Abstract
Autism spectrum disorders are a genetically heterogeneous constellation of syndromes characterized by impairments in reciprocal social interaction. Available somatic treatments have limited efficacy. We have identified inactivating mutations in the gene BCKDK (Branched Chain Ketoacid Dehydrogenase Kinase) in consanguineous families with autism, epilepsy, and intellectual disability. The encoded protein is responsible for phosphorylation-mediated inactivation of the E1α subunit of branched-chain ketoacid dehydrogenase (BCKDH). Patients with homozygous BCKDK mutations display reductions in BCKDK messenger RNA and protein, E1α phosphorylation, and plasma branched-chain amino acids. Bckdk knockout mice show abnormal brain amino acid profiles and neurobehavioral deficits that respond to dietary supplementation. Thus, autism presenting with intellectual disability and epilepsy caused by BCKDK mutations represents a potentially treatable syndrome.
Approximately 25% of individuals with classically defined autism also suffer from epilepsy (1). Although autism is highly heritable (2), the specific genetic causes remain obscure in the majority of cases. Here, we present the results of whole-exome sequencing from two consanguineous families, one of Turkish descent (family 558) and a second of Egyptian ancestry (family 18), presenting with typically developing parents, two siblings with autism, intellectual disability (ID), and either seizure or abnormal electroencephalogram (EEG) and, in one family (558), an unaffected sibling (Fig. 1A, table S1, and supplementary text).
Fig. 1.
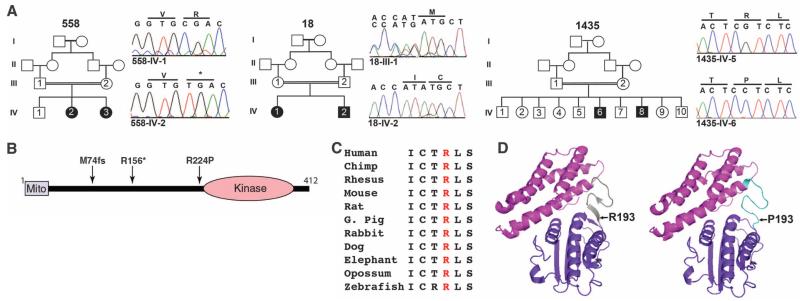
Mutations in the BCKDK gene in patients with autism, ID, and epilepsy. (A) Pedigrees 558, 18, and 1435 display first-cousin consanguinity and two affected patients each (solid symbols), some with unaffected members (open symbols). Sequence chromatograms correspond to noncarrier sibling (for 558 and 1435) or obligate carrier parent (for 18). (B) Schematic of BCKDK protein with mitochondrial localization signal (Mito) and kinase domain, depicting p.M74fs, p.R156*, and p.R224P mutations from pedigrees 18, 558, and 1435, respectively. (C) Sequence conservation of arginine residue throughout evolution (I, Ile; C, Cys; T, Thr; R, Arg; L, Leu; S, Ser). (D) Predicted effect of Arg → Pro substitution leading from a β sheet (left) to an unfolded domain (right). Mutation mapped onto the crystallized rat protein corresponds to amino acid position 193.
We performed whole-exome sequencing in both families as part of parallel efforts among patients with autism and/or epilepsy at Yale and UCSD/Broad Institute. We focused on the identification of homozygous variants predicted to result in loss of protein function, consistent with the presumed mode of recessive inheritance. Using Homozygosity Mapper software (3), we refined the search to identify haplotypes that were identical by descent (fig. S1A). In each of these families, we identified a distinct, null, homozygous mutation in the gene Branched Chain Ketoacid Dehydrogenase Kinase (BCKDK). In family 558, we identified a cytosine-to-thymine substitution in exon 4 (C466T) resulting in a premature stop codon at amino acid position 156, prior to the kinase domain (Fig. 1B). In family 18, a single base deletion (c.G222del) in exon 2 led to a frame-shift terminating the protein at position 74 of 412 amino acids (Fig. 1B and fig. S1B). Both the mutations were in constitutively spliced exons and segregated in the family according to a recessive mode of inheritance. These variants were not present in more than 200 ethnically matched, healthy control chromosomes, nor in our in-house data set of approximately 2200 chromosomes or in publicly available databases. No other homozygous loss-of-function mutations segregating with affected status were identified in either family (table S2).
