Abstract
Osteosarcoma (OS) is the most common primary malignancy of bone. We investigated the roles of insulin-like growth factor binding protein 5 (IGFBP5) domains in modulating OS tumorigenicity and metastasis. The N-terminal (to a lesser extent the C-terminal) domain inhibited cell proliferation and induced apoptosis while the C-terminal domain inhibited cell migration and invasion. The Linker domain had no independent effects. In vivo, the N-terminal domain decreased tumor growth without affecting pulmonary metastases while the C-terminal domain inhibited tumor growth and metastases. In summary, the N- and C-terminal domains modulated OS tumorigenic phenotypes while the C-terminal domain inhibited OS metastatic phenotypes.
Keywords: IGFBP5, insulin-like growth factor binding protein, osteosarcoma, animal model, metastasis
1. INTRODUCTION
Osteosarcoma (OS) is the most common primary malignancy of bone in children and young adults, with a peak incidence in the second decade of life [1]. Although approximately 80% of patients have either macro- or micro-metastatic disease at the time of diagnosis, only 10–15% of these lesions can be identified with current imaging modalities [2,3]. With current treatment protocols, which include wide resection and chemotherapy, the average five year disease-free survival is only 65–70% [4,5]. Despite significant advances in surgical techniques and chemotherapeutic regimens over the past few decades, there has been minimal improvement in patient survival, and there is a critical need to identify molecular markers which provide insight into both the pathogenesis and metastasis of OS. While various genetic and hereditary conditions are associated with OS, the precise molecular mechanisms leading to the development of OS are poorly understood; this may be indicative of the complexity of the disease [6–8].
In previous studies, we have shown that insulin-like growth factor binding protein 5 (IGFBP5) is a critical inhibitor of OS tumorigenesis and metastasis [9,10]. IGFBP5 expression was also decreased in metastatic OS lesions when compared to primary tumors in patient samples [9]. When we examined the role of IGFBP5 in OS pathogenesis and metastasis, we found that it acted as a potent tumor suppressor, and over-expression of IGFBP5 inhibited OS cell proliferation, migration and invasion in vitro, while inducing apoptosis [9]. Conversely, siRNA knockdown of IGFBP5 promoted OS growth and lung metastases in vivo.
IGFBP5 belongs to the IGFBP family of six secreted proteins which play a critical role in regulating insulin-like growth factor 1 (IGF1) and IGF2 signaling, and IGFBP5 is the major IGF binding protein in bone [11]. It contains highly conserved cysteine-rich N- and C-terminal domains and a less conserved central Linker domain [12]. Structural analyses have demonstrated that the N-terminal domain contains a hydrophobic stretch of amino acids serving as the primary IGF binding site [12]. IGFBP5 contains an N-terminal signal peptide that is crucial for protein transport and eventual secretion [12]. The C-terminal domain contains binding sites for several extracellular matrix (ECM) components including heparin and hydroxyapatite and interacts with a wide variety of molecules mediating both IGF-dependent and IGF-independent signaling [12]. The Linker domain contains proteolytic cleavage sites and is the site for several post-translational modifications, including glycosylation and phosphorylation.
In this investigation, we examine the roles of the different IGFBP5 domains in OS pathogenesis and metastasis. Given the unique functions of each domain, we hypothesized that each would modulate different aspects of OS tumorigenicity and metastasis. We found that over-expression of the N-terminal domain inhibited OS cell proliferation and primary tumor growth while promoting apoptosis. Expression of the C domain inhibited OS cell migration, invasion and primary tumor growth, with inhibition of cell proliferation to a lesser extent. Furthermore, we found that the C domain significantly inhibited the formation of spontaneous pulmonary metastases in an orthotopic animal model.
2. MATERIALS AND METHODS
2.1 Tissue Culture
HEK293 and human OS 143B cells were purchased from ATCC (Manassas, VA). We also used a highly tumorigenic MG63.2 OS cell line that was established by serially passaging the metastases of a parental MG63 line, as previously reported [9,10]. Unless otherwise indicated, all chemicals were purchased from Sigma-Aldrich (St Louis, MO) or Fisher Scientific (Pittsburgh, PA).
2.2 Creation of recombinant adenoviral vectors expressing IGFBP5 domains
The cDNA coding regions to the N-terminal, Linker and C-terminal domains of IGFBP5 were PCR amplified and subcloned. We included the native signal peptide of IGFBP5 in each construct to ensure proper protein transport and secretion. The PCR-amplified fragments were confirmed by sequencing. Cloning details and oligo sequences are available upon request. Recombinant adenoviruses were generated using the previously described AdEasy system resulting in adenoviruses expressing the following domains: NL (Adv-NL), N (Adv-N), LC (Adv-LC), L (Adv-L) and C (Adv-C) [13–16]. Each adenoviral construct also expressed RFP to monitor infection efficiency. Identical constructs containing a flag tag were also made. Adenovirus expressing RFP only or full length IGFBP5 (Ad-NLC) were used as controls. Equal efficiency of adenoviral transduction for each construct was achieved by dose titration and direct visualization under fluorescence imaging.
2.3 RNA isolation and semi-quantitative RT-PCR
Total RNA was isolated using TRIZOL Reagents (Invitrogen), and cDNA was generated. The cDNA products were used for semi-quantitative PCR using primers specific to the IGFBP5 domains of interest. All samples were normalized by GAPDH expression. Oligo sequences are available upon request.
2.4 Western blot analysis for construct expression
Western blot analysis was performed as previously described [17,18]. Briefly, the cDNA coding regions of our unique IGFBP5 domains were subcloned into a pSEB-Flag vector. Subsequently, we transiently infected HEK 293 cells, which have a very low endogenous IGFBP5 expression, to verify the production of our constructs. Anti-Flag antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, California) and secondary antibodies conjugated with horseradish peroxidase from Pierce (Rockford, IL).
2.5 Trypan Blue cell proliferation assay
Trypan Blue cell proliferation assays were performed as previously described [19]. Briefly, subconfluent OS cells were infected with the indicated adenoviruses or controls. Cells were replated at 72 hours in 1% FBS complete media. The cells were then collected at the indicated time point and the viable cells counted using Trypan Blue staining. Each assay was performed in triplicate.
2.6 Crystal violet cell proliferation assay
Crystal violet cell proliferation assays were performed as previously described [19]. Briefly, subconfluent OS cells were infected with the indicated adenoviruses or controls. Cells were replated at 72 hours in 1% FBS complete media. Cells were subjected to crystal violet staining 7 days later, and staining intensity was analyzed using ImageJ software. Each assay was performed in triplicate.
2.7 Wound healing cell migration assay
Wound healing migration assays were performed as previously described [17,18]. Briefly, subconfluent OS cells were infected with the indicated adenoviruses or controls. Wounds were made on the monolayers using a pipette tip. Bright field images of three different fields were taken at 0 and 36 hrs. At 36 hrs, the plates were washed, cells fixed with formalin and stained with crystal violet. Quantification of cell migration was done by averaging the greatest gap distances at 3 randomly chosen points. Each assay was performed in triplicate.
2.8 Matrigel cell invasion assay
Matrigel cell invasion assays were performed as previously described [10]. Briefly, subconfluent OS cells were infected with the indicated adenoviruses or controls and plated in the upper chamber of a Matrigel transwell. FCS was used as a chemoattractant and after 72 hrs, the membranes containing the invading cells were fixed in 10% formalin, stained with hematoxylin, cleaned and mounted. Four random high power fields (200×) were counted per insert, and representative images were obtained. Each assay was performed in triplicate.
