Abstract
Despite antiretroviral therapy, proviral latency of human immunodeficiency virus type 1 (HIV-1) remains a principal obstacle to curing the infection [1]. Inducing the expression of latent genomes within resting CD4+ T cells is the primary strategy to clear this reservoir [2]. While histone deacetylase (HDAC) inhibitors such as suberoylanilide hydroxamic acid (SAHA or vorinostat, VOR) can disrupt HIV-1 latency in vitro [3–5], the utility of this approach has never been directly proven in a translational clinical study of HIV-infected patients.
Therefore we isolated the circulating resting CD4+ T cells of patients in whom viremia was fully suppressed by antiretroviral therapy (ART), and directly studied the effect of VOR in this latent reservoir. In each of eight patients studied, a single dose of VOR increased both biomarkers of cellular acetylation, and simultaneously induced an increase in HIV RNA expression in resting CD4+ cells (mean increase 4.8-fold). This is the first demonstration that a molecular mechanism known to enforce HIV latency can be therapeutically targeted in man, provides proof-of-concept for HDAC inhibitors as a new therapeutic class, and defines a precise approach to test novel strategies to directly attack and eradicate latent HIV infection.
Keywords: HDAC inhibitor, antiretroviral therapy, latent infection, resting CD4+ T cell, HIV-1
Among the many important aims of future HIV research is the development of therapies of finite duration capable of eradicating HIV infection. The persistence of quiescent HIV infection within a small population long-lived CD4+ T cells is currently a major obstacle to this goal [1]. HDACs are recruited to the HIV long terminal repeat promoter (LTR), establishing one of several restrictions that can limit LTR expression and maintain viral latency [2]. Deacetylated LTR chromatin appears to play a key contributory role in regulating HIV expression, and especially in maintaining proviral quiescence and latency. In vitro, HDAC inhibitors have been shown to disrupt latent proviral HIV infection in both cell culture models and ex vivo assays using cells from HIV-1-infected patients. While disrupting latency has been proposed as part of a strategy to eradicate HIV infection, previous studies using the weak HDAC inhibitor valproic acid did not consistently demonstrate a marked depletion of resting cell infection [6–10] in patients on ART. However, the effects measured in these studies are significantly downstream of the molecular site of action HDAC inhibitors, and thus the proximal pharmacodynamic measures of HDAC inhibitor activity and HIV-1 expression were not evaluated. Here we show that HDAC inhibitors disrupt the latency of proviral genomes within resting CD4+ T cells, establishing the first class of drugs that could lead to the eradication of HIV infection.
Suberoylanilide hydroxamic acid (vorinostat, VOR) is a potent HDAC inhibitor used to treat human malignancies. At clinically relevant concentrations VOR inhibits the Class I HDACs most important for repression of HIV expression [3, 11], and induces LTR expression and virus production in vitro from the resting CD4+ T cells of HIV+ patients on ART with levels of plasma HIV RNA below the limit of detection (BDL) [4–5,12]. As the most proximal measure of effect on latent infection is expression of HIV-1 RNA, we developed a sensitive assay to enable a direct measurement of unspliced gag HIV RNA within the resting CD4+ T cells of HIV-infected patients. The assay has a limit of detection of 1 copy/million resting CD4+ T cells, and a limit of quantitation of 10 copies/million resting CD4+ T cells.
To evaluate the effect of VOR on latent infection in vivo, HIV-infected patients receiving stable ART with plasma HIV-1 RNA < 50 copies/ml for at least 6 months and a CD4 count >300/µl were enrolled following informed consent. First, to demonstrate that it was ethical to expose patients to an experimental agent with potential risk in a study with no proven clinical benefit for the individual, we validated the ability of this assay of HIV RNA within resting CD4+ T cells to measure HIV expression at baseline, and to detect up-regulation of HIV expression in resting cells from each patient after physiological exposure to VOR.
Patients maintained suppressive ART, and purified populations of resting CD4+ T cells were obtained by continuous-flow leukopheresis and negative selection in an immunomagnetic column [7]. To establish a baseline, we measured the mean quantity of HIV-1 gag RNA in pools of 1 million resting CD4+ T cells immediately after their isolation from patients. To measure validated biomarkers of VOR effect in the PBMCs of patients, we performed parallel assays of total cellular histone acetylation and measured histone acetylation by chromatin immunoprecipitation (ChIP) at the human p21 gene promoter, a gene known to upregulate chromatin acetylation after VOR exposure [13]. Then to model the effect of a clinical dose of VOR, multiple replicate pools of 1 million resting CD4+ T cells were incubated in complete media alone, with 335 nM VOR, or with 3 µg PHA and 60 U IL-2 for 6 hours. VOR conditions were selected to mimic the unbound drug exposure expected after a single 400 mg dose of VOR in vivo [4].
