Abstract
The liver is necessary for survival. Its strategic localisation, blood flow and prominent role in the metabolism of xenobiotics render this organ particularly susceptible to injury by chemicals to which we are ubiquitously exposed. The pathogenesis of most chemical-induced liver injuries is initiated by the metabolic conversion of chemicals into reactive intermediate species, such as electrophilic compounds or free radicals, which can potentially alter the structure and function of cellular macromolecules. Many reactive intermediate species can produce oxidative stress, which can be equally detrimental to the cell. When protective defences are overwhelmed by excess toxicant insult, the effects of reactive intermediate species lead to deregulation of cell signalling pathways and dysfunction of biomolecules, leading to failure of target organelles and eventual cell death. A myriad of genetic factors determine the susceptibility of specific individuals to chemical-induced liver injury. Environmental factors, lifestyle choices and pre-existing pathological conditions also have roles in the pathogenesis of chemical liver injury. Research aimed at elucidating the molecular mechanism of the pathogenesis of chemical-induced liver diseases is fundamental for preventing or devising new modalities of treatment for liver injury by chemicals.
The liver is necessary for survival because it is essential for the coordination of metabolism in the body, including glucose homeostasis, xenobiotic metabolism and detoxification. The liver is also a major site for steroid hormone synthesis and degradation and synthesis of plasma proteins. It continuously provides energy to the whole body by managing the systemic supply of nutrients. In addition, the liver is a mediator of systemic and local innate immunity and an important site of immune regulation (Refs 1, 2).
The liver is a complex organ comprised of parenchymal cells, sinusoidal cells and perisinusoidal cells. Parenchymal cells include hepatocytes, which account for 60% of the adult liver cell population and ~78% of the total liver mass, and bile duct epithelia (cholangiocytes). Sinusoidal cells are composed of hepatic sinusoidal endothelial cells, which account for 20% of total liver cells, and Kupffer cells (hepatic macrophages), which account for 80–90% of the total population of fixed macrophages in the body. Perisinusoidal cells consist of hepatic stellate cells (also known as Ito cells, vitamin-A-storing cells, fat-storing cells or lipocytes), which represent 5–8% of all liver cells, and pit cells, which are the natural killer cells of the liver. The functions of the various classes of liver cells are integrated in the extracellular matrix through direct cell-to-cell communication, paracrine secretion, intracellular signalling, interaction with the extracellular matrix, and generalised response to endocrine and metabolic fluxes (Refs 3, 4). Although histologically the liver appears to be a uniform mass of tissue, it is morphologically, histochemically and functionally heterogeneous. Cells located in the periportal zone differ from those in the perivenous zone in oxygen content, hormones, and cell-to-cell and cell-to-biomatrix interactions. There are also zonal differences in key enzymes, receptors and subcellular structures. Therefore, cells in different regions have different functional capacities for metabolism (Refs 5, 6). This heterogeneity results from the unique blood supply to the different regions of the liver. The liver receives approximately 70% of its blood supply and 40% of its oxygen from the portal vein, with the remainder supplied by the hepatic artery (Ref. 7).
The liver is the initial site of contact for many types of orally ingested therapeutic drugs, alcohol and other xenobiotics from intestinal absorption, thus making this organ particularly susceptible to chemical-induced injury. The spectrum of chemical-induced liver diseases is wide, including dose-dependent hepatotoxicity, other cytopathic toxicities and acute steatosis. Acute and chronic hepatitis, granulomatous hepatitis, cholestasis with bile duct injury, cholestasis with or without hepatitis, steatohepatitis, vascular disorders and tumours are among other known forms of chemical-induced liver diseases. The severity of chemical-induced liver injury varies from minor nonspecific changes in hepatic structure and function to acute liver failure, cirrhosis and liver cancer (Ref. 4). The predominant clinical presentation is acute hepatitis or cholestasis, although almost any clinical pathological pattern of acute or chronic liver disease can occur (Ref. 8). Chemical-induced hepatotoxicity can recapitulate clinicopathological features of several kinds of acute and chronic liver diseases, and thus clinically it presents a diagnostic challenge to health care professionals. However, certain individual drugs tend to have a characteristic clinical signature (Ref. 8).
In this paper, we will summarise our current knowledge of the molecular mechanisms of the pathogenesis of chemical-induced liver injury. More detailed information on the mechanisms of progression of liver disease can be found in other reviews such as those of Henderson and Iredale (Ref. 9) and Guicciardi and Gores (Ref. 10).
Exposure of the liver to xenobiotic chemicals
Xenobiotics are chemicals that are not normally produced or expected to be present in the body. Humans are exposed to many xenobiotics, and several of these xenobiotics are known to have hepatotoxicant potential. Toxic exposure to these chemicals results most often from absorption through the gastrointestinal tract after oral ingestion. Inhalation and absorption through the skin are other common routes of exposure for xenobiotics. There are several hundred industrial chemicals that have been identified as hepatotoxicants; examples include N-nitrosodimethylamine, hydrazine and carbon disulfide (see the Centers for Disease Control and Prevention website for a full list: http://www.cdc.gov/niosh/npg/). Hepatotoxins can come directly from food, such as hepatotoxic mushrooms and fruits, or they can be generated during food processing, such as polycyclic aromatic hydrocarbons (Ref. 11). Fungal mycotoxins (such as aflatoxin B1) can be found in contaminated grains and nuts (Ref. 12). Alcohol consumption can result in serious alcoholic liver diseases, which is a major health care problem in both Europe and the USA (Refs 4, 13). Liver injury also results from environmental exposure to inorganic elements. For example, beryllium is known to produce midzonal liver necrosis as a result of phagocytosis of insoluble beryllium phosphate by Kupffer cells (Ref. 14).
Adverse effects to the liver are one of the most frequently cited reasons for discontinuing the development of drug candidates in the pharmaceutical sector. In addition, hepatotoxicity is recognised during postmarketing phases as one of the main causes for withdrawing medications from the market (Ref. 15). Drug-induced liver necrosis is also one of the leading causes of acute liver failure. Acetaminophen-induced liver injury is most common among all cases of drug-induced liver injuries (Refs 16, 17). The use of vitamins, dietary supplements, herbals and nonproprietary remedies is a popular component of complementary and alternative medicine (Ref. 18). However, many of these dietary supplements, such as Chaparral and MaHuang, are known potent hepatotoxins and their use has resulted in cases of acute liver failure (Ref. 19).
Hepatic biotransformation of xenobiotics
Lipophilic xenobiotics are converted to more hydrophilic forms by biotransformation (xenobiotic metabolism) (Ref. 20). This increase in hydrophilicity of xenobiotics makes their excretion more efficient. Enzymes responsible for xenobiotic metabolism can be found in subcellular compartments such as the endoplasmic reticulum (ER), cytosol, lysosomes, mitochondria, plasma membrane and blood (Ref. 21). The liver is the principal site of xenobiotic metabolism immediately after their absorption from the gastrointestinal tract. The liver also has the highest supply of biotransformation enzymes of all organs in the body. Therefore, it has a key role in xenobiotic detoxification and protection against chemical toxicity.
Biotransformation can be categorised into hydrolysis, reduction, oxidation and conjugation reactions. Depending on the chemical composition of xenobiotics, different types of xenobiotic-metabolising enzymes are involved in these reactions. Enzymatic hydrolysis either produces two molecules or leads to the opening of a ring in the molecule. Reduction is a reaction in which a particular atom, such as oxygen, gains electrons. The reduction of a xenobiotic proceeds either enzymatically or nonenzymatically by interacting with reducing agents such as glutathione (GSH), FADH, FMNH and NAD[P]H. In contrast to reduction reactions, oxidation is a reaction that transfers electrons away from a particular atom (Refs 21, 22). The most important enzyme system for oxidative reactions is cytochrome P450 (CYP). CYP enzymes are mixed-function monooxygenases that catalyse redox and isomerisation reactions in the presence of molecular oxygen (O2). More than 90% of drugs are substrates for the different CYP family of enzymes. The biotransformation of xenobiotics often begins with redox reactions particularly catalysed by CYP enzymes. In addition to biotransformation of xenobiotics, CYPs also have important roles in the synthesis of endogenous compounds such as steroid hormones, bile acids, fat-soluble vitamins and eicosanoids. The highest levels of CYP enzymes involved in xenobiotic biotransformation are found in the liver (Refs 21, 23).
Conjugation refers to the transfer of an endogenous molecule or moiety, usually in the form of a cofactor, to functional chemical groups that either are present on the xenobiotic or are introduced or exposed during oxidation, reduction or hydrolysis reactions. The endogenous molecule or moiety usually has high polarity (hydrophilic). These conjugation reactions usually result in considerable increases in xenobiotic hydrophilicity (with the exception of methylation and acetylation). Conjugation reactions include glucuronidation, sulfation, amino acid conjugation, acetylation, methylation and GSH conjugation (mercapturic acid synthesis). They are catalysed by specific enzymes such as glucuronosyl (or glucuronyl) transferases, sulfotransferases, GSH S-transferases and acetyl and amino acid N-transferases. Some conjugation reactions proceed nonenzymatically. Most conjugation enzymes are located in the cytosol, whereas the family of UDP–glucuronosyltransferases is found in the ER (Refs 21, 24).
Hydroxylation, reduction and oxidation are traditionally called Phase I reactions, which expose or introduce a functional group (–OH, –NH2, –SH or –COOH), usually resulting in increased hydrophilicity of xenobiotics. The functional groups exposed or introduced during Phase I biotransformation are often sites for Phase II conjugation reactions, which usually render the compound even more hydrophilic. The combined effect of Phase I and II reactions makes it easier for xenobiotics to be excreted into urine or bile.
Transporters such as the ABCC (ATP-binding cassette transporter family, subfamily C) efflux proteins and the multidrug resistance proteins (MDRs) have an important role in hepatic xenobiotic elimination by mediating the excretion of parent molecules, metabolites and conjugates into bile or sinusoidal blood in an energy-dependent manner. Transporters can be found in both the canalicular (apical) and sinusoidal membranes of hepatocytes (Refs 25, 26, 27). Transmembrane transporters are important determinants of the overall xenobiotic metabolism and clearance by the liver. The function of these transporters has been described by some investigators as Phase III of the xenobiotic metabolism process. Moreover, coevolution of conjugative enzymes and transporters is believed to have occurred so that the formation of polar conjugates (e.g. glucuronides and GSH derivatives) is coupled to their active excretion from the liver. The net result of these coordinated functions is an efficient removal of xenobiotics and their metabolites from the body (Refs 28, 29, 30) (Fig. 1). For any given compound, one, two or all three of these steps can participate in its elimination.
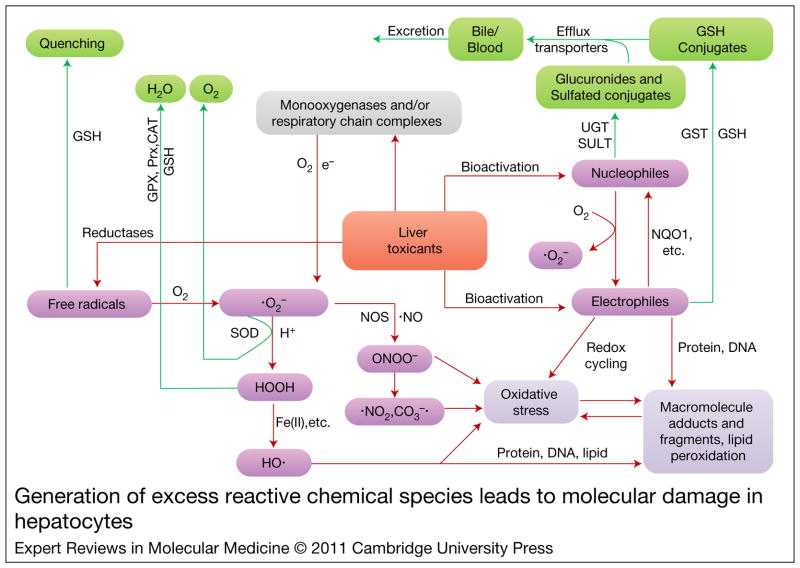
Figure 1. Generation of excess reactive chemical species leads to molecular damage in hepatocytes.
