SUMMARY
In fasted mammals, glucose homeostasis is maintained through induction of the cAMP response element-binding protein (CREB) coactivator transducer of regulated CREB activity 2 (TORC2), which stimulates the gluconeogenic program in concert with the forkhead factor FOXO1. Here we show that starvation also triggers TORC activation in Drosophila, where it maintains energy balance through induction of CREB target genes in the brain. TORC mutant flies have reduced glycogen and lipid stores and are sensitive to starvation and oxidative stress. Neuronal TORC expression rescued stress sensitivity as well as CREB target gene expression in TORC mutants. During refeeding, increases in insulin signaling inhibited TORC activity through the salt-inducible kinase 2 (SIK2)-mediated phosphorylation and subsequent degradation of TORC. Depletion of neuronal SIK2 increased TORC activity and enhanced stress resistance. As disruption of insulin signaling also augmented TORC activity in adult flies, our results illustrate the importance of an insulin-regulated pathway that functions in the brain to maintain energy balance.
INTRODUCTION
Fasting triggers concerted changes in behavior, physical activity, and metabolism that are remarkably well conserved through evolution. In mammals, such responses are often coordinated by transcriptional coactivators that are themselves targets for regulation by environmental cues (Spiegelman and Heinrich, 2004), but the extent to which these coactivators function in model organisms such as Drosophila is less clear.
In the basal state, mammalian transducers of regulated CREB activity (TORCs) are phosphorylated by salt-inducible kinases (SIKs) and sequestered in the cytoplasm via phosphorylation-dependent association with 14-3-3 proteins (Koo et al., 2005; Screaton et al., 2004). During fasting, elevations in circulating pancreatic glucagon promote TORC dephosphorylation via the protein kinase A (PKA)-mediated phosphorylation and inhibition of SIK2.
Increases in intracellular calcium have also been found to stimulate cAMP response element-binding protein (CREB) target gene expression through the activation of calcineurin/PP2B, a calcium/calmodulin-dependent serine/threonine (Ser/Thr) phosphatase that binds directly to and dephosphorylates mammalian TORCs (Koo et al., 2005; Screaton et al., 2004). Indeed, cAMP and calcium signals stimulate TORC dephosphorylation cooperatively through their effects on SIKs and PP2B, respectively. Following their liberation from 14-3-3 proteins, dephosphorylated TORCs shuttle to the nucleus, where they mediate cellular gene expression by associating with CREB over relevant promoters.
TORC2 is thought to function in parallel with FOXO1 to maintain energy balance during fasting. Knockdown and knockout studies support a critical role for both proteins in regulating catabolic programs in the liver (Dentin et al., 2007; Koo et al., 2005; Matsumoto et al., 2007). In Drosophila, starvation promotes the mobilization of glycogen and lipid stores in response to increases in circulating adipokinetic hormone (AKH), the fly homolog of mammalian glucagon (Kim and Rulifson, 2004; Lee and Park, 2004). In parallel, decreases in insulin/IGF signaling (IIS) also stimulate the dephosphorylation and nuclear translocation of Drosophila FOXO (Junger et al., 2003; Puig et al., 2003), which in turn stimulates a wide array of nutrient-regulated genes (Gershman et al., 2007; Zinke et al., 2002).
The accumulation of lipid and glycogen stores in adult flies is highly correlated with resistance to starvation in Drosophila (Djawdan et al., 1998). Indeed, disruption of the IIS pathway promotes lipid accumulation and correspondingly increases resistance to starvation and oxidative stress (Broughton et al., 2005; Clancy et al., 2001). Although FOXO does not appear to be required for starvation resistance in adult flies (Junger et al., 2003), overexpression of FOXO has been found to mimic the starvation phenotype in larvae (Kramer et al., 2003).
Here we address the importance of Drosophila TORC, the single Drosophila homolog of mammalian TORCs, in metabolic regulation. We found that increases in TORC activity during starvation enhance survival through the activation of CREB target genes in the brain. During feeding, increases in insulin signaling inhibit TORC activity through phosphorylation by a Drosophila homolog of mammalian SIK2. These studies indicate that TORC is part of an insulin-regulated pathway that functions in concert with FOXO to promote energy balance and stress resistance.
RESULTS
Activation of TORC by Starvation and Oxidative Stress
Drosophila TORC shares considerable sequence homology with mammalian TORCs in its CREB binding domain (CBD), transactivation domain (TAD), calcineurin (Cn) recognition motif, and regulatory site (Ser157), which is phosphorylated by members of the AMPK family of stress- and energy-sensing Ser/Thr kinases in mammals (Figure 1A, top). Drosophila TORC protein is expressed at low levels during larval and pupal stages, with the highest amounts detected in adults (Figure 1A, bottom). TORC mRNA levels are also increased in adults relative to larvae, although to a lesser extent.
Figure 1. Drosophila TORC Is Activated by Starvation and Oxidative Stress.
(A) Top: schematic of TORC showing the CREB binding domain (CBD), transactivation domain (TAD), nuclear import (NLS) and export (NES) sequences, calcineurin (Cn) recognition motif, and regulatory phosphorylation site (Ser157). Alignment with the mammalian TORC1 sequence and the consensus motif for phosphorylation by salt-inducible kinases (SIKs) and other members of the AMPK family of Ser/Thr kinases is shown. Hydrophobic (Ψ) and basic (B) residues are indicated. Bottom left: immunoblot showing relative amounts of TORC protein in larvae (L1–L3), pupae (P), and adult male flies. Bottom right: qPCR analysis of TORC mRNA amounts at different developmental stages.
(B) Top: immunoblot of Drosophila TORC protein in Drosophila S2 cells exposed to forskolin + isobutylmethylxanthine (F/I), staurosporine (STS), or DMSO vehicle (CON). Amounts of phospho-TORC(Ser157) and total TORC protein recovered from anti-FLAG epitope immunoprecipitates in cells expressing either wild-type (WT) or Ser157Ala mutant Drosophila TORC are shown. Bottom: immunoblot of 14-3-3 proteins recovered from immunoprecipitates of WT or Ser157Ala mutant TORC in HEK293T cells following exposure to forskolin (FSK), STS, or control vehicle. Amounts of phospho-TORC(Ser157) and total TORC shown.
(C) Endogenous Drosophila TORC protein staining by immunofluorescence analysis of Drosophila KC-167 cells under basal conditions (CON) and following exposure to STS. DAPI staining was used to visualize nuclei. Dephosphorylation of TORC by STS in KC-167 cells was verified by immunoblot assay (data not shown).
(D) CRE-luciferase (CRE-luc) reporter activity in HEK293T cells expressing WT or Ser157Ala mutant Drosophila TORC. Effect of FSK or the dominant-negative CREB inhibitor ACREB on CRE-luc activity is shown. n = 6; p < 0.05. Data are given as means ± SD.
(E and F) Top: immunoblots of Drosophila TORC protein in wild-type (w1118) flies exposed to water-only starvation (E) or paraquat (PQT) feeding (F). For starvation assay, time after food withdrawal is shown in hours. Flies were maintained on paraquat-containing food for 24 hr before analysis. Phospho- and dephospho-TORC protein is indicated. Bottom: relative effect of starvation or paraquat feeding on TORC protein and mRNA amounts.