Validation assays were performed in resting CD4+ T cells isolated by leukapheresis from 16 patients with plasma HIV RNA BDL (Fig. 1A). In each patient a total of 48–72 million highly purified resting CD4+ T cells were studied; 12–48 million cells in each condition, depending on cell availability. In 9 patients baseline expression following 6 hours of culture of 16–24 million cells without stimulation in media alone, HIV gag RNA was quantifiable at a mean level of 52 +/− 32 copies/million cells. However, in the other 7 patients in whom 12–24 million cells were studied (Fig. 1A), HIV RNA was not quantifiable at a limit of 10 copies/million cells, although in all but 2 of these patients RNA was detected but not quantifiable (>0 but <10 copies/million cells).
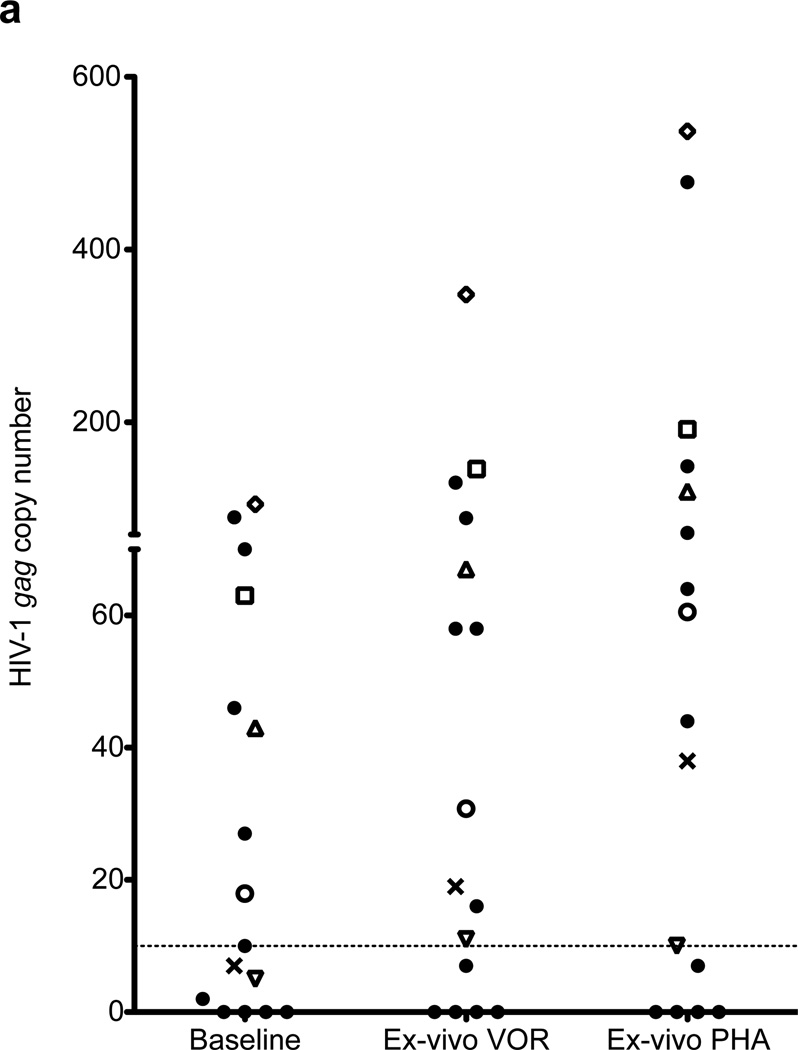
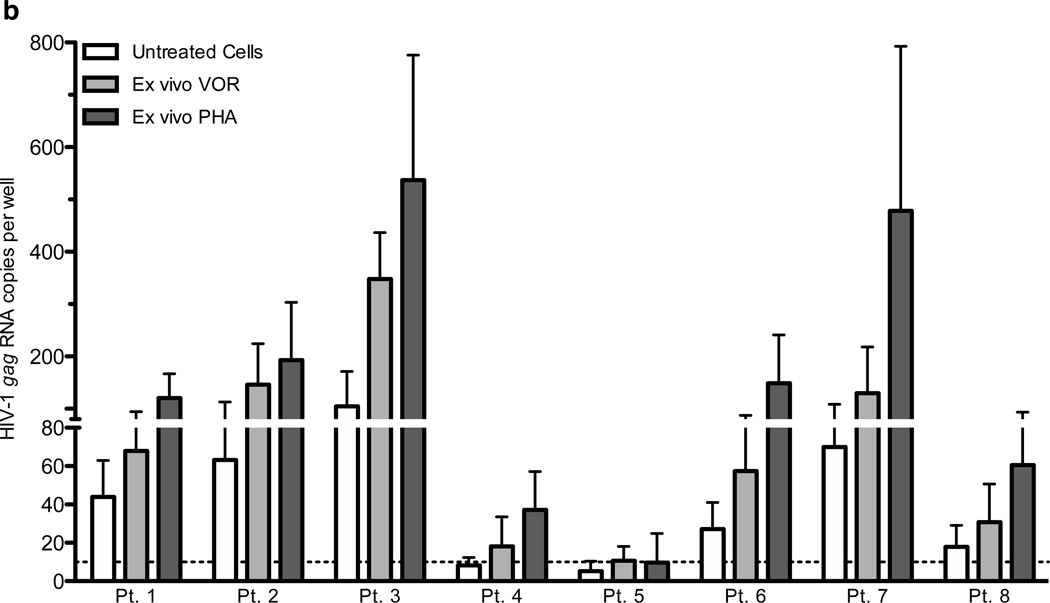
Figure 1. The relative HIV-1 RNA copy number in resting CD4+ T cells of 13 ART-treated HIV+ patients with plasma HIV RNA BDL.
