SUMMARY
Jumonji histone demethylases catalyze removal of methyl marks from lysine residues in histone proteins within nucleosomes. Here, we show that the catalytic domain of demethylase JMJD2A (cJMJD2A) utilizes a distributive mechanism to remove the histone H3 lysine 9 trimethyl mark. By developing a method to assess demethylation of homogeneous, site-specifically methylated nucleosomes, we determined that the kinetic parameters for demethylation of nucleosomes by cJMJD2A are comparable to those of peptide substrates. These findings imply that other domains of the demethylase or its protein partners may contribute to nucleosome recognition in vivo, and in this way, may further regulate demethylation activity and processivity. The quantitative assays of nucleosome demethylation developed in our work provide a platform for future work with complex chromatin substrates and full-length demethylases.
INTRODUCTION
Nucleosomes are comprised of histone proteins that are subject to diverse post-translational modifications. These modifications are predominantly found in the N-terminal tails of histones and have a profound impact on the regulation of transcription. Methylation of lysine 9 in histone H3 (H3 K9) is a conserved eukaryotic modification that demarcates heterochromatin, a transcriptionally silent chromatin state. Trimethylated H3 K9 (H3 K9me3) is required for heritable gene silencing, formation of centromeres, and maintenance of telomere stability (Grewal and Jia, 2007). H3 K9 methylation has also been observed at genes that are transcriptionally silenced in cancer (Kondo et al., 2004). The extent (mono-, di- or tri-) and the half-life of methylation are controlled by opposing actions of histone methyltransferases and demethylases, as well as histone turnover.
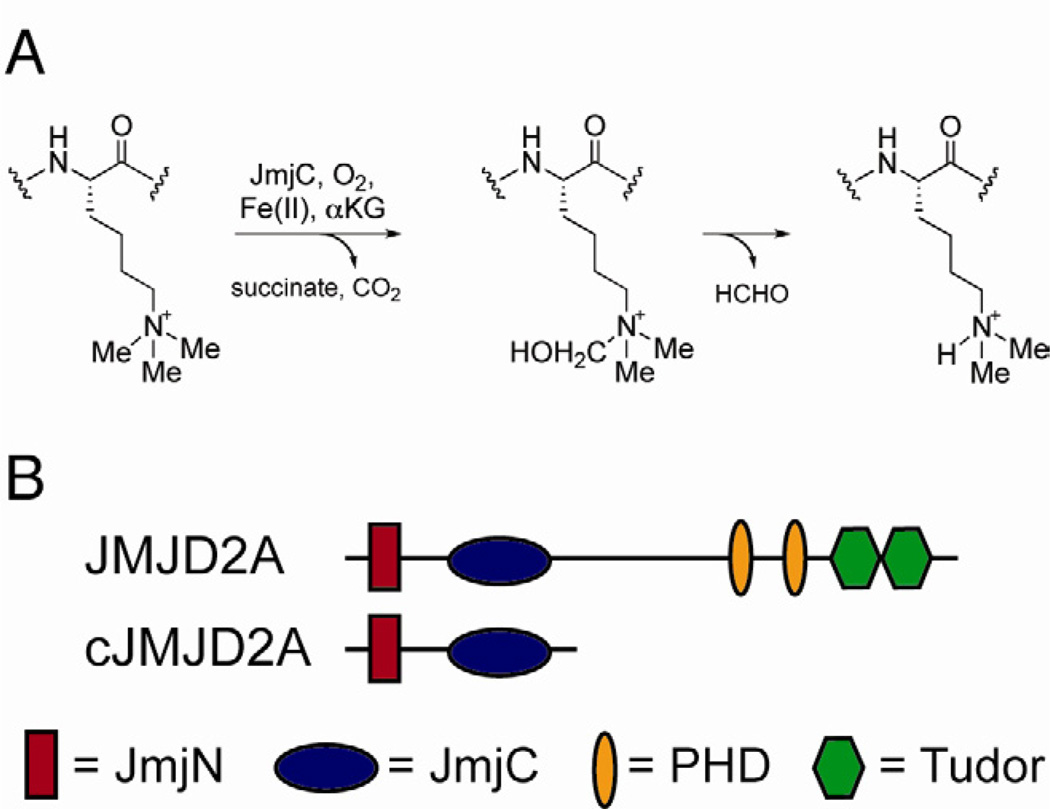
Removal of methyl groups from lysine residues in histones is carried out by two classes of demethylases: the flavin-dependent lysine specific demethylase family consisting of LSD1 and LSD2 (Karytinos et al., 2009; Shi et al., 2004) as well as the iron- and α-ketoglutarate dependent Jumonji C domain-containing demethylases (Klose et al., 2006a; Kooistra and Helin, 2012). The Jumonji family proteins catalyze a wide set of demethylation reactions on histone substrates, including removal of methyl marks from H3 K4, H3 K9, H3 K27, H3 K36 and H4 K20 (Kooistra and Helin, 2012). Common to these proteins is their ability to oxidize methyl groups of methylated lysine substrates to form hemiaminal intermediates. Subsequent release of formaldehyde yields demethylated product (Figure 1A) (Cloos et al., 2006; Ng et al., 2007; Tsukada et al., 2006).