In the basal state, Drosophila TORC is highly phosphorylated at Ser157 and localized to the cytoplasm in Drosophila S2 and KC-167 cells (Figure 1B, top and Figure 1C). Demonstrating the importance of Ser157 phosphorylation in sequestering TORC, Ser157Ala mutant TORC shows only low-level binding to 14-3-3 proteins relative to wild-type TORC in HEK293T cells (Figure 1B, bottom). Exposure to the adenylyl cyclase activator forskolin (FSK) or to staurosporine (STS), an inhibitor of SIKs and other protein kinases (Ravnskjaer et al., 2007; Takemori et al., 2007), promotes TORC dephosphorylation, liberation from 14-3-3 proteins, and nuclear translocation (Figures 1B and 1C).
Consistent with these changes, overexpression of wild-type Drosophila TORC potentiated CRE-luciferase (CRE-luc) reporter activity following exposure of HEK293T cells to FSK, whereas phosphorylation-defective Ser157Ala TORC stimulated CRE-luc activity under basal as well as FSK-induced conditions (Figure 1D). CRE-luc activity is blocked by coexpression of the dominant-negative CREB inhibitor ACREB (Ahn et al., 1998). Taken together, these results indicate that Drosophila TORC modulates CREB target gene expression following its dephosphorylation at Ser157 and nuclear entry in response to cAMP.
Based on the ability of mammalian TORCs to promote fasting metabolism (Koo et al., 2005), we examined whether Drosophila TORC performs a similar function in adult flies. Amounts of dephosphorylated, active TORC increased progressively during water-only starvation (Figure 1E). Feeding adult flies paraquat, a respiratory chain inhibitor that stimulates the production of reactive oxygen species, also promoted the accumulation of dephosphorylated TORC (Figure 1F), suggesting a broader role for this coactivator in stress resistance. Similar to mammalian TORCs (Dentin et al., 2007), the upregulation of TORC in Drosophila appears to reflect an increase in TORC protein stability, as amounts of TORC mRNA did not change significantly in response to fasting or paraquat treatment (Figures 1E and 1F).
Sensitivity of TORC Mutant Flies to Stress
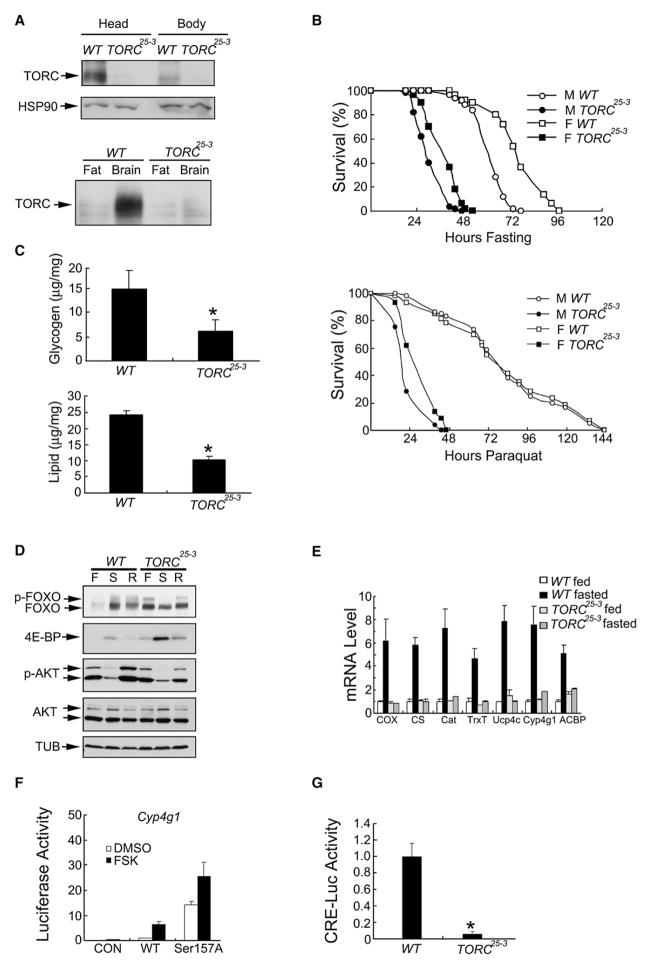
To evaluate the role of TORC in energy homeostasis, we mutated the TORC gene by excising EY00004, a P element insertion from the Berkeley Drosophila Genome Project, located 2.9 kb downstream of the TORC coding region (CG6064; see Figure S1 available online). One line, referred to as TORC25-3, contained a 10 kb deletion that removes the entire transcribed region of TORC. TORC mRNA and protein were not detected in TORC25-3 compared to wild-type controls, where TORC protein is expressed primarily in the brain and at lower levels in other parts of the body (Figure 2A and data not shown). TORC mRNA amounts were more comparable between head and body, however, supporting the idea that TORC activity in different tissues is controlled primarily at the level of protein stability (Figure S2).
Figure 2. TORC Mutant Flies Are Sensitive to Starvation and Oxidative Stress.
(A) Top: immunoblot of TORC in heads and bodies of WT and TORC25-3 flies. Bottom: relative amounts of TORC protein in fat bodies and brains of WT and TORC25-3 larvae.
(B) Relative survival of TORC25-3 and WT flies in response to starvation (top) or paraquat feeding (bottom). Percent survival at different times is shown. n = 50; p < 0.05. Results are representative of three independent experiments.
(C) Total glycogen and lipid content, expressed as μg/mg body weight, in WT and TORC25-3 flies. n = 6; *p < 0.05.
(D) Immunoblot of FOXO proteins in WT and TORC25-3 flies under ad libitum fed (F), starved (S), or refed (R) conditions. Amounts of total and phosphorylated AKT as well as 4E-BP are indicated.
(E) qPCR analysis of head mRNAs from WT and TORC25-3 flies under fasted or fed conditions as indicated. n = 2. Data are representative of two independent experiments.
(F) Effect of WT or Ser157Ala mutant TORC expression on Cyp4g1-luc reporter activity in HEK293T cells, with or without FSK as indicated. n = 3. Data are representative of two independent experiments.
(G) CRE-luc reporter activity in WT and TORC25-3 flies. Comparable amounts and phosphorylation of Drosophila CREB (CREBB-17A) in WT and TORC25-3 flies were confirmed by western blot assay (data not shown). n = 6; *p < 0.05.
Data are given as means ± SD.
Individuals homozygous for TORC25-3 were viable and fertile. However, in response to water-only starvation, TORC25-3 flies lived an average of less than 36 hr, while wild-type flies lived an average of 72 hr (Figure 2B, top). Starvation had similar effects on a second independent TORC mutant line (A4-32; data not shown). TORC25-3 flies were also sensitive to oxidative stress: following exposure to paraquat, their mean survival time was reduced by 50% relative to controls (Figure 2B, bottom). Arguing against a more general “sickly” effect of TORC disruption, TORC25-3 flies had similar food intake as determined using the CAFE method (Ja et al., 2007), and they exhibited comparable or elevated physical activity compared to controls, particularly during fasting (Figure S3 and data not shown).
Respiratory quotients were indistinguishable between TORC mutant and wild-type flies, indicating that glucose oxidation was appropriately induced during feeding, transitioning to fat burning in response to starvation (Figure S3). Consistent with their starvation sensitivity, however, TORC25-3 mutant flies had lower amounts of stored glycogen and lipid relative to wild-type (Figure 2C). These reductions appear specific for adults, because wild-type and TORC25-3 mutant larvae had comparable lipid levels (Figure S4).