Reactive species (pink boxes) are generated during biotransformation of toxicophores, which can then be detoxified by drug-metabolising enzymes and antioxidant systems (green boxes). Excess reactive chemical species react with biomolecules to form molecular adducts, peroxides or broken molecular fragments (purple box), resulting in dysfunction or change in the structure of biomolecules. Green arrows indicate detoxification pathways; red arrows show toxification pathways. CAT, catalase; GSH, glutathione; GPX, glutathione peroxidase; GST, glutathione S-transferase; NOS, nitric oxide synthase; NQO1, NAD(P)H quinone oxidoreductase 1; Prx, peroxiredoxin; SOD, superoxide dismutase; SULT, sulfotransferase; UGT, glucuronosyltransferase. Parts of this figure are adapted from portions of figures from Refs 33, 34.
Harmful reactive chemical species can be produced during biotransformation of xenobiotics and under physiological and pathological conditions
The conversion of xenobiotics from their lipophilic forms to more stable hydrophilic forms favours their excretion from the body and usually results in loss of pharmacological or biological activity. However, biotransformation of certain xenobiotics can also produce short-lived, unstable, highly reactive chemical species that can interact with functional biomolecules and potentially lead to adverse effects. The functional groups or structural motifs in xenobiotics that can produce harmful reactive intermediates have been defined as ‘toxicophores’ (Ref. 31). Toxicophores in drugs and their bioactivation pathways have been empirically catalogued (Ref. 32). Reactive chemical species produced during biotransformation reactions include electrophiles, nucleophiles, free radicals and redox-active reactants (Fig. 1).
Electrophiles are produced by oxidation or reduction reactions. Usually, the parent molecule is changed by the addition of an atom of oxygen or nitrogen, which extracts electrons. This results in a partially or fully positively charged centre in electrophilic molecules. For example, aromatic compounds can be oxidised to epoxides, quinines, quinone imines and methides. Nitroarenes can be reduced to reactive N-oxygenated species and nitrenium ions. Thiolcarbonyl compounds can be oxidised to sulfenes. Electrophiles such as acyl halides, aldehydes, ketones and arene oxides are reactive because their electron-deficient centre allows them to react by sharing electron pairs with electron-rich atoms in nucleophiles. By contrast, nucleophiles such as thiols, amines and hydrazines are species that are strongly attracted to positively charged regions in other chemicals. Nucleophiles can be oxidised to free radicals and electrophiles, whereas electrophiles can be reduced to nucleophiles (Fig. 1). In general, the formation of toxic reactive nucleophiles is rare. The majority of all reactive chemical species are electrophilic (Refs 33, 34).
A free radical is a molecule or molecular fragment that contains one or more unpaired electrons. Radicals are formed when a chemical accepts or loses an electron. Homolytic fission of a covalent bond also generates radicals. Free radical intermediates are known to occur during the metabolism of a wide variety of xenobiotics, such as polyhalogenated alkanes, polyphenols and quinines (Fig. 1). Free radical metabolites can, in principle, be generated during the metabolism of almost any aromatic xenobiotic (Refs 35, 36). Reactive oxygen and nitrogen free radical species can also be generated in response to exposure to toxic concentrations of metals such as iron, copper and arsenic (Ref. 37). The one-electron reduction of nitroaromatic compounds, quinones and other chemicals can be catalysed enzymatically by several of reductases and dehydrogenases. Xenobiotics can also be oxidised to free radicals by peroxidases (Ref. 38). Free radicals usually extract a hydrogen atom from other molecules and can also add to the double bonds of a biomolecule, usually resulting in new free radicals (Ref. 33).
Reactive oxygen species (ROS) are another important type of free radical. This term is used to describe a collection of oxygen-derived free radical and nonradical species such as superoxide anion O2−•), hydroxyl (HO•), peroxyl (RO2•), alkoxyl (RO•) radicals, and hydrogen peroxide (H2O2) (Ref. 39). Superoxide anion is a starting material in the formation of hydrogen peroxide, which can then be converted to hydroxyl radical in the presence of iron. Superoxide is also used in the production of peroxynitrite (ONOO−), which rapidly reacts with CO2 to ultimately form reactive free radicals such as nitrogen dioxide (•NO2) and carbonate anion radical (•CO3−) (Refs 40, 41) (Fig. 1). Superoxide anions are among the least reactive ROS whereas hydroxyl radicals react rapidly with virtually all molecules found in vivo. At low physiological levels, ROS act as ‘redox messengers’ in intracellular signalling and regulation of gene expression. However, when generated in excess amounts, ROS can be harmful to cellular components and tissues by inducing oxidative modifications to cellular macromolecules (Ref. 42). ROS are normal byproducts of metabolism during xenobiotic exposure. Superoxide anion and hydrogen peroxide are released when the cytochrome P450 catalytic cycle is interrupted (uncoupled) following the introduction of the first and the second electron, respectively (Ref. 21). Monooxygenases (e.g. cytochrome P450s) in ER contribute greatly to the increased levels of cellular hydrogen peroxide and superoxide anion, especially in the absence of substrates. More than 60% of electron transfer haemoproteins and about 20–30% of membrane-bound flavoproteins are localised in the ER (Ref. 43). The mitochondrion is another major intracellular source of ROS, where ROS are constantly produced as a byproduct of oxidative respiration under normal conditions. Moreover, mitochondrial ROS production can increase significantly in response to drugs or metabolites that interfere with the respiratory chain (Refs 44, 45). The peroxisome also contributes to cellular hydrogen peroxide production under normal physiological conditions (Ref. 46).
Detoxification of harmful reactive chemical species
During normal biotransformation of endobiotics and nutrients, reactive chemical species such as ROS can also be produced. Depending on the magnitude of their formation, reactive chemical species can be quenched or disposed of by endogenous cytoprotective mechanisms, including Phase II conjugation enzymes and their cofactors, efflux transporters, antioxidant enzymes and small molecules such as vitamin C (Refs 33, 47) (Fig. 1). The tight coupling of bioactivation and detoxification reactions serves to alleviate or eliminate the reactivity of intermediate chemical species (Ref. 48).
Conjugative reactions have evolved from their normal physiological functions to fulfil a variety of cytoprotective functions against potentially harmful xenobiotics (Ref. 49) (Fig. 1). Nucleophiles are generally detoxified by conjugation with a soluble moiety or cofactor at the nucleophilic functional group. Hydroxylated compounds are conjugated by sulfation or glucuronidation, whereas thiols are methylated or glucuronidated. Amines and hydrazines are acetylated. These reactions prevent peroxidase-catalysed conversion of the nucleophiles to free radicals and biotransformation of phenols, aminophenols, catechols and hydroquinones to electrophilic quinones and quinone imines (Ref. 33). Although some GSH conjugates, such as the GSH thiyl radical and GSH disulfide (Ref. 50), are highly reactive, a general mechanism for the detoxification of electrophilic toxicants is conjugation with the thiol group of the nucleophile GSH. This reaction may occur spontaneously or can be facilitated by GSH S-transferases (Refs 51, 52) (Fig. 1). The resulting conjugates are subsequently excreted by efflux transporters into the bile or urine (Refs 28, 29, 30).
To combat oxidative stress and maintain a proper reducing environment, the liver is well equipped with antioxidant mechanisms, including micronutrients such as α-tocopherol (vitamin E) and ascorbic acid (vitamin C). Also, thiol-rich proteins, such as metallothionein, and other metal-sequestering proteins, such as ferritin, are important in preventing the potentially damaging effects of certain metals, such as cadmium and copper. Enzymes such as epoxide hydrolases, catalase, superoxide dismutase (SOD) and GSH peroxidases also have a role in the metabolism and neutralisation of reactive intermediates. For example, superoxide anion, an important ROS, can be converted into hydrogen peroxide by SODs in the cytosol and the mitochondria. Subsequently, hydrogen peroxide is reduced to water by catalase in the peroxisomes, by selenocysteine-containing GSH peroxidases in the cytosol and mitochondria, and by peroxiredoxins in the cytosol, mitochondria and ER. Peroxynitrite, a nitrogen-reactive species, is reduced to nitrite (ONO−) by the selenocysteine-containing GSH peroxidase and peroxiredoxins. In addition, peroxynitrite reacts with oxyhaemoglobin, haem-containing peroxidases or albumin (Ref. 40). The detoxification of superoxide anions, peroxynitrites and peroxides prevents the formation of free radicals produced from the rapid reaction of peroxynitrite with carbon dioxide (CO2) (Ref. 40) and also prevents the formation of extremely reactive hydroxyl radicals, which cannot be quenched enzymatically or by the antioxidants GSH, vitamin E and vitamin C.
The most important antioxidant in the mammalian liver is GSH. Hepatic levels of GSH are usually high and can be readily modulated by regulating its biosynthetic pathway (Refs 53, 54). GSH and redox status are compartmentalised within the hepatocyte, with the highest concentration of GSH found in the cytosol. The relative redox states of the different subcellular compartments from most reducing to most oxidising are mitochondria > nuclei > cytoplasm > ER > extracellular space (Refs 55, 56, 57). GSH is a critical cofactor for several antioxidant pathways, including thiol–disulfide exchange reactions, GSH peroxidase and GSH S-transferases. GSH protects cells from oxidative injury by reducing hydrogen peroxide and scavenging reactive oxygen and nitrogen radicals (Refs 58, 59). The oxidised form of glutathione (GSSG) is reduced back to GSH by glutathione reductase. This reductive reaction needs NADPH, which is generated in the mitochondria in an ATP-dependent manner. Therefore, the ability of the liver to withstand oxidative stress depends on its ability to generate the necessary energy to carry out these vital reactions (Ref. 60). Thus, GSH has an important role in the detoxification of both electrophiles and free radicals.
In addition to regulating redox status, GSH is also an effector of cell signalling. GSH-induced shifts in redox status are known to mediate the dynamic regulation of protein function by the formation of reversible intermolecular, intramolecular and mixed disulfide bonds between protein cysteines and GSH (S-glutathiolation). GSH also stabilises extracellular proteins and protects proteins against irreversible oxidation of critical cysteine residues. The DNA binding of certain transcription factors such as nuclear factor-κB, p53 and activator protein-1 is redox dependent, and redox signalling resulting from oxidative stress can be transduced into altered expression of a wide variety of genes implicated in cell proliferation, differentiation and apoptosis. These processes have been implicated in the pathogenesis of many liver diseases (Refs 58, 59, 61).
Other redox systems, such as thioredoxin, thioredoxin reductase, peroxiredoxin, glutaredoxin and pyridine nucleotide redox couples, regulate the activities of multiple transcription factors, work as growth factors, serve as enzyme cofactors, and also participate in cell signalling and modulation of cell function. In addition, the thioredoxin and glutaredoxin systems have an important role in cellular defences against oxidative stress and in maintaining redox homeostasis by regulating thiol–disulfide exchange. NADPH-dependent thioredoxin reductase, which reduces a broad range of substrates including the oxidised form of thioredoxin, can also directly reduce lipid peroxides, hydrogen peroxide, and dehydroascorbic and lipoic acids (Refs 42, 62).
Modification of biomolecules and shifting of the reducing environment of cells by reactive chemical species
When the balance between bioactivation and detoxification reactions is altered, and the amount of reactive intermediates becomes abnormally elevated, reactive intermediates can overwhelm or bypass the defence systems, resulting in oxidative stress and damage to proteins, nucleic acids and lipids (Fig. 1).