Values are calculated by cycle number, and limit of quantitation of cell-associated RNA is 10 copies. Cells were cultured alone (untreated), with VOR 335 nM (VOR), or activated with 3 µg/ml PHA and 60 U/ml IL2 for 6 hours. Data from 16 patients (a), and those who later received in vivo dosing [1 (Δ), 2 (□), 3 (◊), 4 (X), 5 (▽), 6 (❍), 7 (+), and 8 (⨀)] are shown in detail (b, mean and s.d.).
Following in vitro exposure to 335 nM VOR for 6 hours, HIV RNA expression was significantly upregulated in 8 of 9 patients in whom resting CD4+ T cell HIV RNA was quantifiable without HDAC inhibitor exposure, and also in 3 of 7 patients in whom cell-associated HIV RNA was < 10 copies before HDAC inhibitor exposure. In all 11 patients in whom expression was induced by VOR, HIV RNA expression was also induced after 6 hours of exposure to PHA. Levels of HIV RNA induction were similar after 6 hours of exposure to PHA or VOR, although in other experiments (data not shown) levels of HIV RNA plateaued after 6 hours of exposure to VOR, but continued to increase when PHA exposure was extended for up to 16 hrs.
Of the 11 eligible patients in whom an induction of HIV RNA expression following in vitro exposure to VOR could be measured, eight patients (Fig. 1B; CD4 count 432–1147/µl, mean 713/µl) continued their long-term ART (tenofovir, emtricitabine, and efavirenz in all) and agreed to receive a single dose of VOR at 200 mg to ascertain tolerability. Global cellular histone acetylation in PBMCs was unchanged up to 24 hours after dosing, and histone acetylation at the human p21 gene promoter was modestly increased in only two patients (data not shown).
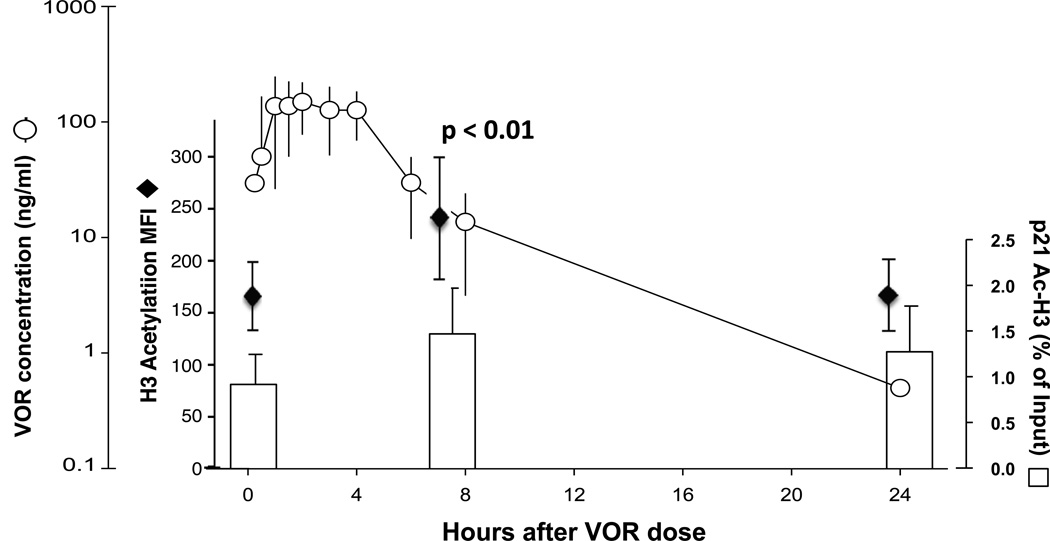
Two to four weeks later the safety and tolerability of a 400 mg dose of VOR was assessed, including acetylation measurements, and VOR plasma concentrations measured serially up to 24 hours after dosing (Fig. 2). Maximum VOR concentrations of 244 ng/ml (median, range 153–301) occurred a median of two hours (range 0.5–4) after dosing. VOR pharmacokinetics were similar to those reported in other populations [14, 15]. This exposure resulted in a significant increase (p < 0.01) in acetylation of total cellular histone H3 (median 1.6-fold) in all eight patients, and trend towards increased acetylation of histones at the human p21 gene as measured by chromatin immunoprecipitation in patients 2–5, and 7 for whom sufficient cells were available (Fig. 2).