We examined effects of TORC disruption on insulin signaling. In line with increases in glucose oxidation, amounts of phosphorylated AKT were comparable between wild-type and TORC25-3 flies during feeding (Figure 2D; Figure S5). By contrast, amounts of dephosphorylated, active FOXO as well as 4E-BP, a FOXO target gene, were actually elevated in starved TORC25-3 flies, likely reflecting a secondary response to the depletion of lipid and glycogen (Figure 2D; Figure S5). While removal of one copy of FOXO had no effect on survival or lipid accumulation in starved TORC25-3 flies, removal of both copies of FOXO was lethal in the TORC mutant background (Figure S6). By contrast with TORC25-3 flies, however, FOXO null (FOXO21/FOXO25) flies had normal lipid levels, and they were as resistant to starvation as wild-type flies (Figure S7) (Junger et al., 2003).
Role of Neuronal TORC in Stress Resistance
We performed gene profiling studies on mRNAs from heads of fasted wild-type and TORC25-3 mutant flies to evaluate the mechanism by which TORC mediates starvation resistance. This analysis revealed a set of 169 genes that were downregulated 4-fold or more in TORC25-3 mutants (Table S1). Many of the genes that were downregulated in TORC25-3 flies appear to function in glucose and lipid metabolism, proteolysis, amino acid transport, and mitochondrial respiration. To identify direct targets for TORC regulation, we selected genes that contain CREB binding sites within 3 kb upstream of the transcription start site and that are induced during starvation for further analysis.
Consistent with the role of mammalian TORCs in promoting mitochondrial gene expression (Wu et al., 2006), mRNAs for mitochondrial respiration (citrate synthase [CS] and cytochrome c oxidase subunit IV [COX]) and reactive oxygen scavenging (UCP4c, catalase [CAT], TrxT, and Cyp4g1) were reduced in TORC25-3 flies. Correspondingly, cytochrome oxidase and citrate synthase enzymatic activities, markers of mitochondrial function, were also lower in TORC mutants compared to wild-type (Figure S8).
We examined whether genes that are downregulated in TORC mutants are directly modulated by TORC. When TORC was over-expressed in a heat-inducible manner using the GAL4/UAS bipartite transactivation system (Brand and Perrimon, 1993), a number of fasting-inducible genes, including COX, CS, and CAT, were upregulated (Figure S9). Indeed, 78 of the 169 genes downregulated in TORC25-3 flies contain CREB binding sites (Figure S10). For example, the Cyp4g1 promoter contains cAMP-responsive elements (CREs) at −375 and −100, and exposure to FSK correspondingly increased Cyp4g1-luciferase reporter activity 10- to 20-fold in human HEK293T cells (Figure 2F). Cyp4g1 reporter activity in these cells was further enhanced by expression of wild-type Drosophila TORC, and to a greater extent by phosphorylation-defective Ser157Ala TORC. Expression of the dominant-negative mouse CREB polypeptide ACREB disrupted effects of TORC on reporter activity, demonstrating the importance of CREB for induction of this gene (Figure S11). CRE-luc reporter activity (Iijima-Ando and Yin, 2005) was effectively eliminated in TORC25-3 compared to wild-type flies, demonstrating the importance of TORC for CREB-mediated transcription in Drosophila (Figure 2G).
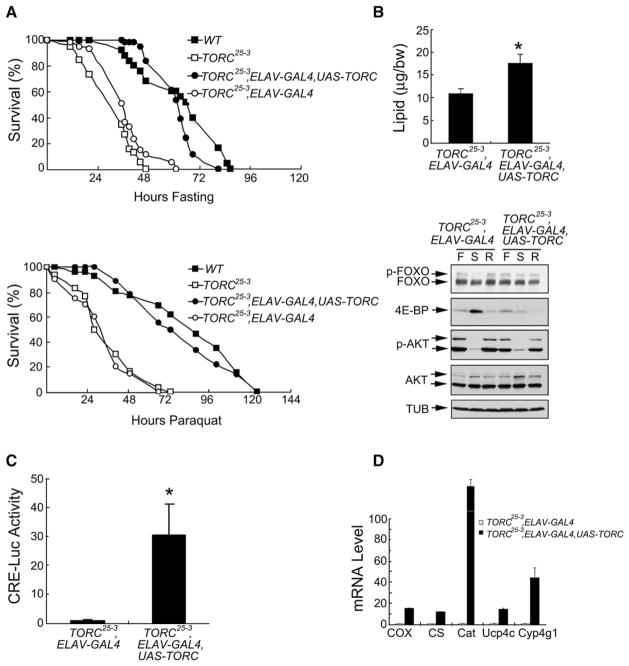
We used the GAL4/UAS transactivation system to supply wild-type TORC in a cell-type-specific manner to TORC25-3 flies. Panneuronal expression of TORC using ELAV-GAL4 to drive expression of UAS-TORC rescued both the starvation- and paraquat-sensitive phenotypes (Figure 3A). Similarly, driving neuronal expression of TORC using SCRATCH-GAL4 also rescued the starvation-sensitive phenotype (Figure S12). In contrast, TORC expression in fat body using r4-GAL4 (Lee and Park, 2004) did not rescue starvation sensitivity (data not shown), arguing for a specific requirement for TORC in neurons.
Figure 3. Neuronal TORC Expression Rescues Sensitivity of TORC25-3 Flies to Starvation and Oxidative Stress.
(A) Relative survival in response to starvation (top) or paraquat feeding (bottom) of WT, TORC25-3, TORC25-3,ELAV-GAL4 control, and TORC25-3,ELAV-GA-L4,UAS-TORC rescued flies. n = 100; p < 0.05.
(B) Top: relative lipid content of TORC25-3,ELAV-GAL4 control and TORC25-3,ELAV-GAL4,UAS-TORC rescued flies. n = 6 flies per group; *p < 0.05. Bottom: immunoblot of FOXO and 4E-BP proteins in TORC mutant TORC25-3,ELAV-GAL4 and TORC25-3,ELAV-GAL4,UAS-TORC rescued flies under fed (F), starved (S), or refed (R) conditions.
(C) CRE-luc activity in TORC25-3,ELAV-GAL4 control and TORC25-3,ELAV-GAL4,UAS-TORC rescued flies. n = 6; *p < 0.05.
(D) qPCR analysis of TORC-regulated genes from head mRNAs of TORC25-3,ELAV-GAL4 control and TORC25-3,ELAV-GAL4,UAS-TORC rescued flies under fasting conditions. Rescue genotype: ELAV-GAL4, w1118/Y;UAS-TORC/+;TORC25-3. Control genotype: ELAV-GAL4, w1118/Y;TORC25-3. Wild-type: w1118.
Data are given as means ± SD.
In keeping with this increase in starvation resistance, total lipid amounts were elevated in TORC rescued flies compared to mutant flies (Figure 3B, top). Interestingly, however, while neuronal TORC fully rescued the starvation sensitivity of TORC mutants, lipid levels were only partially restored, raising the possibility that starvation sensitivity in this setting does not depend exclusively on lipid stores. Consistent with the improvements in these metabolic parameters, FOXO activity and 4E-BP expression in TORC rescued individuals were commensurately downregulated to wild-type levels (Figure 3B, bottom). Moreover, neuronal TORC also rescued CRE reporter activity and fasting-inducible gene expression (Figures 3C and 3D).