Figure 1.
Catalysis and domain architecture of JMJD2A. (A) The abbreviated catalytic mechanism of Jumonji histone demethylases showing the hemiaminal intermediate. (B) Domain organization in JMJD2A.
The removal of transcriptionally repressive H3 K9me3 marks is carried out by the JMJD2 (also known as the KDM4) family of demethylases (Cloos et al., 2006; Fodor et al., 2006; Klose et al., 2006b; Whetstine et al., 2006). Overexpression of several members of the JMJD2 family have been implicated in various diseases (Berry et al., 2012; Cloos et al., 2006; Kawazu et al., 2011; Rui et al., 2010), underscoring the importance of precise regulation of demethylation. Members of the JMJD2 family contain JmjN, JmjC, two PHD domains, and a tandem Tudor domain (Figure 1B). While JmjN and JmjC domains form a composite active site (Couture et al., 2007; Ng et al., 2007), the double Tudor domain is thought to allow recruitment of demethylases to their target sites by binding H3 K4me3 and H4 K20me3 marks (Lee et al., 2008). The function of PHD domains in JMJD2 family is yet to be determined.
The biological significance of this demethylase family has prompted an investigation into its kinetic and structural features (Couture et al., 2007; Hillringhaus et al., 2011; Hopkinson et al., 2010; Krishnan et al., 2012; Ng et al., 2007). In all of these studies, a catalytic construct consisting of JmjN and JmjC domain was used (cJMJD2A). To date, there is no published kinetic data on the full-length enzyme. Additionally, no kinetic studies have evaluated the ability of any demethylase to remove methyl marks from intact and homogenously modified nucleosomes, a necessary first step in the analysis of the demethylation of chromatin substrates.
Herein, we analyzed the intrinsic processivity and nucleosome demethylation ability of cJMJD2A. Our study reveals that the cJMJD2A uses a distributive mechanism to affect multiple demethylations. Successful reconstitution of the demethylation of homogeneous site-specifically methylated nucleosomes has allowed us to compare the kinetic parameters for demethylation of nucleosomes and histone tail peptides. Our findings suggest that nucleosome recognition is not intrinsic to cJMJD2A and may be mediated by other domains in JMJD2A or its interacting partners. These domains and protein partners could provide a regulatory mechanism for demethylase activity and processivity in vivo. To our knowledge, this is the first report of the demethylation kinetics of nucleosomes by any demethylase. Accomplishing this task sets the stage for analysis of the demethylation of more complex chromatin substrates by JMJD2A and other jumonji demethylases.
RESULTS
Kinetic Analysis of Peptide Demethylation by cJMJD2A
To eliminate possible artifacts due to the presence of an affinity tag, we used a tag-less catalytic domain construct where the hexahistidine tag had been removed from cJMJD2A (amino acids 1–350 of JMJD2A) after purification (see Supporting Information). Using a fluorescent assay that follows the formation of the demethylation byproduct formaldehyde (Couture et al., 2007), demethylation of tri- and dimethylated H3 K9 peptides corresponding to amino acids 7–14 of the histone H3 tail (ARKme3STGGK and ARKme2STGGK) by cJMJD2A was analyzed (Figure 2A–B). In these assays, ARKme3STGGK is demethylated more efficiently than ARKme2STGGK. The dimethylated peptide exhibited a six-fold increase in KM and a two-fold decrease in turnover number compared to the trimethylated peptide (KM = 410 µM, kcat = 1.0 min−1 for ARKme2STGGK and KM = 67 µM, kcat = 1.8 min−1 for ARKme3STGGK).
Figure 2.