Figure 2. VOR exposure and histone acetylation.
Median VOR plasma concentrations in patients 1–8 after a single 400 mg oral dose (circles with range shown) is shown in comparison to mean total cellular acetylated histone H3 in PBMCs (diamonds with s.d. shown), and relative levels of acetylated histone H3 at the human p21 gene promoter in resting CD4+ T cells (bars, mean and s.d.). A significant increase (†) in cellular acetylated histone H3 is seen at 8 hours, p < 0.01. A trend towards increased acetylation at the p21 gene is seen in the patients (2–5, 7) in whom sufficient cells were available for analysis.
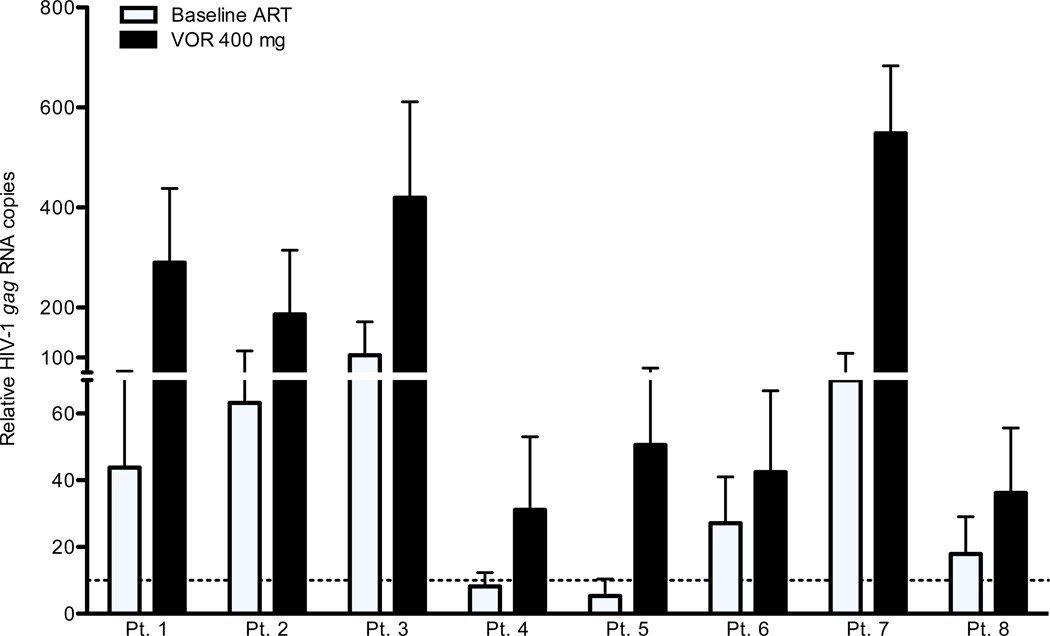
Four to five weeks later a second dose of 400 mg of VOR was administered and resting CD4+ T cells collected 4–7 hours later, after the previously measured peak VOR level. We measured HIV RNA in multiple pools of resting CD4+ T cells within this window of time, when cellular biomarkers of increased acetylation had been previously documented, and induction of HIV LTR expression mediated by HDAC inhibitor activity would be possible. During exposure to VOR, an increase of 1.5- to 10.0-fold (mean 4.8) in expression of unspliced HIV-1 gag RNA within resting CD4+ T cells was measured in all seven patients (Fig. 3). Expression following the 400 mg dose was significantly increased (p<0.01) when compared to baseline levels of RNA expression for all patients. Patients 1 and 2 underwent leukapheresis 3 to 4 months after dosing and protocol completion to provide cells for other research, and in both cases a statistically significant decline in HIV RNA expression per million resting CD4+ T cells after VOR dosing was observed (patient 1: after VOR 290 copies, off-protocol 70 copies/106 cells, p < 0.001; patient 2: after VOR 186 copies, off-protocol 105 copies/106 cells, p = 0.003). These observations support the conclusion that the increase in HIV RNA expression was causally related to VOR exposure. This conclusion assumes that the isolation of RNA from resting CD4+ T cells largely excludes actively infected cells, and reflects RNA expression in cells that are functionally defined as latently infected [16].