Regulation of TORC by Insulin Signaling
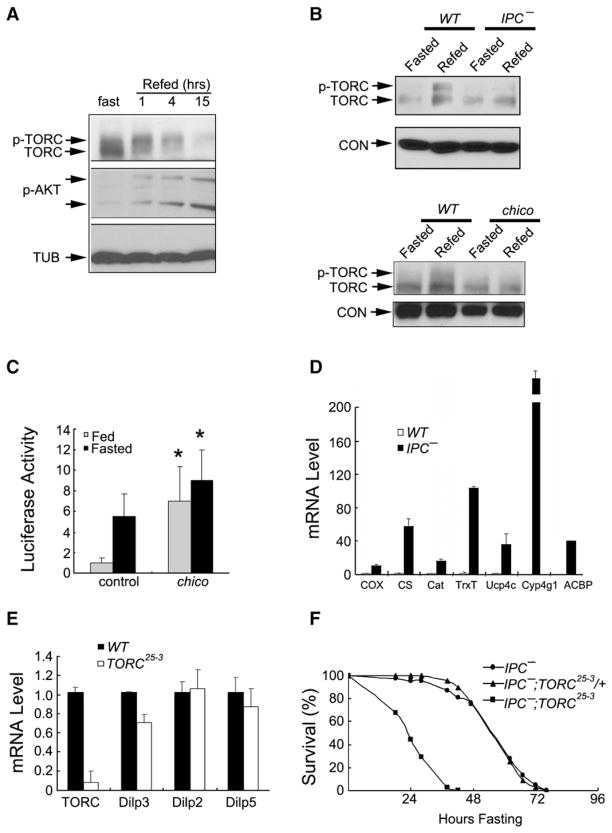
Based on the activation of TORC during starvation, we examined whether increases in insulin signaling inhibit Drosophila TORC activity during refeeding. In contrast to fasting, refeeding triggered TORC phosphorylation after 1 hr and degradation after 4 hr in wild-type flies (Figure 4A). TORC did not undergo phosphorylation during refeeding in flies with defective insulin signaling caused either by mutation of chico, the Drosophila ortholog of the insulin receptor substrate (IRS) protein, or by ablation of the insulin-producing cells (IPCs) (Figure 4B). Indeed, CRE-luc activity and fasting-inducible TORC target gene expression were correspondingly elevated in insulin-signaling mutant flies compared to wild-type (Figures 4C and 4D). These data support the notion that TORC acts downstream of the insulin signaling pathway in Drosophila.
Figure 4. The Insulin Signaling Pathway Regulates TORC Activity in Drosophila.
(A) Immunoblot of phospho-TORC and total TORC protein in fasted or refed flies. Hours after refeeding are indicated. Amounts of phospho-AKT are also shown.
(B) Immunoblots showing amounts of phospho-TORC and total TORC in WT and insulin-producing cell (IPC)-ablated flies (top) and chico mutant flies (bottom) under fasted or refed conditions. Loss of insulin-like peptide (ilp) gene expression in IPC-ablated flies was confirmed by qPCR analysis (data not shown).
(C) Comparison of CRE-luc reporter activity in WT and chico mutant flies under fed or fasted conditions. n = 13 per group; p < 0.05.
(D) qPCR analysis showing relative expression of TORC-regulated genes in WT and IPC-ablated flies.
(E) qPCR analysis of ilp2, ilp3, and ilp5 gene expression in WT and TORC25-3 flies fed ad libitum.
(F) Effect of IPC ablation on starvation sensitivity in TORC25-3 flies.
Data are given as means ± SD.
We considered that neuronal TORC may affect systemic resistance to starvation through feedback inhibition of Drosophila insulin-like peptides (ilps, also known as dilps). In that event, TORC23-5 flies might be predicted to exhibit starvation sensitivity as a consequence of increased ilp expression. However, mRNA amounts for ilp2, ilp3, and ilp5 were either comparable or modestly reduced in TORC23-5 relative to wild-type flies fed ad libitum (Figure 4E). Moreover, IPC ablation did not affect starvation sensitivity in TORC23-5 flies, arguing against a significant role for ilps in this setting (Figure 4F).
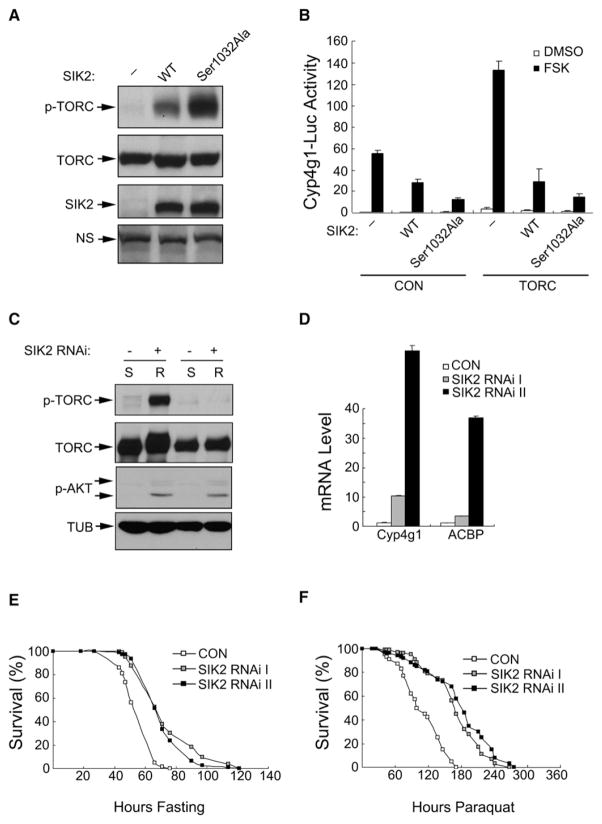
Mammalian SIK2 has been shown to mediate inhibitory effects of insulin on CREB target gene expression in refed mice by phosphorylating TORC2 and promoting its subsequent proteasomal degradation (Dentin et al., 2007). SIK2 kinase activity is inhibited during fasting by the PKA-mediated phosphorylation of SIK2 at Ser587. The presence of a single putative SIK2 homolog (CG4290) in Drosophila (Okamoto et al., 2004) prompted us to test its role in regulating TORC activity. Overexpression of Drosophila SIK2 increased amounts of Ser157-phosphorylated Drosophila TORC in HEK293T cells (Figure 5A). Mutation of the inhibitory PKA phosphorylation site at Ser1032 to Ala in Drosophila SIK2 further increased amounts of phosphorylated TORC. Consistent with these effects, Drosophila SIK2 inhibited Drosophila Cyp4g1-luciferase reporter activity in HEK293T cells expressing Drosophila TORC, and mutant Ser1032Ala SIK2 inhibited reporter activity to a greater extent relative to wild-type (Figure 5B).
Figure 5. The Serine/Threonine Kinase SIK2 Mediates Effects of Insulin Signaling on TORC Activity during Refeeding.
(A) Immunoblot showing effect of WT and constitutively active Ser1032Ala Drosophila SIK2 on amounts of phosphorylated Drosophila TORC in transfected HEK293T cells.
(B) Transient assay of HEK293T cells showing effect of WT and Ser1032Ala mutant SIK2 on Drosophila Cyp4g1-luc reporter activity with or without FSK.