We next searched for other BCKDK variants in patients with similar phenotypes within our in-house exome database enriched for a history of consanguinity. We identified one additional family (family 1435) of Libyan ancestry presenting with autism and ID (table S1 and supplementary text), carrying a missense (c.G671C) mutation (Fig. 1A) mapping within a linkage peak (lod score of 2.2) on chromosome 16 (fig. S1A) and leading to the substitution of a highly conserved arginine with a proline at position 224 in BCKDK (Fig. 1C). Modeling of the p.R224P mutation using the crystal structure of rat Bckdk (4) suggested disruption of the β sheet in a flexible linker domain (Fig. 1D).
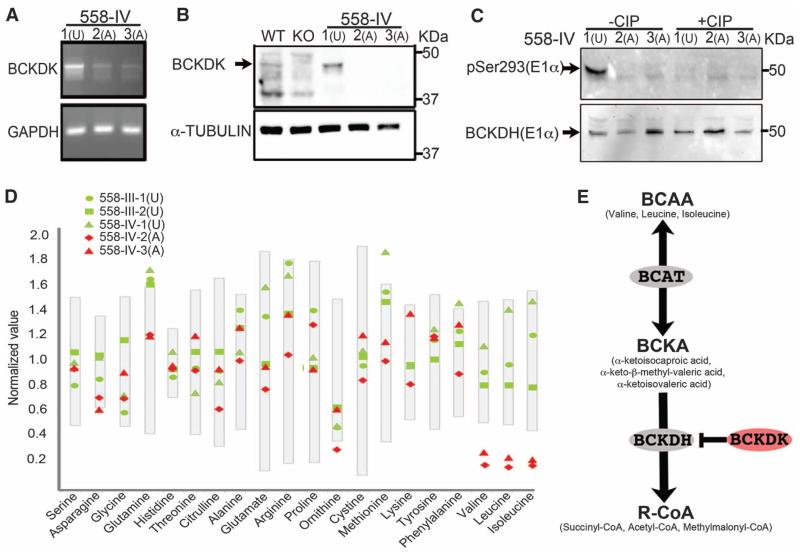
The branched-chain ketoacid dehydrogenase (BCKDH) complex catalyzes the irreversible, rate-limiting step in the catabolism of branched-chain amino acids (BCAAs) (5), and BCKDK encodes a kinase that phosphorylates and thus inactivates the E1α subunit of this complex (6) (Fig. 2E). Mutations in any of the three subunits of the BCKDH complex lead to toxic accumulation of BCAAs and their metabolites and are the cause of maple syrup urine disease (7), characterized by severe neurological complications and treated by dietary restriction of BCAAs, which are essential amino acids (8). BCKDK is expressed widely in mouse and human tissues (fig. S2) (9). Relative to controls, we found reduced mRNA levels in cell lines from the two affected patients in families 558 and 18, suggesting nonsense-mediated mRNA decay (Fig. 2A and fig. S3A), as well as undetectable levels of BCKDK protein on Western blot (Fig. 2B and fig. S3B). Supporting a role of BCKDK in negative regulation of the BCKDH complex, we tested a phospho-specific antibody to the BCKDK phosphorylation site at residue 293 of the E1α BCKDH subunit (10) and found absence of reactivity in dermal fibroblasts of the affected individuals in family 558 (Fig. 2C) and in tissue of the Bckdk−/− mouse (fig. S4). These data suggest that patients with BCKDK mutations may lack basal, negative regulation of BCKDH activity.
Fig. 2.
