Senescent cells extrude fragments of chromatin from the nucleus into the cytoplasm, where they are processed by an autophagic/lysosomal pathway.
Abstract
Cellular senescence is a stable proliferation arrest, a potent tumor suppressor mechanism, and a likely contributor to tissue aging. Cellular senescence involves extensive cellular remodeling, including of chromatin structure. Autophagy and lysosomes are important for recycling of cellular constituents and cell remodeling. Here we show that an autophagy/lysosomal pathway processes chromatin in senescent cells. In senescent cells, lamin A/C–negative, but strongly γ-H2AX–positive and H3K27me3-positive, cytoplasmic chromatin fragments (CCFs) budded off nuclei, and this was associated with lamin B1 down-regulation and the loss of nuclear envelope integrity. In the cytoplasm, CCFs were targeted by the autophagy machinery. Senescent cells exhibited markers of lysosomal-mediated proteolytic processing of histones and were progressively depleted of total histone content in a lysosome-dependent manner. In vivo, depletion of histones correlated with nevus maturation, an established histopathologic parameter associated with proliferation arrest and clinical benignancy. We conclude that senescent cells process their chromatin via an autophagy/lysosomal pathway and that this might contribute to stability of senescence and tumor suppression.
Introduction
Cellular senescence is an irreversible proliferation arrest that is activated in primary human cells by various molecular triggers, including shortened telomeres that result from excessive rounds of cell proliferation (so-called replicative senescence [RS]; Hayflick and Moorhead, 1961; Bodnar et al., 1998); activation of oncogenes, such as B-RAF and N-RAS (so-called oncogene-induced senescence [OIS]; Serrano et al., 1997); and other molecular stresses (Adams, 2009; Kuilman et al., 2010). Both RS and OIS are important tumor suppressor mechanisms in vivo (Braig et al., 2005; Chen et al., 2005; Collado et al., 2005; Michaloglou et al., 2005; Cosme-Blanco et al., 2007; Feldser and Greider, 2007). For example, virtually all benign human nevi harbor activating mutations in N-RAS or its downstream effector, BRAF (Omholt et al., 2002; Pollock et al., 2003; Gray-Schopfer et al., 2007). But, typically, benign nevi do not progress to melanoma. Melanocytes within human nevi display markers of senescence, including cell cycle exit, expression of p16INK4a, senescence-associated β-galactosidase (SA β-gal), and elevated DNA damage signaling (Michaloglou et al., 2005; Gray-Schopfer et al., 2006; Suram et al., 2012; but see also Tran et al., 2012). These data suggest that oncogenic activation of the Ras pathway in melanocytes in vivo ultimately triggers senescence to block melanoma progression.
Senescence-associated proliferation arrest is driven by various interconnected effector pathways, including activation of the p16INK4a/pRB and p53/p21CIP1 tumor suppressor pathways, elevated DNA damage signaling, and chromatin changes (Adams, 2009; Kuilman et al., 2010). In addition, senescence is associated with secretion of various growth regulatory factors, cytokines and chemokines (the so-called senescence-associated secretory phenotype [SASP]; Krtolica et al., 2001). Accumulating evidence indicates that the SASP facilitates control of neoplastic, potentially transformed cells, by the immune system (Xue et al., 2007; Krizhanovsky et al., 2008; Kang et al., 2011; Sagiv et al., 2012).
Evidence also suggests that the lysosomal compartment, being up-regulated in senescence (Lee et al., 2006), plays an important role in senescence-associated cell cycle arrest and generation of the SASP (Young et al., 2009), at least in the context of OIS. Expression of the SASP is dependent on a specific cellular compartment, dubbed the TOR-autophagy spatial coupling compartment (TASCC), where the autophagic catabolic pathway is spatially and temporally linked to the mTOR-associated anabolic pathway, allowing protein degradation to feed raw materials directly into protein synthesis for the SASP (Narita et al., 2011). Flux through linked autophagic (inhibited by mTOR) and protein synthetic (activated by mTOR) pathways is consistent with the ability of rapamycin, an inhibitor of mTOR and activator of autophagy, to both activate and inhibit features of senescence (Demidenko et al., 2009; Cao et al., 2011; Kennedy et al., 2011; Iglesias-Bartolome et al., 2012).