2.9 Caspase 3, 8, and 9 immunohistochemical staining
Immunohistochemical staining for the late apoptotic marker cleaved-caspase 3, caspase 8, and caspase 9 was carried out as previously described [20,21]. Briefly, subconfluent OS cells were infected with the indicated adenoviruses or controls and stained at 72 hrs. The fixed cells were blocked with bovine serum, avidin and biotin blocking solution. Cells were probed with the primary antibody followed by incubation with anti-goat IgG secondary antibody conjugated with horseradish peroxidase (Pierce).
2.10 Flow cytometry
Subconfluent OS cells were infected with the indicated adenoviruses or controls. After 72 hours, the cells were trypsinized and re-plated at subconfluent conditions in 1% FBS complete media. Cells were harvested 48 hrs later, washed with PBS and stained with propidium iodide and AnnexinV. Cells were sorted and categorized as alive, early apoptotic, or late apoptotic based on flow cytometry results.
2.11 Orthotopic xenograft model
The highly tumorigenic and metastatic 143B cell line, with stable expression of luciferase, was used to analyze the in vivo tumorigenic and metastatic effects of our IGFBP5 domains as we have previously described [9]. We chose the 143B cell line because of the relatively short time (4–6 weeks) to tumor formation and metastasis in mice to minimize cost. We have previously demonstrated that both the 143B and MG63.2 cells behave similarly when over-expressing fulllength IGFBP5 [9]. Cells were infected with the indicated adenoviruses or controls. At 36 hours, cells were harvested and prepared for a subperiosteal injection (1.5x106/injection) into the bilateral proximal posterior tibiae of athymic nude mice (male, Harlan Sprague Dawley, 4–6 weeks old) as previously described [9,10]. Although the subperiosteal injection has a lower metastatic efficiency compared to our previously described intratibial injection [10], we now use the subperiosteal injection to avoid the immediate 10% mortality associated with the intratibial injection from marrow embolic events. Five mice were used per group. Tumors were measured every 3–4 days, and the tumor volume and doubling times were calculated as previously reported [22]. Five weeks following implantation of 143B cells, animals were sacrificed, and primary tumors and lungs were harvested for evaluation.
2.12 Xenogen bioluminescence imaging
Bioluminescent imaging was performed every 3–4 days as previously described using the Xenogen IVIS 200 imaging system [9,10]. The pseudoimages were obtained by superimposing the emitted light over the gray-scale photographs of the animal. Quantitative analysis was done with Xenogen’s Living Image V2.50.1 software as previously described [9,10].
2.13 Histologic evaluation
Histologic evaluation was performed as previously described [9,10]. Briefly, the harvested tumors and lungs were fixed with 10% formalin, embedded in paraffin, serially sectioned and stained with hematoxylin and eosin. All samples were assigned a random number and analyzed by a blinded pathologist.
2.14 MicroCT imaging
MicroCT imaging was performed as previously described [14]. Briefly, the harvested tumors were fixed in 10% formalin and subjected to imaging. After the imaging, the data was analyzed via the Amira software program (Visage Imaging, San Diego, California).
2.15 Statistical analysis
A two-tailed Student’s t-test was used for the analysis of cell proliferation, apoptosis, migration, and Matrigel invasion assays as well as the tumor doubling time and spontaneous metastases. The full-length IGFBP5 construct and the mutant constructs were compared with the RFP control using a two-tailed Student’s t-test. All experiments were performed in triplicate and repeated on three separate occasions unless otherwise stated.
3. RESULTS
3.1 Over-expression of IGFBP5 Domains in Osteosarcoma Cell Lines
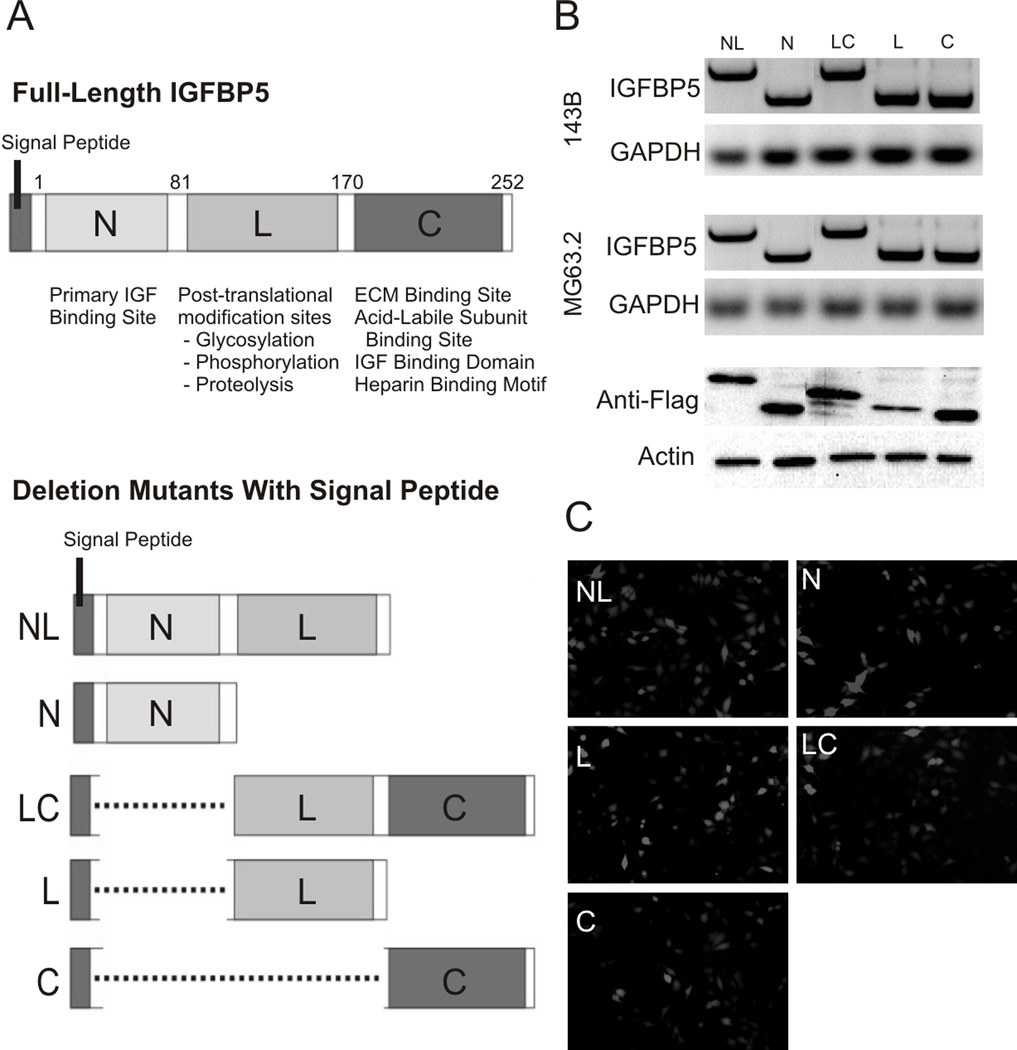
IGFBP5 contains three unique domains with different functions (Figure 1A). The N-terminal domain (N) is an 80 amino acid region that serves as the primary IGF binding site. The Linker domain (L) contains 89 amino acids that undergo post-translational modifications such as glycosylation, phosphorylation and proteolysis. Finally, the C-terminal domain (C) is an 82 amino acid region containing ECM binding sites and secondary IGF binding regions. IGFBP5 also contains an N-terminal signal peptide that is crucial for protein transport and eventual secretion.
Figure 1. IGFBP5 Deletional Mutant Constructs.
