Abstract
The direct sampling feature of liquid sample desorption electrospray ionization (DESI) allows the ionization of liquid samples without adding acids/organic solvents (i.e., without sample pretreatment). As a result, it provides a new approach for probing protein conformation in solution. In this study, it has been observed that native protein ions are generated from proteins in water by DESI. Interestingly, the intensities of the resulting protein ions appear to be higher than those generated by ESI of the proteins in water or in ammonium acetate. For protein solutions that already contain acids/organic solvents, DESI can be used to investigate the influences of these denaturants on protein conformations and the obtained results are in good agreement with spectroscopic data. In addition, online monitoring of protein conformational changes by DESI is feasible; for instance, heat-induced unfolding of ubiquitin can be traced with DESI in water without influences of organic solvents/acids. This DESI method provides a new alternative tool for the study of protein conformation in solution.
The study of protein conformational structure is important as the conformational change can occur to regulate the functions of enzymes and receptors [1]. Besides traditional methods to investigate protein conformations, such as circular dichroism (CD), NMR, and X-Ray, mass spectrometry (MS) is also useful for this purpose due to its high sensitivity [2–4], especially with the advent of soft electrospray ionization (ESI) [5] technique allowing the preservation of noncovalent interactions of proteins during ionization. In the acquired ESI mass spectra of proteins, the charge state distribution (CSD) of protein ions and its width are indicative to the degree of protein unfolding [6–9]. Typically, when a protein is in the folded structure, a narrow CSD in low charge states is observed, while CSD is broadened and shifted to high charge states after unfolding. This is probably because in comparison to a folded protein, the unfolded one has a greater capacity to accommodate a significantly large number of charges on its surface [3, 10, 11]. Therefore, information about the conformational states of the protein can often be extracted based on the structural interpretation of CSDs in ESI-MS, upon controlling other experimental conditions [4].
Ambient mass spectrometry [12] such as desorption electrospray ionization (DESI) [13] and direct analysis in real time (DART) [14] have recently been introduced to provide direct ionization of analytes with little or no sample preparation. It has been shown that DESI is successful in the fast analysis of a variety of different analytes ranging from pharmaceuticals to tissue imaging [15–17]. In addition to being used regularly for solid sample analysis on surface, DESI has been extended to allow the direct analysis of liquid samples [18–22]. In the experiments, ionization of the liquid sample occurs via interactions of the sample with charged microdroplets generated in a pneumatically assisted DESI spray and subsequent desolvation of the resulting secondary microdroplets containing the sample analyte. It has been shown that liquid sample DESI enables convenient on-line coupling of MS with electrochemistry [19, 23] and microfluidics [20], and it can be used for the direct ionization of a wide range of compounds including amino acids, peptides, protein digests and, particularly, high-mass proteins [19] from solution without sample pretreatment. In this sense, liquid sample DESI differs from traditional ESI in that proteins in solution (e.g., in water) can be direct desorbed and ionized without adding acids or organic solvents into samples. Such a direct sampling feature of DESI (i.e., ionizing the sample as it is) is advantageous in studying protein conformation, as those chemical additives or “makeup solvent” used in traditional ESI for obtaining a stable spray or high sensitivity could change protein conformations before ionization (therefore, to preserve the native protein conformation while maintaining high sensitivity, native ESI MS [24] has been introduced in which proteins are ionized from aqueous solutions containing volatile buffers such as ammonium acetate).
In this study, we first examined the DESI ionization of protein in water or ammonium acetate and it turns out that folded protein ions with narrow CSDs in low charge states are generated. With the addition of organic solvents or acids to protein or peptide solutions, the conformational changes can also be detected based on the recorded DESI spectra. In addition, by heating the protein sample solution in the DESI experiment, on-line monitoring of heat-induced protein conformational changes can be performed, providing a simple and fast way to examine the temperature effect on protein conformations.
Experimental
Materials
Cytochrome c, myoglobin, lysozyme, ubiquitin, and human β-endorphin were purchased from Sigma-Aldrich (St. Louis, MO, USA) and used without further purification. The deionized water used for sample preparation was obtained using a Nanopure Diamond Barn-stead purification system (Barnstead International, Dubuque, IA, USA). High-performance liquid chromatography grade methanol was purchased from GFS Chemicals (Columbus, OH, USA), and glacial acetic acid was purchased from Fisher Chemicals (Fair Lawn, NJ, USA).