Kinetic analysis of cJMJD2A-mediated demethylation of peptides. (A–B) Michaelis-Menten plots of initial velocity as a function of the concentration of ARKme3STGGK (A) and ARKme2STGGK (B). Experiments were done in triplicate and are represented as mean ± SEM. (C) Demethylation of ARKme3STGGK (200 µM) by cJMJD2A (1 µM) monitored over time using LC-MSMS. Formation of dimethylated species ([ARKme2STGGK] ~ 150 µM at 15 min) in amounts that largely exceed enzyme concentration supports distributive demethylation. Lines connecting data points are introduced to facilitate visualization and do not represent kinetic fits. For experimental conditions and procedure, see Supporting Information.
We used mass spectrometry (MS) to measure the extent of demethylation of the trimethylated substrate as a function of time (Figure 2C). The rapid disappearance of trimethylated species and accumulation of dimethylated intermediates further support the observation that cJMJD2A demethylates ARKme3STGGK more efficiently than ARKme2STGGK. A slow decay of the dimethyl species and concomitant increase in monomethyl species past the initial four hours of incubation suggest that the enzyme is mostly inactive by this time.
Two consecutive demethylations are required for the formation of monomethylated product from the starting trimethylated peptide. The serial demethylation events can proceed by a processive or distributive mechanism. In a processive demethylation mechanism, the release of the intermediate dimethylated product from the enzyme is substantially slower than its conversion to monomethylated product. In such a scenario, essentially all the dimethylated species exists as bound to the enzyme and thus this intermediate cannot accumulate to amounts exceeding the concentration of the enzyme. In contrast, in a distributive mechanism, release of dimethylated intermediate is substantially faster than its conversion to monomethylated product. In such a scenario the dimethylated intermediate can accumulate in solution to levels that are greater than the concentration of the enzyme. The time course of demethylation by mass spectrometry strongly suggests that the dimethylated species accumulates to amounts greater than that of the enzyme (Figure 2C) implying that demethylation by cJMJD2A proceeds via a distributive mechanism.
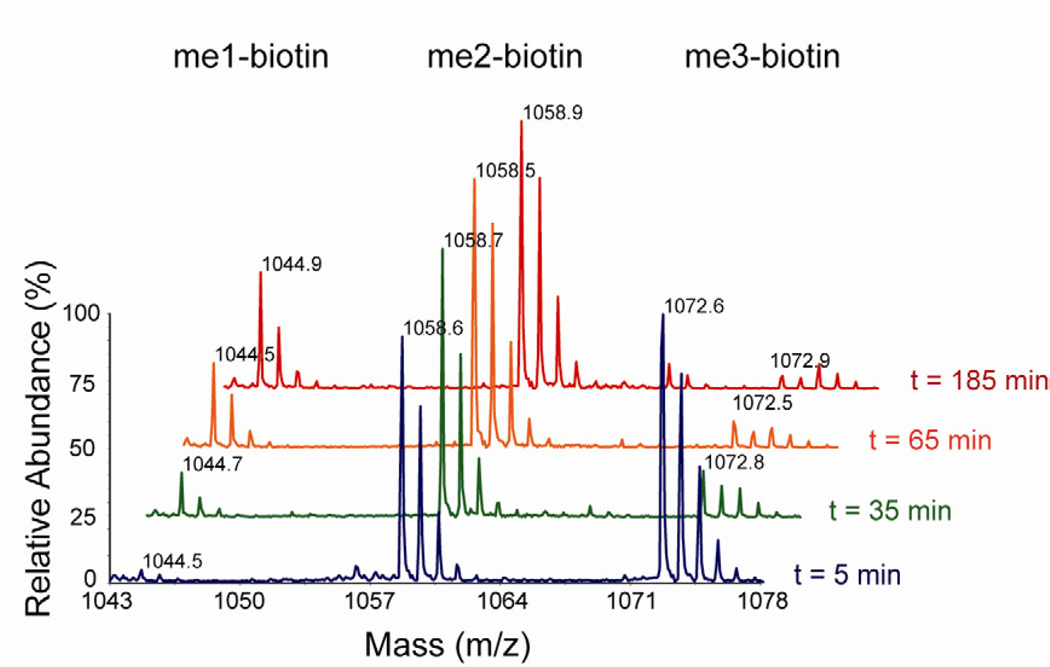
We sought to further confirm this observation in an independent experiment performed with trimethylated peptide ARKme3STGGK with the biotin tag attached to the ε-amino group of the C-terminal lysine (Figure 3). The kinetic parameters for demethylation of this peptide (Figure S1) are similar to that of non-biotinylated peptide (Figure 2). Biotinylated ARKme3STGGK (150 µM) was incubated with the demethylase (5 µM), and the extent of demethylation was monitored over time by MALDI-TOF MS. In analogy to experiments performed with untagged peptide (Figure 2C), accumulation of dimethylated species in excess of enzyme concentration was observed in all time points. This is consistent with the distributive demethylation, where the formation of monomethylated product observed in later time points requires re-binding of dimethylated species. The observation that demethylation of both biotinylated and unlabeled trimethylated peptides results in accumulation of dimethylated intermediate in amounts exceeding the concentration of the enzyme strongly supports the distributive nature of cJMJD2A.