A. A schematic map shows the full-length and deletional mutants generated with the respective functional domains indicated: The N-terminal domain (N) contains an 80 amino acid region, the Linker domain (L) contains 89 amino acids and the C-terminal domain (C) contains 82 amino acids. The reported functions of each of the domains (N-terminal, Linker, and C-terminal) are indicated. Identical constructs with and without Flag tags were made. B. Expression of each construct was confirmed by RT-PCR and Western blot. C. The constructs were cloned into the AdEasy adenoviral expression system co-expressing RFP as an indicator of infectivity. Adenoviral transduction of each of the constructs is shown. Equal efficiency of adenoviral transduction for each construct was achieved by dose titration and direct visualization under fluorescence imaging.
We created the full-length and five different IGFBP5 constructs containing different combinations of the N, L and C domains (Figure 1A, bottom panel). All constructs also contain the N-terminal signal peptide to ensure proper protein transport and secretion. Using adenoviral vectors, we over-expressed these constructs in two highly metastatic OS cell lines, 143B and MG63.2. RNA was harvested at days 3, 5 and 7 and construct expression evaluated by semi-quantitative RT-PCR. As shown in Figure 1B, we observed robust expression of all constructs in both 143B and MG63.2 OS cell lines at 7 days post-infection. Over-expression of identical constructs containing a Flag tag was detectable by Western blot analysis in HEK293 cells, which have very low endogenous IGFBP5 expression (Figure 1B). All adenoviral constructs contained an RFP marker to monitor infection efficiency, and as shown in Figure 1C, we observed equal infectivity under direct fluorescence imaging following titration.
3.2 Cell Proliferation
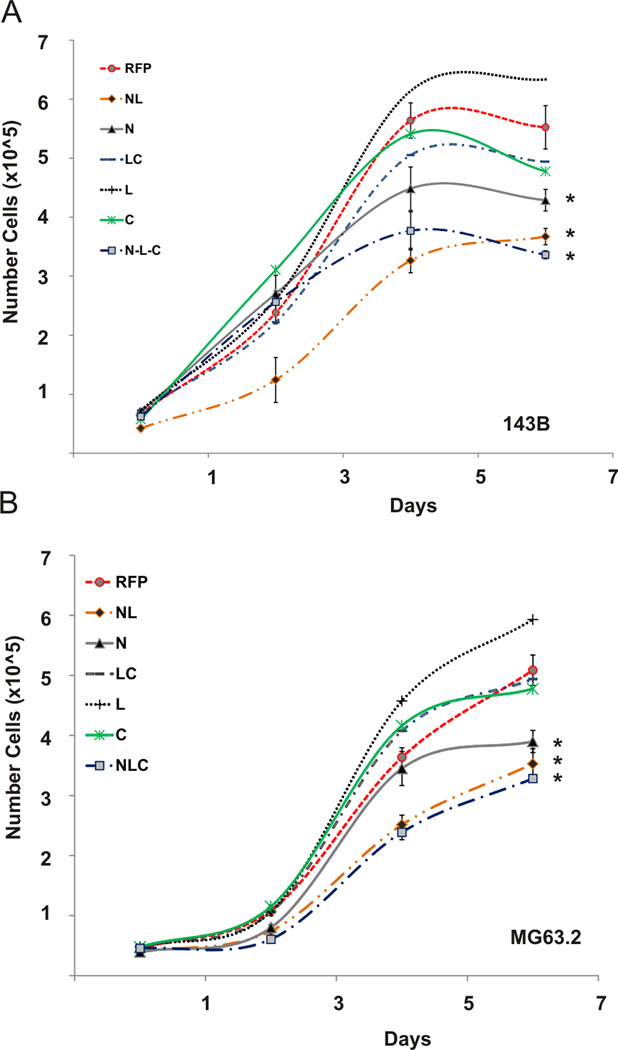
We next assessed the effects of our IGFBP5 constructs on phenotypes important for OS growth and metastasis. Each construct was over-expressed in MG63.2 and 143B OS cell lines, and the proliferative capacity was measured in vitro using Trypan Blue and Crystal Violet cell proliferation assays. The N, NL and NLC domains significantly inhibited proliferation in 143B cells compared to the RFP control (Figure 2A, p < 0.02). To a lesser extent, the C-terminal and LC domains inhibited cell proliferation in 143B cells compared to the RFP control (p < 0.06). Doubling times for the N, NL, NLC, C, LC and RFP groups were 64.1 ± 4.2, 57.9 ± 4.8, 70.5 ± 5.1, 53.7 ± 4.4, 51.8 ± 4.8 and 34.1 ± 4.5 hours, respectively. The N, NL and NLC domains also inhibited cell proliferation in MG63.2 cells when compared to the RFP control (Figure 2B, p<0.03). Doubling times for the N, NL, NLC, C, LC and RFP groups, were 50.7 ± 4.6, 53.1 ± 6.1, 58.8 ± 4.6, 35.0 ± 4.4, 36.4 ± 4.3 and 27.5 ± 4.2 hours in MG63.2 cells, respectively. We observed no statistically significant difference in proliferation between the NL and full-length IGFBP5 (NLC) constructs. Crystal violet staining demonstrated similarly decreased proliferative capacity of OS cells overexpressing the N-terminal domains, and to a lesser extent cells expressing the C-terminal domains, of IGFBP5 in both 143B and MG63.2 cell lines (data not shown). Taken together, these results suggest that the N-terminal domain is largely responsible for the inhibition of OS cell proliferation by IGFBP5, while the C-terminal domain has a lesser effect on OS cell proliferation.
Figure 2. IGFBP5 deletional mutants’ effects on cell proliferation.
A. Cell proliferation: Full length (NLC), deletional mutants and RFP control adenoviruses were used to equally transduce 143B and MG63.2 osteosarcoma cells. Trypan Blue cell proliferation assays were performed at the indicated days. The asterisk (*) indicates a significant difference in doubling time compared to the RFP control (p < 0.02 for 143B, p < 0.03 for MG63.2).