Effect of BCKDK p.R156* mutation in affected and unaffected individuals from family 558. (A) Reverse transcription polymerase chain reaction showing reduced levels of mRNA in affected patients; GAPDH levels are shown as a control. (B) Western blot probed with BCKDK-specific antibody (arrow). Fibroblast lysates from family 558 showing undetectable BCKDK protein in affected patients (A, IV-2, and IV-3) relative to unaffected patients (U, IV-1). Wild-type and Bckdk knockout (KO) mice lysates served as positive and negative controls. (C) Western blot probed with phospho-specific pSer293 antibody of E1α subunit of BCKDH enzyme. In the absence of calf intestinal phospha-tase (CIP), a distinct band is present in the unaffected individual (U), absent in the two affected individuals (A), and nearly abolished after CIP treatment. (D) Reduced levels of plasma BCAAs in affected patients (red symbols) compared with parents and unaffected sibling (green symbols). Gray bars represent normalized standard values. (E) Schematic diagram of the BCAA catabolic pathway. BCAT, branched-chain aminotransferase; BCKA, branched-chain α-keto acid; BCKDH, branched-chain α-keto acid dehydrogenase; R-CoA, succinyl-CoA, acetyl-CoA, and methylmalonyl-CoA.
Mice deficient for Bckdk display increased basal activity of the BCKDH complex as well as reduced BCAAs in various tissues (11). We measured plasma BCAAs in our patients and found that each patient with a homozygous mutation showed notably lower levels of plasma BCAAs than their healthy relatives and with respect to reference ranges (Fig. 2D and tables S3 to S5). Declines in BCAAs in affected patients with null mutations were more substantial (with respect to normal reference ranges) than those in patients with missense variants (tables S3 to S5) (12). There was no accumulation of BCAAs in the urine (table S6), excluding abnormally high BCAA urinary excretion. None of the affected individuals was known to eat a diet deficient in protein, as evidenced by their normal fasting plasma levels of the other essential amino acids (tables S3 to S5 and supplementary text). We did not observe accumulation of organic acids (Fig. 2E) in the blood of our patients (table S7), which suggests that these products were efficiently eliminated in the urine.
To uncover cellular phenotypes of human cells lacking BCKDK, we generated pluripotent stem cells (iPSCs) from the fibroblasts of the healthy brother and the two affected sisters from family 558 by means of episomal reprogramming (supplementary text) (13). All iPSC lines were positive for pluripotent markers (fig. S5B), showed normal karyotype, and demonstrated expected gene expression profiles as a function of these conditions (fig. S6A). We differentiated three iPSC lines per individual into embryoid bodies, then neural rosettes, and finally, neural progenitor cells (NPCs) (fig. S5A). To reduce variability in the differentiation efficiency, we used fluorescence-activated cell sorting (FACS) to generate homogeneous NPC populations. CD184+CD24+CD44−CD271− cells were sorted (14) and passaged for five cycles before analysis, resulting in 99.9% of the cells being positive for Nestin, a marker of neural stem cells. (fig. S7A). There were no notable differences in the morphology or in the proliferation of cells of wild-type and mutant genotypes (fig. S7, A and B).
To uncover potential cell-autonomous phenotypes, we exposed NPCs to reduced levels of BCAAs in the culture media. Typical culture medium is supplemented with a total concentration of ~1200 μM BCAAs, so we reduced these concentrations over several orders of magnitude to nearly zero. We found no genotype-dependent differences in cell survival or proliferation (fig. S7, B and C). We next differentiated the NPCs into mature neurons by basic fibroblast growth factor withdrawal, which resulted in nearly 100% of the cells becoming positive for neuron-specific β-tubulin Tuj1 and the cytoskeletal protein Map2 (fig. S8, A and B). We found no differences in numbers of differentiated cells per well, dendrites per cell, presynaptic punctae per length of dendrite, or area of the cell soma (fig. S8). There were no appreciable differences in gene expression or metabolic profiles of these cultured neurons, arguing against a major cell-autonomous role for BCKDK in the pathogenesis of the disease.