Figure 3.
Demethylation of biotinylated ARKme3STGGK by cJMJD2A. Peptide (150 µM) was incubated with the demethylase (5 µM), and the extent of demethylation over time analyzed by MALDI-TOF MS. See also Figure S1.
Demethylation of Nucleosomes
To assess the ability of cJMJD2A to demethylate nucleosomes, we prepared homogeneous, site-specifically modified nucleosomes using methyllysine analogs (MLAs) (Simon et al., 2007). In this method, a methyl lysine mimic is incorporated into histone H3 via alkylation of a mutant cysteine residue substituting lysine 9 in histone H3, and the modified histone is subsequently incorporated into recombinant nucleosomes (Figure 4A). To quantitatively compare demethylation kinetics between analog-modified and native substrates, we first determined cJMJD2A activity on a trimethyllysine analog peptide ARKCme3STGGK (Figure 4B). We observed an approximate four-fold decrease in kcat (0.48 min−1) and five-fold increase in KM (330 µM) as compared to endogenous trimethylated peptide (Figure 2A), consistent with findings of others (Krishnan et al., 2012).
Figure 4.
Kinetic analysis of cJMJD2A-mediated demethylation of methyllysine analog (MLA)-containing peptides and nucleosomes. (A) Preparation of recombinant homogeneous H3 KC9me3 nucleosomes. Recombinant histone H3 K9C was alkylated with (2-bromoethyl)trimethylammonium bromide to form H3 KC9me3 histones. Subsequent assembly with histone H2A, H2B, H4, and 601 sequence DNA provided homogeneous H3 KC9me3 nucleosomes. (B) Michaelis-Menten plot for demethylation of MLA-containing peptide. (C) Western blot analysis of an experiment performed at 350 µM cJMJD2A. Data analysis and extrapolation of kobs is described in experimental procedures. (D) Kinetics of demethylation of MLA-containing nucleosomes. Data were obtained from three independent experiments and represented as mean ± SEM. See also Figure S2.
Demethylation of nucleosomes by cJMJD2A was carried out under single-turnover conditions with excess enzyme, a method that better enables the measurement of rates of the catalytic step (Johnson, 1992). To allow for sufficient assay sensitivity, demethylation was monitored by quantitative western blotting using antibodies specific for H3 K9me3 and H4 (for normalization). To ensure protein quantification accuracy, the amounts of nucleosomes used in these experiments were within the linear detection range of H3 K9me3 and H4 antibodies (Figure S2A). Importantly, under these conditions, no cross-reactivity was observed between H3 K9me3-specific antibody and mono- and dimethyl recombinant MLA nucleosomes (Figure S2B). By using H3 K9me3-specific antibody to monitor disappearance of starting material and an antibody against H4 as a loading control, we were able to monitor the loss of the trimethylated nucleosome substrate as a function of enzyme concentration. In these experiments, nucleosome concentration was kept constant at 300 nM, while the concentration of cJMJD2A was varied between 50 – 400 µM. For each enzyme concentration, the ratio of H3 KC9me3 to H4 signal was monitored as a function of time, which is used to determine the first-order rate constant for starting material consumption (kobs) (Figure 4C). A catalytic step rate (kmax) of 0.16 min−1 and an apparent KM (K’M) of 240 µM were determined by nonlinear regression analysis of observed first-order rate constants as a function of enzyme concentration (Figure 4D). Due to the presence of thioether moiety, it is possible that the thioether backbone of MLA can undergo oxidation under demethylation conditions, possibly precluding the trimethyl epitope recognition. However, mass spectrometric analysis of MLA peptide indicates negligible oxidation during the reaction (Figure S3).