Reactive intermediates can covalently bind to proteins and form protein adducts. Sulfhydryl groups and primary or secondary amino groups in serine and threonine residues react readily with electrophilic species such as alkyl and benzylic carbonium ions, iminium ions, aldehydes, epoxides, enones, quinone imines and Michael acceptors. Lysine and histidine residues in proteins are similarly targeted by these electrophiles (Refs 63, 64). The identity of many proteins targeted by reactive intermediates of xenobiotics is well documented. Halothane and acetaminophen have been demonstrated to form adducts with specific proteins (Ref. 65). The formation of 3-(cystein-S-yl)-acetaminophen adducts between N-acetyl-p-benzoquinone imine and hepatocellular proteins has been used as a serum biomarker of acetaminophen toxicity in patients with acute liver injury (Refs 66, 67). More detailed information on the identity of proteins modified covalently by xenobiotics can be found in the web-accessible resource known as the reactive metabolite target protein database (Ref. 68). In addition, proteins can be modified by phosphorylation, acylation, methylation, ubiquitylation and sumoylation under electrophilic or oxidative stress conditions (Ref. 69). Adduct formation can lead to several adverse consequences, such as interference with protein function, stimulation of fibrogenesis and induction of immune responses. In alcohol consumers, covalent modification of proteins and other cellular constituents by acetaldehyde has a role in the sequence of events leading to alcoholic liver disease (Ref. 70). The role of protein covalent binding of reactive intermediates in the mechanism of toxicant action has been the subject of extensive investigations for several decades (Ref. 71), yet we do not have a clearly defined role of covalent binding in hepatotoxicity. It is thought that drug–protein adducts contribute to toxicity either through impaired function of the modified proteins or through immune-mediated mechanisms.
DNA can also be modified by reactive chemical species. Oxygen atoms of purine and pyrimidine bases and endocyclic nitrogens of purine bases in DNA are also targets of electrophiles (Ref. 63). This is the case for metabolites of a spectrum of carcinogenic polycyclic hydrocarbons (Refs 72, 73). Diffusible hydroxyl radicals can react either with the sugar units by hydrogen abstraction or with the base moieties by addition (Ref. 74). Metals such as iron and copper also cause various modifications of DNA bases by mediating the formation of free radicals. Lipid peroxides, which result from the reaction between radicals and polyunsaturated fatty acid residues of phospholipids, can further react with metals and finally produce mutagenic and carcinogenic malondialdehyde, 4-hydroxynonenal (HNE) and other exocyclic DNA adducts (etheno and propano adducts) (Refs 37, 75). When DNA damage is poorly repaired, deleterious genetic changes can occur.
Lipid peroxidation is usually the consequence of self-propagating chain reactions of free radicals. Peroxyl and hydroxyl radicals react with unsaturated and mainly polyunsaturated fatty acids in membranes, with major roles in initiating lipid peroxidation (Ref. 76). Lipid peroxidation produces various electrophiles, such as 4-hydroxynonenal (HNE), a major endproduct of peroxidation of membrane N-6-polyunsaturated fatty acids. The reaction of HNE with important signalling proteins might have a role in the progression of liver diseases (Refs 77, 78). The peroxidative breakdown of membrane lipids results in impaired function of mitochondria, ER, lysosomes, plasma membranes and Golgi, and thus has a role in acute liver necrosis and the amplification and progression of liver injury (Ref. 79).
Proper redox homeostasis is crucial for cell survival. The redox status in the cell is maintained by various redox couples, among which GSSG–2GSH is the major one that maintains redox regulation through protein glutathionylation (Refs 56, 80, 81, 82). Normally, the GSH concentration is high in hepatocytes, which is indicative of the reducing environment in the cell. This reductive environment serves as a protective mechanism against oxidative stress. It is also essential for maintaining the integrity of the structure and function of biomolecules (Refs 83, 84). Disturbance in the equilibrium of redox couples by reactive chemical species results in imbalances in redox status. Electrophiles extract electrons from thiols and reduce the amount of GSH. Similarly, free radicals, including reactive oxygen or nitrogen species, abstract hydrogens from the reducing milieu of the cell. These reactions proceed at the expense of reducing equivalents, thus resulting in oxidative stress. Under oxidative stress, redox-dependent proteins are modified, redox signalling pathways are disrupted, and peroxides and free radicals are produced, leading to damage of cell components, including proteins, lipids and DNA (Refs 85, 86). For example, ethanol-induced oxidative stress results in aldehyde–protein adducts in the liver (Ref. 87). Also, liver microsomal malondialdehyde–protein adducts are produced as a result of oxidative stress resulting from chronic dietary iron overload (Ref. 88). In addition, HNE–protein adducts are considered to be good biomarkers of lipid oxidation during liver injury (Refs 77, 87). Redox-dependent post-translational modifications most frequently occur in protein thiols. These are mediated by environmental and endogenously generated reactive species that can trigger cell signalling events leading to toxic effects (Ref. 89). Chloroform, carbon tetrachloride and GSH depletion induce secondary genotoxicity in liver cells through oxidative stress (Ref. 90). In some cases, mitogen-activated protein kinases (MAPKs), extracellular signal-regulated kinase 1/2 (ERK1/2), c-Jun N-terminal kinase (JNK) and the nuclear factor kappa B (NF-κB) pathways are involved in hepatocyte oxidative injury (Refs 91, 92), as seen in alcoholic liver injury. Activating protein 1 (AP-1) is also involved in the deterioration of cell and organ function by toxic xenobiotics (Refs 93, 94).
Adaptation to and repair of damaged biomolecules
In addition to having mechanisms and substances that prevent toxic reactions between reactive chemical species and biomolecules, cells have evolved protective mechanisms that promote repair, removal and replacement of damaged cellular constituents (Fig. 2) (Refs 33, 47). Damaged proteins, chemically altered DNA and peroxidised lipids can be repaired by different mechanisms. The mitochondrial matrix contains repair systems for oxidised proteins, including thioredoxin, thioredoxin reductase, glutaredoxin, GSH and GSH reductase. These systems reduce disulfide bridges or sulfenic acids catalytically. Similarly, methionine sulfoxide reductase reverses methionine sulfoxide back to methionine residues in proteins (Ref. 95). Molecular chaperones such as heat-shock proteins can rescue denatured proteins by ATP-dependent disaggregation and refolding, or assist in their degradation by proteasomes by engaging the chaperone-dependent ubiquitin ligase CHIP (C-terminus of heat-shock protein cognate 70-interacting protein) (Ref. 96). There are distinct proteolytic systems for different physiological tasks under changing environmental and pathophysiological conditions. Damaged proteins can be eliminated by proteolysis in lysosomes (Ref. 97). Proteins damaged by oxidative stress are also removed by the proteasomes in the cytosol and nucleus. Under oxidative stress conditions, there is high proteasome content in the nucleus. This protects the nucleus from formation and accumulation of nondegradable proteins (Ref. 98). Lipid peroxides are repaired by a complex process that operates in concert with a series of reductants along with GSH peroxidase and GSH reductase. Different types of damage to DNA are corrected by specialised mechanisms, each using a different set of repair proteins (Refs 99, 100, 101). Despite its high reactivity with electrophiles and free radicals, nuclear DNA is remarkably stable, in part because it is packaged in chromatin and because of the several repair mechanisms that correct alterations in DNA. Mitochondrial DNA is more prone to damage because of the lack of histones or the type of repair mechanisms present in the nucleus.
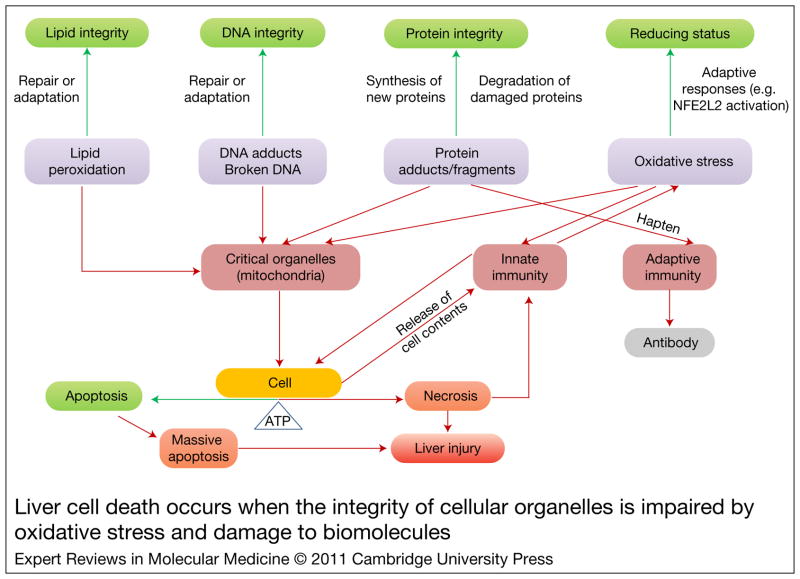
Figure 2. Liver cell death occurs when the integrity of cellular organelles is impaired by oxidative stress and damage to biomolecules.
Damage to biomolecules and oxidative stress can be reversed or tolerated by activation of compensatory responses. Excessive damage or modification of biomolecules by oxidative stress can overwhelm the capacity of repair and adaptive processes, leading to uncorrected changes to critical organelles, among which mitochondria is the most important. This can lead to activation of immune responses, which could further aggravate cellular damage. The interplay between xenobiotic-induced molecular changes, organelle dysfunction and activation of immunity results in cell death and liver injury. Cellular ATP availability is an important determinant of the type of cell death that occurs. Programmed cell death or apoptosis requires ATP, whereas oncotic death (necrosis) predominates when ATP stores are depleted. However, massive apoptosis also leads to apoptotic necrosis. Green boxes indicate normal homeostasis; boxes of other colours show events leading to liver cell injury. Green arrows indicate pathways leading to cell recovery; red arrows show pathways leading to cell damage or death.
Although it has been proposed that biological adaptation to repeated exposure to certain xenobiotics can in some instances result in long-term toxicity (Ref. 102), adaptive responses to acute exposure are generally known to preserve or restore biological homeostasis in the face of chemical insult. These responses involve sensing the initial damage or dysfunction and initiating compensatory changes in gene expression. Adaptive responses are beneficial because they enhance the capacity of different cellular structures to respond more robustly to chemical-induced stress. They are also reversible and critical for preserving cell viability. Disruption of this equilibrium at levels of the molecules, organelle and other cellular organisations leads to an adverse effect or toxicity. Induction of xenobiotic-metabolising enzymes mediated by the xenosensors aryl hydrocarbon receptor (AhR), constitutive androstane receptor (CAR, NR1I3), pregnane X receptor (PXR, NR1I2) and peroxisome proliferator-activated receptor (PPAR) provides examples of adaptive responses to chemical stressors (Refs 103, 104). In response to oxidative stress, KEAP1-NFE2L2 [kelch-like ECH-associated protein 1-nuclear factor (erythroid-derived 2)-like 2] is activated and induces GSH synthesis and alters gene expression for multiple xenobiotic metabolising enzymes and transporters (Refs 105, 106). In those instances when oxidative stress is severe, or when the function of protective enzymes is deeply compromised, cells may ‘sacrifice themselves’ by apoptosis, which protects the surrounding healthy tissue from further damage. Only under the most severe oxidative stress conditions and when adaptation mechanisms fail will cells then undergo necrotic death, which exposes the surrounding nondamaged cells to immune responses (Refs 33, 107).
Oxidative stress and dysfunction of biomolecules result in dysregulation of cell activities, leading to cell death
When damage to cellular biomolecules remains unrepaired or the cellular adaptive responses are overwhelmed by unquenched reactive chemical species, the cell structural components and essential biochemical processes become impaired. These biochemical disorders result in functional and structural damage to certain organelles. Cell death can ensue and, depending on the severity of the damage, clinical manifestations can become evident (Fig. 2). Any intracellular compartment, such as mitochondria, cytoskeleton, lysosome, ER, nucleus or plasma membrane, can be affected by toxicants, initiating different types of cell death. The different pathways mediating modes of cell death could crosstalk through a molecular ‘switch’, such as p53. Inhibition of one particular mode of cell death might activate an alternative pathway to cell death (Refs 108, 109).
