PT, Wang Y. Identification and characterization of proteins interacting with SIRT1 and SIRT3: implications in the anti-aging and metabolic effects of sirtuins. Proteomics. 2009;9:2444–2456. doi: 10.1002/pmic.200800738. [DOI] [PubMed] [Google Scholar]
- Lin SJ, Defossez PA, Guarente L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science. 2000;289:2126–2128. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- Lin WD, Wang CH, Lee CC, Lai CC, Tsai Y, Tsai FJ. Genetic mutation profile of isovaleric acidemia patients in Taiwan. Mol Genet Metab. 2007;90:134–139. doi: 10.1016/j.ymgme.2006.08.011. [DOI] [PubMed] [Google Scholar]
- Lombard DB, Alt FW, Cheng HL, Bunkenborg J, Streeper RS, Mostoslavsky R, Kim J, Yancopoulos G, Valenzuela D, Murphy A, et al. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol Cell Biol. 2007;27:8807–8814. doi: 10.1128/MCB.01636-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Lluch G, Hunt N, Jones B, Zhu M, Jamieson H, Hilmer S, Cascajo MV, Allard J, Ingram DK, Navas P, et al. Calorie restriction induces mitochondrial biogenesis and bioenergetic efficiency. Proc Natl Acad Sci U S A. 2006;103:1768–1773. doi: 10.1073/pnas.0510452103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundby A, Lage K, Weinert BT, Bekker-Jensen DB, Secher A, Skovgaard T, Kelstrup CD, Dmytriyev A, Choudhary C, Lundby C, et al. Proteomic Analysis of Lysine Acetylation Sites in Rat Tissues Reveals Organ Specificity and Subcellular Patterns. Cell Rep. 2012 doi: 10.1016/j.celrep.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattison JA, Roth GS, Beasley TM, Tilmont EM, Handy AM, Herbert RL, Longo DL, Allison DB, Young JE, Bryant M, et al. Impact of caloric restriction on health and survival in rhesus monkeys from the NIA study. Nature. 2012 doi: 10.1038/nature11432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller BF, Robinson MM, Bruss MD, Hellerstein M, Hamilton KL. A comprehensive assessment of mitochondrial protein synthesis and cellular proliferation with age and caloric restriction. Aging Cell. 2012;11:150–161. doi: 10.1111/j.1474-9726.2011.00769.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo HB, Kaiser JT, Chan DC. The mitochondrial transcription and packaging factor Tfam imposes a U-turn on mitochondrial DNA. Nat Struct Mol Biol. 2011;18:1290–U1141. doi: 10.1038/nsmb.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, Walford GA, Sugiana C, Boneh A, Chen WK, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phanstiel DH, Brumbaugh J, Wenger CD, Tian S, Probasco MD, Bailey DJ, Swaney DL, Tervo MA, Bolin JM, Ruotti V, et al. Proteomic and phosphoproteomic comparison of human ES and iPS cells. Nat Meth. 2011;8:821–827. doi: 10.1038/nmeth.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plank M, Wuttke D, van Dam S, Clarke SA, de Magalhaes JP. A meta-analysis of caloric restriction gene expression profiles to infer common signatures and regulatory mechanisms. Mol Biosyst. 2012;8:1339–1349. doi: 10.1039/c2mb05255e. [DOI] [PubMed] [Google Scholar]
- Qiu X, Brown K, Hirschey MD, Verdin E, Chen D. Calorie restriction reduces oxidative stress by SIRT3-mediated SOD2 activation. Cell Metab. 2010;12:662–667. doi: 10.1016/j.cmet.2010.11.015. [DOI] [PubMed] [Google Scholar]
- Richardson DR, Lane DJ, Becker EM, Huang ML, Whitnall M, Suryo Rahmanto Y, Sheftel AD, Ponka P. Mitochondrial iron trafficking and the integration of iron metabolism between the mitochondrion and cytosol. Proc Natl Acad Sci U S A. 2010;107:10775–10782. doi: 10.1073/pnas.0912925107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlicker C, Gertz M, Papatheodorou P, Kachholz B, Becker CF, Steegborn C. Substrates and regulation mechanisms for the human mitochondrial sirtuins Sirt3 and Sirt5. J Mol Biol. 2008;382:790–801. doi: 10.1016/j.jmb.2008.07.048. [DOI] [PubMed] [Google Scholar]