(C) Immunoblot showing effect of neuronal SIK2 RNAi expression on Ser157 phosphorylation of a neuronal TORC-GFP fusion protein in adult flies under starved (S) or refed (R) conditions. Control (−) genotype: ELAV-GAL4/+;UAS-TORC-GFP/+. SIK2-deficient (+) genotype: ELAV-GAL4/+;UAS-TORC-GFP/UAS-SIK2 RNAi.
(D) qPCR analysis of TORC-regulated genes from head mRNAs of control flies and two independent strains expressing SIK2 RNAi in neurons. CON genotype: ELAV-GAL4/Y. SIK RNAi I genotype: ELAV-GAL4/Y;;UAS-SIK2 RNAi/+. SIK RNAi II genotype: ELAV-GAL4/Y;;UAS-SIK2 RNAi/+.
(E and F) Effect of water-only starvation (E) and paraquat (F) on survival of 5-day-old female flies expressing SIK2 RNAi in neurons relative to control. CON genotype: Appl-GAL4/+. SIK RNAi I genotype: Appl-GAL4/+;;UAS-SIK2 RNAi I/+. SIK RNAi II genotype: Appl-GAL4/+;;UAS-SIK2 RNAi II/+. n = 100.
Data are given as means ± SD.
We examined whether depletion of SIK2 increases TORC activity in flies. Neuronal expression of UAS-SIK2 RNAi from an ELAV-GAL4 driver reduced amounts of Ser157-phosphorylated TORC during refeeding, when SIK2 is predicted to be active (Figure 5C; Figure S13). Consistent with the increase in amounts of dephosphorylated, active TORC protein, mRNA amounts for the TORC-regulated genes Cyp4g1 and AcBP were substantially upregulated in SIK2 RNAi flies (Figure 5D). Correspondingly, SIK2 RNAi flies were more resistant to starvation and paraquat feeding relative to controls (Figures 5E and 5F).
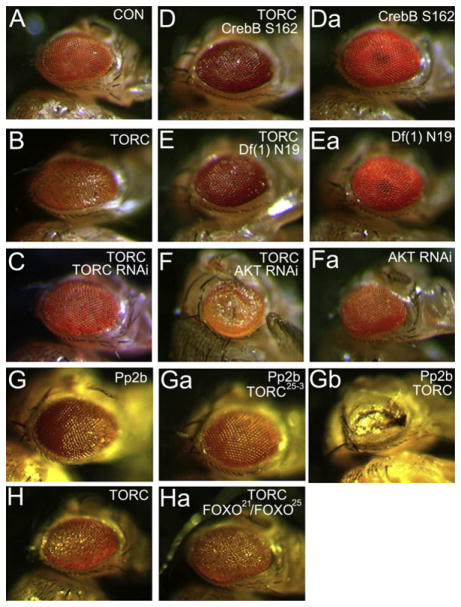
We performed genetic epistasis experiments to evaluate the mechanism by which TORC activity is regulated. Eye-specific overexpression of TORC using a GMR-GAL4 driver led to a rough eye phenotype (Figures 6A–6C). TORC transcriptional activity appeared critical for these effects since reduction of Drosophila CrebB expression using either the chromosomal deficiency Df(1)N19 or the CrebBS162 mutation (Belvin et al., 1999) suppressed the rough eye phenotype in TORC-expressing flies (Figures 6D and 6E).
Figure 6. TORC Acts Downstream of the Serine/Threonine Kinase AKT to Regulate CREB Activity.
(A and B) Effect of TORC overexpression in postmitotic cells of the eye imaginal disc shown relative to control.
(C) Effect of depleting TORC via RNAi-mediated knockdown on eye morphology in TORC-overexpressing flies.
(D and E) Effect of CrebB disruption, either via mutation of the CrebB gene (CrebBS162; [D]) or in flies with a chromosomal deficiency (Df(1)N19; [E]), on eye phenotype in TORC transgenic flies.
(F) Eye morphology in TORC transgenic flies with reduced AKT (AKT RNAi) expression.
(Da–Fa) Effect of each disruption (CrebB, Df(1)N19, and AKT RNAi) on eye phenotype in the absence of TORC overexpression.
(G–Gb) Effect of PP2B overexpression alone (G), in TORC mutant flies (Ga), and in TORC-overexpressing flies (Gb).
(H and Ha) Effect of mutation of FOXO (FOXO21/FOXO25) on the TORC overexpression phenotype (Ha) compared to TORC overexpression alone (H).
Genotypes are GMR-GAL4/+;TM3/+ (A), GMR-GAL4/+;2X UAS-TORC/+ (B), GMR-GAL4/UAS-TORC RNAi;2X UAS-TORC/+ (C) CrebBS162/+;GMR-GAL4/+;2X UAS-TORC/+ (D), CrebBS162/+;GMR-GAL4/+ (Da), Df(1)N19/+; GMR-GAL4/+;2X UAS-TORC/+ (E), Df(1)N19/+;GMR-GAL4/+ (Ea), GMR-GAL4/UAS-AKT RNAi;2X UAS-TORC/+ (F), GMR-GAL4/UAS-AKT RNAi (Fa), GMR-GAL4/UAS-Pp2b-14Dact/+ (G), GMR-GAL4/UAS-Pp2b-14Dact;TORC25-3 (Ga), GMR-GAL4/UAS-Pp2b-14Dact;UAS-TORC/+ (Gb), GMR-GAL4/2X UAS-TORC (H), and GMR-GAL4/2X UAS-TORC;FOXO21/FOXO25 (Ha).
We evaluated the role of the Ser/Thr kinase AKT, which has et al., 2007; Koo et al., 2005; Matsumoto et al., 2007). been shown to inhibit mammalian TORC2 activity through the Depletion of AKT by RNAi-mediated knockdown enhanced the phosphorylation and activation of SIK2 during feeding (Dentin TORC-mediated rough eye phenotype (Figure 6F), arguing that this component of the insulin signaling pathway inhibits TORC activity.
Based on the presence of a conserved calcineurin/PP2B binding motif in Drosophila TORC (Figure 1A), we tested whether this Ser/Thr phosphatase also promotes TORC activation. Supporting this idea, eye-specific expression of constitutively active PP2B alone promoted a rough eye phenotype in wild-type flies, but not in TORC mutant flies, demonstrating the importance of endogenous TORC in this context (Figure 6G). Conversely, overexpression of TORC strongly potentiated effects of PP2B on eye morphology, confirming the ability of PP2B to promote TORC activation.
Because FOXO activity is increased in TORC mutant flies, we tested whether this forkhead protein is required for the TORC eye phenotype. TORC-overexpressing flies in which FOXO was lreduced or eliminated using FOXO21 and FOXO25 alleles (Junger et al., 2003) still exhibited a rough eye phenotype (Figure 6H). FOXO21/FOXO25 null flies also had wild-type levels of TORC protein amounts and activity as measured by immunoblot and CRE-luc reporter assays, indicating that FOXO is not required for TORC activation in this setting (Figure S14).
DISCUSSION
Insulin signaling regulates lipid and glucose metabolism in both C. elegans and Drosophila in part by inhibiting FOXO-dependent transcription (Giannakou and Partridge, 2007). Lipid stores are increased in flies with mutations in the IIS pathway; these animals are resistant to starvation as well as oxidative stress (Broughton et al., 2005; Clancy et al., 2001). We found that TORC enhances survival during starvation in part by stimulating target gene expression in neurons. Although TORC appears to act in parallel with FOXO, the increase in FOXO activity we observed in TORC mutant flies indicates that TORC may also impact on this pathway.