3.3 Apoptosis
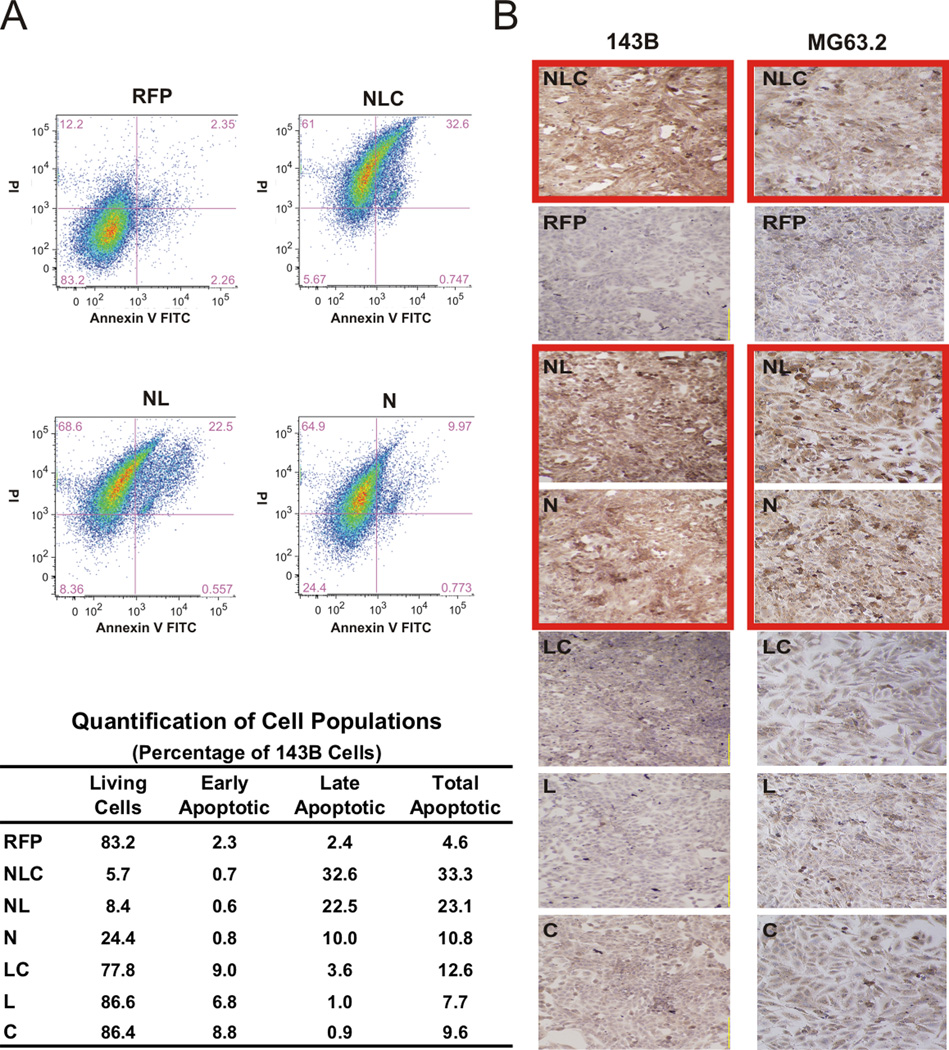
To examine the mechanism underlying the inhibition of OS cell proliferation by our IGFBP5 constructs, we evaluated for the presence of a late apoptotic marker, cleaved caspase-3, and performed flow cytometry to determine the percentage of cells in either early or late stages of apoptosis. As shown in Figure 3A, over-expression of constructs containing the N-terminal domain of IGFBP5 (i.e. N, NL, and NLC) led to a marked increase in necrotic and apoptotic cells compared to the RFP control in 143B cells as determined by flow cytometry. Examples of the sorted populations for the RFP, NLC, NL, and N constructs are shown in Figure 3A, Top Panel and the quantifications for all the constructs are shown in Figure 3A Bottom Panel. Over-expression of the NLC and NL domains resulted in a greater than five-fold higher percentage of apoptotic cells than the RFP control, at 33.3% and 23.1%, respectively (p < 0.01). The N, C and LC domains demonstrated an approximately two-fold increase in apoptosis, at 10.8%, 9.6% and 12.6%, respectively (p < 0.05). Next, we performed immunohistochemical staining with an anti-cleaved caspase-3 antibody. As shown in Figure 3B, we again demonstrated that the most significant induction of apoptosis was by constructs containing the N-terminal domain (outlined red). In the 143B cell line, the NLC, NL and N domains demonstrated the most potent apoptotic induction compared to the RFP control (Figure 3B, left panel). Similarly, in our MG63.2 line the NLC, NL and N domains significantly increased OS cell apoptosis (Figure 3B, right panel). Similar results were seen with immunohistochemical staining for caspase 8 and 9 (Supplemental Figure 1). Taken together, these results suggest that the N-terminal domain of IGFBP5 likely exerts its anti-proliferative effect by inducing OS cell apoptosis. The C-terminal domain had similar effects but to a lesser extent.
Figure 3. IGFBP5 deletional mutants’ effects on apoptosis.
A. Apoptosis by flow cytometry: 143B cells were transduced with the indicated constructs. Cells were stained with propidium iodide & AnnexinV and assessed for apoptosis by flow cytometry. The cells were clustered according to those undergoing early and late phases of apoptosis. Distributions of the populations for RFP, NLC, NL, and N constructs are shown in the top panel. Quantification of the alive, early apoptotic and late apoptotic populations for all constructs is shown in the bottom panel. B. Caspase 3 staining: 143B and MG63.2 cells were transduced with the indicated adenoviruses, and immunohistochemistry staining for cleaved caspase 3 performed as an indicator of apoptosis was performed. Note the increased staining in the N-terminal domain-containing groups (outlined red).
3.4 Cell migration and invasion
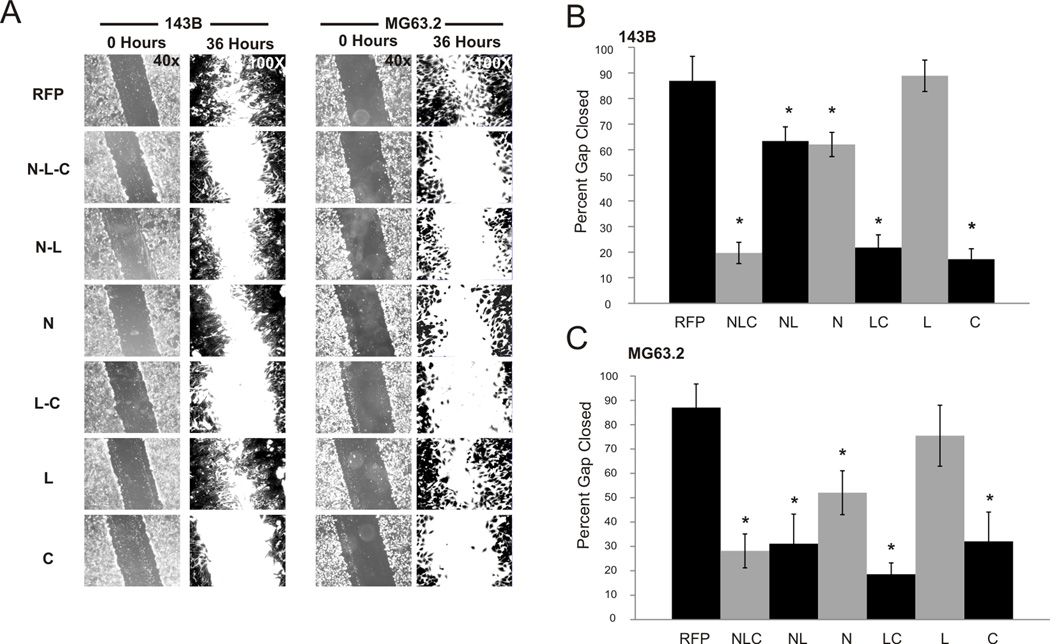
The ability of tumor cells to migrate and invade is considered an important indicator of cell aggressiveness and metastatic ability. The C-terminal domain of IGFBP5 has been found to interact with various ECM components, and we hypothesized that through these interactions, the C domain may inhibit the migratory and invasive capabilities of OS cells. To assess the effect of our IGFBP5 domains on migration in vitro, we performed wound healing assays as previously described [9]. Briefly, a wound was made in a monolayer of adenoviral-transduced OS cells, and the ability of the cells to close the gap was demonstrated qualitatively and measured quantitatively in 143B and MG63.2 OS cells. As shown in Figure 4A (left panel), over-expression of IGFBP5 domains containing the C-terminal region significantly inhibited the migration of 143B cells. When gap closure was quantified (Figure 4B), the NLC, LC and C domains resulted in a 20 ± 4.2%, 21 ± 4.9%, and 17 ± 4.0% closure, respectively, which was significantly lower than the 87 ± 9.1% closure observed in the RFP control (p < 0.01). Similar results were seen in the MG63.2 cell line. As shown in Figure 4A (right panel), the NLC, NL, LC and C domains significantly inhibited cell migration in MG63.2 cells (p < 0.03). The antimigratory effect of the N construct was not as potent as these other domains but still significant compared to the RFP control (p< 0.05). Quantitative gap closure for the NLC, NL, LC and C domains were 28 ± 7.0%, 31 ± 6.1%, 18 ± 4.6% and 32 ± 5.2%, respectively, significantly lower than the 87 ± 9.7% closure obtained in the RFP group (Figure 4C, p < 0.03). For both 143B and MG63.2 cells, the Linker domain (L domain) had no effect on OS cell migration.
Figure 4. IGFBP5 deletional mutants’ effects on cell migration.