Mitochondrial dysfunction is a major mechanism of chemical-induced liver injury. The mitochondrion is a key organelle involved in pyruvate oxidation, fat oxidation and ATP production. It is also a site where external and intracellular stress-response pathways converge to provoke cell death. The opening of permeability transition pores under stress conditions results in sudden increases in permeability of the inner mitochondrial membrane. Depending on the magnitude of this event, mitochondrial membrane permeabilisation can lead to apoptosis by release of cytochrome c, which is a known activator of caspases in the cytosol. Under more serious circumstances, mitochondrial membranes can rupture, leading to ATP depletion and subsequent liver cell necrosis. Abnormal mitochondrial function can also lead to fat accumulation. In the most severe cases of mitochondrial dysfunction, microvesicular steatosis and hypoglycaemia can occur. Macrovacuolar steatosis can also be the result of chemical-induced mitochondrial dysfunction and can often progress to steatohepatitis and cirrhosis (Refs 110, 111, 112, 113, 114, 115, 116).
Apoptosis and necrosis are the most widely recognised forms of hepatocyte cell death. Apoptosis can be initiated by two different pathways. The intracellular signalling pathway is triggered by intracellular stress caused by oxidative stress and damage to proteins, membranes or DNA. The external pathway is initiated by the binding of paracrine molecules such as tumour necrosis factor-α and Fas ligand to death receptors on the cell surface. Each pathway activates caspases and nucleases, resulting in apoptotic cell death. Characteristic changes seen during apoptosis include cell shrinkage, nuclear fragmentation, chromatin condensation and chromosomal DNA fragmentation. In contrast to apoptosis, necrosis can result from extensive damage to the plasma membrane with disturbance of ion transport, dissolution of membrane potential, cell swelling and eventual rupturing of the cell. During necrosis, the cell liberates macromolecular breakdown products, including lipid peroxides, aldehydes and eicosanoids. These molecules can then trigger an inflammatory response in the hepatic parenchyma. Apoptosis is energy dependent and is considered to be a protective mechanism that prevents cells from oncotic necrosis, which often promotes more damage to surrounding cells through the involvement of the immune system. In the presence of ATP, cell death can proceed by apoptosis, but when mitochondria are de-energised, cell death proceeds by necrosis. Apoptosis and necrosis more likely represent the morphological and mechanistic ends of a spectrum of overlapping cell death processes (Fig. 2). Cells on an apoptotic path can be redirected to necrosis or to an overlapping pattern ‘apoptotic necrosis’ (necraptosis) upon depletion of ATP (Fig. 2). Furthermore, with excess apoptosis, the capacity of the tissue for phagocytosis of apoptotic bodies can be exceeded, and ‘secondary’ necrosis can occur (Refs 60, 109, 111, 117). Both necrosis and apoptosis can trigger cytolytic hepatitis.
The interaction between reactive chemical species and cellular biomolecules can also trigger a response by the liver’s immune system. The formation of apoptotic bodies or the release of cell constituents by necrotic hepatocytes can trigger the activation of innate immune cells, producing proinflammatory and cytotoxic mediators. Depending on the magnitude of cell death, this immune response can result in enhanced liver tissue damage. This is postulated to occur in the early stages of alcoholic liver disease (Refs 118, 119). Adaptive immune responses can also be elicited during chemical hepatotoxicity. Covalent binding of reactive metabolites to cellular proteins can lead to the formation of neoantigens (haptens) that can initiate autoantibody production and cytotoxic T-cell responses. In addition, certain drugs such as sulfonamide sulfamethoxazole (Ref. 120) can bind directly to T-cell receptors, mimicking a ligand–receptor interaction. This causes T-cell activation in a major histocompatibility complex-dependent fashion (Refs 121, 122, 123, 124).
Intrahepatic cholestasis induced by drugs such as cyclosporine and anabolic steroids is one of the clinically important forms of acquired cholestatic liver disease. The underlying mechanisms could be associated with canalicular transporters, whose function can be inhibited by the parent chemical or its metabolite. Canalicular transporters are essential for bile formation and for biliary secretion of cholephilic compounds, xenobiotics and their metabolites. Any chemical-mediated functional disturbance of these transport processes can lead to intracellular accumulation of potentially harmful bile constituents and xenobiotics, thus resulting in the development of cholestatic liver cell damage (Refs 125, 126, 127). The bile salt export pump (BSEP) is a known target for drug inhibition in patients who are susceptible to drug-induced cholestasis (Ref. 128). This is the main canalicular transporter involved in the biliary excretion of bile acids. Inhibition of BSEP function leads to accumulation of bile salts in hepatocytes, which can result in liver damage.
Many cases of drug-mediated hepatotoxicityare dose dependent and thus predictable. However, idiosyncratic drug reactions are dose independent and unpredictable. These reactions can result in significant morbidity and mortality (Ref. 129). Considerable evidence indicates that many of these reactions are immune mediated (Ref. 130) and that they are caused by immunogenic conjugates formed from the reaction of reactive metabolites of a drug with cellular proteins. Although this ‘drug–protein antigen response’ concept is well accepted, the precise mechanism of idiosyncratic adverse drug reaction remains unclear. Continued basic research in this area is essential to elucidate the role of drug–protein antigens in hepatotoxicity (Refs 123, 131, 132, 133).
Factors contributing to chemical-induced liver injury
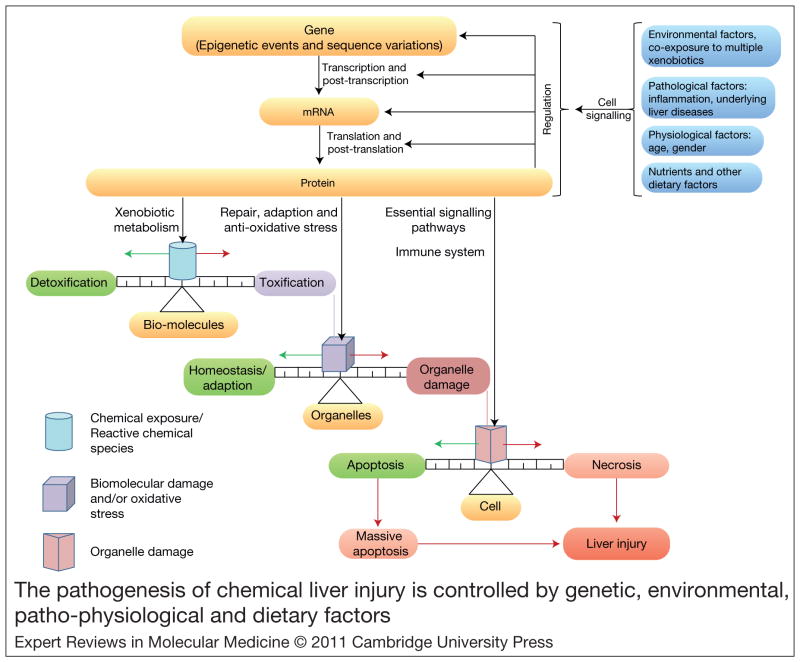
As stated previously, the pathogenesis of most chemical-induced liver injuries is initiated by metabolic activation of the chemical or an imbalance between bioactivation (toxification) and detoxification processes. The propensity of a molecule to form either toxic or chemically reactive metabolites is simply a function of its chemistry. Certain toxicophores present in drugs can be biotransformed into reactive species (Refs 31, 32). Exposure to high doses of a potential toxicant increases the amount of reactive metabolites formed, thus increasing the risk for liver injury (Refs 134, 135). In addition, drugs can either inhibit or induce the expression and function of drug-metabolising enzymes, as seen in the case of human cytochrome P450 3A4. Xenobiotic enzyme induction could lead to altered metabolism of chemicals (Ref. 136), which could greatly affect the toxicity outcome. The progression of chemical liver injury involves competition between cell-damaging events caused by reactive chemical species and protective responses aimed at quenching oxidative stress, repairing macromolecular damage or, ultimately, preserving the integrity of tissue by committing ‘self-sacrificing’ apoptotic cell death. Clinical manifestations of chemical-induced liver injury occur with extensive necrotic cell death and the triggering of an immune response. Therefore, the pathogenesis of chemical-induced liver injury involves many competing processes, shaped by the interplay among genetic determinants, environmental factors and potential underlying pathological processes (Fig. 3).
Figure 3. The pathogenesis of chemical liver injury is controlled by genetic, environmental, patho-physiological and dietary factors.
Other factors such as gender, race, age and lifestyle choices can be risk factors for certain forms of chemical liver injury. The interplay between these factors controls gene transcription and cell signalling events, leading to a myriad of interconnected outcomes such as imbalances between detoxification and toxification metabolic pathways, impaired organelle function, compromised ability to repair molecular damage and alterations in overall cellular homeostasis. With prominent shifts in normal cellular homeostasis, the ability of compensatory and adaptive responses to correct the detrimental effects of toxicants can be hampered. Similarly, repair mechanisms can be overwhelmed and incapable of correcting the cellular chemical injury. Green arrows indicate pathways leading to cell recovery; red arrows show pathways to cell damage or death; black arrows indicate pathways that contribute to chemical-induced liver injury.
Genetic factors have a key role in the initiation of the pathogenesis of chemical-induced liver injury. The toxicant action of a specific xenobiotic chemical in the body is determined by its tissue dosimetry at target sites. The dose of the ultimate toxicant at the site of toxicant action is controlled by the coordination of Phases I–III of xenobiotic metabolism and disposition. The in vivo capacity for xenobiotic metabolism is determined by the combined function of different types of xenobiotic-metabolising enzymes in the body. There is a wild diversity of drug-metabolising enzymes to catalyse the Phase I and II reactions, along with transport proteins (Phase III) involved in drug disposition. For example, there are 57 different active genes encoding CYP enzymes and 58 pseudogenes present in the human genome (Ref. 137). The expression levels of these CYPs are not the same among individual subjects. In addition, individual enzymes show great variation among different individuals. The same is true for Phase II enzymes and drug transporters. Drug-metabolising enzymes and transporters are also known to have a differential and overlapping spectrum of substrates. Therefore, the capacity for metabolism and disposition of chemicals can differ widely from one individual to another.
The catalytic activity of an individual enzyme is determined by its abundance in a given tissue and its intrinsic activity. The amount of xenobiotic-metabolising enzymes in a particular tissue is also dependent on epigenetic regulation, gene transcription, and post-transcriptional, translational and post-translational events. Gene transcription is the most important control among these regulatory steps. Interindividual variation in the expression of xenobiotic-metabolising enzymes could result from polymorphic forms of regulatory systems for gene expression. For example, although the mechanism for controlling constitutive expression of CYP3A4 remains unknown, its gene expression is regulated by a number of transcription factors such as C/EBPα (CCAAT/enhancer binding protein, alpha), C/EBPβ, HNF4α (hepatocyte nuclear factor 4, alpha), HNF3γ, CAR and PXR. These transcription factors recognise regulatory modules in the human CYP3A4 gene (Ref. 138). Changes in PXR expression levels and its amino acid sequence are known to produce alterations in the expression of CYP3A4 (Refs 139, 140, 141). Similarly, expression of the Phase I and II enzymes and transporters is coregulated by a set of nuclear receptors, such as PXR. Therefore, all aspects of xenobiotic metabolism and disposition are regulated by certain transcription factors simultaneously in a coordinated manner. However, the intrinsic activity of individual drug-metabolising enzymes is determined by the amino acid sequences of the peptide and variations thereof. These variations or polymorphic forms have been widely identified and studied. Many polymorphic variants of drug-metabolising enzymes are well known to contribute to the interindividual variations in intrinsic activity, and thus drug biotransformation, even when protein expression levels are the same as for the normal form of the enzyme. About 40% of CYP-dependent metabolic reactions are catalysed by polymorphic enzymes (Refs 139, 142, 143). A collection of genetic information on CYP variants, along with their effects on function, can be found in a web-based resource (Ref. 144). Sequence variations are also found in conjugative enzymes and drug transporters (Ref. 125). In addition, the availability of cofactors and allosteric modificators for xenobiotic-metabolising enzymes and transporters also have an important role in their function and activity. For example, haem is a key cofactor for CYP enzymes. Haem also regulates CYP protein synthesis, assembly, repair and disposal. Cellular haem content itself is tightly controlled by genetic factors (Ref. 145).