We next explored the possibility of cell-nonautonomous mechanisms by returning to the Bckdk−/− mice. Mutants were born at the expected Mendelian ratio and were healthy at birth but showed growth retardation that could be recovered by feeding a BCAA-rich diet (11). Adults developed neurological abnormalities, such as tremors, epileptic seizures, and hindlimb clasping phenotypes observed in some other mouse models of autism spectrum disorders (15, 16). Gross brain histology, however, was normal (11) (fig. S9). To identify possible pathways contributing to the neurological phenotype, we performed a gene expression profile of mouse brain cortex from wild-type and Bckdk−/− mice prior to the onset of seizures (fig. S10A and supplementary text) followed by comprehensive pathway analysis (fig. S10B). There were several relevant perturbed pathways, including the brain-expressed amino acid transporter network (fig. S10C).
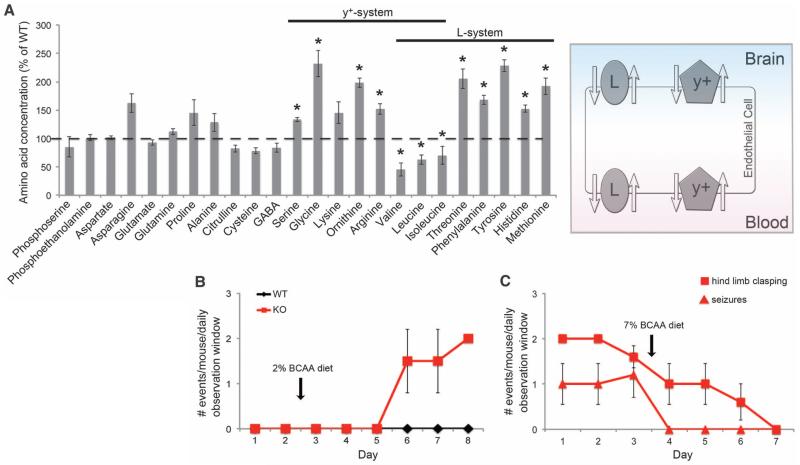
BCAAs are transported across the blood-brain barrier (BBB) mainly by the heterodimeric amino acid transporter SLC7A5/SLC3A2 (or LAT1), a member of the L-type transporter family and, to a lesser extent, by members of the y-type family and the Na+-dependent LNAA transporter (17) (Fig. 3A). In the brain, LAT1 is expressed almost exclusively in the basolateral and apical membranes of the BBB endothelial cells (18) (Fig. 3A). Expression levels of SLC7A5/SLC3A2 change in response to amino acid availability (19, 20). We reasoned that the altered expression of the transporters might be a direct response to the low concentration of plasma BCAAs. At normal plasma amino acid concentrations, these amino acid transporters are fully saturated and shared by a number of large neutral amino acids (LNAAs). Therefore, plasma amino acids compete with each other for transport across the BBB. A decrease in plasma BCAAs could lead to a perturbation in brain concentrations of not only BCAAs but also other LNAAs. Thus, we quantified amino acid concentrations in brain homogenates from post-natal day 14 (P14) wild-type and knockout mice. In addition to the expected reduced brain BCAAs, there were also significantly increased levels of threonine, phenylalanine, tyrosine, histidine, and methionine, the exact LNAAs that are carried by these transporters (Fig. 3A), which suggests imbalanced BBB transport. Because several of these amino acids are precursors for important neurotransmitters, it remains a possibility that the reduced BCAAs and/or the increased LNAAs contribute to the neurological phenotype. In fact, patients with uncontrolled MSUD display reduced brain LNAAs, and the degree to which the reduced or increased amino acids contribute to the phenotype remains controversial (21).
Fig. 3.
Bckdk−/− neurological abnormalities rescued by supplementation with BCAAs. (A) Brain amino acid levels in P14 Bckdk−/− mice. Levels of amino acids were normalized on wet tissue weight and are shown as percentage of wild-type littermates (WT, n = 3; KO, n = 4). Error bars represent SE; *P < 0.05. Right: Schematic of the amino acid transporters expressed in cells of the BBB. The L- and y+-type transporters are responsible for the transport of the BCAAs and other LNAAs. (B) Mice raised on a BCAA-enriched diet (7% BCAA) were assessed for 2 days for hindlimb clasping. Starting on the evening of the second day, the mice were fed a 2% BCAA diet until day 8. Mice were tested for hindlimb clasping twice per day. WT, n = 1; KO, n = 2. Error bars represent SD. The observer was blinded to the genotype. (C) Bckdk−/− mice neurobehavioral tests for animals fed a 2% BCAA diet (day 1 to day 3) or a BCAA-enriched diet (day 3 to day 7). Mice were tested twice per day for hindlimb clasping (1 to 2 months old) and seizures (4 to 6 months old). Error bars represent SD; n = 5 for each test.