Altered chromatin structure also controls senescence. Many senescent cells accumulate specific subnuclear heterochromatic compartments, called senescence-associated heterochromatin foci (SAHF; Narita et al., 2003). These chromatin-dense domains are believed to embed and silence genes responsible for cell cycle progression (Narita et al., 2003; Zhang et al., 2005, 2007), thus facilitating senescence-associated terminal proliferation arrest. Also, it has been shown that the formation of SAHF protects against excessive DNA damage signaling during oncogenic stress (Di Micco et al., 2011). However, the nature of the link, if any, between autophagy and chromatin in senescent cells is unknown.
Here we show that senescent cells extrude fragments of chromatin from the nucleus into the cytoplasm. These cytoplasmic chromatin fragments (CCFs) lack a nuclear envelope and are γ-H2AX–positive and H3K27me3-positive, but 53BP1- and H3K9ac-negative. CCFs are processed in the cytoplasm by an autophagic/lysosomal pathway. Processing of CCFs is associated with reduced histone content in senescent cells and this is prevented by lysosomal inhibitors. Together, these findings reveal a dynamic process of ongoing chromatin metabolism in senescent cells.
Results
Cytoplasmic chromatin fragments in senescent cells
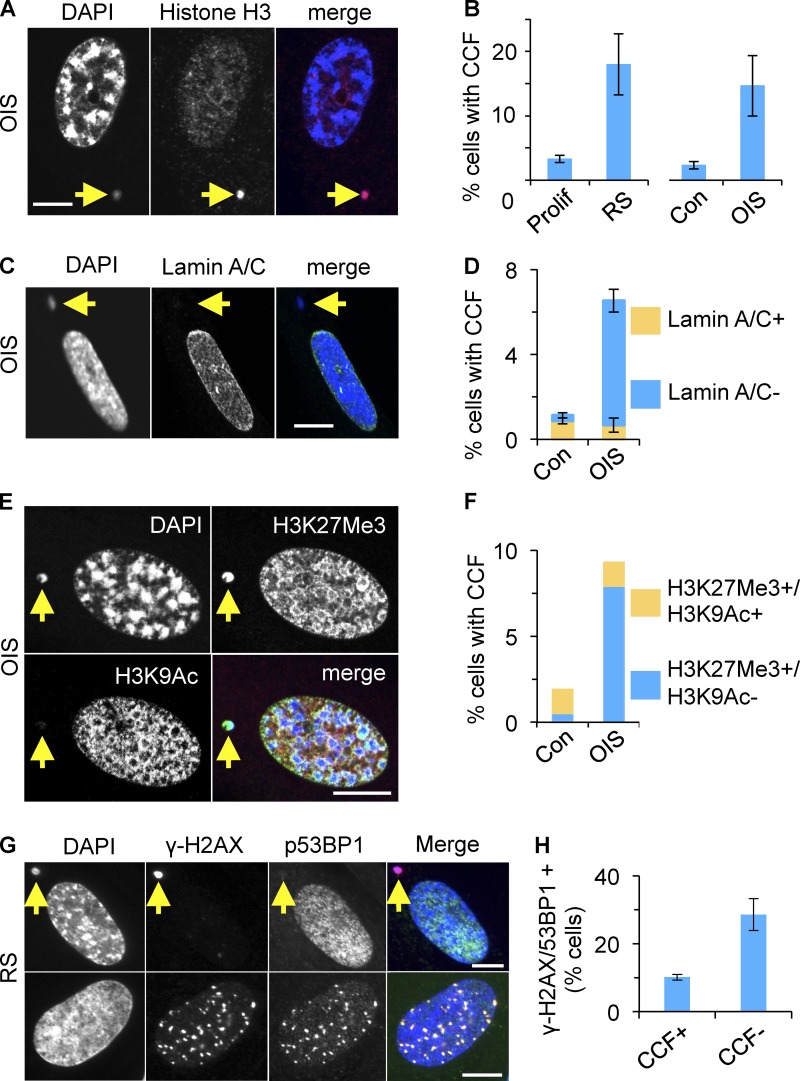
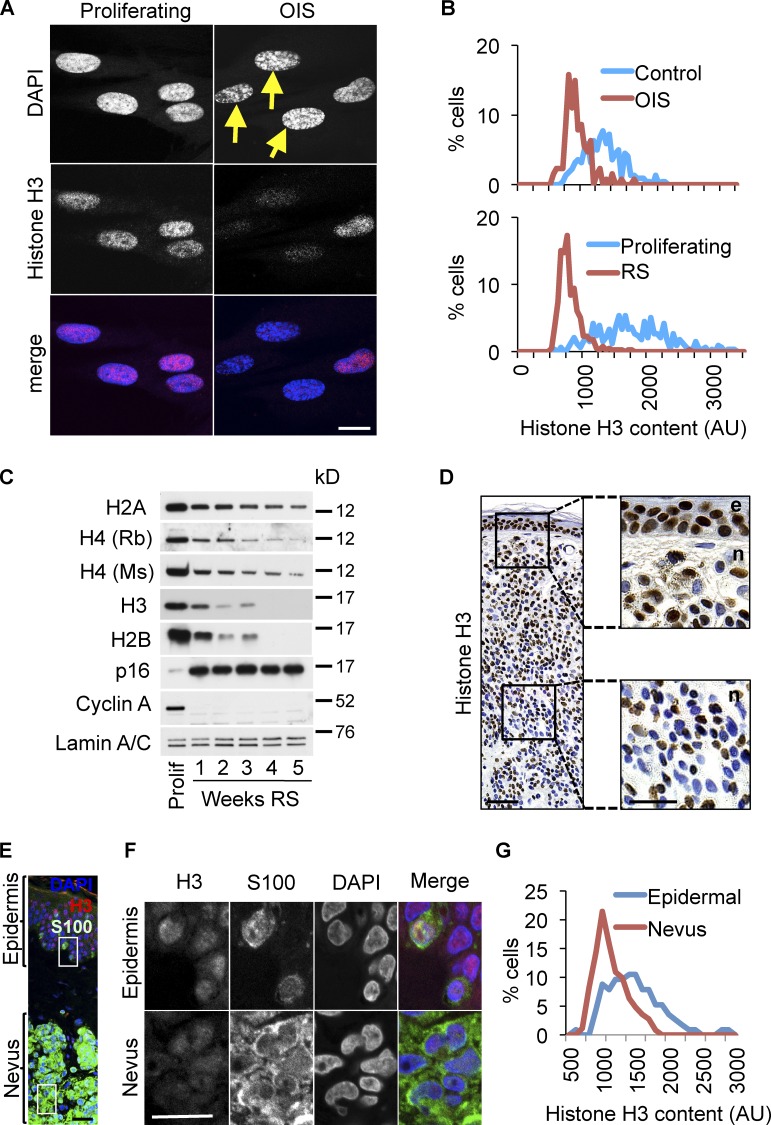
While comparing chromatin of proliferating and senescent cells, we noted chromatin changes in senescent cells in addition to SAHF (Narita et al., 2003), a well-documented feature of senescence. Specifically, in both RS and OIS, we noted an increase in the proportion of cells harboring DAPI- and strongly histone H3-stained CCFs, outside of the main cell nucleus (Fig. 1, A and B). In populations of senescent cells (both RS and OIS), where the majority of the cells were senescent as judged by SA β-gal and cell cycle exit, the proportion of cells containing CCFs typically did not exceed 20% (Fig. S1 A). Among those cells containing CCFs, the mean number of CCFs per cell was 1.65 ± 0.7. Thus, under steady-state conditions, a sizable minority of cells contain 1–3 CCFs.
Figure 1.
Cytoplasmic chromatin fragments in senescent cells. (A) Cytoplasmic chromatin fragments (CCFs) in senescent cells are strongly positive for histone H3. Yellow arrow marks CCF. (B) Increased proportion of cells with CCFs in RS or OIS. Mean ± SEM, n = 3; P < 0.01 (OIS), P < 0.002 (RS). Assays were performed 10 d after induction with tamoxifen (OIS) or 4 wk after the last passage and >90% SA β-gal+ (RS). (C) CCFs in OIS cells are lamin A/C negative. (D) Cells from C were scored for lamin A/C+ and lamin A/C− CCFs. Note that the majority of micronuclei in control cells are lamin A/C positive. Mean ± SEM, n = 3; P < 0.0006 for lamin A/C negative CCF con vs. OIS; P < 0.6 for lamin A/C positive CCF con vs. OIS. (E) CCFs in OIS are strongly positive for heterochromatic histone mark H3K27me3 but not for euchromatic mark H3K9ac. (F) Cells from E were scored for indicated histone modifications in CCFs. A single representative experiment is shown and at least 100 cells were scored per slide. Note the majority of micronuclei in control cells are positive for both marks. (G) CCFs in RS contain γ-H2AX but not 53BP1 (top). Intranuclear γ-H2AX foci colocalize with 53BP1 (bottom). (H) CCF+ and CCF− cells from G were scored for colocalizing intranuclear γ-H2AX and 53BP1 foci. Mean ± SEM, n = 3; P < 0.019. Bars (in IF panels), 10 µm.
Cytoplasmic chromatin fragments generated in senescent cells lacked nuclear lamin A/C (Fig. 1, C and D), in contrast to micronuclei generated by mitotic poisons such as Eg5 inhibitor III, of which 90% were lamin A/C positive (Fig. S1 B and unpublished data); hence designation of the former as CCFs rather than micronuclei. Pointing to a selective mode of formation of these CCFs, they stained positive for heterochromatic histone mark (H3K27me3), negative for euchromatic histone mark (H3K9ac; Fig. 1, E and F), and more intensely for H3 than the main cell nucleus (Fig. 1 A and Fig. S1 C). Indicating the specificity of the immunofluorescence staining, in proliferating cells, H3K27me3 was enriched in the presumptive inactive X-chromosome of these female IMR90 cells, whereas H3K9ac was excluded (Fig. S1 B).
Surprisingly, and again in contrast to micronuclei generated by Eg5 inhibitor III (Fig. S1 B), the vast majority of CCFs stained strongly for γ-H2AX (Fig. 1 G and Fig. S1 E), but not for other DNA damage markers, such as 53BP1 (Fig. 1 G). As expected, nuclear γ-H2AX foci colocalized with 53BP1 (Fig. 1 G). However, we observed that CCF-positive cells contain fewer intranuclear γ-H2AX/53BP1 foci than CCF-negative cells (Fig. 1 H). γ-H2AX–positive CCFs were also detected in RS human melanocytes and RS human mesenchymal progenitor cells (hMPCs), as well as in mouse hepatocytes in vivo (Fig. S1 F). Specifically, in 16-mo-old C57BL/6 mice (n = 3), 2.48 ± 0.32% hepatocytes contained CCFs. In sum, a proportion of senescent cells contain γ-H2AX–positive and H3K27me3-positive CCFs.