TORC appears to be required for the expression of genes that promote lipid and glucose metabolism, amino acid transport, and proteolysis. Consistent with this idea, paralogs for a number of TORC-regulated genes (TrxT, CAT, and UCP4c) appear to be required for starvation and oxidative stress resistance (Chen et al., 2004; Fridell et al., 2005; Mockett et al., 2003; Svensson and Larsson, 2007). Superimposed on these effects, neuronal TORC may also promote systemic resistance to starvation and oxidative stress by modulating the expression of neuropeptide hormones and other circulating factors that regulate peripheral glucose and lipid metabolism.
In mammals, refeeding has been found to increase SIK2 kinase activity during refeeding through the AKT-mediated phosphorylation of SIK2 (Dentin et al., 2007). Phosphorylated TORC2 is subsequently ubiquitinated and degraded through the E3 ligase COP1. Supporting a similar mechanism in Drosophila, RNAi-mediated knockdown of AKT in Drosophila was sufficient to increase TORC activity. Conversely, depletion of neuronal SIK2 enhanced TORC activity and increased resistance to both starvation and paraquat feeding. Although a Drosophila homolog for COP1 has not been identified, we imagine that the ubiquitin-dependent degradation of Drosophila TORC is also critical in modulating its activity in brain as well as other tissues.
Based on its ability to potentiate CREB target gene expression in neurons, TORC may function in other biological settings. Indeed, Drosophila CREB appears to have an important role in learning and memory, circadian rhythmicity, rest homeostasis, and addictive behavior (Belvin et al., 1999; Hendricks et al., 2001; Perazzona et al., 2004; Sakai et al., 2004). Future studies should reveal the extent to which TORC participates in these contexts as well.
EXPERIMENTAL PROCEDURES
Fly Stocks
All Drosophila melanogaster lines were maintained at 25°C on standard food medium. P{EPgy2}EY00004, UAS-rpr, chico1, actin-GAL4, and ELAV-GAL4 were obtained from the Bloomington Drosophila Stock Center. CRE-luc reporter flies, UAS-Pp2b-14Dact, ilp2-GAL4, r4-GAL4, chico2, FOXO21, and FOXO25 have been described previously (Bohni et al., 1999; Iijima-Ando and Yin, 2005; Junger et al., 2003; Lee and Park, 2004; Rulifson et al., 2002; Sullivan and Rubin, 2002). IPC-ablated flies were generated by crossing ilp2-GAL4 with UAS-rpr. r4-GAL4 flies were from J. Park. chico mutants were generated by crossing chico1/Cyo with chico2/Cyo flies. SIK2 RNAi and AKT RNAi flies were obtained from the Vienna Drosophila RNAi Center. TORC RNAi flies were generated by inserting a 600 bp TORC coding region fragment into the pWIZ vector. UAS-TORC-GFP flies were generated as described previously (Bittinger et al., 2004).
Generation of TORC Null Mutant and UAS-TORC Flies
TORC deletion alleles were generated by mobilization of the P{EPgy2} insertion EY00004 with a Δ2-3 source of transposase (Robertson et al., 1988). Potential deletions were screened by PCR. One deletion line, 25-3, was found to have an ~10 kb deletion removing the entire TORC coding sequence; the 25-3 deletion break points were confirmed by sequencing. A second mutant line, A4-32, contained a P{EPgy2} element inserted 1877 bp 3′ to the TORC start codon, resulting in an in-frame stop codon after aa 625 of TORC. For misexpression constructs, the TORC coding region was cloned into pUAST and transgenic lines generated.
Starvation and Oxidative Stress
For starvation assay, 3- to 5-day-old flies were transferred to vials of 1% agar/PBS with filter papers soaked with H2O, and dead flies were scored every 4–8 hr. For oxidative stress, 3- to 5-day-old flies were starved in 1% agar/PBS for 4 hr and then transferred to vials of 20 mM paraquat/10% sucrose/1% agar/PBS, and dead flies were scored every 4–8 hr.
Lipid and Glycogen Measurement
Fly total lipid and glycogen levels were measured as described previously (Van Handel, 1985a, 1985b). For lipids, single flies were crushed in 0.2 ml of chloroform:methanol (1:1). After evaporating solvent, 0.2 ml of sulfuric acid was added, and samples were heated at 37°C for 10 min. After cooling, 2 ml of vanillin reagent (0.12% vanillin in 68% phosphoric acid) was added, and samples were allowed to develop for 5 min. Absorbance was measured at 525 nm. Sesame oil (Sigma, S3547) was used to generate a standard curve. For glycogen content, single flies were crushed in 0.2 ml of 2% sodium sulfate, followed by addition of methanol (1 ml) and centrifugation for 1 min. Supernatants were evaporated, and 2 ml of anthrone reagent (0.14% anthrone in 28% sulfuric acid) was added to each sample. Reactions were incubated at 37°C for 15 min, and absorbance was measured at 625 nm. Purified glycogen (Roche) was used to generate a standard curve.
Respirometry
A Sable Systems International (SSI) TR-2 system was used for flow-through respirometry with a LI-COR LI-6251 CO2 analyzer (resolution < 0.1 ppm CO2), supplemented with an SSI Oxzilla II differential oxygen analyzer and SSI ExpeData data acquisition and analysis software with a UI-2 16-bit measurement interface (basic accuracy 0.03%). Activity was monitored constantly using an SSI AD-2 optical activity detector. Room temperature and air flow rate were also measured and recorded via an A/D converter (UI-2). Bev-A-Line low-permeability tubing (Thermoplastic Processes, Inc.) was used throughout to minimize CO2 and H2O absorbance errors. Room air from a carboy was pulled by an SSI TR-SS3 subsampler through a Drierite/Ascarite/Drierite drying column to remove CO2 and H2O, at a standard temperature and pressure (STP)-corrected flow rate of ~50 ml/min. The prepared air then entered the glass/aluminum respirometry chamber (volume ~ 3 ml; SSI TRRM). During each recording, the CO2 produced and the O2 consumed by the group of 25 flies together with the activity, flow rate, and room temperature were recorded.
During a typical run, a group of 25 flies was cooled for about 10 min at 5°C and transferred to the respirometric chamber. The flies in the chamber were given at least 10 min for temperature acclimatization (room temperature ~ 25°C). Recording was started to establish a 4 min baseline for the CO2 and O2 analyzers with an empty chamber. Each recording consisted of a variable number of data points taken at 1 s intervals using finite impulse response digital filtration to reduce short-term noise (Lighton, 1991).
Data Analysis and Statistics
Recordings were analyzed using SSI ExpeData software. For each recording, the CO2 and O2 baselines were subtracted assuming a linear drift. CO2 in ppm and O2 in % were converted to μl/hr using the recorded flow rate. The activity (measured as V) was transformed to the absolute difference sum (ADS) of the activity. The ADS is the cumulative sum of the absolute difference between all adjacent data points (Lighton and Turner, 2004). The slope and intercept of the linear regression of the ADS values of activity were calculated as a function of time for each recording. The ADS value reached after 10 min was compared across groups. Data are presented as means ± SEM and are representative of at least two independent experiments. Comparisons of different groups were carried out using the two-tailed unpaired Student’s t test. Differences were considered statistically significant at p < 0.05.
qPCR Measurement
Fly heads were collected and RNA was extracted using an RNeasy Mini Kit (QIAGEN). Total RNA (1 μg) was reverse transcribed by SuperScript II transcriptase (Invitrogen), and the generated cDNA was used for real-time PT-PCR (Roche LightCycler 480 Real-Time PCR system, SYBR green) using 2 ng of cDNA template and a 400 nM primer concentration. Values were normalized to rp49.
qPCR Primers
qPCR primers used are listed in Table S2.