Figure 3. VOR upregulates HIV RNA expression.
The relative HIV-1 RNA copy number (mean, s.d.) measured in the resting CD4+ T cells of eight HIV+ patients with plasma HIV RNA BDL is shown on background ART and on ART following a single 400 mg oral dose of VOR. For each subject, the differences are significant (p < 0.01).
Throughout this limited exposure, VOR was well tolerated with no adverse events greater than Grade I, none of which were attributable to VOR. Additionally, we measured single-copy assay viremia [17] on four occasions prior to VOR exposure, and at 8 and 24 hours after doses of 200, 400, and 400 mg of VOR (10 assays in total). Despite the upregulation of HIV RNA expression measured in circulating resting CD4+ T cells, no significant change of low-level viremia was observed. Median plasma HIV RNA/ml by single-copy assay was 13 (range 3 to 23), < 1.0 (range <1 to 3), 2 (range < 1 to 3), < 1 (range <1 to 1), 3 (range 1 to 6), < 1 (range <1 to 5), <1 (range <1 to 1.2), and <1 in all assays, with no consistent trend towards an increase in plasma HIV RNA after VOR exposure.
This study is the first clear demonstration that the quiescence of latent, integrated HIV provirus within resting CD4+ T cells, a significant barrier to the eradication of HIV infection, can be disrupted by an achievable and tolerable exposure to an HDAC inhibitor. A single, clinically tolerable dose of VOR induces the expected biological effect --- histone acetylation --- consistent with HDAC inhibitor exposure within the PBMCs of HIV-infected, ART-treated patients. These effects are temporally associated with increased levels of HIV RNA expression detected within resting CD4+ T cells, demonstrating that, at least for a period of time in some infected cells, all of the restrictions that limit the expression of latent proviral genomes have been overcome. Nevertheless, while histone deacetylation is associated with HIV RNA expression, the precise molecular mechanisms through which VOR mediates this effect remain to be fully explained.
HIV RNA induction in vivo often appeared to be of greater magnitude than that seen in vitro, perhaps due to an underestimation in the modeling of physiological exposure in vitro, or other phenomenon induced by drug exposure in vivo. Further testing will be required to determine if the in vitro assay presented herein is predictive of a subset of patients who do not respond to VOR, or if such pre-screening is unnecessary. Nevertheless, assays of HIV RNA associated with resting CD4+ T cells may be useful in the pre-clinical and clinical testing of new and combination approaches to disrupt HIV latency.
Precise studies will be required to determine which dosing regimens of VOR or other HDAC inhibitors are safe and result in sustained disruption of HIV latency. We did not observe an alteration of low-level viremia, perhaps due to the very low levels of plasma viremia present in these patients at baseline, that our sampling did not capture the brief effect of a single exposure to VOR, or that stable, low-level viremia reflects virion production from sources other than resting CD4+ T cells. Further, a limited evaluation did not reveal a substantial reduction in the frequency of replication-competent HIV within resting CD4+ T cells (data not shown). This is not surprising as such an effect is likely to require more than a single dose of VOR, or additional interventions to clear infected cells. These are important goals for future studies.
VOR, like the HDAC inhibitor in wide clinical use, valproic acid, is deemed a mutagen as predicted by the Ames test in bacteria, although DNA damage induced by VOR is known to be repaired in normal human cells [18, 19]. Mutations in normal cells are not observed following long-term growth in the presence of VOR [20]. Nevertheless, the risks and benefits of attempts to eradicate HIV infection will have to be carefully weighed. While it remains to be seen if the use of VOR in combination with suppressive ART is sufficient to result in the depletion of latently infected resting CD4+ T cells, or whether additional interventions are required, these findings demonstrate that therapy targeted at persistent, latent infection within resting CD4+ T cells is feasible, and open the way for the development of HDAC inhibitors with improved specificity, potency, and safety profiles for the selective targeting of latent proviral genomes.