Apparatus
The liquid sample DESI source used in this study was the same as described previously [19, 23], in which a beam of charged microdroplets from the DESI spray probe was directed onto the outlet of a sample introduction capillary for desorption and ionization. The sample introduction capillary outlet can be placed on a Teflon surface (apparatus a, Figure S–1a, Supporting Information, which can be found in the electronic version of this article) [19], or can be simply put in between the DESI spray probe and the mass spectrometer interface (apparatus b, Figure S–1b, Supporting Information) [23]. The DESI spray probe is virtually an electrosonic spray ionization (ESSI [25], a variant form of ESI) source, in which the high voltage (+5 kV) and high-pressure nebulizing nitrogen gas (160 psi) were used to generate the charged microdroplet beam for sample desorption and ionization. The DESI spray solvent used was methanol/water/acetic acid (50:50:1 by volume) and injected at the flow rate of 10 μL/min. Experiments were mainly carried out using either a LCQ DECA or DECA MAX mass spectrometer (Thermo Finnigan, San Jose, CA, USA) with a heated capillary temperature kept at 150 °C. Protein solutions (6–12 μM) were prepared by dissolving solid protein samples into deionized water and introduced at the flow rate of 5 μL/min for DESI ionization. For on-line monitoring of heat-induced protein conformational changes, a heating tape was used to heat the sample solution flowing through the sample introduction capillary to different temperatures, which were monitored with a digital Omega thermocouple sensor.
In comparison with DESI, ESSI of proteins was also performed, in which proteins in water were mixed with methanol/water/acetic acid (50:50:1 by volume) and then underwent ionization. A high voltage (+5 kV) and high-pressure nebulizing nitrogen gas (160 psi) were used for the ESSI ionization and the MS instrument conditions were kept the same as in the corresponding DESI experiments. In the sensitivity comparison with DESI, ESI experiments were performed using the commercial ESI ion sources of DECA or DECA MAX. The UV absorption experiments were performed on a SpectraMax spectrofluorometer (Molecular Devices, Sunnyvale, CA, USA) and a series of aqueous cytochrome c samples (8 μM) with different pH values adjusted by acetic acid were measured with three replicates at both 394 and 409 nm. The absorbance data were processed using a SoftMax Pro.4.8.
Results and Discussion
In this study, we used DESI to directly sample and ionize various proteins under different conditions, such as in pure water, in aqueous solution containing ammonium acetate, acid, or organic solvent, or at different temperatures. The obtained spectra were analyzed using CSD as indicators to provide protein conformational information in solution.
Generation of Native Protein Ions from the Native Environment
In our experiments, cytochrome c in water was first chosen for DESI analysis. As shown in Figure 1a, when the sample was ionized with the DESI spray solvent containing acetic acid and methanol (MeOH/H2O/HOAc = 50:50:1 by volume), a narrow CSD in low charge states (+7 to +10) of the protein ions was observed, suggesting the generation of folded native protein ions [9, 26, 27] in DESI. In addition, native protein ions were also observed in the DESI of cytochrome c in 100 mM ammonium acetate (Figure S-2, Supporting Information). By contrast, if the spray solvent was mixed with the protein sample and subsequently ionized by ESSI, the spectrum (Figure 1b) displays the appearance of higher charged ions centered at +15, indicating the occurrence of denaturation of the protein due to the high content of denaturants of acetic acid and methanol present in the sample solution (the bimodal distributions in Figure 1b, one centered at +8 and the other at +15, is due to the presence of the two conformer populations, native and denatured cytochrome c [27]). Likely, the difference between the DESI and ESSI spectra is caused by the limited interac tion time of DESI (estimated to be ca. 1 ms, see the discussion in the Supporting Information) between the spray solvent and the protein during DESI ionization, which does not allow the protein to have sufficient time to unfold. Another possibility for the generation of native protein ions in the liquid sample DESI is that the sprayed primary microdroplets might not be completely mixed with the protein solution within the short ionization process so that the protein was not well exposed to the denaturants contained in the microdroplets. However, in the ESSI analysis, the mixing of protein in water with denaturing solvent before ionization allows enough time (practically at least many seconds) for protein denaturation. This contrast between DESI and ESSI analysis is in agreement with a recent study [28] showing that labile aerosol chemicals can be preserved from acid hydrolysis and detected using DESI due to the minimized ionization time while ESI failed in the detection. It is also analogous to an earlier observation of the generation of native protein ions by multiple channel electrospray ionization (MC-ES) [29] in which two sprayed beams containing proteins and methanol/water/acetic acid, respectively, were interacted for ionization.
Figure 1.