Cell Culture and Transfection
HEK293T cells were maintained and transfections were carried out as described previously (Ravnskjaer et al., 2007). For promoter studies, 50 ng of promoter and 50 ng of β-galactosidase plasmids were used per well, and the total amount was kept constant at 300 ng by adding the indicated plasmids or empty pcDNA3 vector. Twenty-four hours after transfection, cells were treated with forskolin (FSK, 10 μM) for 4 hr, and luciferase activity was measured. S2 and KC-167 cells were cultured in Schneider’s medium (Invitrogen) with 10% FCS at room temperature, and transfections were performed using FuGENE 6 (Roche) according to the manufacturer’s instructions. Cells were treated with FSK (10 μM), IBMX (4 μg/ml), or staurosporine (STS, 100 nM).
Western Blotting
Cells or flies were lysed on ice in lysis buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1 mM EDTA, 6 mM EGTA, 20 mM NaF, 1% Triton X-100, and protease inhibitors) for 15–20 min. After centrifugation at 13,000 rpm for 15 min, supernatants were reserved for protein determinations and SDS-PAGE analysis. The antibodies used were phospho-TORC1 (Cell Signaling), FOXO (generous gift of O. Puig), 4E-BP (generous gift of N. Sonenberg), HSP90 (Santa Cruz, sc-7947), tubulin (Upstate, 05-829), FLAG-M2 (Sigma, A8592), phospho-AKT(Ser473) (Cell Signaling Technology, 9271), AKT (Cell Signaling Technology, 9272), and HA (Santa Cruz, sc-7392). TORC antibodies were raised against a synthetic TORC peptide (DYTREIFDSLSLSLG) by Covance Research Products. Sera were collected and purified using a peptide affinity resin.
Supplementary Material
Acknowledgments
We thank O. Puig for FOXO antiserum and N. Sonenberg for 4E-BP antiserum. We also thank J. Yin, U. Heberlein, E. Hafen, T. Aigaki, J. Park, the Bloomington Drosophila Stock Center, and the Vienna Drosophila RNAi Center for fly stocks. We thank Sable Systems International for use of the open-flow respirometry equipment and J.R.B. Lighton and R. Turner for advice. This work was supported by NIH grant GM037828. M.M. is supported by the Kieckhefer Foundation.
Footnotes
ACCESSION NUMBERS
Microarray data reported herein have been deposited in the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) and are accessible through the GEO series accession number GSE10546.
Supplemental Data include two tables and fourteen figures and can be found with this article online at http://www.cellmetabolism.org/cgi/content/full/7/5/434/DC1/.
References
- Ahn S, Olive M, Aggarwal S, Krylov D, Ginty D, Vinson C. A dominant negative inhibitor of CREB reveals that it is a general mediator stimulus-dependent transcription of c-fos. Mol Cell Biol. 1998;18:967–977. doi: 10.1128/mcb.18.2.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belvin MP, Zhou H, Yin JC. The Drosophila dCREB2 gene affects the circadian clock. Neuron. 1999;22:777–787. doi: 10.1016/s0896-6273(00)80736-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittinger MA, McWhinnie E, Meltzer J, Iourgenko V, Latario B, Liu X, Chen CH, Song C, Garza D, Labow M. Activation of cAMP response element-mediated gene expression by regulated nuclear transport of TORC proteins. Curr Biol. 2004;14:2156–2161. doi: 10.1016/j.cub.2004.11.002. [DOI] [PubMed] [Google Scholar]
- Bohni R, Riesgo-Escovar J, Oldham S, Brogiolo W, Stocker H, Andruss BF, Beckingham K, Hafen E. Autonomous control of cell and organ size by CHICO, a Drosophila homolog of vertebrate IRS1–4. Cell. 1999;97:865–875. doi: 10.1016/s0092-8674(00)80799-0. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Broughton SJ, Piper MD, Ikeya T, Bass TM, Jacobson J, Driege Y, Martinez P, Hafen E, Withers DJ, Leevers SJ, Partridge L. Longer lifespan, altered metabolism, and stress resistance in Drosophila from ablation of cells making insulin-like ligands. Proc Natl Acad Sci USA. 2005;102:3105–3110. doi: 10.1073/pnas.0405775102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Rio DC, Haddad GG, Ma E. Regulatory role of dADAR in ROS metabolism in Drosophila CNS. Brain Res Mol Brain Res. 2004;131:93–100. doi: 10.1016/j.molbrainres.2004.08.013. [DOI] [PubMed] [Google Scholar]
- Clancy DJ, Gems D, Harshman LG, Oldham S, Stocker H, Hafen E, Leevers SJ, Partridge L. Extension of life-span by loss of CHICO, a Drosophila insulin receptor substrate protein. Science. 2001;292:104–106. doi: 10.1126/science.1057991. [DOI] [PubMed] [Google Scholar]
- Dentin R, Liu Y, Koo SH, Hedrick S, Vargas T, Heredia J, Yates J, 3rd, Montminy M. Insulin modulates gluconeogenesis by inhibition of the coactivator TORC2. Nature. 2007;449:366–369. doi: 10.1038/nature06128. [DOI] [PubMed] [Google Scholar]
- Djawdan M, Chippindale AK, Rose MR, Bradley TJ. Metabolic reserves and evolved stress resistance in Drosophila melanogaster. Physiol Zool. 1998;71:584–594. doi: 10.1086/515963. [DOI] [PubMed] [Google Scholar]
- Fridell YW, Sanchez-Blanco A, Silvia BA, Helfand SL. Targeted expression of the human uncoupling protein 2 (hUCP2) to adult neurons extends life span in the fly. Cell Metab. 2005;1:145–152. doi: 10.1016/j.cmet.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Gershman B, Puig O, Hang L, Peitzsch RM, Tatar M, Garofalo RS. High-resolution dynamics of the transcriptional response to nutrition in Drosophila: a key role for dFOXO. Physiol Genomics. 2007;29:24–34. doi: 10.1152/physiolgenomics.00061.2006. [DOI] [PubMed] [Google Scholar]
- Giannakou ME, Partridge L. Role of insulin-like signalling in Drosophila lifespan. Trends Biochem Sci. 2007;32:180–188. doi: 10.1016/j.tibs.2007.02.007. [DOI] [PubMed] [Google Scholar]
- Hendricks JC, Williams JA, Panckeri K, Kirk D, Tello M, Yin JC, Sehgal A. A non-circadian role for cAMP signaling and CREB activity in Drosophila rest homeostasis. Nat Neurosci. 2001;4:1108–1115. doi: 10.1038/nn743. [DOI] [PubMed] [Google Scholar]
- Iijima-Ando K, Yin JC. Transgenic cAMP response element reporter flies for monitoring circadian rhythms. Methods Enzymol. 2005;393:302–315. doi: 10.1016/S0076-6879(05)93013-9. [DOI] [PubMed] [Google Scholar]
