Abstract
Dynein is a microtubule-based molecular motor that is involved in various biological functions, such as axonal transport, mitosis, and cilia/flagella movement. Although dynein was discovered 50 years ago, the progress of dynein research has been slow due to its large size and flexible structure. Recent progress in understanding the force-generating mechanism of dynein using x-ray crystallography, cryo-electron microscopy, and single molecule studies has provided key insight into the structure and mechanism of action of this complex motor protein.
It has been 50 years since dynein was discovered and named by Ian Gibbons as a motor protein that drives cilia/flagella bending (Gibbons, 1963; Gibbons and Rowe, 1965). In the mid-1980s, dynein was also found to power retrograde transport in neurons (Paschal and Vallee, 1987). Subsequently, the primary amino acid sequence of the cytoplasmic dynein heavy chain, which contains the motor domain, was determined from the cDNA sequence (Mikami et al., 1993; Zhang et al., 1993). Like other biological motors, such as kinesins and myosins, the amino acid sequence of the dynein motor domain is well conserved. There are 16 putative genes that encode dynein heavy chains in the human genome (Yagi, 2009). Among these is one gene encoding cytoplasmic dynein heavy chain and one encoding retrograde intraflagellar transport dynein heavy chain, while the rest encode for heavy chains of axonemal dyneins. Most of the genes encoding the human dynein heavy chain have a counterpart in Chlamydomonas reinhardtii, which suggests that their functions are conserved from algae to humans.
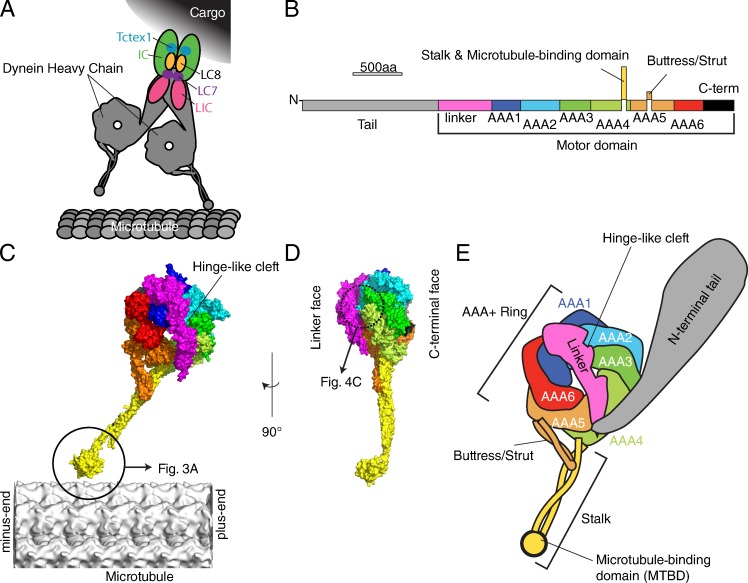
Dynein is unique compared with kinesin and myosin because dynein molecules form large molecular complexes. For example, one axonemal outer arm dynein molecule of C. reinhardtii is composed of three dynein heavy chains, two intermediate chains, and more than ten light chains (King, 2012). Mammalian cytoplasmic dynein consists of two heavy chains and several smaller subunits (Fig. 1 A; Vallee et al., 1988; Allan, 2011). The cargoes of cytoplasmic dynein are various membranous organelles, including lysosomes, endosomes, phagosomes, and the Golgi complex (Hirokawa, 1998). It is likely that one cytoplasmic dynein heavy chain can adapt to diverse cargos and functions by changing its composition.
Figure 1.
Atomic structures of cytoplasmic dynein. (A) Schematic structure of cytoplasmic dynein complex, adapted from Allan (2011). (B) The primary structure of cytoplasmic dynein. (C and D) The atomic model of D. discoideum cytoplasmic dynein motor domain (PDB accession no. 3VKG) overlaid on a microtubule (EMDB-5193; Sui and Downing, 2010) according to the orientation determined by Mizuno et al. (2007) (C) Side view. (D) View from the plus end of microtubule. (E) Schematic domain structure of dynein.
Dynein must have a distinct motor mechanism from kinesin and myosin, because it belongs to the AAA+ family of proteins and does not have the conserved amino acid motifs, called the switch regions, present in kinesins, myosins, and guanine nucleotide-binding proteins (Vale, 1996). Therefore, studying dynein is of great interest because it will reveal new design principles of motor proteins. This review will focus on the mechanism of force generation by cytoplasmic and axonemal dynein heavy chains revealed by recent structural and biophysical studies.
Anatomy of dynein
To understand the chemomechanical cycle of dynein based on its molecular structure, it is important to obtain well-diffracting crystals and build accurate atomic models. Recently, Kon and colleagues determined the crystal structures of Dictyostelium discoideum cytoplasmic dynein motor domain, first at 4.5-Å resolution (Kon et al., 2011), and subsequently at 2.8 Å (without the microtubule binding domain) and 3.8-Å (wild type) resolution (Kon et al., 2012). Carter and colleagues also determined the crystal structures of the Saccharomyces cerevisiae (yeast) cytoplasmic dynein motor domain, first at 6-Å resolution (Carter et al., 2011), and later at 3.3–3.7-Å resolution (Schmidt et al., 2012). According to these crystal structures as well as previous EM studies, the overall structure of the dynein heavy chain is divided into four domains: tail, linker, head, and stalk (Fig. 1, B–E). Simply put, each domain carries out one essential function of a motor protein: the tail is the cargo binding domain, the head is the site of ATP hydrolysis, the linker is the mechanical amplifier, and the stalk is the track-binding domain.
The tail, which is not part of the motor domain and is absent from crystal structures, is located at the N-terminal ∼1,400 amino acid residues and involved in cargo binding (gray in Fig. 1, B and E). The next ∼550 residues comprise the “linker” (pink in Fig. 1, B–E), which changes its conformation depending on the nucleotide state (Burgess et al., 2003; Kon et al., 2005). This linker domain was first observed by negative staining EM in combination with single particle analysis of dynein c, an isoform of inner arm dynein from C. reinhardtii flagella (Burgess et al., 2003). According to the crystal structures, the linker is made of bundles of α-helices and lies across the AAA+ head domain, forming a 10-nm-long rod-like structure (Fig. 1, C and D). Recent class averaged images of D. discoideum cytoplasmic dynein show that the linker domain is stiff along its entire length when undocked from the head (Roberts et al., 2012). The head (motor) domain of dynein is composed of six AAA+ (ATPase associated with diverse cellular activities) modules (Neuwald et al., 1999; color-coded in Fig. 1, B–E). Although many AAA+ family proteins are a symmetric homohexamer (Ammelburg et al., 2006), the AAA+ domains of dynein are encoded by a single heavy chain gene and form an asymmetric heterohexamer. Among the six AAA+ domains, hydrolysis at the first AAA domain mainly provides the energy for dynein motility (Imamula et al., 2007; Kon et al., 2012). The hexameric ring has two distinct faces: the linker face and the C-terminal face. The linker face is slightly convex and the linker domain lies across this side (Fig. 1 D, left side). The other side of the ring has the C-terminal domain (Fig. 1 D, right side).