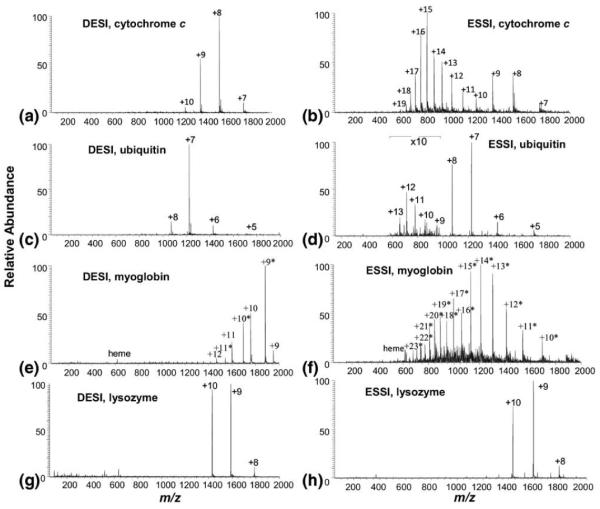
DESI MS spectra of (a) cytochrome c, (c) ubiquitin, (e) myoglobin, and (g) lysozyme; ESSI MS spectra of (b) cytochrome c, (d) ubiquitin, (f) myoglobin, and (h) lysozyme. Experimental conditions: in the DESI experiment, a protein in H2O (6–12 μM) was injected at the flow rate of 5 μL/min and the DESI spray solution was MeOH/H2O/HOAc(50:50:1 by volume) and sprayed at the flow rate of 10 μL/min; in the ESSI experiment, the same protein sample was first mixed with the DESI spray solvent of MeOH/H2O/HOAc(50:50:1 by volume) in the ratio of 1:2 by volume and then directly sprayed by ESSI. The charge numbers labeled with an asterisk correspond to the apomyoglobin ions.
In traditional DESI, in which a solid protein sample on a surface was analyzed, the resulting protein ions were highly dependent on the composition of the spray solvent [30] in which the native-like spray solvent of methanol/water allowed the generation of native protein ions, while the denaturing spray solvent of methanol/water/acetic acid gave rise to denatured protein ions. It is probably because during the traditional solid sample DESI analysis, the ionization commences with the dissolution of the solid protein sample by spray solvent (known as “surface wetting” [31, 32]) so that the native-like solvent can recover the protein conformations but denaturing solvent with methanol and acid can not.
Other proteins were also tested with the liquid sample DESI. As expected, in the case of ubiquitin, DESI generated the native protein ions of +5 to +8 [27] (Figure 1c), while ESSI gave rise to unfolded ions of +9 to +13 (Figure 1d). The relative low abundance of the unfolded ions of +9 to +13 is ascribed to the conformational stability of ubiquitin to denaturation because of its pronounced hydrophobic core and extensive intramolecular hydrogen bonding involving 90% residues [27]. For myoglobin, again, the native myoglobin ions [29, 33] with narrow CSDs from +9 to +12 were observed in DESI (Figure 1e), along with folded apomyoglobin ions (labeled with asterisks in the spectrum). The origin of the latter was probably from in-source collision-induced dissociation (CID) in the region between the outlet of the heated capillary and skimmer of the DECA instrument used, which induced the loss of heme group under the transmission process of the native myoglobin ions in this region [34]. Evidence to support this assumption is that ESI of myoglobin in water using the same instrument also produced apomyoglobin ions (Figure S-4d, Supporting Information). Furthermore, we tested the DESI analysis of myoglobin in water using a different instrument of LTQ-Orbitrap, which mainly generated the native myoglobin ions with negligible amount of apo-form ions (Figure S-3, Supporting Information). By contrast, the high charge state distribution corresponding to the unfolded conformer of apomyoglobin [7, 35] was obtained in ESSI (Figure 1f, the noncovalently bounded heme group was lost in the resulting apomyoglobin ions probably due to acid-induced protein unfolding [36]). For the lysozyme, no apparent difference in CSDs was seen between the ESSI and DESI analysis (Figure 1g and h). Likely, the four disulfide bonds of the protein hold the protein in a folded structure, in which the regular denaturants (methanol or acid) can not break the disulfide bonds so that it often displays the same CSD in both the native and non-native environments [30].
We also examined the sensitivity of the generation of the native protein ions using DESI. The DESI and ESI of proteins such as cytochrome c and myoglobin in water were carried out at the same day for comparison. It was found that the DESI sensitivity is about 60 to 70 times higher than that obtained by ESI analysis of protein in pure water for the generation of native protein ions (Figure S-4, Supporting Information). In DESI, the organic solvent of methanol used in the DESI spray helps the desolvation of the charged microdroplets and the acid used provides sufficient protons for the protein ionization, improving the ionization sensitivity. However, in the ESI analysis of the protein in pure water, in the absence of the methanol or acid, the desolvation or protonation was not sufficient and the spray might not be stable either, thus reducing the ionization efficiency. In addition, we also performed the comparison of liquid DESI of cytochrome c (10 μM) in water with ESI of the protein (10 μM) in 100 μM ammonium acetate; again, the former is more sensitive than the latter by ca. five times. Therefore, liquid sample DESI reported in this study provides a method of choice for generating native protein ions [33, 37], with simplicity and high sensitivity.