Formation of CCF is associated with nuclear-to-cytoplasmic chromatin blebbing
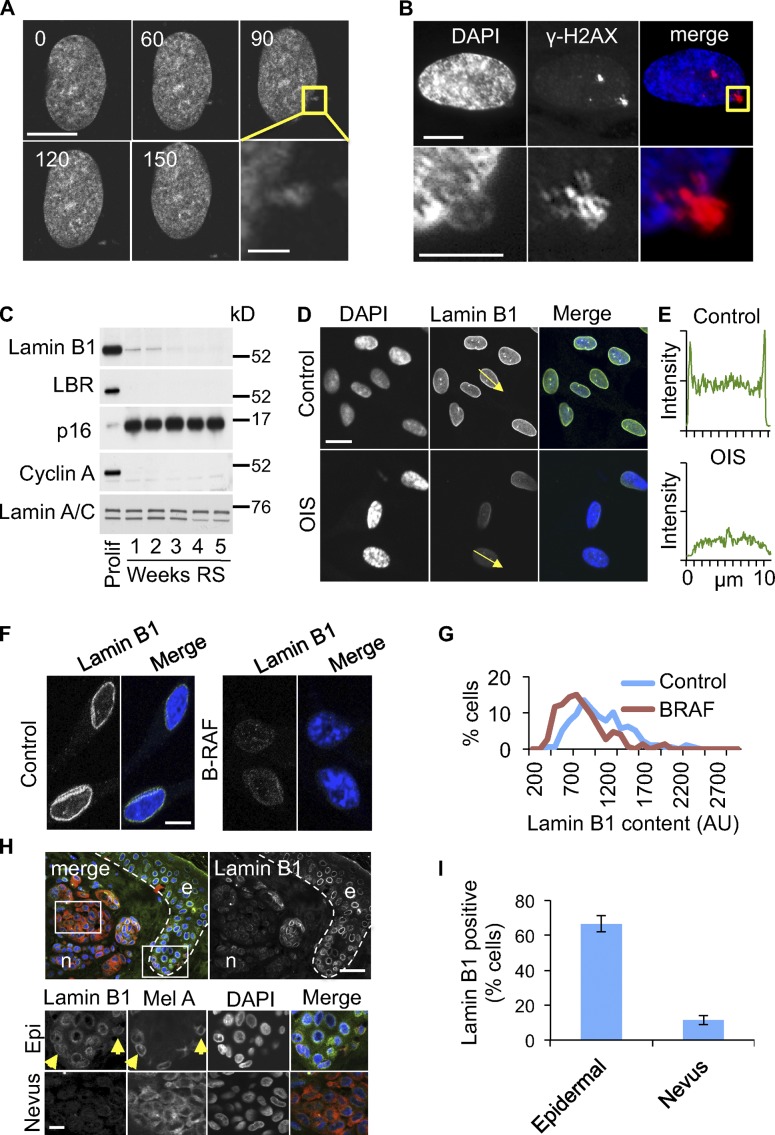
To investigate formation of CCFs in senescence, we transfected near-senescent cells with histone H2B fused at its N terminus to GFP (GFP-H2B) and performed time-lapse fluorescence imaging of live cells. Remarkably, we observed fragments of GFP-H2B “blebbing” off the main cell nucleus of nonmitotic senescent cells (Fig. 2 A; Fig. S2 A; and Video 1). Similarly, we observed apparent nuclear blebbing by staining fixed senescent cells with antibodies to histone H3 (Fig. S2 B) and γ-H2AX (Fig. 2 B). Although nuclear-to-cytoplasmic chromatin blebbing has been reported recently in cancer cell lines (Vargas et al., 2012), the accumulation of cells containing CCFs is consistent with an increase in the frequency of this process in senescent human cells. We conclude that senescent cells lose histone-enriched H3K27me3- and γ-H2AX–positive chromatin to the cytoplasm by extrusion of chromatin through the nuclear envelope.
Figure 2.
Nuclear-to-cytoplasm chromatin blebbing in senescent cells associated with depletion of lamin B1. (A) Extrusion of GFP-H2B–positive chromatin fragment from nonmitotic RS nucleus. Confocal time-lapse microscopy of transiently transfected GFP-H2B–expressing cell, 60× original magnification, 150 nm optical section, time in mins. Bars: 10 µm; (insert) 2 µm. (B) Chromatin fragments blebbing out of nucleus are γ-H2AX positive. Bars: (top) 10 µm; (bottom) 5 µm. (C) Down-regulation of lamin B1 and lamin B receptor (LBR) in RS cells, assayed by Western blot. See Materials and methods for time-course details. (D) Depletion of lamin B1 from OIS cells, assayed by immunofluorescence. (E) A line-scan of lamin B1 fluorescence intensity along the arrows in D. Data shown are from a single representative experiment out of three repeats. (F) Depletion of lamin B1 in BRAFV600E-induced OIS melanocytes in vitro. Bars, 10 µm. (G) Quantitative immunofluorescence of lamin B1 in melanocytes from F. A representative experiment out of two repeats is shown. At least 150 randomly selected cells were assessed. (H) Senescent nevus melanocytes (n) stain less intensely for lamin B1 as compared with nonsenescent melanocytes in epidermis (e). White dotted line shows the basement membrane between the dermis and epidermis. Bottom panels (Epi and Nevus) are expanded from white boxed areas on top. Confocal microscopy for DAPI (blue), MelanA (red), and lamin B1 (green) stained tissue. Bars, 20 µm. (I) Quantitation of results from H for three different nevi. Epifluorescence images of lamin B1– and MelanA-stained nevi were obtained and epidermal or nevus MelanA–positive cells were scored for the presence (positive) of a characteristic lamin B1 “ring.” At least 50 MelanA-expressing epidermal melanocytes and at least 100 nevus melanocytes were scored. Average of three different nevi ± SEM; P < 0.0005.
The observation that chromatin escapes the nucleus of senescent cells suggested that the nuclear envelope might suffer a loss of integrity. To investigate this, we compared expression and localization of the nuclear lamins that underlie the nuclear membrane in proliferating and senescent cells. As recently reported (Shimi et al., 2011; Freund et al., 2012), abundance of lamin B1 was markedly down-regulated in RS and OIS fibroblasts in vitro (Fig. 2, C and D). In particular, lamin B1 was depleted from the perinuclear region, presumably underlying the nuclear envelope (Fig. 2, D and E). Those cells with reduced lamin B1 also exhibited marked SAHF (Fig. S2, C and D). In addition, expression of lamin B receptor (LBR) was also repressed in senescent cells (Fig. 2 C). Similar results were obtained with BRAFV600E-induced senescent melanocytes (Fig. 2, F and G), as well as in melanocytes undergoing RS (Fig. S2 E). Senescent nevus melanocytes in vivo were also deficient in lamin B1, compared with nonsenescent epidermal keratinocytes (Fig. S2 F) and melanocytes (presumably nonsenescent) positioned along the neighboring basement membrane (Fig. 2, H and I). Campisi and coworkers also reported that lamin B1 is down-regulated in senescent cells in vivo, specifically in liver of irradiated mice (Freund et al., 2012). Although expression of lamin A/C was largely unchanged in senescence, we did observe relocalization of lamin A/C in senescent cells, away from the perinuclear region (Fig. S2 G). Together, these results suggest the nuclear envelope is structurally compromised in senescent cells, perhaps facilitating nuclear blebbing.
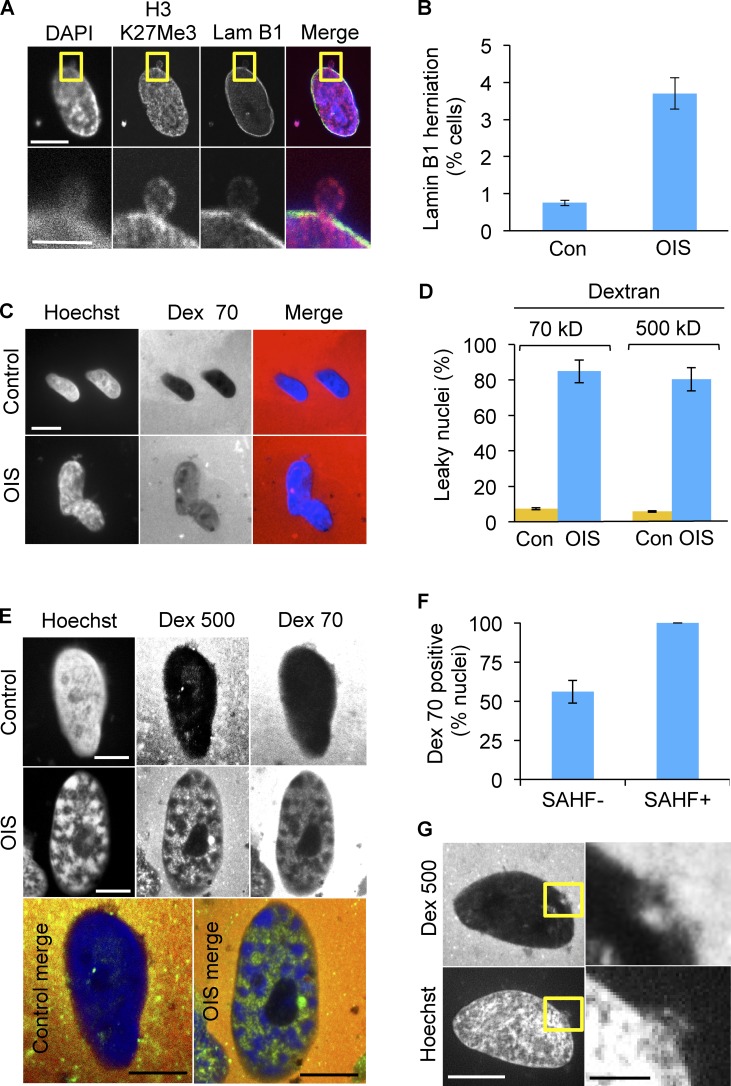
In fact, in some senescent cells, we observed herniation of chromatin through the perinuclear lamin B1 network (Fig. 3, A and B), consistent with the idea that the barrier function of the nuclear membrane of senescent cells is compromised. To directly test this, we harvested nuclei from proliferating and senescent cells, incubated them with fluorescent high molecular weight dextrans, either 70- or 500-kD, and scored uptake by fluorescence microscopy. The nuclei of senescent cells were more permeable to both dextrans (Fig. 3, C and D). Underscoring the link between senescence and envelope permeability, nuclei exhibiting SAHF were invariably permeable to dextrans (Fig. 3, E and F). Significantly, in individual nuclei, we also observed that chromatin extrusion and dextran intrusion occurred in close physical proximity (Fig. 3 G). These results show that down-regulation of lamin B1 in senescent cells is linked to unselective permeability of the nuclear envelope and loss of nuclear chromatin to the cytoplasm.