The stalk domain of dynein was identified as the microtubule-binding domain (MTBD; Gee et al., 1997). It emanates from the C-terminal face of AAA4 and is composed of antiparallel α-helical coiled-coil domain (yellow in Fig. 1, B–E). The tip of the stalk is the actual MTBD. Interestingly, the crystal structures revealed another antiparallel α-helical coiled coil that emerges from AAA5 (orange in Fig. 1, B–E), and this region is called the buttress (Carter et al., 2011) or strut (Kon et al., 2011), which was also observed as the bifurcation of the stalk by negative-staining EM (Burgess et al., 2003; Roberts et al., 2009). The tip of the buttress/strut is in contact with the middle of the stalk and probably works as a mechanical reinforcement of the stalk.
The chemomechanical cycle of dynein
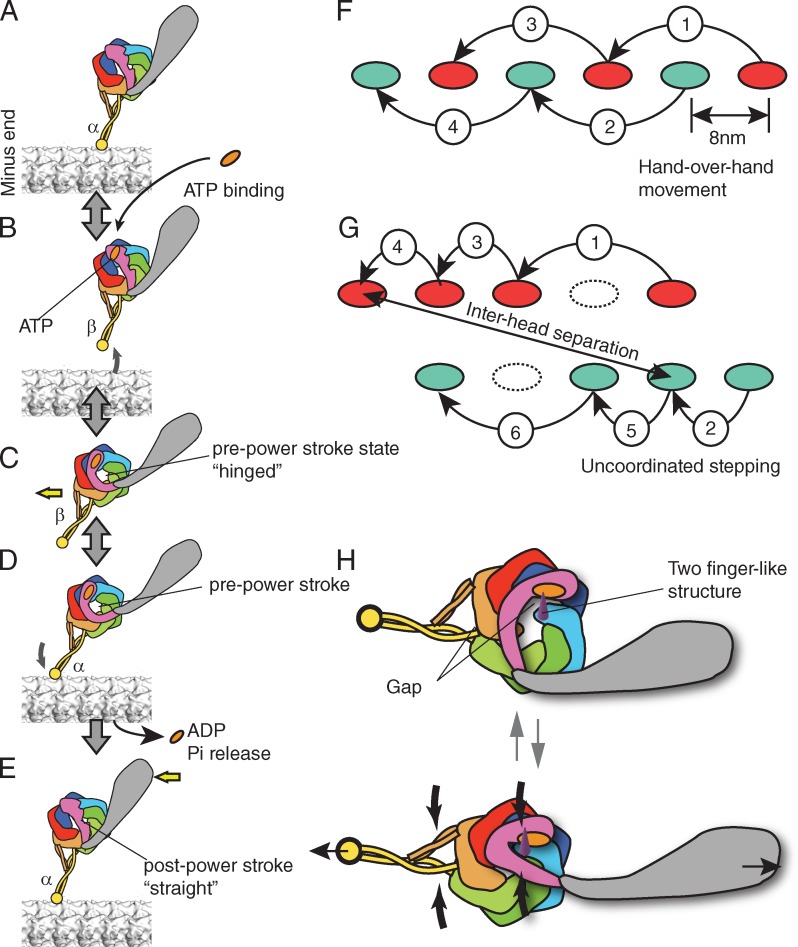
Based on structural and biochemical data, a putative chemomechanical cycle of dynein is outlined in Fig. 2 (A–E). In the no-nucleotide state, dynein is bound to a microtubule through its stalk domain, and its tail region is bound to cargoes (Fig. 2 A). The crystal structures of yeast dynein are considered to be in this no-nucleotide state. When ATP is bound to the AAA+ head, the MTBD quickly detaches from the microtubule (Fig. 2 B; Porter and Johnson, 1983). The ATP binding also induces “hinging” of the linker from the head (Fig. 2 C). According to the biochemical analysis of recombinant D. discoideum dynein (Imamula et al., 2007), the detachment from the microtubule (Fig. 2, A and B) is faster than the later hinging (Fig. 2, B and C). As a result of these two reactions, the head rotates or shifts toward the minus end of the microtubule (for more discussion about “rotate” versus “shift” see the “Dyneins in the axoneme” section) and the MTBD steps forward. The directionality of stepping seems to be mainly determined by the MTBD, because the direction of dynein movement does not change even if the head domain is rotated relative to the microtubule by insertion or deletion of the stalk (Carter et al., 2008). In the presence of ADP and vanadate, dynein is considered to be in this state (Fig. 2 C).
Figure 2.
Presumed chemomechanical cycle and stepping of dynein. (A–E) Chemomechanical cycle of dynein. The pre- and post-power stroke states are also called the primed and unprimed states, respectively. The registries of the stalk coiled coil are denoted as α and β according to Gibbons et al. (2005). (F and G) Processive movement of kinesin (F) and dynein (G). (F) Hand-over-hand movement of kinesin. A step by one head (red) is always followed by the step of another head (green). The stepping of kinesin is on one protofilament of microtubule. (G) Presumed stepping of dynein. The step size varies and the interhead separation can be large. A step by one head (red) is not always flowed by the step of another head (green). (H) A model of strain-based dynein ATPase activation. (G, top) Without strain, the gap between the AAA1 and AAA2 is open and the motor domain cannot hydrolyze ATP. (G, bottom) Under a strain imposed between MTBD and tail (thin black arrows), the gap becomes smaller (thick black arrows) and turns on ATP hydrolysis by dynein.
After the MTBD rebinds to the microtubule at the forward site (Fig. 2 D), release of hydrolysis products from the AAA+ head is activated (Holzbaur and Johnson, 1989) and the hinged linker goes back to the straight conformation (Fig. 2 E; Kon et al., 2005). The crystal structure of D. discoideum dynein is considered to be in the state after phosphate release and before ADP release. This straightening of the linker is considered to be the power-generating step and brings the cargo forward relative to the microtubule.