Probe Protein Conformation in Non-Native Envoirnment
As a direct sampling method, DESI can also be used to probe the protein conformations in a solution that already contains denaturants such as organic solvents or acids so that the effects of these denaturants on protein conformations can be investigated. A series of cytochrome c samples in the different pHs were prepared by adding acetic acid into aqueous samples and then analyzed by DESI. In the Figure 2a–c showing the different CSDs of these samples, one can see that there is a conformation transition of cytochrome c occurring at the pH of 2.6 (i.e., 2% acetic acid in water). This result is exactly in agreement with the UV-absorption measurements of these protein solutions (Figure 2d) in which the abrupt change in UV absorption takes place at 2% acetic acid. The coordination of heme iron with its native His18 and Met80 ligands leads to a Soret absorption maximum at 409 nm. Displacement of these ligands upon acid-induced unfolding of the protein induces a blue shift to 394 nm [26, 38]. The agreement between the two assays suggests that DESI can be used to study protein conformations in different solution environments.
Figure 2.
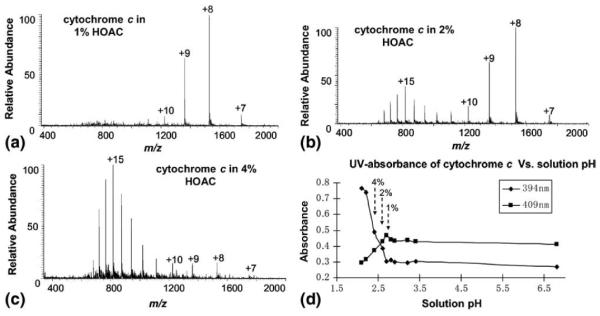
DESI MS spectra of cytochrome c (8 μM) in (a) 1% HOAc, (pH = 2.7), (b) 2% HOAc (pH = 2.6), and (c) 4% HOAc (pH = 2.4); (d) UV spectrum showing the change of absorbances of cytochrome c solution with the solution pH (i.e., the amount of added acetic acid). The 10 data points were obtained from ten 8 μM aqueous cytochrome c samples with different percentages of acetic acid (0%, 0.05%, 0.1%, 0.5%, 0.8%, 1%, 2%, 4%, 6%, and 8%). The converting point for the absorbance lines of 394 and 409 nm corresponds to the sample containing 2% acetic acid.
Another example in this regard is the study of the methanol induced conformational change of a polypeptide β-endorphin, an endogenous opioid that acts as a neuropeptide in the central nervous system [39], by DESI. β-Endorphin has six basic sites consisting of five lysine residues and one N-terminal amino group. With increased methanol component in the β-endorphin solution, the most intense peak shifted from +5 to +4 and the relative abundance of the +6 ions decreased with the general intensity increase trends observed for +2 and +3 ions (Figure 3). Table 1 gives the relative abundance of all ions measured under different amounts of methanol in solution. It is known that the alcohol such as methanol can induce helicity of β-endorphin, which will further reduce the number of accessible basic sites in peptides [39]. In other words, β-endorphin has a more compact structure in methanol/ water than in water. This fact agrees well with the observation of the CSD shift from high to low charge states with the increase of the methanol content in solution, emphasizing that organic solvent-induced peptide conformational changes can also be probed by DESI. Also, the detected conformational changes in this case are exclusively ascribed to methanol, without the influences of other denaturation factors such as pH.
Figure 3.
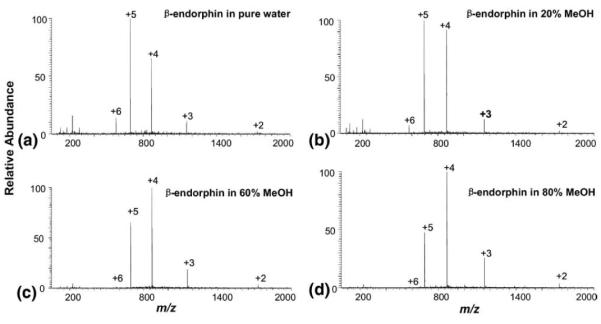
DESI-MS spectra of 10 μM β-endorphin in water containing (a) 0% MeOH, (b) 20%MeOH, (c) 60% MeOH, and (d) 80% MeOH. The DESI spray was composed of MeOH/H2O/HOAc (50:50:1, by volume).
Table 1.