Methods
Patients/Study Procedures
HIV-infected patients receiving stable, standard-of-care ART with plasma HIV-1 RNA < 50 copies/ml and a CD4 count of >300/µl for at least 6 months were enrolled following informed consent. Studies were approved by the UNC institutional biomedical review board and the Food and Drug Administration. Leukapheresis was performed and ex vivo VOR response assayed. Patients with ex vivo VOR response were then administered a 200 mg dose of VOR (donated by Merck Research Laboratories, West Point, PA) to assess safety and tolerability, followed by a 400 mg dose 4 or more weeks later during which pharmacokinetic parameters, including maximum concentration (Cmax) peak and apparent oral clearance (CL/F), as well as levels of global cellular histone and human p21 promoter histone H3 acetylation. A second 400 mg dose of VOR was administered 4 or more weeks later, and apheresis spanning the time point of measured Cmax VOR concentrations was performed to assay in vivo VOR response.
Measurement of Resting CD4+ T cell-associated HIV-1 RNA
Leukapheresis and isolation of resting CD4+ T cells have been described elsewhere [4]. To measure the ex vivo response of resting CD4+ T cells to VOR, cells were incubated in media containing 335 nM VOR (Merck Research Laboratories) or 3 µg/ml PHA (Remel, Lenexa, KS) and 60 U/ml IL-2 or media alone for 6 hrs. Cells were washed and plated at 1 million/ well in a 96-well plate and pelleted. Cell pellets were snap frozen and stored at −80°C. To measure the in vivo response to VOR, immediately following leukapheresis, resting CD4+ T-cells were isolated and plated at 1 million cells/ well, pelleted, snap frozen, and stored at −80°C.
Total RNA was isolated from 12 to 48 pools of 1 million resting cells using the Magmax 96 Total RNA isolation kit (Ambion, Austin, TX) following the manufacturer’s protocol. Duplicate pools of cDNA were synthesized from DNase-treated, isolated RNA using the SuperScript® III First-Strand Synthesis SuperMix kit (Invitrogen, Carlsbad, CA) according to the manufacturer’s procedures. Two additional duplicates wells from each treatment condition did not include reverse transcriptase and served as control for DNA contamination. Duplicate PCR amplification of duplicate cDNA was performed using the ABI7500 Fast Real-Time PCR machine and previously published primers and probe [20]. A standard curve was generated for each PCR reaction using cDNA synthesized from in-vitro transcribed RNA where the p5’ plasmid served as template [22]. Results of the four PCR replicates representing each of the original 12 to 48 pools of RNA were averaged and the standard deviation determined for each condition. Inputs for samples in which basal and VOR-induced HIV RNA expression were measured (Fig. 3) were compared by the quantitation of TATA Binding Protein RNA [23] and the quantitation of total HIV DNA, and showed no trend towards higher values in the post-VOR samples.
We determined that our HIV RNA PCR assay could detect the difference between 1 copy and ≥10 copies using dilutions of an HIV RNA internal standard [24]. Detectable PCR signal less than 10 copies (1–9 copies) was treated in all analyses as 5 copies. No PCR signal or < 1 copy was treated in all analyses as 0 copies. As the entire pool of cDNA was not amplified, and individual PCR amplifications are therefore subject to stochastic sampling effects, we have termed the result “relative HIV-1 gag RNA copies.” The Wilcoxon rank sum test was used to calculate the statistical significance of all comparisons between conditions.
Measurement of total histone acetylation by flow cytometry
PBMCs collected before and after 400 mg VOR dosing were fixed and permeabilized using Phosflow fix buffer I and Phosphflow permeabilization buffer II (BD Biosciences, San Diego, CA) according to the manufacturer’s protocol. Cells were then washed in stain buffer (2% FBS, 0.09 % sodium azide), blocked with 8% normal goat serum (Invitrogen), and incubated with anti-acetyl histone H3 (1:100 dilution, catalog #06-599, Millipore, Billerica, MA) or control rabbit IgG, in blocking solution for 60 minutes at room temperature. Cells were then washed and incubated with goat-anti-Rabbit IgG FITC conjugated secondary antibody (1: 250 dilution, Millipore, Billerica, MA) in stain buffer for 30 min at 25°C in the dark. Following a final wash, cells were analyzed by flow cytometry using a CyAn ADP flow cytometer and Summit 4.3 software (Beckman Coulter Inc, Miami, FL).
Chromatin Immunoprecipitation (ChIP)
ChIP assays were performed as previously described [8] with the following modifications: PBMCs were crossed-linked with 1% Formaldehyde and nuclei extracted using the appropriate buffers (Diagenode, Denville, NJ). Nuclei were lysed using an SDS lysis buffer containing mammalian protease cocktail inhibitor (Sigma, St Louis, MO) and sonicated to fragment chromatin to 500–1000 base pairs using a Bioruptor standard sonicator (Diagenode, Denville, NJ). 1 × 106 sonicated cells were used to set up each immunoprecipitation reaction using 5 µg of anti-acetyl-histone H3 (Ac-H3 catalogue #17–615, Millipore) or rabbit pre-immune immunoglobulin G (Sigma). PCR of immunoprecipitates or input DNA was performed using primers targeting the human p21 promoter [25]. The percent of immunoprecipitated p21 promoter DNA was determined by comparing the cycle threshold values of each reaction to a standard curve generated from input DNA and is reported as the percent of input.