- Ja WW, Carvalho GB, Mak EM, de la Rosa NN, Fang AY, Liong JC, Brummel T, Benzer S. Prandiology of Drosophila and the CAFE assay. Proc Natl Acad Sci USA. 2007;104:8253–8256. doi: 10.1073/pnas.0702726104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junger MA, Rintelen F, Stocker H, Wasserman JD, Vegh M, Radimerski T, Greenberg ME, Hafen E. The Drosophila forkhead transcription factor FOXO mediates the reduction in cell number associated with reduced insulin signaling. J Biol. 2003;2:20. doi: 10.1186/1475-4924-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SK, Rulifson EJ. Conserved mechanisms of glucose sensing and regulation by Drosophila corpora cardiaca cells. Nature. 2004;431:316–320. doi: 10.1038/nature02897. [DOI] [PubMed] [Google Scholar]
- Koo SH, Flechner L, Qi L, Zhang X, Screaton RA, Jeffries S, Hedrick S, Xu W, Boussouar F, Brindle P, et al. The CREB coactivator TORC2 is a key regulator of fasting glucose metabolism. Nature. 2005;437:1109–1111. doi: 10.1038/nature03967. [DOI] [PubMed] [Google Scholar]
- Kramer JM, Davidge JT, Lockyer JM, Staveley BE. Expression of Drosophila FOXO regulates growth and can phenocopy starvation. BMC Dev Biol. 2003;3:5. doi: 10.1186/1471-213X-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G, Park JH. Hemolymph sugar homeostasis and starvation-induced hyperactivity affected by genetic manipulations of the adipokinetic hormone-encoding gene in Drosophila melanogaster. Genetics. 2004;167:311–323. doi: 10.1534/genetics.167.1.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lighton JR. Measurements on insects. In: Payne PA, editor. Concise Encyclopedia on Biological and Biomedical Measurement Systems. Oxford: Pergamon Press; 1991. pp. 201–208. [Google Scholar]
- Lighton JR, Turner RJ. Thermolimit respirometry: an objective assessment of critical thermal maxima in two sympatric desert harvester ants, Pogonomyrmex rugosus and P. californicus. J Exp Biol. 2004;207:1903–1913. doi: 10.1242/jeb.00970. [DOI] [PubMed] [Google Scholar]
- Matsumoto M, Pocai A, Rossetti L, Depinho RA, Accili D. Impaired regulation of hepatic glucose production in mice lacking the forkhead transcription factor foxo1 in liver. Cell Metab. 2007;6:208–216. doi: 10.1016/j.cmet.2007.08.006. [DOI] [PubMed] [Google Scholar]
- Mockett RJ, Bayne AC, Kwong LK, Orr WC, Sohal RS. Ectopic expression of catalase in Drosophila mitochondria increases stress resistance but not longevity. Free Radic Biol Med. 2003;34:207–217. doi: 10.1016/s0891-5849(02)01190-5. [DOI] [PubMed] [Google Scholar]
- Okamoto M, Takemori H, Katoh Y. Salt-inducible kinase in steroidogenesis and adipogenesis. Trends Endocrinol Metab. 2004;15:21–26. doi: 10.1016/j.tem.2003.11.002. [DOI] [PubMed] [Google Scholar]
- Perazzona B, Isabel G, Preat T, Davis RL. The role of cAMP response element-binding protein in Drosophila long-term memory. J Neurosci. 2004;24:8823–8828. doi: 10.1523/JNEUROSCI.4542-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig O, Marr MT, Ruhf ML, Tjian R. Control of cell number by Drosophila FOXO: downstream and feedback regulation of the insulin receptor pathway. Genes Dev. 2003;17:2006–2020. doi: 10.1101/gad.1098703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravnskjaer K, Kester H, Liu Y, Zhang X, Lee D, Yates JR, 3rd, Montminy M. Cooperative interactions between CBP and TORC2 confer selectivity to CREB target gene expression. EMBO J. 2007;26:2880–2889. doi: 10.1038/sj.emboj.7601715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson HM, Preston CR, Phillis RW, Johnson-Schlitz DM, Benz WK, Engels WR. A stable genomic source of P element transposase in Drosophila melanogaster. Genetics. 1988;118:461–470. doi: 10.1093/genetics/118.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rulifson EJ, Kim SK, Nusse R. Ablation of insulin-producing neurons in flies: growth and diabetic phenotypes. Science. 2002;296:1118–1120. doi: 10.1126/science.1070058. [DOI] [PubMed] [Google Scholar]
- Sakai T, Tamura T, Kitamoto T, Kidokoro Y. A clock gene, period, plays a key role in long-term memory formation in Drosophila. Proc Natl Acad Sci USA. 2004;101:16058–16063. doi: 10.1073/pnas.0401472101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Screaton RA, Conkright MD, Katoh Y, Best JL, Canettieri G, Jeffries S, Guzman E, Niessen S, Yates JR, 3rd, Takemori H, et al. The CREB coactivator TORC2 functions as a calcium- and cAMP-sensitive coincidence detector. Cell. 2004;119:61–74. doi: 10.1016/j.cell.2004.09.015. [DOI] [PubMed] [Google Scholar]
- Spiegelman BM, Heinrich R. Biological control through regulated transcriptional coactivators. Cell. 2004;119:157–167. doi: 10.1016/j.cell.2004.09.037. [DOI] [PubMed] [Google Scholar]
- Sullivan KM, Rubin GM. The Ca(2+)-calmodulin-activated protein phosphatase calcineurin negatively regulates EGF receptor signaling in Drosophila development. Genetics. 2002;161:183–193. doi: 10.1093/genetics/161.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson MJ, Larsson J. Thioredoxin-2 affects lifespan and oxidative stress in Drosophila. Hereditas. 2007;144:25–32. doi: 10.1111/j.2007.0018-0661.01990.x. [DOI] [PubMed] [Google Scholar]
- Takemori H, Kanematsu M, Kajimura J, Hatano O, Katoh Y, Lin XZ, Min L, Yamazaki T, Doi J, Okamoto M. Dephosphorylation of TORC initiates expression of the StAR gene. Mol Cell Endocrinol. 2007;265–266:196–204. doi: 10.1016/j.mce.2006.12.020. [DOI] [PubMed] [Google Scholar]
- Van Handel E. Rapid determination of glycogen and sugars in mosquitoes. J Am Mosq Control Assoc. 1985a;1:299–301. [PubMed] [Google Scholar]
- Van Handel E. Rapid determination of total lipids in mosquitoes. J Am Mosq Control Assoc. 1985b;1:302–304. [PubMed] [Google Scholar]
- Wu Z, Huang X, Feng Y, Handschin C, Gullicksen PS, Bare O, Labow M, Spiegelman B, Stevenson SC. Transducer of regulated CREB-binding proteins (TORCs) induce PGC-1alpha transcription and mitochondrial biogenesis in muscle cells. Proc Natl Acad Sci USA. 2006;103:14379–14384. doi: 10.1073/pnas.0606714103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinke I, Schutz CS, Katzenberger JD, Bauer M, Pankratz MJ. Nutrient control of gene expression in Drosophila: microarray analysis of starvation and sugar-dependent response. EMBO J. 2002;21:6162–6173. doi: 10.1093/emboj/cdf600. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.