Figure 3.
Blebbing of chromatin from nucleus to cytoplasm in senescent cells is associated with loss of nuclear envelope integrity. (A) Herniation of H3K27me3-positive chromatin into the cytoplasm of OIS cells. Yellow boxed area is magnified in bottom panels. Bars: (top panels) 10 µm; (bottom panels) 5 µm. (B) Quantitation of lamin B1 gaps or herniations in OIS. Mean ± SEM, n = 2; P < 0.01. (C) Isolated nuclei of senescent cells are permeable to both 70- and 500-kD dextrans, indicating impairment of the barrier function of nuclear envelope. Dark nuclei exclude fluorescent dextran. Bars, 10 µm. (D) Quantitation of results from C. Mean ± SD, n = 4; P < 0.000001 for both 70- and 500-kD dextrans. (E) Representative OIS SAHF+ nucleus showing dextran permeability. Bars, 10 µm. (F) Quantitation of dextran 70 uptake in SAHF+ and SAHF− OIS cells. P < 1.9 × 10−5. (G) In individual nuclei, chromatin extrusion and dextran intrusion occurred in close physical proximity. Right-hand panels are expanded from yellow boxed areas on the left. Bars: (left) 10 µm; (right) 2 µm.
Lysosomal/autophagic processing of histones in senescence
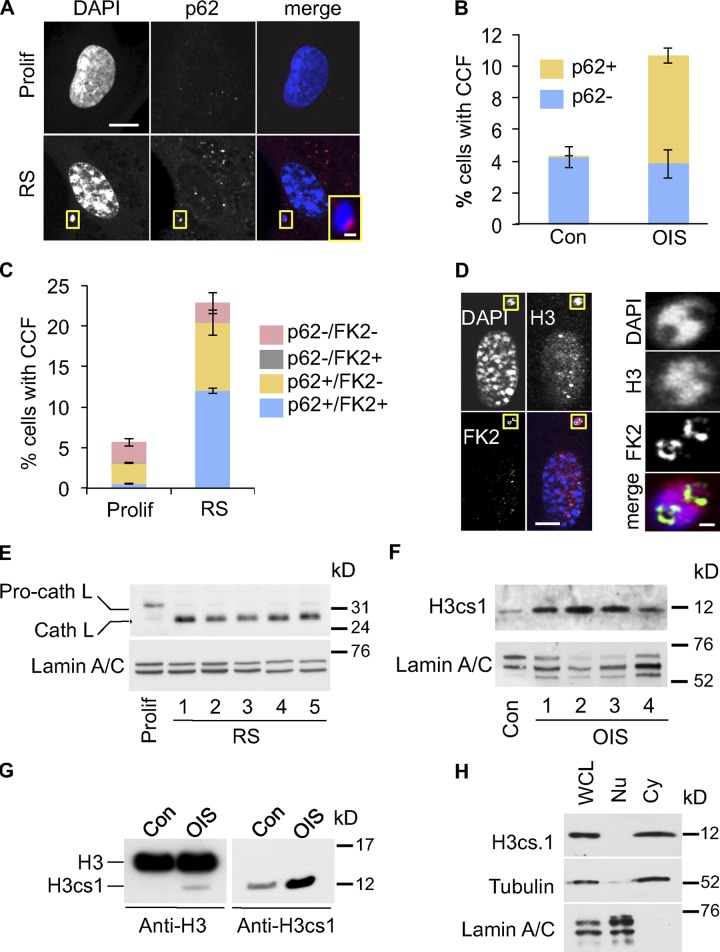
Next, we considered the fate of the CCFs in senescent cells. Senescent cells up-regulate autophagy, and this recycling process is required for secretion of inflammatory cytokines by senescent cells (Young et al., 2009). Hence, we hypothesized that CCFs might be substrates for the autophagy/lysosomal processing pathway. To test this, we compared the localization of the CCFs to p62SQSTM (p62), an adaptor protein that directs protein aggregates into the autophagy pathway (Pankiv et al., 2007). Consistent with an increase in autophagy, in senescent cells we observed increased staining of both presumptive lysosomes (with Lysotracker red) and p62, the latter in the form of characteristic focal “p62 bodies” (Lamark et al., 2003; Fig. S3, A and B). Strikingly, we invariably observed that CCFs in senescent cells were closely juxtaposed to a p62 body (Fig. 4, A and B; and Fig. S3 C). Indeed, frequently the p62 body appeared to nestle within an inclusion in the chromatin fragment (Fig. S3 C). Importantly, sites of CCFs and p62 juxtaposition are distinct from the TASCC previously described in senescent cells (Narita et al., 2011), as less than 0.5% of CCFs overlapped with TASCC (Fig. S3 D). In the autophagy pathway, p62 serves as an adaptor between LC3-II in the autophagosome and ubiquitinated target proteins or aggregates (Pankiv et al., 2007). Therefore, we asked whether CCFs in senescent cells are ubiquitinated. Indeed, p62 and conjugated ubiquitin (detected by FK2 antibody) often partially colocalized in CCFs (Fig. 4 C; and Fig. S3, E and F). Moreover, consistent with ubiquitin being an adaptor between the chromatin and p62, ubiquitinated proteins were seemingly most enriched at the chromatin surface (Fig. 4 D). The p62–ubiquitin interaction directs substrates to the autophagic/lysosomal proteolysis pathway and, concordant with this, in senescent cells we observed increased expression of the proteolytically cleaved, activated form of cathepsin L, a lysosomal protease (Fig. 4 E and Fig. S3 G; Ishidoh et al., 1998). This was accompanied by accumulation of histone H3cs1, a modified form of H3 that results from N-terminal proteolysis of H3 by cathepsin L (Fig. 4, F and G; Duncan et al., 2008). As expected from lysosomal degradation of CCFs, H3cs1 was present predominantly in the cytoplasm (Fig. 4 H).
Figure 4.
Cytoplasmic histone is processed by a lysosomal/autophagy pathway. (A) Close juxtaposition of senescence-associated CCF (RS) with p62 nuclear bodies. Bars: 10 µm; (inset) 1 µm. (B) Quantitation of control and OIS cells with CCF overlapping p62 (p62+) or not (p62−). Mean ± SEM, n = 3. (C) Quantitation of proliferating and RS cells with CCF overlapping p62 (p62+) or not (p62−) and protein ubiquitination (FK2+) or not (FK2−). Mean ± SEM, n = 3; P < 0.0008 for FK2+/p62+ CCF, comparing proliferating and RS cells. (D) Ubiquitinated proteins appear to line chromatin surface in CCF. Yellow boxed areas are magnified in right-hand panels. Bars: (left panels) 10 µm; (right panels) 1 µm. (E) Apparent activation of cathepsin L in RS IMR90 cells. In contrast to RS cells, the majority of cathepsin L in proliferating control cells exists in its inactive pro-cathepsin L form. Loading was normalized by cell number and lamin A/C was used as a loading control. See Materials and methods for details of time course. (F) Accumulation of H3cs.1, a specific N-terminal cleavage product of H3, in OIS cells. See Materials and methods for details of time course. Western blotting was performed using the same cellular lysates as in Fig. S4 B, and the same lamin A/C panel is shown in both figures. (G) Accumulation of H3cs.1 and corresponding high mobility H3 in OIS. (H) Predominant cytoplasmic localization of H3cs.1 in senescent cells. α-Tubulin and lamin A/C were used as fractionation controls for cytoplasmic and nuclear fractions, respectively.