Relative abundance of β-endorphin as a function of concentration of methanol
| Methanol concentration (%) | % Relative abundance of charge states |
||||
|---|---|---|---|---|---|
| +6 | +5 | +4 | +3 | +2 | |
| 0 | 7.22 | 51.55 | 34.02 | 6.19 | 1.03 |
| 20 | 3.73 | 46.58 | 42.85 | 5.68 | 1.16 |
| 60 | 1.05 | 34.65 | 52.49 | 9.71 | 2.1 |
| 80 | 0.56 | 26.82 | 55.87 | 14.53 | 2.23 |
In another similar experiment, it was found that methanol-induced conformational change of ubiquitin in aqueous solution can also be monitored by DESI. In the liquid sample DESI spectrum of 12 μM aqueous ubiquitin, the narrow CSD from +5 to +8 representing the native conformation was observed (Figure S-5a, Supporting Information). With the increasing percent of methanol in the solution to 20%, 50%, there was no pronounced change for the CSD (Figure S-5b and S-5c, Supporting Information). This observation is in line with the previous study which shows the generation of the folded ubiquitin ions from ubiquitin in aqueous solution containing 60% methanol at pH 7.2 by ESI [40]. It is also in good agreement with the recent study of electrospray-assisted laser desorption ionization (ELDI) [37]. When the concentration of the methanol increased to 80%, ubiquitin unfolded with the CSD shifted to the higher charge state distribution centered at +12 (Figure S-5d, Supporting Information).
Online Monitoring Heat-Induced Protein Conformational Changes
Liquid sample DESI is also ideal for on-line monitoring of protein conformational changes in solution and the instrumentation involved in the experiments is simple. As a demonstration, we chose to examine the denaturing effect of heat on protein conformations. In our study, the experiments were carried out simply by using a modified sample introduction capillary wrapped with a heating tape. As the protein sample was injected through the capillary for DESI ionization, it was heated up to a certain temperature and the corresponding MS spectrum was recorded. Figure 4a shows the acquired DESI mass spectrum of ubiquitin in water (pH = 6.0) at 25 °C and the native ions of +6 to +8 were observed. As the temperature of the protein sample increased to 73 °C, another peak distribution of +9 to +13 appeared, corresponding to the unfolded protein ions (Figure 4b). However, the relative intensities of the ions of +9 to +13 are low, which is consistent with the observation of the protein conformational stability toward acids/organic solvents shown in Figure 1d and indicates the high unfolding energy of this protein. With the further increase of the temperature, the intensities of these unfolded protein ions increased accordingly. At 94 °C, the unfolded protein ions dominated in the spectrum (Figure 4c), suggesting substantial unfolding of the protein at this temperature [27]. The heat denaturation transition curve for ubiquitin, a plot of [F]/([F] + [U]) as a function of temperature [27], is also displayed in Figure 4d (F designates the tightly folded form of ubiquitin and U designates the unfolded form of the protein; the values of F and U were calculated by summing the heights of the spectral peaks corresponding to ions +6 to +8 and +9 to +13, respectively). From this plot, the transition temperature was estimated to be 80 °C. These results showed that the liquid sample DESI can be used for on-line monitoring of protein conformational changes with simple instrumentation. In a previous report of heat-induced ubiquitin conformational changes using ESI [27], as protein solutions were prepared in different pHs, the conformational changes observed were contributed to a combination of heat and acid effects. In this DESI experiment, since proteins in pure water were used, the conformational changes arose exclusively from heat.
Figure 4.
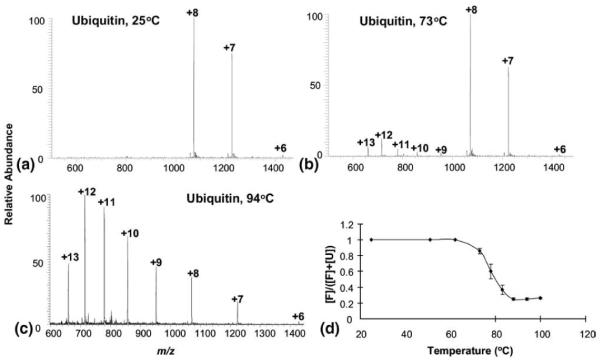
DESI MS spectra of 10 μM ubiquitin in water at different temperatures of: (a) 25 °C; (b) 73 °C, and (c) 94 °C (background subtracted); (d) denaturation transition curves for ubiquitin plotted as a function of temperature.
Conclusions
In conclusion, we have shown the ionization characteristics of liquid sample DESI for protein samples and have demonstrated novel applications of the liquid sample DESI for the protein conformation study by taking advantage of its direct sampling nature. It has been found that native protein ions can be generated from the aqueous solution with high sensitivity using DESI. In addition, DESI can be used to examine protein conformations in non-native environments and to probe conformation changes in solution, such as organic solvent or heat-induced protein unfolding. Given the simplicity and fast ionization nature of liquid DESI as well as the importance of protein conformation study, this work would be able to find useful applications in bioanalysis, such as the detection of labile protein complexes, metalloenzymes, and multiprotein assemblies, which are sensitive to acids or organic solvents.