DISCUSSION
Histone methylation, a post-translational modification critical to the regulation of transcription, is controlled by the opposing actions of histone methyltransferases and demethylases. Tight control of the site and extent (mono-, di-, or tri-) of methylation is crucial for proper cellular function. Jumonji histone demethylases are critical regulators of nucleosomal methylation, and their misregulation is associated with cancer and neurological disorders (Kooistra and Helin, 2012). By antagonizing repressive H3 K9 di- and trimethylation, enzymes belonging to JMJD2 family of demethylases have a crucial impact on transcription, an effect well studied in the context of nuclear hormone receptor mediated transcription (Wissmann et al., 2007). Herein, we report our findings on the processivity and kinetic analysis of nucleosome demethylation by cJMJD2A. Determining the intrinsic behavior of the catalytic domain of JMJD2A provides a better understanding of how this demethylase may be regulated.
The ability of cJMJD2A to catalyze demethylation was first analyzed on methylated histone tail peptides. The kinetic parameters obtained show high turnover for this family of enzymes (kcat(ARKme3STGGK) = 1.8 min−1; kcat(ARKme2STGGK) = 1.0 min−1) (Figure 2), indicating that enzyme obtained in this manner is well behaved and comparable to the most active preparations published (Krishnan et al., 2012). Our findings also indicate that the trimethylated peptide is a better substrate than the dimethylated variant for cJMJD2A, predominantly due to differences in KM (KM(ARKme3STGGK) = 67 µM; KM(ARKme2STGGK) = 410 µM). The preference for trimethylated substrate is consistent with previous reports using His-tagged catalytic constructs of JMJD2A (Couture et al., 2007; Hillringhaus et al., 2011). Additionally, mass spectrometric monitoring of demethylation of the trimethylated peptide over time showed the rapid disappearance of trimethylated species and the accumulation of the intermediate dimethylated peptide, which slowly decays to a monomethyl product (Figure 2C). These findings are consistent with our kinetic measurements indicating strong preference for trimethylated peptide.
Mechanistically, multiple methyl marks can be demethylated either distributively or processively, depending on whether peptide substrates dissociate or remain enzyme-bound between consecutive demethylations. Our findings indicate that the catalytic domain of JMJD2A by itself is distributive in its action. The release of the dimethyl intermediate may have a significant impact on the regulation of transcription. For example, HP1 is recruited to heterochromatic loci by specific association with H3 K9me2/3, while sites containing H3 K9me1/0 generally lack HP1 and remain euchromatic (Bannister et al., 2001; Canzio et al., 2011; Fischle et al., 2008; Grewal and Jia, 2007; Lachner et al., 2001). Thus in one possible scenario, the release of the dimethyl peptide may serve as an additional check-point before full commitment to the functional output signaled by the monomethylated state. Moreover, the observation that the dimethylated peptide is a ten-fold poorer substrate than the trimethylated peptide raises the possibility that demethylation of dimethylated substrates may, in some instances, be carried out by other demethylases. For example, in the context of androgen receptor-mediated transcription, JMJD2C associates with a flavoprotein demethylase LSD1, and in this complex, LSD1 acts on mono- and dimethyl H3 K9 marks (Metzger et al., 2005; Wissmann et al., 2007). Like JMJD2C, JMJD2A is also known to associate with LSD1 in an androgen receptor-dependent fashion (Kauffman et al., 2011).
Our data further imply that cJMJD2A does not significantly discriminate between histone tail peptide and nucleosomal substrates. The kcat/KM values for these two substrates are within 2.5-fold of one another (kcat/KM (ARKC9me3STGGK) = 1.5 × 10−3 µM−1min−1 and kmax/K’M (H3 KC9me3 Nuc) = 0.67 × 10−3 µM−1min−1). This result implies that the catalytic domain of JMJD2A predominantly recognizes residues immediately surrounding the H3 K9, and does not recognize additional features on the nucleosome. Recognition of other chromatin features or other modifications by the double Tudor or PHD domains of the full-length demethylase may result in tighter association and an increase in demethylase activity. Such additional interactions may also increase the processivity of the catalytic domain. By analogy, protein interacting partners may also contribute to the regulation of activity and processivity. In vivo, it is plausible that in the presence of the relevant chromatin marks and/or protein partners, full-length JMJD2A may demethylate in a processive manner yielding monomethylated product. Such regulation of processivity would have significant implications for controlling the specific output of JMJD2A proteins in a context dependent manner (Bua et al., 2009; Collins et al., 2008; Iwase et al., 2007; Kim et al., 2006; Rottach et al., 2010).