A. Wound healing assay: Full length (NLC), deletional mutants and RFP control adenoviruses were used to equally transduce 143B and MG63.2 osteosarcoma cells. A wound was made across a monolayer of cells and closure of the wound was observed. At 36 hours, the cells were fixed in formalin and stained with crystal violet. Representative images taken at 0 hrs and 36 hrs are shown to demonstrate closure of the wound. B. Closure of the wound was measured, percentage of wound closure calculated and measurements from three random fields averaged. The graph depicts the percentage of the gap closed for each of the constructs. The asterisk (*) indicates a significant difference in wound closure compared to the RFP control (p < 0.01 for 143B, p < 0.03 for MG63.2).
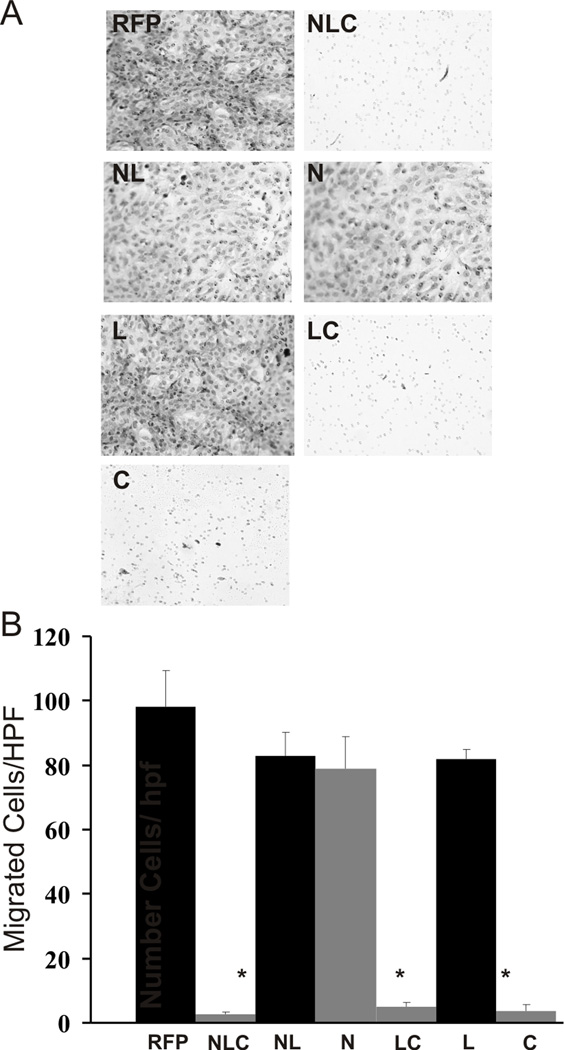
Next, we determined the invasive potential of OS cells using a previously described Matrigel invasion assay [9]. Adenoviral-transduced MG63.2 and 143B cells were plated in the upper chamber of a Matrigel-coated transwell insert, and their ability to invade through a porous Matrigel matrix was measured. As shown in Figure 5A for 143B cells, we found significant cell invasion in the RFP, NL, N and L domain treatment groups, while there was minimal invasion in the NLC, LC and C domain constructs. When quantifying the number of cells invaded (Figure 5B), we found the NLC, LC and C domains exhibited the most significant inhibitory effect on cell invasion (p < 0.01). In fact, the inhibition by the LC and C domains was equally as potent as the NLC construct. We observed no significant inhibitory effect on cell invasion by the NL, N and L domain constructs. Similar results were seen with MG63.2 cells (data not shown). While the N-domain had some inhibitory effects on cell migration, its inhibitory effects on cell invasion was minimal. These results suggest that the C domain of IGFBP5, likely through its interactions with ECM components, is responsible for the inhibition of cell migration and cell invasion in these osteosarcoma lines.
Figure 5. IGFBP5 deletional mutants’ effects on cell invasion.
A. Matrigel invasion assay: Full length (NLC), deletional mutants and RFP control adenoviruses were used to equally transduce 143B and MG63.2 osteosarcoma cells. The cells were plated on Matrigel coated polycarbonate membranes containing pores. Fetal calf serum was used as a chemoattractant. Representative images of 143B cells that migrated through the coated membrane are shown (MG63.2 data not shown). B. Random fields of migrated cells per treatment group were counted, averaged and depicted on the graph shown; the asterisk (*) indicates a significant difference in cell invasion compared to the RFP control (p < 0.01).
3.5 Primary tumor growth
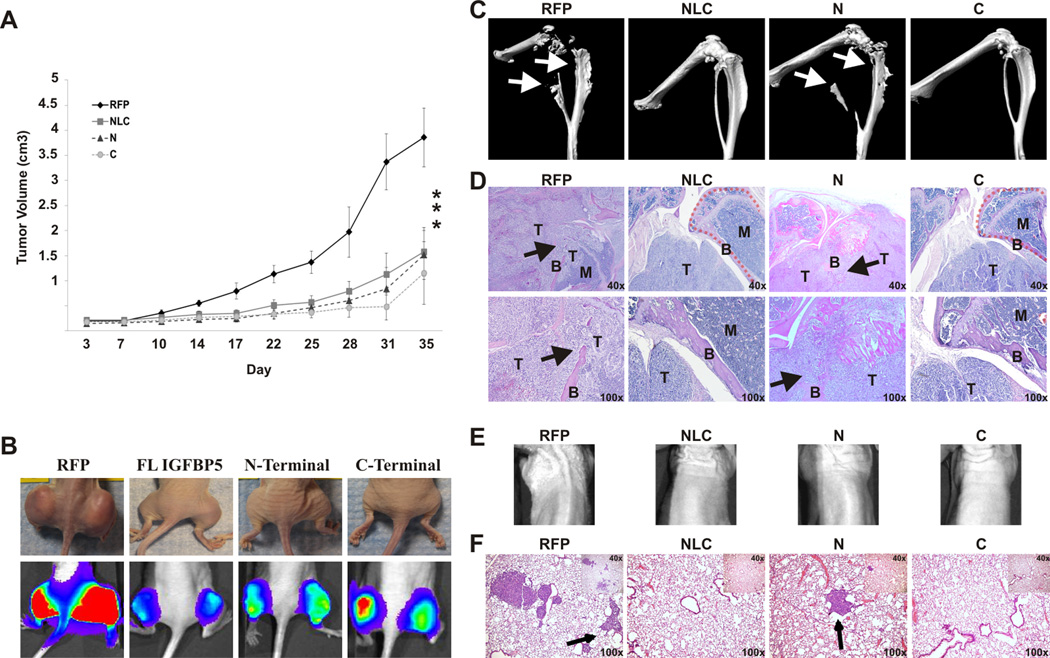
We next sought to investigate the role of our IGFBP5 domains on primary OS tumor growth in an orthotopic animal model. Given the unique functions of the N- and C-terminal domains demonstrated in our in vitro studies, we chose to use IGFBP5 constructs expressing only the N and C domains for the in vivo studies. The NLC and RFP constructs were used as positive and negative controls, respectively. The luciferase tagged 143B cells were equally infected with adenovirus expressing RFP only (Ad-RFP), Ad-NLC, Ad-N or Ad-C and injected subperiosteally in the posterior tibia. The animals were subjected to biweekly Xenogen bioluminescent imaging. As seen in Figure 6A, over-expression of the N and C domains by adenoviral transduction in the 143B cell line resulted in decreased primary tumor growth compared with the RFP control (p<0.01). The effect of these individual domains was equivalent to the anti-tumor effect of the full-length IGFBP5 construct. Over-expression of the N and C domains increased tumor doubling times by 166% and 117%, respectively compared to the RFP control group. The doubling times during the log phase of growth for the N domain, C domain, full length IGFBP5 and RFP control groups are shown in Table 1. When the primary tumors were grossly examined, there was a marked difference in tumor size between the N domain, C domain, full length and RFP treatment groups (Figure 6B, top row). Live animal bioluminescent imaging four days prior to tumor harvesting further demonstrated the difference in tumor sizes between these four groups (Figure 6B, bottom row). We found that by the second week, there was consistently increased luciferase activity visualized by live animal biweekly Xenogen bioluminescent imaging in the RFP group and a marked decrease in the NLC, N domain and C domain treatment groups (data not shown). Taken together, the full length, N, and C domains had inhibitory effects on tumor growth.