The data suggest that the neurological phenotype may be treated by dietary supplementation with BCAAs. To test this hypothesis, we studied the effect of a chow diet containing 2% BCAAs or a BCAA-enriched diet, consisting of 7% BCAAs, on the neurological phenotypes of the Bckdk−/− mice. Mice raised on the BCAA-enriched diet were phenotypically normal. On the 2% BCAA diet, however, Bckdk−/− mice had clear neurological abnormalities not seen in wild-type mice, such as seizures and hindlimb clasping, that appeared within 4 days of instituting the 2% BCAA diet (Fig. 3B). These neurological deficits were completely abolished within a week of the Bckdk−/− mice starting the BCAA-enriched diet, which suggests that they have an inducible yet reversible phenotype (Fig. 3C).
Our experiments have identified a Mendelian form of autism with comorbid ID and epilepsy that is associated with low plasma BCAAs. Although the incidence of this disease among patients with autism and epilepsy remains to be determined, it is probably quite a rare cause of this condition. We have shown that murine Bckdk−/− brain has a disrupted amino acid profile, suggesting a role for the BBB in the pathophysiology of this disorder. The mechanism by which abnormal brain amino acid levels lead to autism, ID, and epilepsy remains to be investigated. We have shown that dietary supplementation with BCAAs reverses some of the neurological phenotypes in mice. Finally, by supplementing the diet of human cases with BCAAs, we have been able to normalize their plasma BCAA levels (table S10), which suggests that it may be possible to treat patients with mutations in BCKDK with BCAA supplementation.
Supplementary Material
Acknowledgments
We thank the families for their participation; R. Weavers and E. Morava (Nijmegen Medical Center, Netherlands) and B. Barshop for providing patients; M. Kara’s colleague for arranging shipment during the Libyan revolution; C. Lynch for the pSer293 antibody; the S. Taylor lab (UCSD) for help in protein modeling; M. Seashore, W. MacLean Jr., T. Cowan, and A. El-Gharabawy for suggestions; N. Wright-Davis, M. Murtha, M. Raubeson, N. DiLullo, M. Walker, Y. Song, N. Lifton, K. Bilguvar, A. Caglayan, Z. Omay, M. Choi, N. Carriero, R.D. Bjornson, P. Ventola, K. Koenig, and A. Bozik for technical assistance; the Yale Center for Genome Analysis (S. Mane); and the Sanford Burnham Institute. Supported by NIH grants P01HD070494, R01NS048453, and P30NS047101 (J.G.G.), RC2MH089956 (M.W.S.), K08MH087639 (A.R.G.), T32MH018268 and (P.E.-F.); Broad Institute grant U54HG003067 (E.L.); the Center for Inherited Disease Research for genotyping; the Simons Foundation Autism Research Initiative (J.G.G. and M.W.S.); a Veterans Administration Merit Award (R.A.H.); the German Research Foundation (G.N.); the American Academy of Child and Adolescent Psychiatry Pilot Research Award/Elaine Schlosser Lewis Fund (P.E.-F.); and the American Psychiatric Association/Lilly Research Fellowship (P.E.-F.). J.G.G. is an investigator of the Howard Hughes Medical Institute. Data have been deposited into dbGap (phs000288) and the NCBI Sequence Read Archive (SRS351252) (whole exome sequencing) and into GEO (GSE39447) (microarrays). J.G.G. is a consultant for Halozyme Therapeutics, a biopharmaceutical company that develops products targeting the extracellular matrix. M.W.S. is a consultant to Synapdx, that is developing diagnostic tests for autism, and to Pfizer Pharmaceuticals that is working to develop autism therapeutics. Bckdk gene trap mice are available for noncommercial research from R.A.H. under MTA agreement with Lexicon. J.G.G. and M.W.S. are inventors on a patent application filed 17 July 2012 by UCSD covering diagnostic and therapeutic strategies for patients with autism and epilepsy.