Several environmental factors have a role in chemical-induced liver injury by regulating xenobiotic metabolism and disposition through various signalling pathways. Transcription factors are cellular mediators of gene regulation. Some of these sense the presence of exogenous stressors and regulate gene expression accordingly as an attempt to counteract any of their potential damaging effects. For example, a variety of structurally diverse endobiotics and xenobiotics, such as bile acids, pharmaceuticals, and environmental, dietary and occupational chemicals, activate PXR and CAR to induce Phase I and II drug-metabolising enzymes and transporters (Refs 146, 147). Induction or suppression of expression of drug-metabolising enzyme results in changes in the normal metabolism and disposition of drugs, which might in turn promote adverse responses (Ref. 148).
Predisposition by pathological and physiological factors also has a role in the initiation of pathogenesis of chemical liver injury. The induction and downregulation of CYP3A4 under pathophysiological conditions, such as inflammatory responses, are key processes involved in determining whether the toxic or therapeutic effects of drugs predominate (Ref. 149). Significant changes in the expression of drug-metabolising enzymes occur during ontogeny. The CYP3A subfamily is the major drug-metabolising CYP enzyme expressed at all stages of development. The expression of different members of this subfamily, however, undergoes significant switches from the fetal predominant form CYP3A7 to the major adult form CYP3A4 (Refs 150, 151). A higher expression of CYP3A4 mRNA in women has also been reported (Ref. 152). In addition to affecting drug metabolism and disposition capacity, genetic determinants, environmental factors and pre-existing pathological conditions can also modulate the function of repair systems for damaged proteins, DNA and lipids, and also immune responses in the liver. For example, human leukocyte antigen (HLA)-B*5701 genotype is a major determinant of antimicrobial agent flucloxacillin-induced liver injury (Ref. 153).
In summary, the initiation of pathogenesis of chemical liver injury is the outcome of interactions between xenobiotics and genetic factors, environmental factors, pre-existing pathobiological conditions and physiological factors. The dynamic changes, uncertainty and diversity of these factors pose significant difficulties in predicting and managing liver injury induced by xenobiotics.
Perspective
Integrity of the function and structure of the liver is necessary for preserving human health. Notably, the liver is particularly susceptible to chemical injury because of its strategic location, prominent blood supply and prevalent role in biotransformation of xenobiotics. Xenobiotic-induced liver diseases represent a major health care problem and a significant source of morbidity and mortality. Unfortunately, the underlying pathophysiological mechanisms of most hepatocellular forms of chemical-induced hepatic injury are still poorly understood. Without a proper understanding of the underlying cellular and molecular mechanisms involved, diseases such as alcoholic liver disease and idiosyncratic drug responses remain a challenge for both basic research scientists and clinicians. A better understanding of the molecular mechanisms of drug-induced liver injury and the interplay among genetic determinants, environmental factors and pre-existing pathological conditions will help in the prevention and development of new forms of treatment for acute and chronic liver diseases.
Chemical-induced liver injury is an acquired disease, and therefore it can be prevented. The ideal approach, in the case of therapeutic drugs, would be to avoid the use of medications with a hepatotoxic liability in patients prone to develop idiosyncratic reactions. This would require a thorough knowledge of genetic traits and environmental factors that render certain individuals vulnerable to drug liver toxicity. Another approach would be the use and development of new drugs that lack known toxicophores, while retaining the desired pharmacological and therapeutic activity (Refs 154, 155, 156, 157).
Proper diagnosis is critical for the correct management of chemical liver disease. Early identification of hepatotoxicity may avert serious complications associated with liver injury. Accurate diagnosis of chemical-induced liver injury is based on a combination of appropriate history, clinical presentation, biochemical tests and exclusion of other potential causes (Ref. 158). The information required for accurate diagnosis may not always be available or might not be complete. Certain subclinical statuses, such as an adaptive response, insidiously developing liver tumours or hepatic fibrosis, might not be detected by conventional biochemical tests of liver function (Ref. 4). Sometimes liver diseases can result from exposure to unknown chemical species. However, liver toxicity by certain individual chemicals tends to have a characteristic clinical signature (Ref. 8). Macromolecular adducts can provide reliable indicators of exposure to certain chemicals and can help in predicting toxicological outcomes (Refs 71, 159). Future research endeavours on the molecular mechanisms of chemical liver injury, including the identification of reactive intermediates, macromolecular adducts and altered cell signalling pathways, could lead to the development and standardisation of novel clinical or subclinical biomarkers of liver toxicant exposure, initiation of injury, progression of disease and overall prognosis.
Collectively, the susceptibility to chemical liver injury is thought to be determined in great part by genetic factors. The Human Genome Program, the HapMap Program (Ref. 160), the 1000 Genomes Project (Ref. 161) and the ENCODE program (Ref. 162) have been instrumental in decoding the role of genetics and gene regulation networks in many pathological conditions, as well as adverse reactions induced by drugs. The accomplishment of these efforts will help in understanding the susceptibility and responses of individuals to chemical exposure in order to develop or choose appropriate therapies for specific patients (personalised medicine) while minimising the possibility of adverse reactions.
Acknowledgments
We recognise many outstanding published contributions of investigators in the field that could not be included in this review owing to space limitations. We express our thanks to the peer reviewers of the article. J.E.M. was the Marlene L. Cohen and Jerome H. Fleisch Scholar in the Department of Pharmaceutical Sciences at the University of Connecticut from 2007 to 2010. X.G., postdoctoral fellow in José Manautou’s laboratory, was supported by the National Institutes of Health Grant DK069557.
References
- 1.Nemeth E, Baird AW, O’Farrelly C. Microanatomy of the liver immune system. Seminars in Immunopathology. 2009;31:333–343. doi: 10.1007/s00281-009-0173-4. [DOI] [PubMed] [Google Scholar]
- 2.Dienes HP, Drebber U. Pathology of immune-mediated liver injury. Digestive Diseases. 2010;28:57–62. doi: 10.1159/000282065. [DOI] [PubMed] [Google Scholar]
- 3.Misdraji J. Embryology, anatomy, histology, and developmental anomalies of the liver. In: Sleisenger MH, Feldman M, et al., editors. Sleisenger and Fordtran’s Gastrointestinal and Liver Disease: Pathophysiology, Diagnosis, Management. 9. Saunders/Elsevier; Philadelphia, PA: 2010. pp. 1201–1206. [Google Scholar]
- 4.Carithers RL, Jr, Mcclain CJ. Alcoholic liver disease. In: Sleisenger MH, Feldman M, et al., editors. Sleisenger and Fordtran’s Gastrointestinal and Liver Disease: Pathophysiology, Diagnosis, Management. 9. Saunders/Elsevier; Philadelphia, PA: 2010. pp. 1383–1400. [Google Scholar]
- 5.Jungermann K, Kietzmann T. Zonation of parenchymal and nonparenchymal metabolism in liver. Annual Review of Nutrition. 1996;16:179–203. doi: 10.1146/annurev.nu.16.070196.001143. [DOI] [PubMed] [Google Scholar]
- 6.Gebhardt R. Metabolic zonation of the liver: regulation and implications for liver function. Pharmacology and Therapeutics. 1992;53:275–354. doi: 10.1016/0163-7258(92)90055-5. [DOI] [PubMed] [Google Scholar]
- 7.Malarkey DE, et al. New insights into functional aspects of liver morphology. Toxicologic Pathology. 2005;33:27–34. doi: 10.1080/01926230590881826. [DOI] [PubMed] [Google Scholar]
- 8.Gunawan B, Kaplowitz N. Clinical perspectives on xenobiotic-induced hepatotoxicity. Drug Metabolism Reviews. 2004;36:301–312. doi: 10.1081/dmr-120034148. [DOI] [PubMed] [Google Scholar]
- 9.Henderson NC, Iredale JP. Liver fibrosis: cellular mechanisms of progression and resolution. Clinical Science. 2007;112:265–280. doi: 10.1042/CS20060242. [DOI] [PubMed] [Google Scholar]
- 10.Guicciardi ME, Gores GJ. Apoptosis as a mechanism for liver disease progression. Seminars in Liver Disease. 2010;30:402–410. doi: 10.1055/s-0030-1267540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hochstein C, Arnesen S, Goshorn J. Environmental health and toxicology resources of the United States National Library of Medicine. Medical Reference Services Quarterly. 2007;26:21–45. doi: 10.1300/J115v26n03_02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kensler TW, et al. Aflatoxin: a 50-year odyssey of mechanistic and translational toxicology. Toxicological Sciences. 2011;120 (Suppl 1):S28–S48. doi: 10.1093/toxsci/kfq283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stubbs MA, Morgan MY. Managing alcohol dependence and alcohol-related liver disease: a problem for the hepatologist, psychiatrist or economist? Clinical Medicine. 2011;11:189–193. doi: 10.7861/clinmedicine.11-2-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zimmerman HJ. Hepatotoxicity: The Adverse Effects of Drugs and Other Chemicals on the Liver. 2. Lippincott Williams & Wilkins; Philadelphia: 1999. [Google Scholar]
- 15.Goldkind L, Laine L. A systematic review of NSAIDs withdrawn from the market due to hepatotoxicity: lessons learned from the bromfenac experience. Pharmacoepidemiology and Drug Safety. 2006;15:213–220. doi: 10.1002/pds.1207. [DOI] [PubMed] [Google Scholar]
- 16.Reuben A, Koch DG, Lee WM. Drug-induced acute liver failure: results of a U.S. multicenter, prospective study. Hepatology. 2010;52:2065–2076. doi: 10.1002/hep.23937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chun LJ, et al. Acetaminophen hepatotoxicity and acute liver failure. Journal of Clinical Gastroenterology. 2009;43:342–349. doi: 10.1097/MCG.0b013e31818a3854. [DOI] [PubMed] [Google Scholar]
- 18.Rhee SM, Garg VK, Hershey CO. Use of complementary and alternative medicines by ambulatory patients. Archives of Internal Medicine. 2004;164:1004–1009. doi: 10.1001/archinte.164.9.1004. [DOI] [PubMed] [Google Scholar]
- 19.Seeff LB. Herbal hepatotoxicity. Clinics in Liver Disease. 2007;11:577–596. vii. doi: 10.1016/j.cld.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 20.Lehman-McKeeman LD. Absorption, distribution, and excretion of toxicants. In: Casarett LJ, Doull J, et al., editors. Casarett and Doull’s Toxicology: The Basic Science of Poisons. 7. McGraw-Hill; New York: 2008. pp. 131–159. [Google Scholar]