- Schwer B, Bunkenborg J, Verdin RO, Andersen JS, Verdin E. Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc Natl Acad Sci U S A. 2006;103:10224–10229. doi: 10.1073/pnas.0603968103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B, Eckersdorff M, Li Y, Silva JC, Fermin D, Kurtev MV, Giallourakis C, Comb MJ, Alt FW, Lombard DB. Calorie restriction alters mitochondrial protein acetylation. Aging Cell. 2009;8:604–606. doi: 10.1111/j.1474-9726.2009.00503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimazu T, Hirschey MD, Hua L, Dittenhafer-Reed KE, Schwer B, Lombard DB, Li Y, Bunkenborg J, Alt FW, Denu JM, et al. SIRT3 deacetylates mitochondrial 3-hydroxy-3-methylglutaryl CoA synthase 2 and regulates ketone body production. Cell Metab. 2010;12:654–661. doi: 10.1016/j.cmet.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith BC, Settles B, Hallows WC, Craven MW, Denu JM. SIRT3 substrate specificity determined by peptide arrays and machine learning. ACS Chem Biol. 2011;6:146–157. doi: 10.1021/cb100218d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohal RS, Weindruch R. Oxidative stress, caloric restriction, and aging. Science. 1996;273:59–63. doi: 10.1126/science.273.5271.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Someya S, Yu W, Hallows WC, Xu J, Vann JM, Leeuwenburgh C, Tanokura M, Denu JM, Prolla TA. Sirt3 mediates reduction of oxidative damage and prevention of age-related hearing loss under caloric restriction. Cell. 2010;143:802–812. doi: 10.1016/j.cell.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibbetts AS, Appling DR. Compartmentalization of Mammalian Folate-Mediated One-Carbon Metabolism. Annu Rev Nutr. 2010;30:57–81. doi: 10.1146/annurev.nutr.012809.104810. [DOI] [PubMed] [Google Scholar]
- Tiffany KA, Roberts DL, Wang M, Paschke R, Mohsen AW, Vockley J, Kim JJ. Structure of human isovaleryl-CoA dehydrogenase at 2.6 A resolution: structural basis for substrate specificity. Biochemistry-Us. 1997;36:8455–8464. doi: 10.1021/bi970422u. [DOI] [PubMed] [Google Scholar]
- Vedrenne V, Galmiche L, Chretien D, de Lonlay P, Munnich A, Rotig A. Mutation in the mitochondrial translation elongation factor EFTs results in severe infantile liver failure. J Hepatol. 2012;56:294–297. doi: 10.1016/j.jhep.2011.06.014. [DOI] [PubMed] [Google Scholar]
- Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Cotney J, Shadel GS. Human mitochondrial ribosomal protein MRPL12 interacts directly with mitochondrial RNA polymerase to modulate mitochondrial gene expression. J Biol Chem. 2007;282:12610–12618. doi: 10.1074/jbc.M700461200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:W214–220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weindruch R, Walford RL, Fligiel S, Guthrie D. The retardation of aging in mice by dietary restriction: longevity, cancer, immunity and lifetime energy intake. J Nutr. 1986;116:641–654. doi: 10.1093/jn/116.4.641. [DOI] [PubMed] [Google Scholar]
- Wenger CD, Lee MV, Hebert AS, McAlister GC, Phanstiel DH, Westphall MS, Coon JJ. Gas-phase purification enables accurate, multiplexed proteome quantification with isobaric tagging. Nat Meth. 2011a;8:933–935. doi: 10.1038/nmeth.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenger CD, Phanstiel DH, Lee MV, Bailey DJ, Coon JJ. COMPASS: a suite of pre- and post-search proteomics software tools for OMSSA. Proteomics. 2011b;11:1064–1074. doi: 10.1002/pmic.201000616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf-Yadlin A, Hautaniemi S, Lauffenburger DA, White FM. Multiple reaction monitoring for robust quantitative proteomic analysis of cellular signaling networks. Proceedings of the National Academy of Sciences. 2007;104:5860–5865. doi: 10.1073/pnas.0608638104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R, Dephoure N, Haas W, Huttlin EL, Zhai B, Sowa ME, Gygi SP. Correct Interpretation of Comprehensive Phosphorylation Dynamics Requires Normalization by Protein Expression Changes. Molecular & Cellular Proteomics. 2011;10 doi: 10.1074/mcp.M111.009654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Dittenhafer-Reed KE, Denu JM. SIRT3 protein deacetylates isocitrate dehydrogenase 2 (IDH2) and regulates mitochondrial redox status. J Biol Chem. 2012;287:14078–14086. doi: 10.1074/jbc.M112.355206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H, et al. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.