Consistent with histone proteolysis in senescent cells, we observed that senescent cells were depleted of total histone content, based on immunofluorescence assays (Fig. 5, A and B; and Fig. S4 A). Moreover, we observed by Western blot that senescent fibroblasts generated either through RS or OIS showed a progressive decline in histone content over a period of weeks (Fig. 5 C and Fig. S4 B) after they were judged senescent by expression of SA β-gal, p16INK4a, and repression of cyclin A (Fig. 5 C; and Fig. S4, B and C). Underscoring the reduced histone content of senescent cells, cells exhibiting SAHF stained less intensely for histone H3 than did cells without SAHF (Fig. 5 A), even though SAHF are, in part, defined by their intense staining for heterochromatic histone marks (Narita et al., 2003; Zhang et al., 2005).
Figure 5.
Senescent cells in vitro and in vivo contain reduced histone content. (A) Immunofluorescent confocal images of histone H3 in control or OIS cells (11 d after activation of ER-RASG12V). Yellow arrows indicate cells with most pronounced SAHF. (B) Quantitative immunofluorescence analysis of histone H3 in RS and OIS cells. Representative of two independent experiments. (C) Progressive loss of core histones in senescent IMR90 cells in replicative senescence (RS). Lysates were normalized by cell number and an equal number of cells was loaded per each lane. Lamin A/C was used as a confirmatory loading control. See Materials and methods for details of time course. (D) Immunohistochemistry for histone H3 in human dermal nevus (n) and adjacent epidermis (e). Right-hand panels are expanded from boxed areas on the left. Note decreased nevus staining for H3 (right bottom) as compared with epidermis (right top). Bars: (left) 100 µm; (right) 50 µm. (E) Immunofluorescence of histone H3 (red) and S100 (melanocytes [green]) in human benign nevus and adjacent epidermis. Bars, 20 µm. (F) Higher magnification of boxed regions in E shows reduced staining for H3 in nevus (bottom panels), compared with epidermal melanocytes (top panels). Bars, 20 µm. (G) Quantitative immunofluorescence of H3 in epidermal and nevus melanocytes. Images were obtained in blue (DAPI), green (S100), and red (H3) channels and then H3 intensity was measured in either S100+ epidermal cells, strictly adjacent to the basal membrane, or in S100+ nevus melanocytes. H3 intensity histograms represent fluorescence intensity distribution, combined from three individual nevi. At least 150 epidermal (50 per nevus) and 300 nevus (100 per nevus) melanocytes were assessed.
To confirm that senescent cells become depleted of histone content, we established conditions to measure DNA in whole-cell lysates prepared in buffer containing 2% SDS. Using fluorometric DNA quantitation assay we found that measurements of DNA concentration were linear with respect to input-purified human genomic DNA or dilution of cell crude lysate over the relevant concentration range (Fig. S4, D and E). This assay was used to Western blot whole-cell lysates from proliferating and senescent cells, normalized for equal DNA content. This confirmed that total histone content is reduced in senescent cells (Fig. S4 F). Our ability to detect depletion of histones using multiple antibodies (including two different antibodies to histone H4; Fig. 5 C) reduces the likelihood that the observation depends on altered epitope exposure (antigenicity) between proliferating and senescent cells. Previous studies showed that senescent yeast and fibroblasts in vitro contain reduced amounts of histone and/or harbor histone-depleted regions of the genome (Dang et al., 2009; Feser et al., 2010; O’Sullivan et al., 2010). Here we conclude that senescent human cells are progressively depleted of histones, and, importantly, this phenomenon occurs, at least partly, after onset of senescence judged by cell cycle exit and expression of SA β-gal.
To test whether senescent cells are depleted of histones in vivo, we stained nevi for histones. On average, nevus melanocytes stained less intensely for histone H3, compared with nonsenescent epidermal keratinocytes and melanocytes (Fig. 5, D–G). Importantly, in nevi, we observed that the proportion of nuclei with low histone content increased in the deeper portion of the neoplasm, away from the skin surface (Fig. 5 D and Fig. S5, A–C). In contrast, expression of p16INK4a, a commonly applied marker of senescence, stained heterogeneously through the nevus as reported previously (Michaloglou et al., 2005), in no obvious pattern (Fig. S5 D). The deeper portion of a nevus is considered more “mature,” a phenomenon that is poorly defined at the molecular level but is associated with a lower proliferative index and, importantly, is inversely linked to malignancy (Smolle et al., 1989a,b; Barnhill et al., 2004; Glatz et al., 2010; Ruhoy et al., 2011).
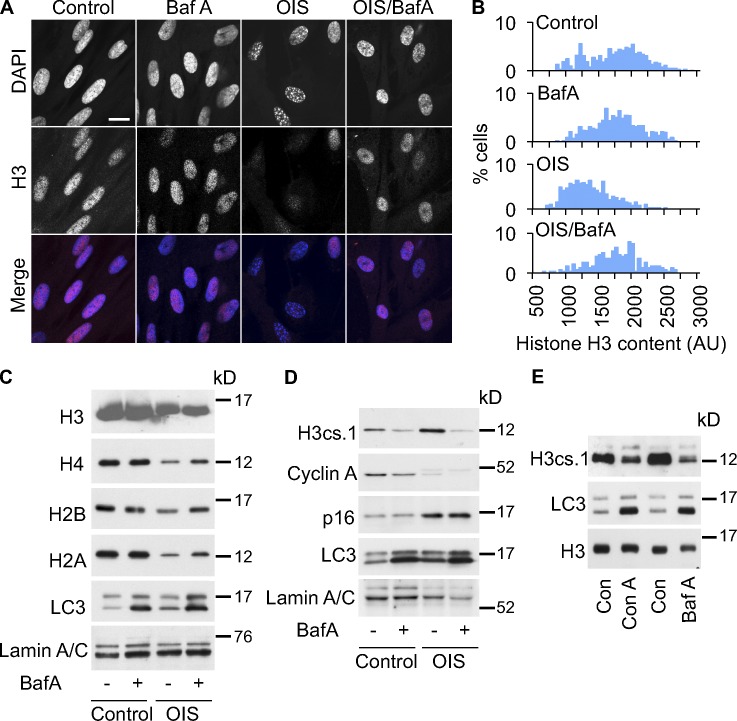
To test whether lysosomes are involved in processing of histones in senescent cells, we treated the cells with bafilomycin A or concanamycin A, inhibitors of lysosome acidification (Klionsky et al., 2008). This treatment antagonized the decrease in histone content (Fig. 6, A–C) and production of H3cs1 (Fig. 6, D and E). Importantly, bafilomycin A inhibited the autophagic/lysosomal machinery as indicated by accumulation of cleaved/lipidated LC3-II (Fig. 6, C–E; Klionsky et al., 2008), but did not affect the wider senescence program, as reflected in expression of p16INK4a and repression of cyclin A (Fig. 6 D; Kuilman et al., 2010). This suggests that the effect of bafilomycin A reflects a direct effect of autophagy/lysosomes on histone dynamics. Taken together, these results indicate that autophagy and lysosomes contribute to proteolytic processing of histones in senescence.
Figure 6.
Senescence-associated loss of histones is V-ATPase dependent. (A) Bafilomycin A1 (BafA1) blocks the loss of nuclear histone H3 content in cells undergoing OIS. BafA1 was added to cells at 50 nM on day 5 after RASG12V induction. Cells were harvested 24 h later, and stained for histone H3. Bars, 10 µm. (B) Quantitative histone H3 immunofluorescence in cells from A. Single representative experiment out of three repeats. (C) RASG12V-induced OIS cells were treated with BafA1 as described in A and then assessed for histones by Western blot. LC3 I/II has been used as a control for BafA1 activity. Lysates from 10,000 cells were loaded per well. (D) BafA1 blocks the accumulation of H3cs.1 in cells undergoing OIS. (E) BafA1 and concanamycin A (Con A) block the accumulation of H3cs.1 in RS cells.