Overall, our ability to quantitatively assess the removal of methyl marks from site-specifically methylated nucleosomes by a demethylase is the necessary first step in further detailed investigations of chromatin modification by demethylases and demethylase-containing protein complexes. These studies will also address the importance of the cross-talk between chromatin marks in the regulation of histone methylation.
SIGNIFICANCE
Histone lysine methylation within nucleosomes is a reversible process catalyzed by opposing actions of histone methyltransferases and histone demethylases. As precise control of the methylation extent of nucleosomes is required for proper gene expression, nucleosome methylation must be tightly regulated. Jumonji histone demethylases are a class of Fe(II) and α-ketoglutarate-dependent dioxygenases that catalyze methyl removal from a number of methylated lysine residues, and their misregulation is correlated with diseases. By quantitatively analyzing the demethylation activity of a catalytic construct of JMJD2A, a member of the human Jumonji demethylases, we provide insight into the ability of a prototypic member of the family to remove a methyl mark. The results point to two important features of the cJMJD2A: an intrinsic distributive nature and its predominant recognition of the histone tail segment surrounding the methyl lysine residue. Our studies directly raise the possibility that the activity and processivity of the catalytic domain may be regulated in a context-specific manner by domains outside the catalytic domain. The method developed here will enable future quantitative studies on mechanistic roles of the non-catalytic regions as well as interacting proteins of demethylases in regulating demethylation of nucleosomes.
EXPERIMENTAL PROCEDURES
Demethylation of Nucleosomes
Demethylations of nucleosomes were performed by incubation of nucleosomes containing H3 KC9me3 (300 nM) with α-ketoglutarate (1 mM), ascorbate (1 mM), varying concentrations of cJMJD2A (50 – 400 µM), and Fe(NH4)2(SO4)2 (twice the concentration of cJMJD2A) in 50 mM Hepes (pH 7.5) at room temperature. Reactions were initiated by addition of methylated nucleosomes. Time points were quenched using 3:1 mixture of 6× SDS sample loading buffer and 0.5 M EDTA pH 8.0 and boiled at 100 °C for 2 min. Samples were run on 15% Criterion gels at 200 V for 40 min and transferred onto Immun-blot PVDF membranes (Bio-Rad) at 4 °C for 2 hr at 600 mAmp. Membranes were blocked at room temperature in 10 mL Odyssey Blocking Buffer (LI-COR Biosciences) for 75 min, and then incubated overnight with H3 K9me3 antibody (Upstate 07-442, Lot: 2017310) and H4 antibody (Active Motif 39269, Lot: 11908001) at 1:800 and 1:1000 dilutions, respectively in 1:1 mixture of 1× PBS and Odyssey Blocking Buffer containing 0.02% Tween-20 at 4 °C. Membranes were subsequently washed with 1× TBS and 0.05% Tween-20 (4 times, 4 min each), and then incubated with IRDye 680LT Goat Anti-Rabbit secondary antibody (LI-COR 926-68021, Lot: C10628-01) at 1:20,000 dilutions in 1:1 mixture of 1× PBS and Odyssey Blocking Buffer containing 0.02% Tween-20 for 40 min at room temperature. Following this, membranes were washed as described above and analyzed using Odyssey Application Software (LI-COR Biosciences).