The study reported in this paper was originally presented at the ASMS conference in 2009 [41]. During the revision of this paper, we also noticed a latest publication [42] reporting the generation of native ions using another ambient ionization method of extractive electrospray ionization.
Supplementary Material
Acknowledgments
The authors acknowledge support for this work by NSF (CHE-0911160). The authors gratefully acknowledge Professor Michael L. Gross at Washington University in St. Louis for the access to the NIH/NCRR Mass Spectrometry Resources (grant number 2P41RR000954) and are also thankful to Mr. Hao Zhang for his kind help.
Footnotes
Appendix A Supplementary Material Supplementary material associated with this article may be found in the online version at doi:10.1016/j.jasms.2010.06.003.
References
- 1.Zhang Y. Progress and Challenges in Protein Structure Prediction. Curr. Opin. Struct. Biol. 2008;18:342–348. doi: 10.1016/j.sbi.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Przybylski M, Glocker MO. Electrospray Mass Spectrometry of Biomacromolecular Complexes with Noncovalent Interactions—New Analytical Perspectives for Supramolecular Chemistry and Molecular Recognition Processes. Angew. Chem. Int. Ed. 1996;35:806–826. [Google Scholar]
- 3.Kaltashov IA, Abzalimov RR. Do Ionic Charges in ESI MS Provide Useful Information on Macromolecular Structure? J. Am. Soc. Mass Spectrom. 2008;19:1239–1246. doi: 10.1016/j.jasms.2008.05.018. [DOI] [PubMed] [Google Scholar]
- 4.Grandori R. Electrospray-Ionization Mass Spectrometry for Protein Conformational Studies. Curr. Org. Chem. 2003;7:1589–1603. [Google Scholar]
- 5.Fenn JB, Mann M, Meng CK, Wong SF, Whitehouse CM. Electrospray Ionization for Mass Spectrometry of Large Biomolecules. Science. 1989;246:64–71. doi: 10.1126/science.2675315. [DOI] [PubMed] [Google Scholar]
- 6.Konermann L, Collings BA, Douglas DJ. Cytochrome c Folding Kinetics Studied by Time-Resolved Electrospray Ionization Mass Spectrometry. Biochemistry. 1997;36:5554–5559. doi: 10.1021/bi970046d. [DOI] [PubMed] [Google Scholar]
- 7.Katta V, Chait BT. Observation of the Hemoglobin Complex in Native Myoglobin by Electrospray-Ionization Mass Spectrometry. J. Am. Chem. Soc. 1991;113:8534–8535. [Google Scholar]
- 8.Loo JA, Edmonds CG, Udseth HR, Smith RD. Effect of Reducing Disulfide-Containing Proteins on Electrospray Ionization Mass Spectra. Anal. Chem. 1990;62:693–698. doi: 10.1021/ac00206a009. [DOI] [PubMed] [Google Scholar]
- 9.Chowdhury SK, Katta V, Chait BT. Probing Conformational Changes in Proteins by Mass Spectrometry. J. Am. Chem. Soc. 1990;112:9012–9013. [Google Scholar]
- 10.Nemirovskiy O, Giblin DE, Gross ML. Electrospray Ionization Mass Spectrometry and Hydrogen/Deuterium Exchange for Probing the Interaction of Calmodulin with Calcium. J. Am. Soc. Mass Spectrom. 1999;10:711–718. doi: 10.1016/S1044-0305(99)00036-7. [DOI] [PubMed] [Google Scholar]
- 11.Simmons DA, Konermann L. Characterization of Transient Protein Folding Intermediates during Myoglobin Reconstitution by Time-Resolved Electrospray Mass Spectrometry with On-Line Isotopic Pulse Labeling. Biochemistry. 2002;41:1906–1914. doi: 10.1021/bi011697j. [DOI] [PubMed] [Google Scholar]
- 12.Cooks RG, Ouyang Z, Takats Z, Wiseman JM. Ambient Mass Spectrometry. Science. 2006;311:1566–1570. doi: 10.1126/science.1119426. [DOI] [PubMed] [Google Scholar]
- 13.Takats ZW, J. M., Gologan B, Cooks RG. Mass Spectrometry Sampling Under Ambient Conditions with Desorption Electrospray Ionization. Science. 2004;306:471–473. doi: 10.1126/science.1104404. [DOI] [PubMed] [Google Scholar]