Footnotes
www.sciencemag.org/cgi/content/full/science.1224631/DC1
Materials and Methods
Supplementary Text
Figs. S1 to S10
Tables S1 to S10
References and Notes
- 1.Tuchman R, Rapin I. Lancet Neurol. 2002;1:352. doi: 10.1016/s1474-4422(02)00160-6. [DOI] [PubMed] [Google Scholar]
- 2.Bailey A, et al. Psychol. Med. 1995;25:63. doi: 10.1017/s0033291700028099. [DOI] [PubMed] [Google Scholar]
- 3.Seelow D, Schuelke M, Hildebrandt F, Nürnberg P. Nucleic Acids Res. 2009;37:W593. doi: 10.1093/nar/gkp369. Web Server issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Machius M, Chuang JL, Wynn RM, Tomchick DR, Chuang DT. Proc. Natl. Acad. Sci. U.S.A. 2001;98:11218. doi: 10.1073/pnas.201220098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harris RA, Joshi M, Jeoung NH, Obayashi M. J. Nutr. 2005;135(suppl.):1527S. doi: 10.1093/jn/135.6.1527S. [DOI] [PubMed] [Google Scholar]
- 6.Suryawan A, et al. Am. J. Clin. Nutr. 1998;68:72. doi: 10.1093/ajcn/68.1.72. [DOI] [PubMed] [Google Scholar]
- 7.Menkes JH, Hurst PL, Craig JM. Pediatrics. 1954;14:462. [PubMed] [Google Scholar]
- 8.Snyderman SE, Norton PM, Roitman E, Holt LE., Jr. Pediatrics. 1964;34:454. [PubMed] [Google Scholar]
- 9.Doering CB, Coursey C, Spangler W, Danner DJ. Gene. 1998;212:213. doi: 10.1016/s0378-1119(98)00182-6. [DOI] [PubMed] [Google Scholar]
- 10.Lynch CJ, et al. Am. J. Physiol. Endocrinol. Metab. 2003;285:E854. doi: 10.1152/ajpendo.00153.2003. [DOI] [PubMed] [Google Scholar]
- 11.Joshi MA, et al. Biochem. J. 2006;400:153. doi: 10.1042/BJ20060869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lepage N, McDonald N, Dallaire L, Lambert M. Clin. Chem. 2397;43:1997. [PubMed] [Google Scholar]
- 13.Okita K, et al. Nat. Methods. 2011;8:409. doi: 10.1038/nmeth.1591. [DOI] [PubMed] [Google Scholar]
- 14.Yuan SH, et al. PLoS ONE. 2011;6:e17540. doi: 10.1371/journal.pone.0017540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chao HT, et al. Nature. 2010;468:263. doi: 10.1038/nature09582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmeisser MJ, et al. Nature. 2012;486:256. doi: 10.1038/nature11015. [DOI] [PubMed] [Google Scholar]
- 17.Hawkins RA, O’Kane RL, Simpson IA, Viña JR. J. Nutr. 2006;136(suppl.):218S. doi: 10.1093/jn/136.1.218S. [DOI] [PubMed] [Google Scholar]
- 18.Duelli R, Enerson BE, Gerhart DZ, Drewes LR. J. Cereb. Blood Flow Metab. 2000;20:1557. doi: 10.1097/00004647-200011000-00005. [DOI] [PubMed] [Google Scholar]
- 19.Wagner CA, Lang F, Bröer S. Am. J. Physiol. Cell Physiol. 2001;281:C1077. doi: 10.1152/ajpcell.2001.281.4.C1077. [DOI] [PubMed] [Google Scholar]
- 20.Verrey F. Pflugers Arch. 2003;445:529. doi: 10.1007/s00424-002-0973-z. [DOI] [PubMed] [Google Scholar]
- 21.Zinnanti WJ, et al. Brain. 2009;132:903. doi: 10.1093/brain/awp024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.