Measurement of Vorinostat PK
Five milliliters of blood was collected without anticoagulant for pharmacokinetic assays before dose and at 0.25, 0.50, 1.0, 1.5, 2.0, 3.0, 4.0, 6.0, 8.0, and 24 hours after dosing. Blood samples were allowed to clot at room temperature for 30 minutes, centrifuged at 2,000× g for 15 minutes at 4°C, and stored at −70°C until analysis. Vorinostat concentrations were measured from human serum using a modification of the method by Du et al. [14]. Individual serum concentrations were used to estimate vorinostat pharmacokinetic variables AUC0–24h, Cmax, Tmax, CL/F, and apparent terminal t1/2. WinNonlin Phoenix was used for the calculations. The apparent terminal t1/2 was estimated from the best-fit variables of a single exponential to the log-linear portion of the serum concentration/time curve using unweighted linear regression. AUC0–24h was calculated using the linear up/log down method. Cmax and Tmax were obtained by inspection of the concentration-time data.
Acknowledgements
This study was supported by National Institutes of Health grants AI084553, AI095052, and AI096113, and a grant from Merck & Co. to DMM, National Institutes of Health grants RR024383 to the UNC TRaCS Institute, AI50410 to the UNC Center for AIDS Research, and an equipment grant from the James B. Pendleton Charitable Trust. Merck & Co. provided VOR. We thank NP Dahl, R Sackmann, M Cheema, A Wiegand, and N White for technical support, Y. Park and the staff the UNC Blood Bank, R.J. Bedimo and C.M. van der Horst for study oversight, and J. Scepanski and A. Sugarbaker for patient follow-up and study coordination. We thank MS Cohen, JV Garcia-Martinez, WC Greene, J Karn, RF Siliciano for helpful discussions. Finally, we are grateful for the contribution of the patients who have participated in these studies.
Competing financial interests are declared as DJ Hazuda is an employee of Merck and Co., and JJ Eron and DM Margolis have received grants and consulting honoraria from Merck and Co. NM Archin, DJ Hazuda, and DM Margolis are co-inventors of a patent for the use of VOR in HIV infection, but to lead this study DM Margolis has remanded ownership of this patent to UNC Chapel Hill.
Footnotes
Author Contributions: NM Archin, AL Liberty, SK Choudhary, DJ Hazuda and DM Margolis designed and performed primary study assays and performed study analysis. JD Kuruc, AD Crooks, JJ Eron, and DM Margolis designed, implemented, and oversaw the clinical protocol. MG Hudgens and RJ Bosch designed the data analysis. AD Kashuba oversaw PK assays and analyzed this data. MF Kearney, E Anderson, and JM Coffin designed and performed single-copy assays.
REFERENCES
- 1.Blankson JN, Persaud D, Siliciano RF. The challenge of viral reservoirs in HIV-1 infection. Annu Rev Med. 2002;53:557. doi: 10.1146/annurev.med.53.082901.104024. [DOI] [PubMed] [Google Scholar]
- 2.Margolis DM. Histone deacetylase inhibitors and HIV latency. Curr Opin HIV AIDS. 2011;6:25. doi: 10.1097/COH.0b013e328341242d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Archin NM, et al. Expression of Latent Human Immunodeficiency Type-1 is Induced by Novel and Selective Histone Deacetylase Inhibitors. AIDS. 2009;23:1799. doi: 10.1097/QAD.0b013e32832ec1dc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Archin NM, et al. Expression of Latent HIV Induced by the Potent HDAC Inhibitor Suberoylanilide Hydroxamic Acid. AIDS Res Hum Retroviruses. 2009;25:207. doi: 10.1089/aid.2008.0191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Contreras X, et al. Suberoylanilide hydroxamic acid reactivates HIV from latently infected cells. J Biol Chem. 2009;284:6782. doi: 10.1074/jbc.M807898200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lehrman G, et al. Depletion of latent HIV infection in vivo: a proof of concept study. Lancet. 2005;366:549. doi: 10.1016/S0140-6736(05)67098-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Archin NM, et al. Valproic acid without intensified antiviral therapy has limited impact on persistent HIV infection of resting CD4+ T cells. AIDS. 2008;22:1131. doi: 10.1097/QAD.0b013e3282fd6df4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siliciano JD, et al. Stability of the latent reservoir for HIV-1 in patients receiving valproic acid. J Infect Dis. 2007;195:833. doi: 10.1086/511823. [DOI] [PubMed] [Google Scholar]