Figure 6. In vivo tumor and metastases.
A. Primary tumor growth of 143B cells expressing control RFP, the full-length NLC, N-terminal or C-terminal constructs: At 36 hours, cells were harvested and injected subperiosteally into the bilateral proximal posterior tibiae of athymic nude mice. Tumors were measured and subjected to live bioluminescent imaging every 3–4 days. The tumor volume and doubling time were calculated. At five weeks, animals were sacrificed and primary tumors and lungs were harvested for evaluation. The asterisk (*) indicates a significant difference in doubling time compared to the RFP control (p < 0.01).
B. Representative photographic and bioluminescent images of the primary tumors of mice for each of the indicated groups: Bioluminescent imaging and quantitative analysis was performed every 3–4 days. The photographs and bioluminescent images seen are at four days prior to tumor harvesting. C. MicroCT of the primary tumors: Note the bone destruction of the tibia and fibula (arrows) in the RFP and N-terminal constructs. Conversely, the full-length NLC and C-terminal constructs had normal appearing tibias and fibulas. D. Hematoxylin and eosin staining of the primary tumors: Note that the cortex of the tibia is intact in the full-length NLC and C-terminal groups (dashed lines), whereas that bone structure is lost in the RFP and N-terminal groups due to tumor invasion. In the RFP and N-terminal groups, the bone cortex (B) is destroyed by the tumor (T), and the bone marrow space (M) is being replaced by tumor. E. Representative bioluminescent images of the thorax of the mice four days prior to sacrifice. F. Lung metastases: Representative hematoxylin and eosin stained sections of the lungs of animals from the indicated groups are shown. Note the metastases seen in the RFP and the N-terminal groups (arrow), whereas the full-length NLC and C-terminal groups show minimal metastases.
Table 1.
Tumor Growth & Metastases
| Treatment Groups | ||||
|---|---|---|---|---|
| RFP | NLC | N-Term | C-Term | |
| Tumor Double Time (days) | 4.7 ± 0.9 | 9.8 ±2.1 | 12.5 ±3.2 | 10.2 ± 3.3 |
| Lung Metastasis/hpf | 16.7 ± 8.7 | 3.2 ± 2.6 | 11.3 ± 4.9 | 1.4 ± 2.9 |
3.6 Tumor invasion and spontaneous pulmonary metastases
We next characterized the role of the IGFBP5 domains on OS primary tumor invasion and spontaneous pulmonary metastasis. MicroCT analysis was conducted on primary tumors to evaluate for bone invasion and destruction at the primary tumor site. The proximal tibias of mice in the RFP and N domain groups demonstrated significant destruction of the bone architecture (Figure 6C), with bone destruction extending to the fibulas in these groups. However, the tibias and fibulas of mice in the C domain and NLC over-expression groups remained entirely intact, with no bone invasion or destruction. Random sagittal sections dividing the tibia and tumors were generated and stained with hematoxylin and eosin (Figure 6D). The RFP and the N domain groups showed significant destruction and invasion through the posterior cortex and into the bone marrow, while the tumors in the C domain and full-length NLC groups had little or no bone destruction or invasion on histologic analysis.
Next, we examined for the presence of spontaneous pulmonary metastases. The lungs were subjected to histologic evaluation to evaluate for the presence of micro-metastatic disease. Random sections at the maximal width of the lungs were stained with hematoxylin and eosin (Figure 6F). We found significant micro-metastatic disease in both the RFP and N domain treatment groups; however, treatment with the C domain and NLC group significantly inhibited the formation of spontaneous pulmonary metastases (p < 0.01). The average number of metastases per high power field for the RFP, full length IGFBP5, N domain and C domain groups are shown in Table 1. As expected, the pulmonary micro-metastatic disease did not have sufficient tumor burden and bioluminescent signal to be detected by bioluminescent imaging of the animal (Figure 6E). Overall, our evaluations suggest the C-terminal domain of IGFBP5 is responsible for the suppression of both OS tumor invasion and pulmonary metastases.
4. DISCUSSION
We have previously reported the inhibition of osteosarcoma tumorigenicity and metastasis by IGFBP5 [9]. Analysis demonstrated that IGFBP5 expression was down-regulated in metastatic pulmonary lesions taken from OS patients. Upon characterizing the role of IGFBP5 in four OS cell lines, we found that over-expression of IGFBP5 inhibited cell proliferation and primary tumor growth, while inducing apoptosis. Our studies also showed that IGFBP5 inhibited cell migration and invasion in vitro, while inhibiting the formation of spontaneous pulmonary metastases in vivo. In this study, we functionally characterize the three domains of IGFBP5 and their effects on OS tumorigenicity and metastasis. We find that the N domain of IGFBP5 inhibits cell proliferation while inducing the apoptotic cascade. The C domain inhibits OS cell migration and invasion in vitro with a lesser effect on the inhibition of cell proliferation than the N domain. In an orthotopic tumor model, we find that both N and C domains inhibit primary tumor growth, but only the C domain significantly inhibits primary tumor invasion and the formation of spontaneous pulmonary metastases. The L domain has no independent effects on OS tumorigenesis or metastasis.
Little is currently known about the role of IGFBP5 in OS, but the IGF pathway has been implicated in a number of human cancers [23,24]. Furthermore, studies elucidating the role of IGFBP5 in tumor progression have provided conflicting results. For example, IGFBP5 is down-regulated in renal and cervical tumors and induces caspase expression and apoptosis in breast cancer cells [25–30]. However, other studies have shown that IGFBP5 is up-regulated in highgrade ovarian carcinoma and thyroid papillary carcinomas [25–30]. These results may reflect the complexity of IGFBP5 signaling as well as its ability to modulate both IGF-dependent and IGFindependent effects [31]. To our knowledge, there have been no studies functionally characterizing the domains of IGFBP5 in osteosarcoma, and our work is the first of such reported in the literature.
In our study, we find that the N domain of IGFBP5 primarily suppresses cell proliferation and tumor growth while inducing apoptosis. These findings are supported by the reported functions of the N-terminal domain; studies have shown that IGFBP5 contains a high-affinity IGF binding site located in the N-terminal region termed ‘mini-IGFBP5’ [32]. Deletion mutagenesis studies have shown that this N-terminal fragment binds IGF-I and, with reduced affinity, IGF-II, exhibiting an inhibitory effect on IGF-I signaling [32]. IGF-I activation results in downstream activation of the mitogen-activated protein kinase (MAPK) pathway and phosphatidylinositol-3-kinase (PI3K) pathway, resulting in cellular proliferation and inhibition of apoptosis [33]. Given the inhibitory effect of the N domain on IGF-I signaling, it is likely that the IGFBP5 N-terminal domain exhibits its anti-proliferative and pro-apoptotic effects by modulating the IGF-I signaling axis in OS cells.