- 21.Parkinson A, Ogilvie BW. Biotransformation of xenobiotics. In: Casarett LJ, Doull J, et al., editors. Casarett and Doull’s Toxicology: The Basic Science of Poisons. 7. McGraw-Hill; New York: 2008. pp. 161–304. [Google Scholar]
- 22.Testa B, Kramer SD. The biochemistry of drug metabolism – an introduction: part 3. Reactions of hydrolysis and their enzymes. Chemistry and Biodiversity. 2007;4:2031–2122. doi: 10.1002/cbdv.200790169. [DOI] [PubMed] [Google Scholar]
- 23.Testa B, Kramer SD. The biochemistry of drug metabolism – an introduction: part 2. Redox reactions and their enzymes. Chemistry and Biodiversity. 2007;4:257–405. doi: 10.1002/cbdv.200790032. [DOI] [PubMed] [Google Scholar]
- 24.Testa B, Kramer SD. The biochemistry of drug metabolism – an introduction: part 4. Reactions of conjugation and their enzymes. Chemistry and Biodiversity. 2008;5:2171–2336. doi: 10.1002/cbdv.200890199. [DOI] [PubMed] [Google Scholar]
- 25.Trauner M, Boyer JL. Bile salt transporters: molecular characterization, function, and regulation. Physiological Reviews. 2003;83:633–671. doi: 10.1152/physrev.00027.2002. [DOI] [PubMed] [Google Scholar]
- 26.Alrefai WA, Gill RK. Bile acid transporters: structure, function, regulation and pathophysiological implications. Pharmaceutical Research. 2007;24:1803–1823. doi: 10.1007/s11095-007-9289-1. [DOI] [PubMed] [Google Scholar]
- 27.Kosters A, Karpen SJ. Bile acid transporters in health and disease. Xenobiotica. 2008;38:1043–1071. doi: 10.1080/00498250802040584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nies AT, Schwab M, Keppler D. Interplay of conjugating enzymes with OATP uptake transporters and ABCC/MRP efflux pumps in the elimination of drugs. Expert Opinion on Drug Metabolism and Toxicology. 2008;4:545–568. doi: 10.1517/17425255.4.5.545. [DOI] [PubMed] [Google Scholar]
- 29.Pang KS, Maeng HJ, Fan J. Interplay of transporters and enzymes in drug and metabolite processing. Molecular Pharmaceutics. 2009;6:1734–1755. doi: 10.1021/mp900258z. [DOI] [PubMed] [Google Scholar]
- 30.Zamek-Gliszczynski MJ, et al. Integration of hepatic drug transporters and phase II metabolizing enzymes: mechanisms of hepatic excretion of sulfate, glucuronide, and glutathione metabolites. European Journal of Pharmaceutical Sciences. 2006;27:447–486. doi: 10.1016/j.ejps.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 31.Williams DP, Naisbitt DJ. Toxicophores: groups and metabolic routes associated with increased safety risk. Current Opinion in Drug Discovery and Development. 2002;5:104–115. [PubMed] [Google Scholar]
- 32.Kalgutkar AS, et al. A comprehensive listing of bioactivation pathways of organic functional groups. Current Drug Metabolism. 2005;6:161–225. doi: 10.2174/1389200054021799. [DOI] [PubMed] [Google Scholar]
- 33.Gregus Z. Mechenism of toxicity. In: Casarett LJ, Doull J, et al., editors. Casarett and Doull’s Toxicology: The Basic Science of Poisons. 7. McGraw-Hill; New York: 2008. pp. 45–106. [Google Scholar]
- 34.Testa B, Kramer SD. The biochemistry of drug metabolism – an introduction: part 5. Metabolism and bioactivity. Chemistry and Biodiversity. 2009;6:591–684. doi: 10.1002/cbdv.200900022. [DOI] [PubMed] [Google Scholar]
- 35.Mason RP, Chignell CF. Free radicals in pharmacology and toxicology – selected topics. Pharmacological Reviews. 1981;33:189–211. [PubMed] [Google Scholar]
- 36.Mason RP. Free radical intermediates in the metabolism of toxic chemicals. In: Pryor WA, editor. Free Radicals in Biology. Vol. 5. Academic Press; New York: 1982. pp. 161–222. [Google Scholar]
- 37.Valko M, Morris H, Cronin MT. Metals, toxicity and oxidative stress. Current Medicinal Chemistry. 2005;12:1161–1208. doi: 10.2174/0929867053764635. [DOI] [PubMed] [Google Scholar]
- 38.Aust SD, et al. Free radicals in toxicology. Toxicology and Applied Pharmacology. 1993;120:168–178. doi: 10.1006/taap.1993.1100. [DOI] [PubMed] [Google Scholar]
- 39.Halliwell B, Cross CE. Oxygen-derived species: their relation to human disease and environmental stress. Environmental Health Perspectives. 1994;102 (Suppl 10):5–12. doi: 10.1289/ehp.94102s105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ferrer-Sueta G, Radi R. Chemical biology of peroxynitrite: kinetics, diffusion, and radicals. ACS Chemical Biology. 2009;4:161–177. doi: 10.1021/cb800279q. [DOI] [PubMed] [Google Scholar]
- 41.Gutteridge JM. Biological origin of free radicals, and mechanisms of antioxidant protection. Chemico-biological Interactions. 1994;91:133–140. doi: 10.1016/0009-2797(94)90033-7. [DOI] [PubMed] [Google Scholar]
- 42.Circu ML, Aw TY. Reactive oxygen species, cellular redox systems, and apoptosis. Free Radical Biology and Medicine. 2010;48:749–762. doi: 10.1016/j.freeradbiomed.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zangar RC, Davydov DR, Verma S. Mechanisms that regulate production of reactive oxygen species by cytochrome P450. Toxicology and Applied Pharmacology. 2004;199:316–331. doi: 10.1016/j.taap.2004.01.018. [DOI] [PubMed] [Google Scholar]
- 44.Kovacic P, et al. Mechanism of mitochondrial uncouplers, inhibitors, and toxins: focus on electron transfer, free radicals, and structure–activity relationships. Current Medicinal Chemistry. 2005;12:2601–2623. doi: 10.2174/092986705774370646. [DOI] [PubMed] [Google Scholar]
- 45.Murphy MP. How mitochondria produce reactive oxygen species. Biochemical Journal. 2009;417:1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fritz R, et al. Compartment-dependent management of H(2)O(2) by peroxisomes. Free Radical Biology and Medicine. 2007;42:1119–1129. doi: 10.1016/j.freeradbiomed.2007.01.014. [DOI] [PubMed] [Google Scholar]
- 47.Davies KJ. Oxidative stress, antioxidant defenses, and damage removal, repair, and replacement systems. IUBMB Life. 2000;50:279–289. doi: 10.1080/713803728. [DOI] [PubMed] [Google Scholar]
- 48.Williams DP, et al. Are chemically reactive metabolites responsible for adverse reactions to drugs? Current Drug Metabolism. 2002;3:351–366. doi: 10.2174/1389200023337423. [DOI] [PubMed] [Google Scholar]
- 49.McCarver DG, Hines RN. The ontogeny of human drug-metabolizing enzymes: phase II conjugation enzymes and regulatory mechanisms. Journal of Pharmacology and Experimental Therapeutics. 2002;300:361–366. doi: 10.1124/jpet.300.2.361. [DOI] [PubMed] [Google Scholar]
- 50.van Bladeren PJ. Glutathione conjugation as a bioactivation reaction. Chemico-Biological Interactions. 2000;129:61–76. doi: 10.1016/s0009-2797(00)00214-3. [DOI] [PubMed] [Google Scholar]
- 51.Whalen R, Boyer TD. Human glutathione S-transferases. Seminars in Liver Disease. 1998;18:345–358. doi: 10.1055/s-2007-1007169. [DOI] [PubMed] [Google Scholar]
- 52.Dragovic S, et al. Role of human glutathione S-transferases in the inactivation of reactive metabolites of clozapine. Chemical Research in Toxicology. 2010;23:1467–1476. doi: 10.1021/tx100131f. [DOI] [PubMed] [Google Scholar]
- 53.Forman HJ, Zhang H, Rinna A. Glutathione: overview of its protective roles, measurement, and biosynthesis. Molecular Aspects of Medicine. 2009;30:1–12. doi: 10.1016/j.mam.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lu SC. Regulation of glutathione synthesis. Molecular Aspects of Medicine. 2009;30:42–59. doi: 10.1016/j.mam.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hansen JM, Go YM, Jones DP. Nuclear and mitochondrial compartmentation of oxidative stress and redox signaling. Annual Review of Pharmacology and Toxicology. 2006;46:215–234. doi: 10.1146/annurev.pharmtox.46.120604.141122. [DOI] [PubMed] [Google Scholar]
- 56.Jones DP. Redefining oxidative stress. Antioxidants and Redox Signaling. 2006;8:1865–1879. doi: 10.1089/ars.2006.8.1865. [DOI] [PubMed] [Google Scholar]
- 57.Circu ML, Aw TY. Glutathione and apoptosis. Free Radical Research. 2008;42:689–706. doi: 10.1080/10715760802317663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yuan L, Kaplowitz N. Glutathione in liver diseases and hepatotoxicity. Molecular Aspects of Medicine. 2009;30:29–41. doi: 10.1016/j.mam.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 59.Han D, et al. Mechanisms of liver injury. III. Role of glutathione redox status in liver injury. American Journal of Physiology. Gastrointestinal and Liver Physiology. 2006;291:G1–G7. doi: 10.1152/ajpgi.00001.2006. [DOI] [PubMed] [Google Scholar]
- 60.Kaplowitz N. Mechanisms of liver cell injury. Journal of Hepatology. 2000;32:39–47. doi: 10.1016/s0168-8278(00)80414-6. [DOI] [PubMed] [Google Scholar]
- 61.Biswas S, Chida AS, Rahman I. Redox modifications of protein-thiols: emerging roles in cell signaling. Biochemical Pharmacology. 2006;71:551–564. doi: 10.1016/j.bcp.2005.10.044. [DOI] [PubMed] [Google Scholar]
- 62.Kalinina EV, Chernov NN, Saprin AN. Involvement of thio-, peroxi-, and glutaredoxins in cellular redox-dependent processes. Biochemistry. 2008;73:1493–1510. doi: 10.1134/s0006297908130099. [DOI] [PubMed] [Google Scholar]
- 63.Srivastava A, et al. Role of reactive metabolites in drug-induced hepatotoxicity. Handbook of Experimental Pharmacology. 2010;196:165–194. doi: 10.1007/978-3-642-00663-0_7. [DOI] [PubMed] [Google Scholar]
- 64.Zhou S, et al. Drug bioactivation, covalent binding to target proteins and toxicity relevance. Drug Metabolism Reviews. 2005;37:41–213. doi: 10.1081/dmr-200028812. [DOI] [PubMed] [Google Scholar]
- 65.Cohen SD, et al. Selective protein covalent binding and target organ toxicity. Toxicology and Applied Pharmacology. 1997;143:1–12. doi: 10.1006/taap.1996.8074. [DOI] [PubMed] [Google Scholar]
- 66.Davern TJ, 2nd, et al. Measurement of serum acetaminophen-protein adducts in patients with acute liver failure. Gastroenterology. 2006;130:687–694. doi: 10.1053/j.gastro.2006.01.033. [DOI] [PubMed] [Google Scholar]
- 67.James LP, et al. Pharmacokinetics of acetaminophen-protein adducts in adults with acetaminophen overdose and acute liver failure. Drug Metabolism and Disposition. 2009;37:1779–1784. doi: 10.1124/dmd.108.026195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hanzlik RP, et al. The reactive metabolite target protein database (TPDB) – a web-accessible resource. BMC Bioinformatics. 2007;8:95. doi: 10.1186/1471-2105-8-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liebler DC. Protein damage by reactive electrophiles: targets and consequences. Chemical Research in Toxicology. 2008;21:117–128. doi: 10.1021/tx700235t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Thiele GM, Klassen LW, Tuma DJ. Formation and immunological properties of aldehyde-derived protein adducts following alcohol consumption. Methods in Molecular Biology. 2008;447:235–257. doi: 10.1007/978-1-59745-242-7_17. [DOI] [PubMed] [Google Scholar]
- 71.Rubino FM, et al. Toward an ‘omic’ physiopathology of reactive chemicals: thirty years of mass spectrometric study of the protein adducts with endogenous and xenobiotic compounds. Mass Spectrometry Reviews. 2009;28:725–784. doi: 10.1002/mas.20207. [DOI] [PubMed] [Google Scholar]
- 72.Benigni R. Structure-activity relationship studies of chemical mutagens and carcinogens: mechanistic investigations and prediction approaches. Chemical Reviews. 2005;105:1767–1800. doi: 10.1021/cr030049y. [DOI] [PubMed] [Google Scholar]