Discussion
In this study we show that down-regulation of lamin B1 in senescent cells is linked to permeability of the nuclear envelope and dispersal of chromatin fragments (CCFs) from the nucleus to the cytoplasm, where they are targeted by the lysosomal/autophagy machinery. Total histone content is progressively decreased in senescent cells in vitro, and in vivo this appears to be linked to enhanced tumor suppression in melanocytic neoplasia. The lysosomal compartment plays a role in this senescence-associated degradation of histones.
Lamin B1 has been previously shown to be down-regulated in senescent cells (Shimi et al., 2011; Freund et al., 2012). Here we extend these observations to show that lamin B1 is dramatically down-regulated in vivo in senescent melanocytes of benign human nevi. We also show in vitro that this down-regulation is linked to permeabilization of the nuclear envelope and loss of chromatin from the nucleus to the cytoplasm. Significantly, Yang et al. (2011) previously reported an increased frequency of nuclear blebbing in in vitro cultured lamin B1/B2-null keratinocytes. On the other hand, lamin B1 and lamin B2 knockout mice die shortly after birth (Vergnes et al., 2004; Coffinier et al., 2010). Clearly, B-type lamins do have essential functions, but this is context dependent. We also note that senescent cells retain expression of lamin B2 (Shimi et al., 2011; Freund et al., 2012).
Interestingly, defects in nuclear envelope morphology and integrity, including herniations of the nuclear envelope (Bridger and Kill, 2004; Goldman et al., 2004; Scaffidi and Misteli, 2005), are also a feature of the progeroid aging syndrome Hutchinson-Gilford progeria syndrome (HGPS), frequently caused by a splicing mutation in the lamin A gene. This mutation generates a cryptic splice site in exon 11, thereby deleting a proteolytic cleavage site in the variant lamin A protein (progerin). This, in turn, leads to incomplete protein processing and accumulation of mislocalized progerin in the nuclear envelope (Butin-Israeli et al., 2012). Other mutations in the lamin A gene have also been linked to the aberrant nucleocytoplasmic compartmentalization, apparently due to ruptures in the nuclear envelope (De Vos et al., 2011; Houben et al., 2013). Together, this suggests that defects in the nuclear lamina are linked to impaired containment of chromatin in cell senescence, premature aging, and other laminopathies. The A- and B-type lamins are thought to form separate, but interacting, fibrillar networks (Shimi et al., 2008) underlying the nuclear envelope, and their functions and regulation have been suggested to be relatively nonredundant (Butin-Israeli et al., 2012). Hence, it is conceivable that impairment of either the A- or B-type lamin network can lead to a loss of the nuclear envelope integrity.
We found that lamin A/C–negative CCFs are strongly positive for both heterochromatic histone mark H3K27me3 and γ-H2AX. Conceivably, enrichment of γ-H2AX in CCFs may reflect a role in routing of damaged genome fragments out of the nucleus and targeting them to the autophagy machinery, as reflected by a close association of γ-H2AX–positive ubiquitinated CCFs with p62 bodies. Indeed, the fact that CCF-positive cells have a tendency to contain less intranuclear DDR foci indirectly supports this hypothesis. Alternatively, the fact that CCFs are negative for 53BP1 might also suggest a non-DDR role for at least a proportion of γ-H2AX in senescent cells. Interestingly, formation of γ-H2AX foci unlinked to downstream DDR signaling has been reported previously in cellular senescence induced by some non-DNA damaging agents (Pospelova et al., 2009). Other studies have also shown that phosphorylation of H2AX can occur in the absence of DNA damage (Soutoglou and Misteli, 2008; Ziegler-Birling et al., 2009). Dissociation of H2AX phosphorylation from DNA damage has also been reported for normal mitotic (Ichijima et al., 2005; McManus and Hendzel, 2005) and heat shock–treated cells (Hunt et al., 2007; Laszlo and Fleischer, 2009). Taken together, these reports suggest that DNA damage–independent formation of γ-H2AX is quite common, although the mechanism and function of this phenomenon is yet to be defined.
We observed that core histones are progressively decreased during senescence. Genome site-specific and/or overall loss of core histones in senescence has been previously reported for yeast (Dang et al., 2009; Feser et al., 2010) and fibroblasts (O’Sullivan et al., 2010). Our results extend these observations to show that this phenomenon continues to occur after onset of senescence, as typically defined by molecular markers such as cell cycle exit, expression of p16INK4a and SA β-gal. Although cellular senescence is commonly viewed as a static endpoint, previous reports have suggested that this may not be the case. These reports have characterized “early” senescence and “late” or “deep” senescence, based on analyses of DNA damage signaling and expression of repetitive elements (Chen and Ozanne, 2006; Passos et al., 2010; De Cecco et al., 2013). Proteolytic processing of histones in senescent cells in vitro is linked to lysosomal activity, based on its association with activated cathepsin L (a lysosomal protease) and accumulation of H3.cs1, a cathepsin L cleavage product of full-length histone H3 (Duncan et al., 2008; Adams-Cioaba et al., 2011). Accumulation of H3.cs1 and depletion of histones was suppressed by lysosomal vacuolar-type ATPase inhibitors, showing their dependence on lysosomal activity. Interestingly, because H3.cs1 is broadly associated with gene activation (Santos-Rosa et al., 2009), it is tempting to speculate that H3cs.1 might also have a role in changing the gene expression program in senescence.
Depletion of histones from senescent cells was not accompanied by a corresponding decrease in DNA content. So, even though histones appeared concentrated in CCFs (see Fig. 1 A), it seems likely that depletion of histones also occurs via CCF-independent mechanisms. Depletion of histones relative to DNA is expected to have profound consequences for the cell. Recently, Sedivy and colleagues reported “opening” and decompaction of chromatin at intergenic regions and pericentromeric satellite sequences of senescent cells (De Cecco et al., 2013). Moreover, “late” senescent cells exhibit increased expression and transposition of LINE-1 retrotransposons (De Cecco et al., 2013). Expression of Alu retrotransposons is also increased in senescent cells (Wang et al., 2011). Opening of chromatin structure caused by depletion of histones in senescent cells might contribute to this apparent dysregulation of the epigenome. Loss of heterochromatin and altered genome function has been previously proposed to be a cause of cellular senescence and, ultimately, tissue aging (Howard, 1996; Villeponteau, 1997; Imai and Kitano, 1998). Of course, the cell might also counter the effects of histone depletion on chromatin structure through mechanisms that remain to be determined. Consistent with this idea, Sedivy and coworkers have also shown that nuclear protein content actually increases in senescent cells (De Cecco et al., 2011), and some heterochromatin proteins accumulate in physiologically aged tissues (Kreiling et al., 2011). On balance, it appears that cellular senescence is associated with a tendency to opening of chromatin due to depletion of histones, but this is offset by compensatory mechanisms likely designed to maintain control of the genome. As a specific example, histone H1 is depleted from senescent cells and apparently replaced by HMGA proteins (Funayama et al., 2006; Narita et al., 2006). To the extent that senescence contributes to physiological aging, histone depletion and chromatin opening might be considered “degenerative” and compensatory changes as “protective.” A wealth of evidence, including recent genetic studies in model organisms, shows that chromatin features are altered with age and exert an important influence on aging and longevity (Fraga et al., 2005; Michishita et al., 2008; Dang et al., 2009; Feser et al., 2010; Greer et al., 2010; Peleg et al., 2010; Siebold et al., 2010; Wood et al., 2010; Kreiling et al., 2011; Maures et al., 2011; Kanfi et al., 2012).