Our observed inhibition of OS cell migration, cell invasion and pulmonary metastases by the Cterminal domain is supported by previous work characterizing the functional role of the C domain; this domain has been shown to interact with extracellular matrix (ECM) components such as fibronectin, vitronectin, thrombospondin-1 and hydroxyapatite [12,31]. In addition, IGFBP5 contains three heparin binding motifs within the C-terminal and Linker domains [34,35]. Deletion mutagenesis studies indicate that the C domain is the primary heparin binding site, with the Linker domain only effective in the absence of the C-terminal region [35]. The ability of the C domain to bind ECM components may regulate OS cell-cell and cell-matrix interactions. In addition, the C-terminal region contains a unique nuclear localization sequence (NLS) that regulates intracellular IGFBP5 transport [31]. We believe the observed inhibition of cell migration, cell invasion and spontaneous pulmonary metastases likely arises from the C domain’s interactions with various ECM components and its ability to modulate intracellular signaling. Interestingly, our in vivo studies demonstrated a significant inhibition of primary tumor growth by both the C-terminal and N-terminal domains. Our in vitro studies showed the N-terminal domain to have significant inhibition of proliferation, while the C-terminal domain had a lesser but not statistically significant (p < 0.06) anti-proliferative effect. Previous studies have also shown that the C-terminal region contains secondary IGF-I binding sites, and we believe the observed inhibition of primary tumor growth, and to a lesser extent in vitro cell proliferation, by C domain-containing constructs may be a result of its actions in the IGF-1 signaling pathway [31]. It is possible that these secondary IGF-I binding sites modulate cell proliferation and thus primary tumor growth to a greater degree in vivo than in a less physiologic monolayer culture in vitro.
In summary, we have functionally characterized the unique domains of IGFBP5 in the inhibition of OS tumorigenicity and metastasis. We have demonstrated that the N domain of IGFBP5 suppresses cell proliferation and primary tumor growth while inducing apoptosis. The C domain primarily inhibits cell migration and cell invasion while also preventing primary tumor growth and spontaneous pulmonary metastases. We believe that further studies are warranted to investigate both IGF dependent and IGF independent signaling pathways that may be regulated by the IGFBP5 domains. Furthermore, identification of the anti-metastatic C domain can be used to develop novel recombinant proteins for therapeutic interventions in the clinical arena.
Supplementary Material
The 143B and MG63.2 cells were transduced with the indicated IGFBP5 construct adenoviruses. Immunohistochemistry staining for caspase 8 and 9 were performed to assess for apoptosis. Note the increased staining in the N-terminal domain-containing groups (outlined red).
ACKNOWLEDGEMENT
This work was supported in part by research grants from The American Cancer Society (HHL, TCH), the Brinson Foundation (HHL, TCH, RCH), The National Institute of Health (HHL, TCH, RCH), and the Orthopaedic Research and Education Foundation (HHL, RCH).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICT OF INTEREST: NONE
REFERENCES
- 1.Whelan JS. Osteosarcoma. Eur J Cancer. 1997;33:1611–1618. doi: 10.1016/s0959-8049(97)00251-7. discussion 1618–9. [DOI] [PubMed] [Google Scholar]
- 2.Kaste SC, Pratt CB, Cain AM, Jones-Wallace DJ, Rao BN. Metastases detected at the time of diagnosis of primary pediatric extremity osteosarcoma at diagnosis: imaging features. Cancer. 1999;86:1602–1608. doi: 10.1002/(sici)1097-0142(19991015)86:8<1602::aid-cncr31>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 3.Yonemoto T, Tatezaki S, Ishii T, Satoh T, Kimura H, Iwai N. Prognosis of osteosarcoma with pulmonary metastases at initial presentation is not dismal. Clin Orthop. 1998:194–199. doi: 10.1097/00003086-199804000-00024. [DOI] [PubMed] [Google Scholar]
- 4.Davis AM, Bell RS, Goodwin PJ. Prognostic factors in osteosarcoma: a critical review. J Clin Oncol. 1994;12:423–431. doi: 10.1200/JCO.1994.12.2.423. [DOI] [PubMed] [Google Scholar]
- 5.Mankin HJ, Hornicek FJ, Rosenberg AE, Harmon DC, Gebhardt MC. Survival data for 648 patients with osteosarcoma treated at one institution. Clin Orthop. 2004:286–291. doi: 10.1097/01.blo.0000145991.65770.e6. [DOI] [PubMed] [Google Scholar]
- 6.Letson GD, Muro-Cacho CA. Genetic and molecular abnormalities in tumors of the bone and soft tissues. Cancer Control. 2001;8:239–251. doi: 10.1177/107327480100800304. [DOI] [PubMed] [Google Scholar]
- 7.Ragland BD, Bell WC, Lopez RR, Siegal GP. Cytogenetics and molecular biology of osteosarcoma. Lab Invest. 2002;82:365–373. doi: 10.1038/labinvest.3780431. [DOI] [PubMed] [Google Scholar]
- 8.Fuchs B, Pritchard DJ. Etiology of osteosarcoma. Clin Orthop. 2002:40–52. doi: 10.1097/00003086-200204000-00007. [DOI] [PubMed] [Google Scholar]
- 9.Su Y, Wagner ER, Luo Q, Huang J, Chen L, He BC, Zuo GW, Shi Q, Zhang BQ, Zhu G, Bi Y, Luo J, Luo X, Kim SH, Shen J, Rastegar F, Huang E, Gao Y, Gao JL, Yang K, Wietholt C, Li M, Qin J, Haydon RC, He TC, Luu HH. Insulin-like growth factor binding protein 5 suppresses tumor growth and metastasis of human osteosarcoma. Oncogene. 2011;30:3907–3917. doi: 10.1038/onc.2011.97. [DOI] [PubMed] [Google Scholar]
- 10.Su Y, Luo X, He BC, Wang Y, Chen L, Zuo GW, Liu B, Bi Y, Huang J, Zhu GH, He Y, Kang Q, Luo J, Shen J, Chen J, Jin X, Haydon RC, He TC, Luu HH. Establishment and characterization of a new highly metastatic human osteosarcoma cell line. Clin Exp Metastasis. 2009;26:599–610. doi: 10.1007/s10585-009-9259-6. [DOI] [PubMed] [Google Scholar]
- 11.Rosenzweig SA. What's new in the IGF-binding proteins? Growth Horm IGF Res. 2004;14:329–336. doi: 10.1016/j.ghir.2004.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mukherjee A, Rotwein P. Insulin-like growth factor binding protein-5 in osteogenesis: facilitator or inhibitor? Growth Horm IGF Res. 2007;17:179–185. doi: 10.1016/j.ghir.2007.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He TC, Zhou S, da Costa LT, Yu J, Kinzler KW, Vogelstein B. A simplified system for generating recombinant adenoviruses. Proc Natl Acad Sci U S A. 1998;95:2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kang Q, Song WX, Luo Q, Tang N, Luo J, Luo X, Chen J, Bi Y, He BC, Park JK, Jiang W, Tang Y, Huang J, Su Y, Zhu GH, He Y, Yin H, Hu Z, Wang Y, Chen L, Zuo GW, Pan X, Shen J, Vokes T, Reid RR, Haydon RC, Luu HH, He TC. A comprehensive analysis of the dual roles of BMPs in regulating adipogenic and osteogenic differentiation of mesenchymal progenitor cells. Stem Cells Dev. 2009;18:545–559. doi: 10.1089/scd.2008.0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kang Q, Sun MH, Cheng H, Peng Y, Montag AG, Deyrup AT, Jiang W, Luu HH, Luo J, Szatkowski JP, Vanichakarn P, Park JY, Li Y, Haydon RC, He TC. Characterization of the distinct orthotopic bone-forming activity of 14 BMPs using recombinant adenovirus-mediated gene delivery. Gene Ther. 2004;11:1312–1320. doi: 10.1038/sj.gt.3302298. [DOI] [PubMed] [Google Scholar]