- 14.Cody RB, Laramee JA, Durst HD. Versatile New Ion Source for the Analysis of Materials in Open Air under Ambient Conditions. Anal. Chem. 2005;77:2297–2302. doi: 10.1021/ac050162j. [DOI] [PubMed] [Google Scholar]
- 15.Denes J, Katona M, Hosszu A, Czuczy N, Takats Z. Analysis of Biological Fluids by Direct Combination of Solid Phase Extraction and Desorption Electrospray Ionization Mass Spectrometry. Anal. Chem. 2009;81:1669–1675. doi: 10.1021/ac8024812. [DOI] [PubMed] [Google Scholar]
- 16.Kauppila T, Wiseman JM, Ketola RA, Kotiaho T, Cooks RG, Kostiainen R. Desorption Electrospray Ionization Mass Spectrometry for the Analysis of Pharmaceuticals and Metabolites. Rapid Commun. Mass Spectrom. 2006;20:387–392. doi: 10.1002/rcm.2304. [DOI] [PubMed] [Google Scholar]
- 17.Cotte-Rodriguez I, Chen H, Cooks RG. Rapid Trace Detection of Triacetone Triperoxide (TATP) by Complexation Reactions during Desorption Electrospray Ionization. Chem. Commun. 2006:953–955. doi: 10.1039/b515122h. [DOI] [PubMed] [Google Scholar]
- 18.Miao Z, Chen H. Analysis of Continuous-Flow Liquid Samples by Desorption Electrospray Ionization-Mass Spectrometry (DESI-MS). Proceedings of the 56th Annual American Society for Mass Spectrometry Conference on Mass Spectrometry; Denver, CO. June, 2008; [DOI] [PubMed] [Google Scholar]
- 19.Miao Z, Chen H. Direct Analysis of Liquid Samples by Desorption Electrospray Ionization-Mass Spectrometry (DESI-MS) J. Am. Soc. Mass Spectrom. 2009;20:10–19. doi: 10.1016/j.jasms.2008.09.023. [DOI] [PubMed] [Google Scholar]
- 20.Ma X, Zhao M, Lin Z, Zhang S, Yang C, Zhang X. Versatile Platform Employing Desorption Electrospray Ionization Mass Spectrometry for High-Throughput Analysis. Anal. Chem. 2008;80:6131–6136. doi: 10.1021/ac800803x. [DOI] [PubMed] [Google Scholar]
- 21.Chipuk JE, Brodbelt JS. Transmission Mode Desorption Electrospray Ionization. J. Am. Soc. Mass Spectrom. 2008;19:1612–1620. doi: 10.1016/j.jasms.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y, Chen H. Detection of Saccharides by Reactive Desorption Electrospray Ionization (DESI) Using Modified Phenylboronic Acids. Int. J. Mass Spectrom. 2010;289:98–107. [Google Scholar]
- 23.Li J, Dewald HD, Chen H. Online Coupling of Electrochemical Reactions with Liquid Sample Desorption Electrospray Ionization-Mass Spectrometry. Anal. Chem. 2009;81:9716–9722. doi: 10.1021/ac901975j. [DOI] [PubMed] [Google Scholar]
- 24.Van den Heuvel RHH, Heck AJR. Native Protein Mass Spectrometry: From Intact Oligomers to Functional Machineries. Curr. Opin. Chem. Biol. 2004;8:519–526. doi: 10.1016/j.cbpa.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 25.Takáts Z, Wiseman JM, Gologan B, Cooks RG. Electrosonic Spray Ionization—a Gentle Technique or Generating Folded Proteins and Protein Complexes in the Gas Phase and Studying Ion-Molecule Reactions at Atmospheric Pressure. Anal. Chem. 2004;76:4050–4058. doi: 10.1021/ac049848m. [DOI] [PubMed] [Google Scholar]
- 26.Liu J, Konermann L. Irreversible Thermal Denaturation of Cytochrome c Studied by Electrospray Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2009;20:819–828. doi: 10.1016/j.jasms.2008.12.016. [DOI] [PubMed] [Google Scholar]
- 27.Mirza UA, Cohen SL, Chait BT. Heat-Induced Conformational Changes in Proteins Studied by Electrospray Ionization Mass Spectrometry. Anal. Chem. 1993;65:1–6. doi: 10.1021/ac00049a003. [DOI] [PubMed] [Google Scholar]
- 28.Laskin J, Laskin A, Roach PJ, Slysz GW, Anderson GA, Nizkorodov SA, Bones DL, Nguyen LQ. High-Resolution Desorption Electrospray Ionization Mass Spectrometry for Chemical Characterization of Organic Aerosols. Anal. Chem. 2010;82:2048–2058. doi: 10.1021/ac902801f. [DOI] [PubMed] [Google Scholar]