- 73.Kho R, et al. Ring systems in mutagenicity databases. Journal of Medicinal Chemistry. 2005;48:6671–6678. doi: 10.1021/jm050564j. [DOI] [PubMed] [Google Scholar]
- 74.Dizdaroglu M, et al. Free radical-induced damage to DNA: mechanisms and measurement. Free Radical Biology and Medicine. 2002;32:1102–1115. doi: 10.1016/s0891-5849(02)00826-2. [DOI] [PubMed] [Google Scholar]
- 75.Burcham PC. Genotoxic lipid peroxidation products: their DNA damaging properties and role in formation of endogenous DNA adducts. Mutagenesis. 1998;13:287–305. doi: 10.1093/mutage/13.3.287. [DOI] [PubMed] [Google Scholar]
- 76.Dix TA, Aikens J. Mechanisms and biological relevance of lipid peroxidation initiation. Chemical Research in Toxicology. 1993;6:2–18. doi: 10.1021/tx00031a001. [DOI] [PubMed] [Google Scholar]
- 77.Poli G, Biasi F, Leonarduzzi G. 4-Hydroxynonenal-protein adducts: a reliable biomarker of lipid oxidation in liver diseases. Molecular Aspects of Medicine. 2008;29:67–71. doi: 10.1016/j.mam.2007.09.016. [DOI] [PubMed] [Google Scholar]
- 78.Smathers RL, et al. Overview of lipid peroxidation products and hepatic protein modification in alcoholic liver disease. Chemico-biological Interactions. 2011;192:107–112. doi: 10.1016/j.cbi.2011.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Poli G, Albano E, Dianzani MU. The role of lipid peroxidation in liver damage. Chemistry and Physics of Lipids. 1987;45:117–142. doi: 10.1016/0009-3084(87)90063-6. [DOI] [PubMed] [Google Scholar]
- 80.Ghezzi P, Bonetto V, Fratelli M. Thiol-disulfide balance: from the concept of oxidative stress to that of redox regulation. Antioxidants and Redox Signaling. 2005;7:964–972. doi: 10.1089/ars.2005.7.964. [DOI] [PubMed] [Google Scholar]
- 81.Gallogly MM, Mieyal JJ. Mechanisms of reversible protein glutathionylation in redox signaling and oxidative stress. Current Opinion in Pharmacology. 2007;7:381–391. doi: 10.1016/j.coph.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 82.Schafer FQ, Buettner GR. Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. Free Radical Biology and Medicine. 2001;30:1191–1212. doi: 10.1016/s0891-5849(01)00480-4. [DOI] [PubMed] [Google Scholar]
- 83.Farout L, Friguet B. Proteasome function in aging and oxidative stress: implications in protein maintenance failure. Antioxidants and Redox Signaling. 2006;8:205–216. doi: 10.1089/ars.2006.8.205. [DOI] [PubMed] [Google Scholar]
- 84.Klatt P, Lamas S. Regulation of protein function by S-glutathiolation in response to oxidative and nitrosative stress. European Journal of Biochemistry. 2000;267:4928–4944. doi: 10.1046/j.1432-1327.2000.01601.x. [DOI] [PubMed] [Google Scholar]
- 85.Farber JL. Mechanisms of cell injury by activated oxygen species. Environmental Health Perspectives. 1994;102 (Suppl 10):17–24. doi: 10.1289/ehp.94102s1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Turrens JF. Superoxide production by the mitochondrial respiratory chain. Bioscience Reports. 1997;17:3–8. doi: 10.1023/a:1027374931887. [DOI] [PubMed] [Google Scholar]
- 87.Smathers RL, et al. Overview of lipid peroxidation products and hepatic protein modification in alcoholic liver disease. Chemico-biological Interactions. 2011;192:107–112. doi: 10.1016/j.cbi.2011.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Valerio LG, Jr, Petersen DR. Formation of liver microsomal MDA-protein adducts in mice with chronic dietary iron overload. Toxicology Letters. 1998;98:31–39. doi: 10.1016/s0378-4274(98)00100-3. [DOI] [PubMed] [Google Scholar]
- 89.Rudolph TK, Freeman BA. Transduction of redox signaling by electrophile-protein reactions. Science Signaling. 2009;2:re7. doi: 10.1126/scisignal.290re7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Beddowes EJ, Faux SP, Chipman JK. Chloroform, carbon tetrachloride and glutathione depletion induce secondary genotoxicity in liver cells via oxidative stress. Toxicology. 2003;187:101–115. doi: 10.1016/s0300-483x(03)00058-1. [DOI] [PubMed] [Google Scholar]
- 91.Czaja MJ. Cell signaling in oxidative stress-induced liver injury. Seminars in Liver Disease. 2007;27:378–389. doi: 10.1055/s-2007-991514. [DOI] [PubMed] [Google Scholar]
- 92.Allen RG, Tresini M. Oxidative stress and gene regulation. Free Radical Biology and Medicine. 2000;28:463–499. doi: 10.1016/s0891-5849(99)00242-7. [DOI] [PubMed] [Google Scholar]
- 93.Zima T, Kalousova M. Oxidative stress and signal transduction pathways in alcoholic liver disease. Alcoholism, Clinical and Experimental Research. 2005;29:110S–115S. doi: 10.1097/01.alc.0000189288.30358.4b. [DOI] [PubMed] [Google Scholar]
- 94.Wu D, Cederbaum AI. Oxidative stress and alcoholic liver disease. Seminars in Liver Disease. 2009;29:141–154. doi: 10.1055/s-0029-1214370. [DOI] [PubMed] [Google Scholar]
- 95.Ugarte N, Petropoulos I, Friguet B. Oxidized mitochondrial protein degradation and repair in aging and oxidative stress. Antioxidants and Redox Signaling. 2010;13:539–549. doi: 10.1089/ars.2009.2998. [DOI] [PubMed] [Google Scholar]
- 96.Lee S, Tsai FT. Molecular chaperones in protein quality control. Journal of Biochemistry and Molecular Biology. 2005;38:259–265. doi: 10.5483/bmbrep.2005.38.3.259. [DOI] [PubMed] [Google Scholar]
- 97.Zhang Y, et al. Lysosomal proteolysis is the primary degradation pathway for cytosolic ferritin and cytosolic ferritin degradation is necessary for iron exit. Antioxidants and Redox Signaling. 2010;13:999–1009. doi: 10.1089/ars.2010.3129. [DOI] [PubMed] [Google Scholar]
- 98.Breusing N, Grune T. Regulation of proteasome-mediated protein degradation during oxidative stress and aging. Biological Chemistry. 2008;389:203–209. doi: 10.1515/BC.2008.029. [DOI] [PubMed] [Google Scholar]
- 99.Christmann M, et al. Mechanisms of human DNA repair: an update. Toxicology. 2003;193:3–34. doi: 10.1016/s0300-483x(03)00287-7. [DOI] [PubMed] [Google Scholar]
- 100.Camenisch U, Naegeli H. Role of DNA repair in the protection against genotoxic stress. Exs. 2009;99:111–150. doi: 10.1007/978-3-7643-8336-7_5. [DOI] [PubMed] [Google Scholar]
- 101.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Molecular Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Barouki R. Linking long-term toxicity of xeno-chemicals with short-term biological adaptation. Biochimie. 2010;92:1222–1226. doi: 10.1016/j.biochi.2010.02.026. [DOI] [PubMed] [Google Scholar]
- 103.Xie W, et al. Orphan nuclear receptor-mediated xenobiotic regulation in drug metabolism. Drug Discovery Today. 2004;9:442–449. doi: 10.1016/S1359-6446(04)03061-2. [DOI] [PubMed] [Google Scholar]
- 104.Ramadoss P, Marcus C, Perdew GH. Role of the aryl hydrocarbon receptor in drug metabolism. Expert Opinion on Drug Metabolism and Toxicology. 2005;1:9–21. doi: 10.1517/17425255.1.1.9. [DOI] [PubMed] [Google Scholar]
- 105.Copple IM, et al. The keap1-nrf2 cellular defense pathway: mechanisms of regulation and role in protection against drug-induced toxicity. Handbook of Experimental Pharmacology. 2010;196:233–266. doi: 10.1007/978-3-642-00663-0_9. [DOI] [PubMed] [Google Scholar]
- 106.Shen G, Kong AN. Nrf2 plays an important role in coordinated regulation of phase II drug metabolism enzymes and phase III drug transporters. Biopharmaceutics and Drug Disposition. 2009;30:345–355. doi: 10.1002/bdd.680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Williams GM, Iatropoulos MJ. Alteration of liver cell function and proliferation: differentiation between adaptation and toxicity. Toxicologic Pathology. 2002;30:41–53. doi: 10.1080/01926230252824699. [DOI] [PubMed] [Google Scholar]
- 108.Green DR, Kroemer G. Cytoplasmic functions of the tumour suppressor p53. Nature. 2009;458:1127–1130. doi: 10.1038/nature07986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Orrenius S, Nicotera P, Zhivotovsky B. Cell death mechanisms and their implications in toxicology. Toxicological Sciences. 2011;119:3–19. doi: 10.1093/toxsci/kfq268. [DOI] [PubMed] [Google Scholar]
- 110.Pessayre D, et al. Mitochondrial involvement in drug-induced liver injury. Handbook of Experimental Pharmacology. 2010;196:311–365. doi: 10.1007/978-3-642-00663-0_11. [DOI] [PubMed] [Google Scholar]
- 111.Jones DP, et al. Mechanisms of pathogenesis in drug hepatotoxicity putting the stress on mitochondria. Molecular Interventions. 2010;10:98–111. doi: 10.1124/mi.10.2.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kass GE. Mitochondrial involvement in drug-induced hepatic injury. Chemico-biological Interactions. 2006;163:145–159. doi: 10.1016/j.cbi.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 113.Malhi H, Guicciardi ME, Gores GJ. Hepatocyte death: a clear and present danger. Physiological Reviews. 2010;90:1165–1194. doi: 10.1152/physrev.00061.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wei MC, et al. Proapoptotic BAX and BAK: a requisite gateway to mitochondrial dysfunction and death. Science. 2001;292:727–730. doi: 10.1126/science.1059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Joza N, et al. Essential role of the mitochondrial apoptosis-inducing factor in programmed cell death. Nature. 2001;410:549–554. doi: 10.1038/35069004. [DOI] [PubMed] [Google Scholar]
- 116.Jones BE, et al. Hepatocytes sensitized to tumor necrosis factor-alpha cytotoxicity undergo apoptosis through caspase-dependent and caspase-independent pathways. Journal of Biological Chemistry. 2000;275:705–712. doi: 10.1074/jbc.275.1.705. [DOI] [PubMed] [Google Scholar]
- 117.Lemasters JJ. V. Necrapoptosis and the mitochondrial permeability transition: shared pathways to necrosis and apoptosis. American Journal of Physiology. 1999;276:G1–G6. doi: 10.1152/ajpgi.1999.276.1.G1. [DOI] [PubMed] [Google Scholar]
- 118.Nagy LE. Recent insights into the role of the innate immune system in the development of alcoholic liver disease. Experimental Biology and Medicine (Maywood, NJ) 2003;228:882–890. doi: 10.1177/153537020322800803. [DOI] [PubMed] [Google Scholar]
- 119.Nath B, Szabo G. Alcohol-induced modulation of signaling pathways in liver parenchymal and nonparenchymal cells: implications for immunity. Seminars in Liver Disease. 2009;29:166–177. doi: 10.1055/s-0029-1214372. [DOI] [PubMed] [Google Scholar]
- 120.Depta JP, et al. Drug interaction with T-cell receptors: T-cell receptor density determines degree of cross-reactivity. Journal of Allergy and Clinical Immunology. 2004;113:519–527. doi: 10.1016/j.jaci.2003.11.030. [DOI] [PubMed] [Google Scholar]
- 121.Kaplowitz N. Biochemical and cellular mechanisms of toxic liver injury. Seminars in Liver Disease. 2002;22:137–144. doi: 10.1055/s-2002-30100. [DOI] [PubMed] [Google Scholar]
- 122.Holt MP, Ju C. Mechanisms of drug-induced liver injury. AAPS Journal. 2006;8:E48–E54. doi: 10.1208/aapsj080106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ju C, Uetrecht JP. Mechanism of idiosyncratic drug reactions: reactive metabolite formation, protein binding and the regulation of the immune system. Current Drug Metabolism. 2002;3:367–377. doi: 10.2174/1389200023337333. [DOI] [PubMed] [Google Scholar]
- 124.Adams DH, et al. Mechanisms of immune-mediated liver injury. Toxicological Sciences. 2010;115:307–321. doi: 10.1093/toxsci/kfq009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Pauli-Magnus C, Meier PJ. Hepatobiliary transporters and drug-induced cholestasis. Hepatology. 2006;44:778–787. doi: 10.1002/hep.21359. [DOI] [PubMed] [Google Scholar]