Histone loss was also observed in nevus melanocytes in vivo. Interestingly, in contrast to lamin B1 that was largely decreased in both the less (superficial) and more mature (deep) regions of the nevus, depletion of histones was most marked in the more mature component. Thus, both in vitro and in vivo, down-regulation of lamin B1 is associated with senescence-associated proliferation arrest, whereas progressive depletion of histones in part reflects a more advanced stage of senescence, perhaps reflecting senescence “maturation” or “deep senescence.” Accordingly, the more mature region of a nevus might be considered to be in a state of deep senescence. Consistent with this idea, the rare mitotic cells observed within nevi are found at lower frequency in the more mature lower portion of the nevus (Ruhoy et al., 2011). Importantly, nevus maturation is inversely linked to malignancy (Smolle et al., 1989a,b; Barnhill et al., 2004; Glatz et al., 2010; Ruhoy et al., 2011). Hence, deep senescence might equate to enhanced senescence-associated proliferation arrest and tumor suppression.
Materials and methods
Cell lines and tissue culture
Human fetal lung IMR90 diploid fibroblasts were purchased from the American Type Culture Collection and cultured at 2–4% oxygen in DMEM (phenol red free) supplemented with 20% FBS, l-glutamine, and antibiotics. For ER-RAS induction, ER-RAS–IMR90 cells were supplemented with 100 nM of 4-hydroxytamoxifen (4-OHT) and maintained in 4-OHT–containing medium until harvesting.
Lightly pigmented neonatal human epidermal melanocytes (Invitrogen) were cultured in medium 254 with human melanocyte growth supplement (HMGS), 100 U/ml penicillin, and 100 µg/ml streptomycin (all from Invitrogen).
In experiments where proliferating and RS cells or control and OIS cells were directly compared at single time points, the assays were performed between days 10 and 15 after ER-RASG12V induction for OIS and between weeks 4 and 5 after last split for RS (and at least 90% SA β-gal+ and absence of observable proliferation). In time-course experiments (Figs. 2 C, 4 E, and 5 C), cells harvested 3 weeks after the last split (and at least 90% SA β-gal+ and absence of observable proliferation) were considered week 1. In OIS time-course experiments using ER-RASG12V (Fig. 4 F and Fig. S4 B), week 1 was harvested 1 week after the addition of tamoxifen. For both RS and OIS, subsequent time points were collected at weekly intervals.
Vectors, transfections, and viral infections
The following vectors were used in this study: pLNC-ER-H-RASG12V, encoding a fusion protein of the estrogen receptor (ER) ligand-binding domain and H-RASG12V (described in Barradas et al., 2009); pBOS-H2B-GFP (BD); and pPA-H2B-TagRFP (Evrogen). Retroviral-mediated gene transfer was performed using Phoenix packaging cells and polyethylenimine (PEI; Linear MW 25,000; Polysciences, Inc.; Kennedy et al., 2011). Fibroblasts were drug selected with 1 µg/ml puromycin or 500 µg/ml neomycin and kept in drug for duration of experiments. For transient transfection of plasmids encoding GFP- or RFP-histone H2B, vectors were delivered by Amaxa nucleofection (solution R, X001 program). After 48 h, cells were seeded onto 3-cm glass-bottom dishes (Iwaki America, Inc.) for subsequent time-lapse microscopy.
Melanocyte infection
Vesicular stomatitis virus G pseudotyped lentivectors encoding the oncogene BRAF600E under the transcriptional control of the cytomegalovirus initial early promoter and puromycin resistance from the simian virus 40 promoter were generated using standard methods (Kennedy et al., 2011). Melanocytes were infected overnight in normal culture medium supplemented with 2 µg/ml polybrene (EMD Millipore) and thereafter cultured in the presence of 1 µg/ml puromycin for 13 d.
Inhibition of V-ATPase
To inhibit V-ATPase activity, 50 nM bafilomycin A1 or 1 nM concanamycin A (both from Sigma-Aldrich) was added to cells for 24 h before harvesting.
Immunofluorescence
For immunofluorescence, cells growing on coverslips were fixed with methanol at −20°C for 20 min and then rehydrated in TBS-HCl, pH 7.6. After two 5-min washes in TBS/0.05% (vol/vol) Tween 20 (TBS-T), cells were blocked in TBS-T containing 2% (wt/vol) goat serum for 20 min. After that, cells were incubated in appropriately diluted (in TBS-T/2% [wt/vol] goat serum) primary antibodies for 1 h at room temperature. Primary antibodies used in this study were: histone H3 (39163, 1:1,000; Active Motif); γ-H2A.X (phosphor-S139, ab11174, 1 µg/ml; Abcam), 53BP1 (4937, 1:500; Cell Signaling Technology), H3K27Me3 (39536, 1:5,000; Active Motif); H3K9Ac (39585, 1:1,000; Active Motif); lamin A/C (ab13910, 1 µg/ml; Abcam); lamin B1 (ab16048, 1 µg/ml; Abcam); p62SQSTM (SC-25575, 1 µg/ml; Santa Cruz Biotechnology, Inc.); and anti-ubiquitinated proteins, clone FK2 (04–263, 1 µg/ml; EMD Millipore). As negative controls, the following IgG-matched antibodies were used: anti-HA tag (mouse IgG1, 2367; Cell Signaling Technology); anti–rabbit IgG (M7023; Sigma-Aldrich); and normal rabbit serum (S-5000; Vector Laboratories). After subsequent TBS-T washes, cells were incubated with Alexa 488– or Alexa 594–conjugated secondary antibodies (Molecular Probes). Nuclei were counterstained with DAPI at 1 µg/ml.
Confocal microscopy
Confocal images have been obtained using a confocal microscope (model A1R; Nikon) and 60×/1.4 NA Plan Apo VC objective lens (Nikon). Images were obtained and brightness and contrast were adjusted using NIS-Elements AR3 software (Nikon) in compliance with the journal image policy.
Confocal time-lapse microscopy
For live-cell imaging, H2B-RFP– or H2B-GFP–expressing IMR-90 cells were seeded onto 3-cm glass-bottom dishes (Iwaki America, Inc.) 24 h before time lapse. Live-cell imaging was then performed in a humidified chamber at 37°C in a CO2-enriched atmosphere using a confocal microscope (model A1R; Nikon) equipped with a PanFluor 40×/1.30 NA objective lens (Nikon). Images were acquired and processed using NIS-Elements AR3 software (Nikon).
Dextran nuclear permeability assay
After harvesting, 2.5 × 106 cells were washed with 10 ml of cold PBS and then resuspended in 5 ml of cold hypotonic lysis buffer (1 mM KCl, 1.5 mM MgCl2, 1 mM DTT, and 10 mM Tris-Cl, pH 8), supplemented with protease and phosphatase inhibitors. After 15 min incubation on ice, 10–20 strokes with a Dounce homogenizer were made. Intact nuclei, as monitored by phase-contrast microscopy, were pelleted at 500 g for 5 min and then resuspended in sucrose buffer (10 mM Tris-Cl, 5 mM EGTA, 80 mM KCl, 20 mM NaCl, and 250 mM sucrose, pH 7.6). Texas red–conjugated 70-kD (D1829; Molecular Probes) and FITC-conjugated 500-kD (D7136; Molecular Probes) dextrans were added at 1:500, nuclei were carefully pipetted and, after 15 min incubation, transferred into 3-cm glass bottom dishes (Iwaki America, Inc.). Images were captured using either a fluorescence (Eclipse 80i; Nikon) or confocal (model A1R; Nikon) microscope. For scoring, images of cells were obtained based on Hoechst staining and then all cells were scored as positive or negative for dextran by visual assessment of images obtained in green (500-kD dextran) or red (70-kD dextran) channels. At least 100 cells were scored in each of 4 replicates.
Immunohistochemistry
Immunohistochemistry was performed on paraffin-embedded human nevi obtained from the Fox Chase Cancer Center (Philadelphia, PA). Formalin-fixed paraffin-embedded sections were deparaffinized and rehydrated by passage through xylene and a graded alcohol series. Endogenous peroxidase activity was inactivated by treatment with 3% hydrogen peroxide, after which antigen retrieval was performed by incubation in boiling citrate buffer. Sections were blocked in 5% serum for 1 h, and then incubated with primary antibody for 1 h at room temperature or overnight at 4°C (for lamin B1). The following antibodies were used: anti-histone H3 (Clone 96C10, 3680; Cell Signaling Technology); anti-histone H2B (ab1790; Abcam); anti-histone H2A (ab18255; Abcam); anti-lamin B1 (ab16048; Abcam); and CINtec Histology kit for p16 (9511; MTM). Sections were then incubated in secondary antibody for 1 h (Envision+ kit [Dako] or Vectastain ABC system [Vector Laboratories]) and the staining was visualized with DAB.