The MTBD of dynein
As outlined in Fig. 2, the nucleotide state of the head domain may control the affinity of the MTBD to the microtubule. Conversely, the binding of the MTBD to the microtubule should activate the ATPase activity of the head domain. This two-way communication is transmitted through the simple ∼17-nm-long α-helical coiled-coil stalk and the buttress/strut, and its structural basis has been a puzzling question.
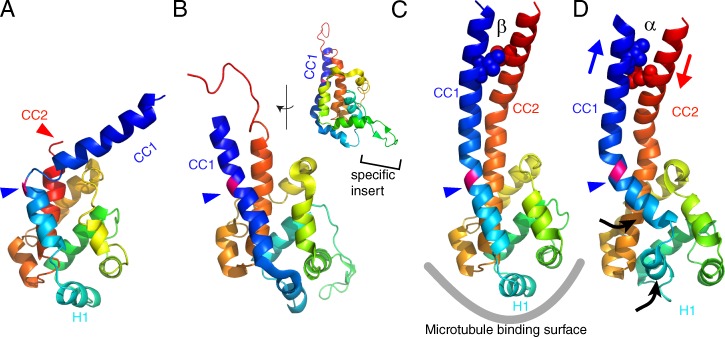
Currently there are three independent MTBD atomic structures in the Protein Data Bank (PDB): One of the crystal structures of the D. discoideum dynein motor domain contains the MTBD (Fig. 3 A), and Carter et al. (2008) crystallized the MTBD of mouse cytoplasmic dynein fused with a seryl tRNA-synthetase domain (Fig. 3 C). The MTBD structure of C. reinhardtii axonemal dynein was solved using nuclear magnetic resonance (PDB accession no. 2RR7; Fig. 3 B). The MTBD is mostly composed of α-helices and the three structures are quite similar to each other within the globular MTBD (Fig. 3). Note that dynein c has an additional insert at the MTBD–microtubule interface (Fig. 3 B, inset), whose function is not yet clear. The three structures start to deviate from the junction between the MTBD and the coiled-coil region of the stalk (Fig. 3, A–C, blue arrowheads). Particularly, one of the stalk α-helix (CC2) in D. discoideum dynein motor domain appears to melt at the junction with the MTBD (Fig. 3 A, red arrowhead). This structural deviation suggests that the stalk coiled coil at the junction is flexible, which is consistent with the observation by EM (Roberts et al., 2009).
Figure 3.
Atomic models of the MTBD of dynein. (A) D. discoideum cytoplasmic dynein (PDB accession no. 3VKH). (B) C. reinhardtii dynein c (PDB accession no. 2RR7). The inset shows the side view, highlighting the dynein c–specific insert. (C) Mouse cytoplasmic dynein (PDB accession no. 3ERR). (D) Mouse cytoplasmic dynein fit to the MTBD–microtubule complex derived from cryo-EM (PDB accession no. 3J1T). All the MTBD structures were aligned using least square fits and color-coded with a gradient from the N to C terminus. CC1, coiled coil helix 1; CC2, coiled coil helix 2. The blue arrowheads points to the junction between MTBD and the stalk, where a well-conserved proline residue (colored pink) is located. In C and D, two residues (isoleucine 3269 and leucine 3417) are shown as spheres. The two residues form hydrophobic contacts in the β-registry (C), whereas they are separated in the α-registry (D) because of the sliding between the two α-helices (blue and red arrows). Conformational changes observed in the mouse dynein MTBD in complex with a microtubule by cryo-EM are shown by black arrows. Note that the cryo-EM density map does not have enough resolution to observe sliding between CC1 and CC2. The sliding was modeled based on targeted molecular dynamics (Redwine et al., 2012).
Various mechanisms have been proposed to explain how the affinity between the MTBD and a microtubule is controlled. Gibbons et al. (2005) proposed “the helix-sliding hypothesis” (for review see Cho and Vale, 2012). In brief, this hypothesis proposes that the sliding between two α-helices CC1 and CC2 (Fig. 3, C and D; blue and red arrows) may control the affinity of this domain to a microtubule. When Gibbons’s classification (Gibbons et al., 2005) of the sliding state is applied to the three MTBD structures, the stalk in the D. discoideum dynein motor domain is in the “α-registry” state (not visible in Fig. 3 A because of the melting of CC2), which corresponds to the strong binding state. However, the mouse cytoplasmic and C. reinhardtii axonemal MTBDs have the “β-registry” stalk (Fig. 3 C), which corresponds to the weak binding state.
To observe conformational changes induced by the α-registry and/or microtubule binding, Redwine et al. (2012) solved the structure of mouse dynein MTBD in complex with a microtubule at 9.7-Å resolution using cryo-EM and single particle analysis. The MTBD was coupled with seryl tRNA-synthetase to fix the stalk helix in the α-registry. At this resolution, α-helices are visible, and they used molecular dynamics to fit the crystal structure of mouse MTBD (β-registry) to the cryo-EM density map. According to this result, the first helix H1 moves ∼10 Å to a position that avoids a clash with the microtubule (Fig. 3 D, black arrows). This also induces opening of the stalk helix (CC1). Together with mutagenesis and single-molecule motility assays, Redwine et al. (2012) proposed that this new structure represents the strong binding state. Currently, it is not clear why the MTBD structure of D. discoideum dynein motor domain (α-registry, Fig. 3 A) is not similar to the new α-registry mouse dynein MTBD, and this problem needs to be addressed by further studies.
Structures around the first ATP binding site
Another central question about motor proteins is how Ångstrom-scale changes around the nucleotide are amplified to generate steps >8 nm. For dynein, the interface between the first nucleotide-binding pocket and the linker seem to be the key force-generating element (Fig. 4). The crystal structures of dynein give us clues about how nucleotide-induced conformational changes may be transmitted to and amplified by the linker domain.
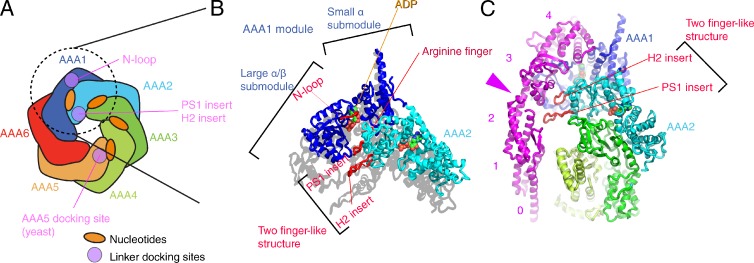
Figure 4.