- 16.Luo J, Deng ZL, Luo X, Tang N, Song WX, Chen J, Sharff KA, Luu HH, Haydon RC, Kinzler KW, Vogelstein B, He TC. A protocol for rapid generation of recombinant adenoviruses using the AdEasy system. Nat Protoc. 2007;2:1236–1247. doi: 10.1038/nprot.2007.135. [DOI] [PubMed] [Google Scholar]
- 17.Luo X, Sharff KA, Chen J, He TC, Luu HH. S100A6 expression and function in human osteosarcoma. Clin Orthop Relat Res. 2008;466:2060–2070. doi: 10.1007/s11999-008-0361-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luu HH, Zhou L, Haydon RC, Deyrup AT, Montag AG, Huo D, Heck R, Heizmann CW, Peabody TD, Simon MA, He TC. Increased expression of S100A6 is associated with decreased metastasis and inhibition of cell migration and anchorage independent growth in human osteosarcoma. Cancer Lett. 2005;229:135–148. doi: 10.1016/j.canlet.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 19.Luu HH, Kang Q, Park JK, Si W, Luo Q, Jiang W, Yin H, Montag AG, Simon MA, Peabody TD, Haydon RC, Rinker-Schaeffer CW, He TC. An orthotopic model of human osteosarcoma growth and spontaneous pulmonary metastasis. Clin Exp Metastasis. 2005;22:319–329. doi: 10.1007/s10585-005-0365-9. [DOI] [PubMed] [Google Scholar]
- 20.Luo Q, Kang Q, Si W, Jiang W, Park JK, Peng Y, Li X, Luu HH, Luo J, Montag AG, Haydon RC, He TC. Connective tissue growth factor (CTGF) is regulated by Wnt and bone morphogenetic proteins signaling in osteoblast differentiation of mesenchymal stem cells. J Biol Chem. 2004;279:55958–55968. doi: 10.1074/jbc.M407810200. [DOI] [PubMed] [Google Scholar]
- 21.Peng Y, Kang Q, Luo Q, Jiang W, Si W, Liu BA, Luu HH, Park JK, Li X, Luo J, Montag AG, Haydon RC, He TC. Inhibitor of DNA binding/differentiation helix-loop-helix proteins mediate bone morphogenetic protein-induced osteoblast differentiation of mesenchymal stem cells. J Biol Chem. 2004;279:32941–32949. doi: 10.1074/jbc.M403344200. [DOI] [PubMed] [Google Scholar]
- 22.Luu HH, Zagaja GP, Dubauskas Z, Chen SL, Smith RC, Watabe K, Ichikawa Y, Ichikawa T, Davis EM, Le Beau MM, Rinker-Schaeffer CW. Identification of a novel metastasis-suppressor region on human chromosome 12. Cancer Res. 1998;58:3561–3565. [PubMed] [Google Scholar]
- 23.Manara MC, Landuzzi L, Nanni P, Nicoletti G, Zambelli D, Lollini PL, Nanni C, Hofmann F, Garcia-Echeverria C, Picci P, Scotlandi K. Preclinical in vivo study of new insulin-like growth factor-I receptor--specific inhibitor in Ewing's sarcoma. Clin Cancer Res. 2007;13:1322–1330. doi: 10.1158/1078-0432.CCR-06-1518. [DOI] [PubMed] [Google Scholar]
- 24.Avnet S, Sciacca L, Salerno M, Gancitano G, Cassarino MF, Longhi A, Zakikhani M, Carboni JM, Gottardis M, Giunti A, Pollak M, Vigneri R, Baldini N. Insulin receptor isoform A and insulin-like growth factor II as additional treatment targets in human osteosarcoma. Cancer Res. 2009;69:2443–2452. doi: 10.1158/0008-5472.CAN-08-2645. [DOI] [PubMed] [Google Scholar]
- 25.Stolf BS, Carvalho AF, Martins WK, Runza FB, Brun M, Hirata R, Jr, Jordao Neves E, Soares FA, Postigo-Dias J, Kowalski LP, Reis LF. Differential expression of IGFBP-5 and two human ESTs in thyroid glands with goiter, adenoma and papillary or follicular carcinomas. Cancer Lett. 2003;191:193–202. doi: 10.1016/s0304-3835(02)00679-1. [DOI] [PubMed] [Google Scholar]
- 26.Cheung C, Vesey D, Cotterill A, Douglas M, Gobe G, Nicol D, Johnson D. Altered messenger RNA and protein expressions for insulin-like growth factor family members in clear cell and papillary renal cell carcinomas. Int J Urol. 2005;12:17–28. doi: 10.1111/j.1442-2042.2004.00993.x. [DOI] [PubMed] [Google Scholar]
- 27.Takahashi M, Papavero V, Yuhas J, Kort E, Kanayama HO, Kagawa S, Baxter RC, Yang XJ, Gray SG, Teh BT. Altered expression of members of the IGF-axis in clear cell renal cell carcinoma. Int J Oncol. 2005;26:923–931. [PubMed] [Google Scholar]
- 28.Wang H, Rosen DG, Fuller GN, Zhang W, Liu J. Insulin-like growth factor-binding protein 2 and 5 are differentially regulated in ovarian cancer of different histologic types. Mod Pathol. 2006;19:1149–1156. doi: 10.1038/modpathol.3800637. [DOI] [PubMed] [Google Scholar]
- 29.Butt AJ, Dickson KA, Jambazov S, Baxter RC. Enhancement of tumor necrosis factoralpha- induced growth inhibition by insulin-like growth factor-binding protein-5 (IGFBP-5), but not IGFBP-3 in human breast cancer cells. Endocrinology. 2005;146:3113–3122. doi: 10.1210/en.2004-1408. [DOI] [PubMed] [Google Scholar]
- 30.Butt AJ, Dickson KA, McDougall F, Baxter RC. Insulin-like growth factor-binding protein-5 inhibits the growth of human breast cancer cells in vitro and in vivo. J Biol Chem. 2003;278:29676–29685. doi: 10.1074/jbc.M301965200. [DOI] [PubMed] [Google Scholar]
- 31.Akkiprik M, Feng Y, Wang H, Chen K, Hu L, Sahin A, Krishnamurthy S, Ozer A, Hao X, Zhang W. Multifunctional roles of insulin-like growth factor binding protein 5 in breast cancer. Breast Cancer Res. 2008;10:212. doi: 10.1186/bcr2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kalus W, Zweckstetter M, Renner C, Sanchez Y, Georgescu J, Grol M, Demuth D, Schumacher R, Dony C, Lang K, Holak TA. Structure of the IGF-binding domain of the insulin-like growth factor-binding protein-5 (IGFBP-5): implications for IGF and IGF-I receptor interactions. Embo J. 1998;17:6558–6572. doi: 10.1093/emboj/17.22.6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.LeRoith D, Roberts CT., Jr The insulin-like growth factor system and cancer. Cancer Lett. 2003;195:127–137. doi: 10.1016/s0304-3835(03)00159-9. [DOI] [PubMed] [Google Scholar]
- 34.Beattie J, Allan GJ, Lochrie JD, Flint DJ. Insulin-like growth factor-binding protein-5 (IGFBP-5): a critical member of the IGF axis. Biochem J. 2006;395:1–19. doi: 10.1042/BJ20060086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song H, Shand JH, Beattie J, Flint DJ, Allan GJ. The carboxy-terminal domain of IGFbinding protein-5 inhibits heparin binding to a site in the central domain. J Mol Endocrinol. 2001;26:229–239. doi: 10.1677/jme.0.0260229. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The 143B and MG63.2 cells were transduced with the indicated IGFBP5 construct adenoviruses. Immunohistochemistry staining for caspase 8 and 9 were performed to assess for apoptosis. Note the increased staining in the N-terminal domain-containing groups (outlined red).