- 29.Shiea J, Wang CH. Applications of Multiple Channel Electrospray Ionization Sources for Biological Sample Analysis. J. Mass Spectrom. 1997;32:247–250. [Google Scholar]
- 30.Myung S, Wiseman JM, Valentine SJ, Takats Z, Cooks RG, Clemmer DE. Coupling Desorption Electrospray Ionization with Ion Mobility/Mass Spectrometry for Analysis of Protein Structure: Evidence for Desorption of Folded and Denatured States. J. Phys. Chem. B. 2006;110:5045–5051. doi: 10.1021/jp052663e. [DOI] [PubMed] [Google Scholar]
- 31.Costa AB, Cooks RG. Simulated Splashes: Elucidating the Mechanism of Desorption Electrospray Ionization Mass Spectrometry. Chem. Phys. Lett. 2008;464:1–8. [Google Scholar]
- 32.Lane A, Nyadong L, Galhena AS, Shearer TL, Stout EP, Parry RM, Kwasnik M, Wang MD, Hay ME, Fernandez FM, Kubanek J. Desorption Electrospray Ionization Mass Spectrometry Reveals Surface-Mediated Antifungal Chemical Defense of a Tropical Seaweed. Proc. Natl. Acad. Sci. U.S.A. 2009;106:7314–7319. doi: 10.1073/pnas.0812020106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shiea J, Yuan C-H, Huang M-Z, Cheng S-C, Ma Y-L, Tseng W-L, Chang H-C, Hung W-C. Detection of Native Protein Ions in Aqueous Solution under Ambient Conditions by Electrospray Laser Desorption/Ionization Mass Spectrometry. Anal. Chem. 2008;80:4845–4852. doi: 10.1021/ac702108t. [DOI] [PubMed] [Google Scholar]
- 34.Mao D, Ding C, Douglas DJ. Hydrogen/Deuterium Exchange of Myoglobin Ions in a Linear Quadrupole Ion Trap. Rapid Commun. Mass Spectrom. 2002;16:1941–1945. doi: 10.1002/rcm.810. [DOI] [PubMed] [Google Scholar]
- 35.Babu KR, Douglas DJ. Methanol-Induced Conformations of Myoglobin at pH 4.0. Biochemistry. 2000;39:14702–14710. doi: 10.1021/bi001265t. [DOI] [PubMed] [Google Scholar]
- 36.Konermann L, Rosell FI, Mauk AG, Douglas DJ. Acid-Induced Denaturation of Myoglobin Studied by Time-Resolved Electrospray Ionization Mass Spectrometry. Biochemistry. 1997;36:6448–6454. doi: 10.1021/bi970353j. [DOI] [PubMed] [Google Scholar]
- 37.Peng IX, Loo RO, Shiea J, Loo JA. Reactive-Electrospray-Assisted Laser Desorption/Ionization for Characterization of Peptides and Proteins. Anal. Chem. 2008;80:6995–7003. doi: 10.1021/ac800870c. [DOI] [PubMed] [Google Scholar]
- 38.Konermann L, Douglas DJ. Acid-Induced Unfolding of Cytochrome c at Different Methanol Concentrations: Electrospray Ionization Mass Spectrometry Specifically Monitors Changes in the Tertiary Structure. Biochemistry. 1997;36:12296–12302. doi: 10.1021/bi971266u. [DOI] [PubMed] [Google Scholar]
- 39.Lin H, Dass C. Conformational Changes in β-endorphin as Studied by Electrospray Ionization Mass Spectrometry. Rapid Commun. Mass Spectrom. 2001;15:2341–2346. doi: 10.1002/rcm.513. [DOI] [PubMed] [Google Scholar]
- 40.Konermann L, Douglas DJ. Unfolding of Proteins Monitored by Electrospray Ionization Mass Spectrometry: A Comparison of Positive and Negative Ion Modes. J. Am. Soc. Mass Spectrom. 1998;9:1248–1254. doi: 10.1016/S1044-0305(98)00103-2. [DOI] [PubMed] [Google Scholar]
- 41.Miao Z, Wu Q, Wu S, Chen H. Generation of Native Protein Ions and Ultrafast H/D Exchange in Liquid Sample Desorption Electrospray Ionization Mass Spectrometry (DESI-MS). Proceedings of the 57th Annual American Society for Mass Spectrometry Conference on Mass Spectrometry; Philadelphia, PA. May, 2009. [Google Scholar]
- 42.Chen H, Yang S, Li M, Hu B, Li J, Wang J. Sensitive Detection of Native Proteins Using Extractive Electrospray Ionization Mass Spectrometry. Angew. Chem. Int. Ed. 2010;49:3053–3056. doi: 10.1002/anie.200906886. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