Structures around the first ATP binding site. (A) Schematic domain structure of the head domain. Regions contacting the linker domain are colored purple. (B) AAA submodules surrounding the first nucleotide-binding pocket (PDB accession no. 3VKG, chain A). The linker is connected to AAA1 domain by the “N-loop.” To highlight that the two finger-like structures are protruding, the shadow of the atomic structure has been cast on the plane parallel to the head domain. (C) Interaction between the linker and the two finger-like structures. The pink arrowhead points to the hinge-like structure of the linker. The pink numbers indicates the subdomain of the linker.
The main ATP catalytic site is located between AAA1 and AAA2 (Fig. 4, A and B). There are four ADP molecules in the D. discoideum dynein crystal structures, but the first ATP binding site alone drives the microtubule-activated ATPase activity, based on biochemical experiments on dyneins whose ATP binding sites were mutated (Kon et al., 2012).
One AAA+ module is composed of a large submodule and a small α submodule (Fig. 4 B). The large α/β submodule is located inside of the ring and the small α submodule is located outside. The large submodule bulges toward the linker face, and the overall ring forms a dome-like shape (Fig. 1 D).
The main ATP catalytic site is surrounded by three submodules: AAA1 large α/β, AAA1 small α, and AAA2 large α/β (Fig. 4, A and B). Based on the structural changes of other AAA+ proteins (Gai et al., 2004; Suno et al., 2006; Wendler et al., 2012), the gap between AAA1 and AAA2 modules is expected to open and close during the ATPase cycle.
In fact, the size of the gap varies among the dynein crystal structures. The crystal structures of yeast dyneins show a larger gap between AAA1 and AAA2, which might be the reason why no nucleotide was found in the binding pocket. Although Schmidt et al. (2012) soaked the crystals in a high concentration of various nucleotides (up to 25 mM of ATP), no electron densities corresponding to the nucleotide were observed at the first ATP binding site. Among dynein crystal structures, one of D. discoideum dynein (PDB accession no. 3VKH, chain A) has the smallest gap, but it is still considered to be in an “open state” because the arginine finger in the AAA2 module (Fig. 4 B, red) is far from the phosphates of ADP. Because the arginine finger is essential for ATP hydrolysis in other AAA+ proteins (Ogura et al., 2004), the gap is expected to close and the arginine finger would stabilize the negative charge during the transition state of ATP hydrolysis.
The presumed open/close conformational change between AAA1 and AAA2 would result in the movement of two “finger-like” structures protruding from the AAA2 large α/β submodule (Fig. 4 B). The two finger-like structures are composed of the H2 insert β-hairpin and preSensor I (PS-I) insert. In D. discoideum dynein crystal structure, the two finger-like structures are in contact with the “hinge-like cleft” of the linker (Fig. 4 C, pink arrowhead). The hinge-like cleft is one of the thinnest parts of the linker, where only one α-helix is connecting between the linker subdomains 2 and 3.
In the yeast crystal structures, which have wider gaps between AAA1 and AAA2, the two finger-like structures are not in direct contact with the linker and separated by 18 Å. Instead, the N-terminal region of the linker is in contact with the AAA5 domain (Fig. 4 A). To test the functional role of the linker–AAA5 interaction, Schmidt et al. (2012) mutated a residue involved in the interaction (Phe3446) and found that the mutation resulted in severe motility defects, showing strong microtubule binding and impaired ATPase activities. In D. discoideum dynein crystals, there is no direct interaction between AAA5 and the linker, which suggests that the gap between AAA1 and AAA2 may influence the interaction between the head and linker domain. The contact between the linker and AAA5 may also influence the gap around AAA5, because the gap between AAA5 and AAA6 is large in yeast dynein crystal, whereas the one between AAA4 and AAA5 is large in D. discoideum dynein.
The movement of two finger-like structures would induce remodeling of the linker. According to the recent cryo-EM 3D reconstructions of cytoplasmic dynein and axonemal dynein c (Roberts et al., 2012), the linker is visible across the head and there is a large gap between AAA1 and AAA2 in the no-nucleotide state. This linker structure is considered to be the “straight” state (Fig. 2, A and E). In the presence of ADP vanadate, the gap between AAA1 and AAA2 is closed and the N-terminal region of linker is near AAA3, which corresponds to the pre-power stroke “hinged” state (Fig. 2, C and D). The transition from the hinged state to the straight state of the linker is considered to be the force-generating step of dynein.
Processivity of dynein
As the structure of dynein is different from other motor proteins, dynein’s stepping mechanism is also distinct. Both dynein and kinesin are microtubule-based motors and move processively. Based on the single molecule tracking experiment with nanometer accuracy (Yildiz et al., 2004), it is widely accepted that kinesin moves processively by using its two motor domain alternately, called the “hand-over-hand” mechanism. To test whether dynein uses a similar mechanism to kinesin or not, recently Qiu et al. (2012) and DeWitt et al. (2012) applied similar single-molecule approaches to dynein.
To observe the stepping, the two head domains of yeast recombinant cytoplasmic dynein were labeled with different colors and the movement of two head domains was tracked simultaneously. If dynein walks by the hand-over-hand mechanism, the step size would be 16 nm and the stepping of one head domain would always be followed by the stepping of another head domain (alternating pattern), and the trailing head would always take a step (Fig. 2 F). Contrary to this prediction, both groups found that the stepping of the head domains is not coordinated when the two head domains are close together. These observations indicated that the chances of a leading or trailing head domain stepping are not significantly different (Fig. 2 G; DeWitt et al., 2012; Qiu et al., 2012).
This stepping pattern predicts that the distance between the head domains can be long. In fact, the distance between the two head domains is on average ∼18 ± 11 nm (Qiu et al., 2012) or 28.4 ± 10.7 nm (DeWitt et al., 2012), and as large as ∼50 nm (DeWitt et al., 2012). When the two head domains are separated, there is a tendency where stepping of the trailing head is preferred over that of the forward head.