Data were analyzed by dividing fluorescent antibody signal of H3 KC9me3 by that of H4. These ratios were normalized to the H3 KC9me3/H4 ratio at time zero, graphed as a function of time, and fitted to the equation [H3 KC9me3] = [H3 KC9me3]t=0e−kobst. Obtained kobs values were then graphed against cJMJD2A concentrations and fit to kobs = (kmaxX)/(X+K’M) with nonlinear regression to determine kmax and K’M parameters, where X is the concentration of cJMJD2A.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Lim, Shokat, and Wells laboratories for use of their instruments. We thank Lindsey Pack and the past and present Fujimori lab members for helpful discussions. We also thank Drs. Lisa Racki and Daniele Canzio for their help in nucleosome reconstitutions. This work is supported by the American Heart Association Predoctoral Fellowship (to C.S.), UCSF Program for Breakthrough Biomedical Research, Sidney Kimmel Foundation for Cancer Research, Basil O'Connor Starter Scholar Research Award, and Searle Scholars Program (to D.G.F.). Mass Spectrometry was provided by the Bio-Organic Biomedical Mass Spectrometry Resource at UCSF supported by NIH NIGMS P41GM103481 and 1S10RR026662.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- Berry WL, Shin S, Lightfoot SA, Janknecht R. Oncogenic features of the JMJD2A histone demethylase in breast cancer. Int. J. Oncol. 2012;41:1701–1706. doi: 10.3892/ijo.2012.1618. [DOI] [PubMed] [Google Scholar]
- Bua DJ, Kuo AJ, Cheung P, Liu CL, Migliori V, Espejo A, Casadio F, Bassi C, Amati B, Bedford MT, et al. Epigenome microarray platform for proteome-wide dissection of chromatin-signaling networks. PLoS ONE. 2009;4:e6789. doi: 10.1371/journal.pone.0006789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canzio D, Chang EY, Shankar S, Kuchenbecker KM, Simon MD, Madhani HD, Narlikar GJ, Al-Sady B. Chromodomain-Mediated Oligomerization of HP1 Suggests a Nucleosome-Bridging Mechanism for Heterochromatin Assembly. Mol. Cell. 2011;41:67–81. doi: 10.1016/j.molcel.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloos PAC, Christensen J, Agger K, Maiolica A, Rappsilber J, Antal T, Hansen KH, Helin K. The putative oncogene GASC1 demethylates tri- and dimethylated lysine 9 on histone H3. Nature. 2006;442:307–311. doi: 10.1038/nature04837. [DOI] [PubMed] [Google Scholar]
- Collins RE, Northrop JP, Horton JR, Lee DY, Zhang X, Stallcup MR, Cheng X. The ankyrin repeats of G9a and GLP histone methyltransferases are mono- and dimethyllysine binding modules. Nat. Struct. Mol. Biol. 2008;15:245–250. doi: 10.1038/nsmb.1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couture J-F, Collazo E, Ortiz-Tello PA, Brunzelle JS, Trievel RC. Specificity and mechanism of JMJD2A, a trimethyllysine-specific histone demethylase. Nat. Struct. Mol. Biol. 2007;14:689–695. doi: 10.1038/nsmb1273. [DOI] [PubMed] [Google Scholar]
- Fischle W, Franz H, Jacobs SA, Allis CD, Khorasanizadeh S. Specificity of the chromodomain Y chromosome family of chromodomains for lysine-methylated ARK(S/T) motifs. J. Biol. Chem. 2008;283:19626–19635. doi: 10.1074/jbc.M802655200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fodor BD, Kubicek S, Yonezawa M, O'Sullivan RJ, Sengupta R, Perez-Burgos L, Opravil S, Mechtler K, Schotta G, Jenuwein T. Jmjd2b antagonizes H3K9 trimethylation at pericentric heterochromatin in mammalian cells. Genes Dev. 2006;20:1557–1562. doi: 10.1101/gad.388206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SI, Jia S. Heterochromatin revisited. Nat. Rev. Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- Hillringhaus L, Yue WW, Rose NR, Ng SS, Gileadi C, Loenarz C, Bello SH, Bray JE, Schofield CJ, Oppermann U. Structural and evolutionary basis for the dual substrate selectivity of human KDM4 histone demethylase family. J. Biol. Chem. 2011;286:41616–41625. doi: 10.1074/jbc.M111.283689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkinson RJ, Hamed RB, Rose NR, Claridge TD, Schofield CJ. Monitoring the activity of 2-oxoglutarate dependent histone demethylases by NMR spectroscopy: direct observation of formaldehyde. Chembiochem. 2010;11:506–510. doi: 10.1002/cbic.200900713. [DOI] [PubMed] [Google Scholar]
- Iwase S, Lan F, Bayliss P, de la Torre-Ubieta L, Huarte M, Qi HH, Whetstine JR, Bonni A, Roberts TM, Shi Y. The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell. 2007;128:1077–1088. doi: 10.1016/j.cell.2007.02.017. [DOI] [PubMed] [Google Scholar]