- 126.Kosters A, Karpen SJ. The role of inflammation in cholestasis: clinical and basic aspects. Seminars in Liver Disease. 2010;30:186–194. doi: 10.1055/s-0030-1253227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Geier A, et al. Principles of hepatic organic anion transporter regulation during cholestasis, inflammation and liver regeneration. Biochimica et Biophysica Acta. 2007;1773:283–308. doi: 10.1016/j.bbamcr.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 128.Pauli-Magnus C, Meier PJ, Stieger B. Genetic determinants of drug-induced cholestasis and intrahepatic cholestasis of pregnancy. Seminars in Liver Disease. 2010;30:147–159. doi: 10.1055/s-0030-1253224. [DOI] [PubMed] [Google Scholar]
- 129.Kaplowitz N. Idiosyncratic drug hepatotoxicity. Nature Reviews. Drug Discovery. 2005;4:489–499. doi: 10.1038/nrd1750. [DOI] [PubMed] [Google Scholar]
- 130.Zhang X, et al. Involvement of the immune system in idiosyncratic drug reactions. Drug Metabolism and Pharmacokinetics. 2011;26:47–59. doi: 10.2133/dmpk.dmpk-10-rv-085. [DOI] [PubMed] [Google Scholar]
- 131.Uetrecht J. Idiosyncratic drug reactions: past, present, and future. Chemical Research in Toxicology. 2008;21:84–92. doi: 10.1021/tx700186p. [DOI] [PubMed] [Google Scholar]
- 132.Li J, Uetrecht JP. The danger hypothesis applied to idiosyncratic drug reactions. Handbook of Experimental Pharmacology. 2010;196:493–509. doi: 10.1007/978-3-642-00663-0_18. [DOI] [PubMed] [Google Scholar]
- 133.Daly AK. Drug-induced liver injury: past, present and future. Pharmacogenomics. 2010;11:607–611. doi: 10.2217/pgs.10.24. [DOI] [PubMed] [Google Scholar]
- 134.Smith DA, Obach RS. Metabolites in safety testing (MIST): considerations of mechanisms of toxicity with dose, abundance, and duration of treatment. Chemical Research in Toxicology. 2009;22:267–279. doi: 10.1021/tx800415j. [DOI] [PubMed] [Google Scholar]
- 135.Uetrecht JP. New concepts in immunology relevant to idiosyncratic drug reactions: the ‘danger hypothesis’ and innate immune system. Chemical Research in Toxicology. 1999;12:387–395. doi: 10.1021/tx980249i. [DOI] [PubMed] [Google Scholar]
- 136.Zhou SF. Drugs behave as substrates, inhibitors and inducers of human cytochrome P450 3A4. Current Drug Metabolism. 2008;9:310–322. doi: 10.2174/138920008784220664. [DOI] [PubMed] [Google Scholar]
- 137.Nelson DR. The cytochrome p450 homepage. Human Genomics. 2009;4:59–65. doi: 10.1186/1479-7364-4-1-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Martinez-Jimenez CP, et al. Transcriptional regulation and expression of CYP3A4 in hepatocytes. Current Drug Metabolism. 2007;8:185–194. doi: 10.2174/138920007779815986. [DOI] [PubMed] [Google Scholar]
- 139.Liu Y, et al. The effects of splicing variant of PXR PAR-2 on CYP3A4 and MDR1 mRNA expressions. Clinica Chimica Acta. 2009;403:142–144. doi: 10.1016/j.cca.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 140.Koyano S, et al. Functional characterization of four naturally occurring variants of human pregnane X receptor (PXR): one variant causes dramatic loss of both DNA binding activity and the transactivation of the CYP3A4 promoter/enhancer region. Drug Metabolism and Disposition. 2004;32:149–154. doi: 10.1124/dmd.32.1.149. [DOI] [PubMed] [Google Scholar]
- 141.Tompkins LM, Sit TL, Wallace AD. Unique transcription start sites and distinct promoter regions differentiate the pregnane X receptor (PXR) isoforms PXR 1 and PXR 2. Drug Metabolism and Disposition. 2008;36:923–929. doi: 10.1124/dmd.107.018317. [DOI] [PubMed] [Google Scholar]
- 142.Ingelman-Sundberg M. The human genome project and novel aspects of cytochrome P450 research. Toxicology and Applied Pharmacology. 2005;207:52–56. doi: 10.1016/j.taap.2005.01.030. [DOI] [PubMed] [Google Scholar]
- 143.Zanger UM, et al. Functional pharmacogenetics/genomics of human cytochromes P450 involved in drug biotransformation. Analytical and Bioanalytical Chemistry. 2008;392:1093–1108. doi: 10.1007/s00216-008-2291-6. [DOI] [PubMed] [Google Scholar]
- 144.Sim SC, Ingelman-Sundberg M. The human cytochrome P450 (CYP) allele nomenclature website: a peer-reviewed database of CYP variants and their associated effects. Human Genomics. 2010;4:278–281. doi: 10.1186/1479-7364-4-4-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Correia MA, Sinclair PR, De Matteis F. Cytochrome P450 regulation: the interplay between its heme and apoprotein moieties in synthesis, assembly, repair, and disposal. Drug Metabolism Reviews. 2011;43:1–26. doi: 10.3109/03602532.2010.515222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hernandez JP, Mota LC, Baldwin WS. Activation of CAR and PXR by dietary, environmental and occupational chemicals alters drug metabolism, intermediary metabolism, and cell proliferation. Current Pharmacogenomics and Personalized Medicine. 2009;7:81–105. doi: 10.2174/187569209788654005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zollner G, et al. Role of nuclear receptors in the adaptive response to bile acids and cholestasis: pathogenetic and therapeutic considerations. Molecular Pharmaceutics. 2006;3:231–251. doi: 10.1021/mp060010s. [DOI] [PubMed] [Google Scholar]
- 148.Lynch T, Price A. The effect of cytochrome P450 metabolism on drug response, interactions, and adverse effects. American Family Physician. 2007;76:391–396. [PubMed] [Google Scholar]
- 149.Pelkonen O, et al. Inhibition and induction of human cytochrome P450 enzymes: current status. Archives of Toxicology. 2008;82:667–715. doi: 10.1007/s00204-008-0332-8. [DOI] [PubMed] [Google Scholar]
- 150.Hakkola J, Tanaka E, Pelkonen O. Developmental expression of cytochrome P450 enzymes in human liver. Pharmacology and Toxicology. 1998;82:209–217. doi: 10.1111/j.1600-0773.1998.tb01427.x. [DOI] [PubMed] [Google Scholar]
- 151.Hines RN. Ontogeny of human hepatic cytochromes P450. Journal of Biochemical and Molecular Toxicology. 2007;21:169–175. doi: 10.1002/jbt.20179. [DOI] [PubMed] [Google Scholar]
- 152.Wolbold R, et al. Sex is a major determinant of CYP3A4 expression in human liver. Hepatology. 2003;38:978–988. doi: 10.1053/jhep.2003.50393. [DOI] [PubMed] [Google Scholar]
- 153.Daly AK, et al. HLA-B*5701 genotype is a major determinant of drug-induced liver injury due to flucloxacillin. Nature Genetics. 2009;41:816–819. doi: 10.1038/ng.379. [DOI] [PubMed] [Google Scholar]
- 154.Evans DC, et al. Drug-protein adducts: an industry perspective on minimizing the potential for drug bioactivation in drug discovery and development. Chemical Research in Toxicology. 2004;17:3–16. doi: 10.1021/tx034170b. [DOI] [PubMed] [Google Scholar]
- 155.Coles B. Effects of modifying structure on electrophilic reactions with biological nucleophiles. Drug Metabolism Reviews. 1984;15:1307–1334. doi: 10.3109/03602538409029962. [DOI] [PubMed] [Google Scholar]
- 156.Kalgutkar AS. Handling reactive metabolite positives in drug discovery: what has retrospective structure-toxicity analyses taught us? Chemico-biological Interactions. 2011;192:46–55. doi: 10.1016/j.cbi.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 157.Thompson RA, et al. Risk assessment and mitigation strategies for reactive metabolites in drug discovery and development. Chemico-biological Interactions. 2011;192:65–71. doi: 10.1016/j.cbi.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 158.Halegoua-De Marzio D, Navarro VJ. Drug-induced hepatotoxicity in humans. Current Opinion in Drug Discovery and Development. 2008;11:53–59. [PubMed] [Google Scholar]
- 159.Chang LW, et al. Macromolecular adducts: biomarkers for toxicity and carcinogenesis. Annual Review of Pharmacology and Toxicology. 1994;34:41–67. doi: 10.1146/annurev.pa.34.040194.000353. [DOI] [PubMed] [Google Scholar]
- 160.Consortium TIH. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Durbin RM, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Birney E, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
Further reading, resources and contacts
- Gregus Z. Mechanism of toxicity. In: Casarett LJ, Doull J, et al., editors. Casarett and Doull’s Toxicology: The Basic Science of Poisons. 7. McGraw-Hill; New York: 2008. pp. 45–106. This book chapter provides the most comprehensive and detailed information on the molecular and cellular mechanism of toxicity by xenobiotics. [Google Scholar]
- Kaplowitz N. Biochemical and cellular mechanisms of toxic liver injury. Seminars in Liver Disease. 2002;22:137–144. doi: 10.1055/s-2002-30100. This review focuses on the mechanisms of different forms of cell death. [DOI] [PubMed] [Google Scholar]
- Abboud G, Kaplowitz N. Drug-induced liver injury. Drug Safety. 2007;30:277–294. doi: 10.2165/00002018-200730040-00001. This review provides practical information on the diagnosis and management of patients with suspected drug-related liver disease. Pathogenesis is also discussed briefly. [DOI] [PubMed] [Google Scholar]
- Testa B, Kramer SD. The biochemistry of drug metabolism – an introduction: parts 2–5. Redox, hydrolysis, conjugation reactions and their enzymes, metabolism and bioactivity. Chemistry and Biodiversity. 2007–2009;4456:257–405. 2031–2122, 2171–2336, 591–684. doi: 10.1002/cbdv.200790032. [DOI] [PubMed] [Google Scholar]
- Parkinson A, Ogilvie BW. Biotransformation of xenobiotics. In: Casarett LJ, Doull J, et al., editors. Casarett and Doull’s Toxicology: The Basic Science of Poisons. 7. McGraw-Hill; New York: 2008. pp. 161–304. The reviews by Test and Kramer and the book chapter by Parkinson and Ogilvie demonstrate comprehensive and detailed information on detoxification and bio-activation of xenobiotics. [Google Scholar]
- Hinson JA, Roberts DW, James LP. Mechanisms of acetaminophen-induced liver necrosis. Handbook of Experimental Pharmacology. 2010;196:369–405. doi: 10.1007/978-3-642-00663-0_12. This review introduces the molecular mechanism of chemical-induced liver injury using a model compound acetaminophen, which is a leading cause of drug-induced liver injury. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun LJ, et al. Acetaminophen hepatotoxicity and acute liver failure. Journal of Clinical Gastroenterology. 2009;43:342–349. doi: 10.1097/MCG.0b013e31818a3854. This review provides information useful for clinical management of acetaminophen liver injury. [DOI] [PubMed] [Google Scholar]
- Daly AK. Drug-induced liver injury: past, present and future. Pharmacogenomics. 2010;11:607–611. doi: 10.2217/pgs.10.24. This review introduces research on the association between certain genetic polymorphisms of the immune system and drug metabolism enzymes and drug-induced liver injury. [DOI] [PubMed] [Google Scholar]
- Gu X, et al. Role of NF-kappaB in regulation of PXR-mediated gene expression: a mechanism for the suppression of cytochrome P-450 3A4 by proinflammatory agents. Journal of Biological Chemistry. 2006;281:17882–17889. doi: 10.1074/jbc.M601302200. This research paper provides an example for a possible molecular mechanism by which the expression of a specific drug metabolism enzyme is regulated under inflammation conditions. [DOI] [PubMed] [Google Scholar]