In addition, even though the recombinant cytoplasmic dynein is a homodimer, the two heavy chains do not function equally. While walking along the microtubule, the leading head tends to walk on the right side, whereas the trailing head walks on the left side (DeWitt et al., 2012; Qiu et al., 2012). This arrangement suggests that the stepping mechanism is different between the two heads. In fact, when one of the two dynein heavy chains is mutated to abolish the ATPase activity at AAA1, the heterodimeric dynein still moves processively (DeWitt et al., 2012), with the AAA1-mutated dynein heavy chain remaining mostly in the trailing position. This result clearly demonstrates that allosteric communication between the two AAA1 domains is not required for processivity of dynein. It is likely that the mutated head acts as a tether to the microtubule, as it is known that wild-type dynein can step processively along microtubules under external load even in the absence of ATP (Gennerich et al., 2007).
These results collectively show that dynein moves by a different mechanism from kinesin. It is likely that the long stalk and tail allow dynein to move in a more flexible manner.
Dyneins in the axoneme
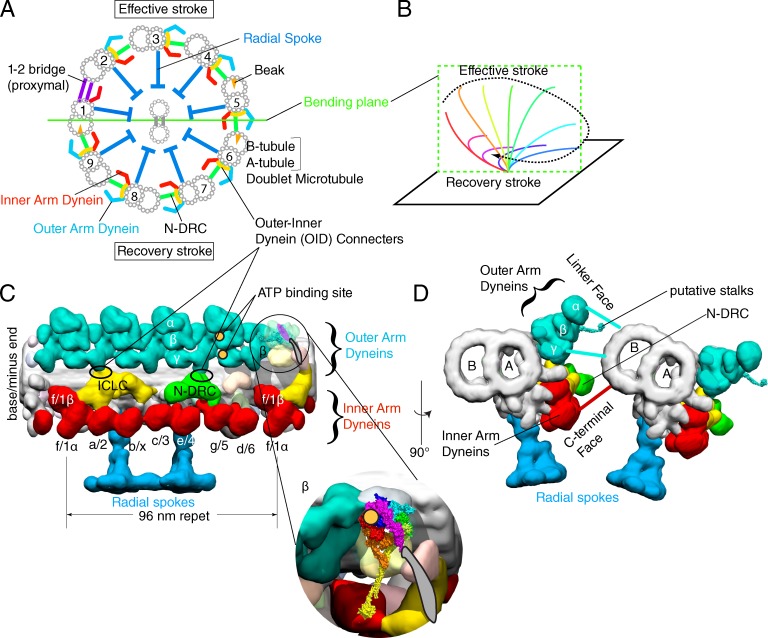
As mentioned in the introduction, >10 dyneins work in motile flagella and cilia. The core of flagella and cilia is the axoneme, which is typically made of nine outer doublet microtubules and two central pair microtubules (“9 + 2,” Fig. 5 A). The axonemes are found in various eukaryotic cells ranging from the single-cell algae C. reinhardtii to human. Recent extensive cryo-electron tomography (cryo-ET) in combination with genetics revealed the highly organized and complex structures of axonemes that are potentially important for regulating dynein activities (Fig. 5, C and D; Nicastro et al., 2006; Bui et al., 2008, 2009, 2012; Heuser et al., 2009, 2012; Movassagh et al., 2010; Lin et al., 2012; Carbajal-González et al., 2013; Yamamoto et al., 2013).
Figure 5.
Arrangement of axonemal dyneins. (A) The schematic structure of the motile 9 + 2 axoneme, viewed from the base of flagella. (B) Quasi-planar asymmetric movement of the 9 + 2 axoneme typically observed in trachea cilia or in C. reinhardtii flagella. (C and D) 3D structure of a 96-nm repeat of doublet microtubules in the distal/central region of C. reinhardtii flagella (EMDB-2132; Bui et al., 2012). N-DRC, the nexin-dynein regulatory complex; ICLC, intermediate chain/light chain complex. Inner arm dynein subspecies are labeled according to Bui et al. (2012) and Lin et al. (2012). To avoid the confusion with the linker domain of dynein, the structures connecting between outer and inner arm dyneins are labeled as “connecters,” which are normally called “linkers.” Putative ATP binding sites of outer arm dynein determined by biotin-ADP (Oda et al., 2013) are indicated by orange circles. The atomic structure of cytoplasmic dynein is placed into the β-heavy chain of outer arm dynein and its enlarged view is shown in the inset. (D) Two doublet microtubules, viewed from the base of flagella.
The basic mechanochemical cycles of axonemal dyneins are believed to be shared with cytoplasmic dynein. Dynein c is an inner arm dynein of C. reinhardtii and used extensively to investigate the conformational changes of dynein, as shown in Fig. 2 (A–E), by combining EM and single-particle analysis (Burgess et al., 2003; Roberts et al., 2012). Structural changes of axonemal dyneins complexed with microtubules are also observed by quick-freeze and deep-etch EM (Goodenough and Heuser, 1982; Burgess, 1995), cryo-EM (Oda et al., 2007), negative-staining EM (Ueno et al., 2008), and cryo-ET (Movassagh et al., 2010). According to these studies, the AAA+ head domains are constrained near the A-tubule in the no-nucleotide state. In the presence of nucleotide, the head domains move closer to the B-tubule and/or the minus end of microtubule, and their appearance becomes heterogeneous, which is consistent with the observation of isolated dynein c that shows greater flexibility between tail and stalk in the ADP/vanadate state (Burgess et al., 2003).
One of the controversies about the structural changes of axonemal dyneins is whether their stepping involves “rotation” or “shift” of the head (Fig. 2, B to D). The stalk angle relative to the microtubule seems to be a constant ∼60° irrespective of the nucleotide state (Ueno et al., 2008; Movassagh et al., 2010). This angle is similar to the angle obtained from cryo-EM study of the MTBD–microtubule complex (Redwine et al., 2012). Based on these observations, Ueno et al. (2008) and Movassagh et al. (2010) hypothesize that the “shift” of the head pulls the B-microtubule toward the distal end. However, Roberts et al. (2012) propose that the “rotation” of head and stalk is involved in the stepping based on the docking of dynein c head into an averaged flagella tomogram obtained by Movassagh et al. (2010). This issue needs to be resolved by more reliable and high-resolution data, but these two models may not be mutually exclusive. For example, averaged tomograms may be biased toward the microtubule-bound stalk because tomograms are aligned using microtubules.