- Johnson KA. Transient-State Kinetic Analysis of Enzyme Reaction Pathways. In: Sigman DS, editor. The Enzymes. Vol. 20. Academic Press; 1992. pp. 1–61. [Google Scholar]
- Karytinos A, Forneris F, Profumo A, Ciossani G, Battaglioli E, Binda C, Mattevi A. A novel mammalian flavin-dependent histone demethylase. J. Biol. Chem. 2009;284:17775–17782. doi: 10.1074/jbc.M109.003087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffman EC, Robinson BD, Downes MJ, Powell LG, Lee MM, Scherr DS, Gudas LJ, Mongan NP. Role of androgen receptor and associated lysine-demethylase coregulators, LSD1 and JMJD2A, in localized and advanced human bladder cancer. Mol. Carcinog. 2011;50:931–944. doi: 10.1002/mc.20758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawazu M, Saso K, Tong KI, McQuire T, Goto K, Son DO, Wakeham A, Miyagishi M, Mak TW, Okada H. Histone demethylase JMJD2B functions as a co-factor of estrogen receptor in breast cancer proliferation and mammary gland development. PLoS ONE. 2011;6:e17830. doi: 10.1371/journal.pone.0017830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Daniel J, Espejo A, Lake A, Krishna M, Xia L, Zhang Y, Bedford MT. Tudor, MBT and chromo domains gauge the degree of lysine methylation. EMBO Rep. 2006;7:397–403. doi: 10.1038/sj.embor.7400625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Kallin EM, Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nat. Rev. Genet. 2006a;7:715–727. doi: 10.1038/nrg1945. [DOI] [PubMed] [Google Scholar]
- Klose RJ, Yamane K, Bae Y, Zhang D, Erdjument-Bromage H, Tempst P, Wong J, Zhang Y. The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature. 2006b;442:312–316. doi: 10.1038/nature04853. [DOI] [PubMed] [Google Scholar]
- Kondo Y, Shen L, Yan PS, Huang TH, Issa JP. Chromatin immunoprecipitation microarrays for identification of genes silenced by histone H3 lysine 9 methylation. Proc. Natl. Acad. Sci. U S A. 2004;101:7398–7403. doi: 10.1073/pnas.0306641101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooistra SM, Helin K. Molecular mechanisms and potential functions of histone demethylases. Nat. Rev. Mol. Cell. Biol. 2012;13:297–311. doi: 10.1038/nrm3327. [DOI] [PubMed] [Google Scholar]
- Krishnan S, Collazo E, Ortiz-Tello PA, Trievel RC. Purification and assay protocols for obtaining highly active Jumonji C demethylases. Anal. Biochem. 2012;420:48–53. doi: 10.1016/j.ab.2011.08.034. [DOI] [PubMed] [Google Scholar]
- Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- Lee J, Thompson JR, Botuyan MV, Mer G. Distinct binding modes specify the recognition of methylated histones H3K4 and H4K20 by JMJD2A-tudor. Nat. Struct. Mol. Biol. 2008;15:109–111. doi: 10.1038/nsmb1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger E, Wissmann M, Yin N, Müller JM, Schneider R, Peters AHFM, Günther T, Buettner R, Schüle R. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- Ng SS, Kavanagh KL, McDonough MA, Butler D, Pilka ES, Lienard BMR, Bray JE, Savitsky P, Gileadi O, von Delft F, et al. Crystal structures of histone demethylase JMJD2A reveal basis for substrate specificity. Nature. 2007;448:87–91. doi: 10.1038/nature05971. [DOI] [PubMed] [Google Scholar]
- Rottach A, Frauer C, Pichler G, Bonapace IM, Spada F, Leonhardt H. The multi-domain protein Np95 connects DNA methylation and histone modification. Nucl. Acids Res. 2010;38:1796–1804. doi: 10.1093/nar/gkp1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rui L, Emre NCT, Kruhlak MJ, Chung H-J, Steidl C, Slack G, Wright GW, Lenz G, Ngo VN, Shaffer AL, et al. Cooperative epigenetic modulation by cancer amplicon genes. Cancer Cell. 2010;18:590–605. doi: 10.1016/j.ccr.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Simon MD, Chu F, Racki LR, De La Cruz CC, Burlingame AL, Panning B, Narlikar GJ, Shokat KM. The Site-Specific Installation of Methyl-Lysine Analogs into Recombinant Histones. Cell. 2007;128:1003–1012. doi: 10.1016/j.cell.2006.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada Y-i, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- Whetstine JR, Nottke A, Lan F, Huarte M, Smolikov S, Chen Z, Spooner E, Li E, Zhang G, Colaiacovo M, et al. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell. 2006;125:467–481. doi: 10.1016/j.cell.2006.03.028. [DOI] [PubMed] [Google Scholar]
- Wissmann M, Yin N, Müller JM, Greschik H, Fodor BD, Jenuwein T, Vogler C, Schneider R, Günther T, Buettner R, et al. Cooperative demethylation by JMJD2C and LSD1 promotes androgen receptor-dependent gene expression. Nat. Cell Biol. 2007;9:347–353. doi: 10.1038/ncb1546. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.