- 9.Sagot-Lerolle N, et al. Prolonged valproic acid treatment does not reduce the size of latent HIV reservoir. AIDS. 2008;22:1125. doi: 10.1097/QAD.0b013e3282fd6ddc. [DOI] [PubMed] [Google Scholar]
- 10.Archin NM, et al. Antiretroviral intensification and valproic acid have limited effect on residual HIV-1 viremia or resting CD4+ cell infection. PLoS One. 2010;5:e9390. doi: 10.1371/journal.pone.0009390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keedy KS, et al. A limited group of class I histone deacetylases act to repress human immunodeficiency virus type-1 expression. J Virol. 2009;83:4749. doi: 10.1128/JVI.02585-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edelstein LC, Micheva-Viteva S, Phelan BD, Dougherty JP. Activation of latent HIV type 1 gene expression by suberoylanilide hydroxamic acid (SAHA), an HDAC inhibitor approved for use to treat cutaneous T cell lymphoma. AIDS Res Hum Retroviruses. 2009;25:883. doi: 10.1089/aid.2008.0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gui CY, et al. Histone deacetylase (HDAC) inhibitor activation of p21WAF1 involves changes in promoter-associated proteins, including HDAC1. Proc Natl Acad Sci U S A. 2004;101:1241. doi: 10.1073/pnas.0307708100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Du L, Musson DG, Wang AQ. Stability studies of vorinostat and its two metabolites in human plasma, serum and urine. Rapid Commun Mass Spectrom. 2005;19:1779. doi: 10.1002/rcm.1984. [DOI] [PubMed] [Google Scholar]
- 15.Rubin EH, et al. A study to determine the effects of food and multiple dosing on the pharmacokinetics of vorinostat given orally to patients with advanced cancer. Clin Cancer Res. 2006;12:7039. doi: 10.1158/1078-0432.CCR-06-1802. [DOI] [PubMed] [Google Scholar]
- 16.Hermankova M, et al. Analysis of human immunodeficiency virus type 1 gene expression in latently infected resting CD4+ T lymphocytes in vivo. J Virol. 2003;77:7383. doi: 10.1128/JVI.77.13.7383-7392.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palmer S, et al. New real-time reverse transcriptase-initiated PCR assay with single-copy sensitivity for human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol. 2003;41:4531. doi: 10.1128/JCM.41.10.4531-4536.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee JH, Choy ML, Ngo L, Foster SS, Marks PA. Histone deacetylase inhibitor induces DNA damage, which normal but not transformed cells can repair. Proc Natl Acad Sci U S A. 2010;107:14639. doi: 10.1073/pnas.1008522107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee JH, Choy ML, Ngo L, Venta-Perez G, Marks PA. Mechanisms of Resistance to Histone Deacetylase Inhibitors: Role of Chk1. Proc Natl Acad Sci U S A. 2011;108:19629. doi: 10.1073/pnas.1117544108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marks PA. personal communication.
- 21.Israel-Ballard K, et al. TaqMan RT-PCR and VERSANT HIV-1 RNA 3.0 (bDNA) assay Quantification of HIV-1 RNA viral load in breast milk. J Clin Virol. 2005;34:253. doi: 10.1016/j.jcv.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 22.Robinson LH, Gale CV, Kleim JP. Inclusion of full-length human immunodeficiency virus type 1 (HIV-1) gag sequences in viral recombinants applied to drug susceptibility phenotyping. J Virol Methods. 2002;104:147. doi: 10.1016/s0166-0934(02)00059-9. [DOI] [PubMed] [Google Scholar]
- 23.Radonić A, et al. Guideline to reference gene selection for quantitative real-time PCR. Biochem Biophys Res Commun. 2004;313:856. doi: 10.1016/j.bbrc.2003.11.177. [DOI] [PubMed] [Google Scholar]
- 24.May RC, et al. Change-Point Models to Estimate the Limit of Detection. University of North Carolina at Chapel Hill Biostatistics Technical Report Series. Working Paper 26. 2012 Mar; http://biostatsbepresscom/uncbiostat/papers/art26.
- 25.Tonelli R, et al. G1 cell-cycle arrest and apoptosis by histone deacetylase inhibition in MLL-AF9 acute myeloid leukemia cells is p21 dependent and MLL-AF9 independent. Leukemia. 2006;20:1307. doi: 10.1038/sj.leu.2404221. [DOI] [PubMed] [Google Scholar]