To interpret these structural changes of axonemal dyneins, docking atomic models of dynein is necessary. According to Roberts et al. (2012), the linker face of inner arm dynein c is oriented outside of axoneme (Fig. 5 D). For outer arm dyneins, we used cryo-EM in combination with biotin-ADP-streptavidin labeling and showed that the ATP binding site, most likely AAA1, is on the left side of the AAA+ head (Fig. 5 C; Oda et al. (2013)). Assuming that the stalks extend out of the plane toward the viewer, the linker face of outer arm dynein is oriented outside of axoneme (Fig. 5 C, inset; and Fig. 5 D). If it were the opposite, the AAA1 would be located on the right side of the AAA+ head. In summary, both inner and outer arm dynein seem to have the same arrangement, with their linker face oriented outside of the axoneme (Fig. 5 D).
A unique characteristic of axonemal dyneins is that these dyneins are under precise temporal and spatial control. To generate a planer beating motion (Fig. 5 B), dyneins should be asymmetrically controlled, because the dyneins located on doublets 2–4 drive the effective stroke, whereas the ones on doublets 6–8 drive the recovery stroke (Fig. 5 A). Based on the cryo-ET observation of axonemes, Nicastro et al. (2006) proposed that “linkers” between dyneins provide hard-wiring to coordinate motor activities. Because the linkers in axonemes are distinct structures from the linker domain of dynein, for clarity, here we call them “connecters.” According to the recent cryo-ET of proximal region of C. reinhardtii flagella (Bui et al., 2012), there are in fact asymmetries among nine doublets that are localized to the connecters between outer and inner arm dynein, called the outer-inner dynein (OID) connecters (Fig. 5, A and C). Recently we identified that the intermediate chain 2 (IC2) of outer arm dynein is a part of the OID connecters, and a mutation of the N-terminal region of IC2 affects functions of both outer and inner arm dyneins (Oda et al., 2013), which supports the idea that the connecters between dyneins are involved in axonemal dynein regulation.
Closing remarks
Thanks to the crystal structures, we can now design and interpret experiments such as single molecule assays and EM based on the atomic models of dynein. Our understanding of the molecular mechanism and cellular functions of dyneins will be significantly advanced by these experiments in the near future.
One important direction of dynein research is to understand the motor mechanisms closer to the in vivo state. For example, the step sizes of cytoplasmic dynein purified from porcine brain is ∼8 nm independent of load (Toba et al., 2006). This result suggests that intermediate and light chain bound to the dynein heavy chain may modulate the motor activity of dynein. To address such questions, Trokter et al. (2012) reconstituted human cytoplasmic dynein complex from recombinant proteins, although the reconstituted dynein did not show robust processive movement. Further studies are required to understand the movement of cytoplasmic dynein. Similarly, axonemal dyneins should also be studied using mutations in a specific gene that does not affect the overall flagella structure, rather than depending on null mutants that cause the loss of large protein complexes.
Detailed full chemomechanical cycle of dynein and its regulation are of great importance. Currently, open/closed states of the gap between AAA1 and AAA2 are not clearly correlated with the chemomechanical cycle of dynein. Soaking dynein crystal with nucleotides showed that the presence of ATP alone is not sufficient to close the gap, at least in the crystal (Schmidt et al., 2012). This result suggests that other factors such as a conformational change of the linker are required. For other motors, ATP hydrolysis is an irreversible chemical step, which is often “gated” by strain. In the case of kinesin, ATP is hydrolyzed by a motor domain only when a forward strain is applied by the other motor domain through the neck linker (Cross, 2004; Kikkawa, 2008). A similar strain-based gating mechanism may play important roles in controlling the dynein ATPase. Upon MTBD binding to the forward binding site, a strain between MTBD and tail would be applied to the dynein molecule. The Y-shaped stalk and strut/buttress under the strain would force the head domain to close the gap between AAA4 and AAA5 (Fig. 2 H). Similarly, the linker under the strain would be hooked onto the two finger-like structures and close the gap between AAA1 and AAA2 (Fig. 2 H). The gap closure then triggers ATP hydrolysis by dynein. This strain-based gating of dynein is consistent with the observation that the rate of nonadvancing backward steps, which would depend on ATP hydrolysis, is increased by load applied to dynein (Gennerich et al., 2007). To explain cilia and flagella movement, the geometric clutch hypothesis has been proposed (Lindemann, 2007), which contends that the forces transverse (t-force) to the axonemal axis act on the dynein to regulate dynein activities. In the axoneme, dynein itself can be the sensor of the t-force by the strain-based gating mechanism. Further experiments are required to test this idea, but the strain-based gating could be a shared property of biological motors.
Acknowledgments
We thank Toshiki Yagi for discussion and critical reading of the manuscript.
This work was supported by the Funding Program for Next Generation World-Leading Researchers (LS027) and a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports Science and Technology of Japan, the Uehara Memorial Foundation, and the Takeda Science Foundation.
Footnotes
Abbreviations used in this paper:
- cryo-ET
- cryo-electron tomography
- MTBD
- microtubule-binding domain
References
- Allan V.J. 2011. Cytoplasmic dynein. Biochem. Soc. Trans. 39:1169–1178 10.1042/BST0391169 [DOI] [PubMed] [Google Scholar]
- Ammelburg M., Frickey T., Lupas A.N. 2006. Classification of AAA+ proteins. J. Struct. Biol. 156:2–11 10.1016/j.jsb.2006.05.002 [DOI] [PubMed] [Google Scholar]
- Bui K.H., Sakakibara H., Movassagh T., Oiwa K., Ishikawa T. 2008. Molecular architecture of inner dynein arms in situ in Chlamydomonas reinhardtii flagella. J. Cell Biol. 183:923–932 10.1083/jcb.200808050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui K.H., Sakakibara H., Movassagh T., Oiwa K., Ishikawa T. 2009. Asymmetry of inner dynein arms and inter-doublet links in Chlamydomonas flagella. J. Cell Biol. 186:437–446 10.1083/jcb.200903082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui K.H., Yagi T., Yamamoto R., Kamiya R., Ishikawa T. 2012. Polarity and asymmetry in the arrangement of dynein and related structures in the Chlamydomonas axoneme. J. Cell Biol. 198:913–925 10.1083/jcb.201201120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess S.A. 1995. Rigor and relaxed outer dynein arms in replicas of cryofixed motile flagella. J. Mol. Biol. 250:52–63 10.1006/jmbi.1995.0357 [DOI] [PubMed] [Google Scholar]
- Burgess S.A., Walker M.L., Sakakibara H., Knight P.J., Oiwa K. 2003. Dynein structure and power stroke. Nature. 421:715–718 10.1038/nature01377 [DOI] [PubMed] [Google Scholar]
- Carbajal-González B.I., Heuser T., Fu X., Lin J., Smith B.W., Mitchell D.R., Nicastro D. 2013. Conserved structural motifs in the central pair complex of eukaryotic flagella. Cytoskeleton (Hoboken). 70:101–120 10.1002/cm.21094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter A.P., Garbarino J.E., Wilson-Kubalek E.M., Shipley W.E., Cho C., Milligan R.A., Vale R.D., Gibbons I.R. 2008. Structure and functional role of dynein’s microtubule-binding domain. Science. 322:1691–1695 10.1126/science.1164424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter A.P., Cho C., Jin L., Vale R.D. 2011. Crystal structure of the dynein motor domain. Science. 331:1159–1165 10.1126/science.1202393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho C., Vale R.D. 2012. The mechanism of dynein motility: insight from crystal structures of the motor domain. Biochim. Biophys. Acta. 1823:182–191 10.1016/j.bbamcr.2011.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross R.A. 2004. The kinetic mechanism of kinesin. Trends Biochem. Sci. 29:301–309 10.1016/j.tibs.2004.04.010 [DOI] [PubMed] [Google Scholar]
- DeWitt M.A., Chang A.Y., Combs P.A., Yildiz A. 2012. Cytoplasmic dynein moves through uncoordinated stepping of the AAA+ ring domains. Science. 335:221–225 10.1126/science.1215804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gai D., Zhao R., Li D., Finkielstein C.V., Chen X.S. 2004. Mechanisms of conformational change for a replicative hexameric helicase of SV40 large tumor antigen. Cell. 119:47–60 10.1016/j.cell.2004.09.017 [DOI] [PubMed] [Google Scholar]
- Gee M.A., Heuser J.E., Vallee R.B. 1997. An extended microtubule-binding structure within the dynein motor domain. Nature. 390:636–639 10.1038/37663 [DOI] [PubMed] [Google Scholar]
- Gennerich A., Carter A.P., Reck-Peterson S.L., Vale R.D. 2007. Force-induced bidirectional stepping of cytoplasmic dynein. Cell. 131:952–965 10.1016/j.cell.2007.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons I.R. 1963. Studies on the protein components of cilia from Tetrahymena pyriformis. Proc. Natl. Acad. Sci. USA. 50:1002–1010 10.1073/pnas.50.5.1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons I.R., Rowe A.J. 1965. Dynein: A protein with adenosine triphosphatase activity from cilia. Science. 149:424–426 10.1126/science.149.3682.424 [DOI] [PubMed] [Google Scholar]
- Gibbons I.R., Garbarino J.E., Tan C.E., Reck-Peterson S.L., Vale R.D., Carter A.P. 2005. The affinity of the dynein microtubule-binding domain is modulated by the conformation of its coiled-coil stalk. J. Biol. Chem. 280:23960–23965 10.1074/jbc.M501636200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough U.W., Heuser J.E. 1982. Substructure of the outer dynein arm. J. Cell Biol. 95:798–815 10.1083/jcb.95.3.798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuser T., Raytchev M., Krell J., Porter M.E., Nicastro D. 2009. The dynein regulatory complex is the nexin link and a major regulatory node in cilia and flagella. J. Cell Biol. 187:921–933 10.1083/jcb.200908067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuser T., Barber C.F., Lin J., Krell J., Rebesco M., Porter M.E., Nicastro D. 2012. Cryoelectron tomography reveals doublet-specific structures and unique interactions in the I1 dynein. Proc. Natl. Acad. Sci. USA. 109:E2067–E2076 10.1073/pnas.1120690109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N. 1998. Kinesin and dynein superfamily proteins and the mechanism of organelle transport. Science. 279:519–526 10.1126/science.279.5350.519 [DOI] [PubMed] [Google Scholar]
- Holzbaur E.L., Johnson K.A. 1989. Microtubules accelerate ADP release by dynein. Biochemistry. 28:7010–7016 10.1021/bi00443a034 [DOI] [PubMed] [Google Scholar]
- Imamula K., Kon T., Ohkura R., Sutoh K. 2007. The coordination of cyclic microtubule association/dissociation and tail swing of cytoplasmic dynein. Proc. Natl. Acad. Sci. USA. 104:16134–16139 10.1073/pnas.0702370104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikkawa M. 2008. The role of microtubules in processive kinesin movement. Trends Cell Biol. 18:128–135 10.1016/j.tcb.2008.01.002 [DOI] [PubMed] [Google Scholar]
- King S.M. 2012. Integrated control of axonemal dynein AAA(+) motors. J. Struct. Biol. 179:222–228 10.1016/j.jsb.2012.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kon T., Mogami T., Ohkura R., Nishiura M., Sutoh K. 2005. ATP hydrolysis cycle-dependent tail motions in cytoplasmic dynein. Nat. Struct. Mol. Biol. 12:513–519 10.1038/nsmb930 [DOI] [PubMed] [Google Scholar]
- Kon T., Sutoh K., Kurisu G. 2011. X-ray structure of a functional full-length dynein motor domain. Nat. Struct. Mol. Biol. 18:638–642 10.1038/nsmb.2074 [DOI] [PubMed] [Google Scholar]
- Kon T., Oyama T., Shimo-Kon R., Imamula K., Shima T., Sutoh K., Kurisu G. 2012. The 2.8 Å crystal structure of the dynein motor domain. Nature. 484:345–350 10.1038/nature10955 [DOI] [PubMed] [Google Scholar]
- Lin J., Heuser T., Song K., Fu X., Nicastro D. 2012. One of the nine doublet microtubules of eukaryotic flagella exhibits unique and partially conserved structures. PLoS ONE. 7:e46494 10.1371/journal.pone.0046494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindemann C.B. 2007. The geometric clutch as a working hypothesis for future research on cilia and flagella. Ann. N. Y. Acad. Sci. 1101:477–493 10.1196/annals.1389.024 [DOI] [PubMed] [Google Scholar]
- Mikami A., Paschal B.M., Mazumdar M., Vallee R.B. 1993. Molecular cloning of the retrograde transport motor cytoplasmic dynein (MAP 1C). Neuron. 10:787–796 10.1016/0896-6273(93)90195-W [DOI] [PubMed] [Google Scholar]
- Mizuno N., Narita A., Kon T., Sutoh K., Kikkawa M. 2007. Three-dimensional structure of cytoplasmic dynein bound to microtubules. Proc. Natl. Acad. Sci. USA. 104:20832–20837 10.1073/pnas.0710406105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Movassagh T., Bui K.H., Sakakibara H., Oiwa K., Ishikawa T. 2010. Nucleotide-induced global conformational changes of flagellar dynein arms revealed by in situ analysis. Nat. Struct. Mol. Biol. 17:761–767 10.1038/nsmb.1832 [DOI] [PubMed] [Google Scholar]
- Neuwald A.F., Aravind L., Spouge J.L., Koonin E.V. 1999. AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 9:27–43 [PubMed] [Google Scholar]
- Nicastro D., Schwartz C., Pierson J., Gaudette R., Porter M.E., McIntosh J.R. 2006. The molecular architecture of axonemes revealed by cryoelectron tomography. Science. 313:944–948 10.1126/science.1128618 [DOI] [PubMed] [Google Scholar]
- Oda T., Hirokawa N., Kikkawa M. 2007. Three-dimensional structures of the flagellar dynein-microtubule complex by cryoelectron microscopy. J. Cell Biol. 177:243–252 10.1083/jcb.200609038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda T., Yagi T., Yanagisawa H., Kikkawa M. 2013. Identification of the outer-inner dynein linker as a hub controller for axonemal Dynein activities. Curr. Biol. 23:656–664 10.1016/j.cub.2013.03.028 [DOI] [PubMed] [Google Scholar]
- Ogura T., Whiteheart S.W., Wilkinson A.J. 2004. Conserved arginine residues implicated in ATP hydrolysis, nucleotide-sensing, and inter-subunit interactions in AAA and AAA+ ATPases. J. Struct. Biol. 146:106–112 10.1016/j.jsb.2003.11.008 [DOI] [PubMed] [Google Scholar]
- Paschal B.M., Vallee R.B. 1987. Retrograde transport by the microtubule-associated protein MAP 1C. Nature. 330:181–183 10.1038/330181a0 [DOI] [PubMed] [Google Scholar]
- Porter M.E., Johnson K.A. 1983. Transient state kinetic analysis of the ATP-induced dissociation of the dynein-microtubule complex. J. Biol. Chem. 258:6582–6587 [PubMed] [Google Scholar]
- Qiu W., Derr N.D., Goodman B.S., Villa E., Wu D., Shih W., Reck-Peterson S.L. 2012. Dynein achieves processive motion using both stochastic and coordinated stepping. Nat. Struct. Mol. Biol. 19:193–200 10.1038/nsmb.2205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redwine W.B., Hernández-López R., Zou S., Huang J., Reck-Peterson S.L., Leschziner A.E. 2012. Structural basis for microtubule binding and release by dynein. Science. 337:1532–1536 10.1126/science.1224151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A.J., Numata N., Walker M.L., Kato Y.S., Malkova B., Kon T., Ohkura R., Arisaka F., Knight P.J., Sutoh K., Burgess S.A. 2009. AAA+ Ring and linker swing mechanism in the dynein motor. Cell. 136:485–495 10.1016/j.cell.2008.11.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A.J., Malkova B., Walker M.L., Sakakibara H., Numata N., Kon T., Ohkura R., Edwards T.A., Knight P.J., Sutoh K., et al. 2012. ATP-driven remodeling of the linker domain in the dynein motor. Structure. 20:1670–1680 10.1016/j.str.2012.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt H., Gleave E.S., Carter A.P. 2012. Insights into dynein motor domain function from a 3.3-Å crystal structure. Nat. Struct. Mol. Biol. 19:492–497: S1 10.1038/nsmb.2272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui H., Downing K.H. 2010. Structural basis of interprotofilament interaction and lateral deformation of microtubules. Structure. 18:1022–1031 10.1016/j.str.2010.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suno R., Niwa H., Tsuchiya D., Zhang X., Yoshida M., Morikawa K. 2006. Structure of the whole cytosolic region of ATP-dependent protease FtsH. Mol. Cell. 22:575–585 10.1016/j.molcel.2006.04.020 [DOI] [PubMed] [Google Scholar]
- Toba S., Watanabe T.M., Yamaguchi-Okimoto L., Toyoshima Y.Y., Higuchi H. 2006. Overlapping hand-over-hand mechanism of single molecular motility of cytoplasmic dynein. Proc. Natl. Acad. Sci. USA. 103:5741–5745 10.1073/pnas.0508511103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trokter M., Mücke N., Surrey T. 2012. Reconstitution of the human cytoplasmic dynein complex. Proc. Natl. Acad. Sci. USA. 109:20895–20900 10.1073/pnas.1210573110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno H., Yasunaga T., Shingyoji C., Hirose K. 2008. Dynein pulls microtubules without rotating its stalk. Proc. Natl. Acad. Sci. USA. 105:19702–19707 10.1073/pnas.0808194105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale R.D. 1996. Switches, latches, and amplifiers: common themes of G proteins and molecular motors. J. Cell Biol. 135:291–302 10.1083/jcb.135.2.291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallee R.B., Wall J.S., Paschal B.M., Shpetner H.S. 1988. Microtubule-associated protein 1C from brain is a two-headed cytosolic dynein. Nature. 332:561–563 10.1038/332561a0 [DOI] [PubMed] [Google Scholar]
- Wendler P., Ciniawsky S., Kock M., Kube S. 2012. Structure and function of the AAA+ nucleotide binding pocket. Biochim. Biophys. Acta. 1823:2–14 10.1016/j.bbamcr.2011.06.014 [DOI] [PubMed] [Google Scholar]
- Yagi T. 2009. Bioinformatic approaches to dynein heavy chain classification. Methods Cell Biol. 92:1–9 10.1016/S0091-679X(08)92001-X [DOI] [PubMed] [Google Scholar]
- Yamamoto R., Song K., Yanagisawa H.A., Fox L., Yagi T., Wirschell M., Hirono M., Kamiya R., Nicastro D., Sale W.S. 2013. The MIA complex is a conserved and novel dynein regulator essential for normal ciliary motility. J. Cell Biol. 201:263–278 10.1083/jcb.201211048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz A., Tomishige M., Vale R.D., Selvin P.R. 2004. Kinesin walks hand-over-hand. Science. 303:676–678 10.1126/science.1093753 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Tanaka Y., Nonaka S., Aizawa H., Kawasaki H., Nakata T., Hirokawa N. 1993. The primary structure of rat brain (cytoplasmic) dynein heavy chain, a cytoplasmic motor enzyme. Proc. Natl. Acad. Sci. USA. 90:7928–7932 10.1073/pnas.90.17.7928 [DOI] [PMC free article] [PubMed] [Google Scholar]