Fluorescent-based immunohistochemistry
After dehydration, antigen retrieval, and protein blocking, tissue sections were incubated with primary antibodies, appropriately diluted with PBS-Tween 20. Samples were then washed 3× for 5 min each in PBST and then secondary Alexa Fluor 488 or 594 antibodies (1:500) were applied for 30 min. After washing, samples were stained with 1 µg/ml DAPI and embedded in VectaMount mounting medium (Vector Laboratories). Antibodies used for this study are as follows: lamin B1 (ab16048, 1:1,000; Abcam), histone H3 (3680, 1:500; Cell Signaling Technology), MelanA (M7196, 1:50; Dako), and S100 (X0311, 1:400; Dako).
Automated quantitative fluorescence microscopy
After staining slides for immunofluorescence as described previously, random microscopy fields were captured at 60× original magnification (Plan Apo 60×/1.4 NA oil lens; Nikon) and with a fluorescence microscope (Eclipse 80i; Nikon) equipped with an Orca-ER CCV digital camera (Hamamatsu Photonics). Average nuclear fluorescence was measured within the linear fluorescence range, using MetaMorph software and integrated morphometry analysis module. Identical preparation, acquisition, and analysis parameters were used for comparative microscopy. At least 300 cells from at least 25 random fields were analyzed per sample.
Immunoblotting
Cells were lysed in 1× Laemmli sample buffer and 5–20 µg of protein was resolved by SDS-PAGE followed by transfer onto PVDF membrane and probing with antibodies. Antibodies used in this study are as follows: anti-histone H3 (39163; Active Motif); anti-histone H2B (ab1790; Abcam); anti-histone H2A (ab18255; Abcam); anti-histone H4 (39269; Active Motif); anti–histone H4 (2935; Cell Signaling Technology); anti–lamin B1 (ab16048; Abcam); anti–lamin A/C (ab13910; Abcam); anti–LC3-B (ab48394; Abcam); anti–cyclin A (SC-754; Santa Cruz Biotechnology, Inc.); anti-p16 (clone G175-405, 51-1325GR; BD); anti–cathepsin L (ab6314; Abcam); anti-p53 (Ab-6, Pantropic, OP43; EMD Millipore); anti-H3cs.1 (39573; Active Motif); anti–α-tubulin (T5168; Sigma-Aldrich); anti–rabbit IgG, HRP linked (NA934; GE Healthcare); and anti–mouse IgG, HRP linked (P0447; Dako).
Quantitation of DNA
DNA concentration was measured in 1 µl of cellular lysate in 1× Laemmli buffer using a Qubit fluorometer and a broad range dsDNA quantitation kit.
Statistical analysis
Results shown are means ± SD or means ± SEM as indicated. P-value was calculated by student’s type-2 two-tailed t test.
Online supplemental material
Fig. S1 supports Fig. 1 and shows the proportion of lamin A/C–negative CCFs in relation to other markers of cellular senescence (Fig. S1 A), and lamin A/C staining control for bona fide micronuclei induced by Eg5 inhibitor III (Fig. S1 B). It also provides fluorescence in situ cytometry data showing higher content of histone H3 and γ-H2AX in CCFs in comparison to the main nuclei (Fig. S1, C and E). Fig. S1 D is related to Fig. 1 E and is a specificity control for antibodies against H3K27Me3 and H3K9Ac. Fig. 1 F shows the presence of γ-H2AX–positive CCFs in other biological models in vitro and in vivo. Fig. S2 is related to Fig. 2 and provides additional information on the looping of chromatin from the nuclei of senescent cells as well as on the loss of lamin B1 during RS and OIS in vitro and in benign nevi in vivo. Fig. S3 is related to Fig. 4 and shows up-regulated lysosomal compartment in cells undergoing OIS (Fig. S3, A and B), close association of CCFs with the p62 bodies (Fig. S3, D and E), IgG control for FK2 antibody (Fig. S3 F), and activation of cathepsin L during OIS (Fig. S3 G). Fig. S4 is related to Fig. 5 and provides additional information on the loss of histones in cells undergoing RS (Fig. S4 A) and OIS (Fig. S4 B). Fig. S4, C–E are methodological and provide SA β-Gal control for the onset of RS shown in Fig. 5 C and DNA measurements for subsequent normalization of loading by DNA content (Fig. S4 F). Fig. S5 is related to Fig. 5 and shows the loss of histones H3, H2A, and H2B in human nevi applying either DAB (Fig. S5, A and B) or fluorescence-related (Fig. S5 C) IHC methods. Fig. S5 A also shows MelanA, H&E, and IgG controls for Fig. S2 F. A common immunohistochemical p16 staining pattern in nevi is shown in Fig. S5 D. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201212110/DC1.
Supplementary Material
Acknowledgments
The authors would like to acknowledge microscopy core facility at the Beatson Institute.
Work in the laboratory of P.D. Adams is funded by NIA program project P01 AG031862 and CR-UK program C10652/A10250. Work in the laboratory of S.L. Berger was funded by P01 AG031862. J.F. Passos is funded by a BBSRC David Phillips Fellowship. G. Hewitt is funded by a BBSRC studentship.
Footnotes
Abbreviations used in this paper:
- CCF
- cytoplasmic chromatin fragment
- OIS
- oncogene-induced senescence
- RS
- replicative senescence
- SA β-gal
- senescence-associated β-galactosidase
- SAHF
- senescence-associated heterochromatin foci
References
- Adams P.D. 2009. Healing and hurting: molecular mechanisms, functions, and pathologies of cellular senescence. Mol. Cell. 36:2–14 10.1016/j.molcel.2009.09.021 [DOI] [PubMed] [Google Scholar]
- Adams-Cioaba M.A., Krupa J.C., Xu C., Mort J.S., Min J. 2011. Structural basis for the recognition and cleavage of histone H3 by cathepsin L. Nat Commun. 2:197 10.1038/ncomms1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnhill R., Piepkorn M., Busam K., 2004. Pathology of Melanocytic Nevi and Malignant Melanoma. Springer; 2nd ed, XIV, 406 pp [Google Scholar]
- Barradas M., Anderton E., Acosta J.C., Li S., Banito A., Rodriguez-Niedenführ M., Maertens G., Banck M., Zhou M.M., Walsh M.J., et al. 2009. Histone demethylase JMJD3 contributes to epigenetic control of INK4a/ARF by oncogenic RAS. Genes Dev. 23:1177–1182 10.1101/gad.511109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodnar A.G., Ouellette M., Frolkis M., Holt S.E., Chiu C.P., Morin G.B., Harley C.B., Shay J.W., Lichtsteiner S., Wright W.E. 1998. Extension of life-span by introduction of telomerase into normal human cells. Science. 279:349–352 10.1126/science.279.5349.349 [DOI] [PubMed] [Google Scholar]
- Braig M., Lee S., Loddenkemper C., Rudolph C., Peters A.H., Schlegelberger B., Stein H., Dörken B., Jenuwein T., Schmitt C.A. 2005. Oncogene-induced senescence as an initial barrier in lymphoma development. Nature. 436:660–665 10.1038/nature03841 [DOI] [PubMed] [Google Scholar]
- Bridger J.M., Kill I.R. 2004. Aging of Hutchinson-Gilford progeria syndrome fibroblasts is characterised by hyperproliferation and increased apoptosis. Exp. Gerontol. 39:717–724 10.1016/j.exger.2004.02.002 [DOI] [PubMed] [Google Scholar]
- Butin-Israeli V., Adam S.A., Goldman A.E., Goldman R.D. 2012. Nuclear lamin functions and disease. Trends Genet. 28:464–471 10.1016/j.tig.2012.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao K., Graziotto J.J., Blair C.D., Mazzulli J.R., Erdos M.R., Krainc D., Collins F.S. 2011. Rapamycin reverses cellular phenotypes and enhances mutant protein clearance in Hutchinson-Gilford progeria syndrome cells. Sci. Transl. Med. 3:89ra58 10.1126/scitranslmed.3002346 [DOI] [PubMed] [Google Scholar]
- Chen J.H., Ozanne S.E. 2006. Deep senescent human fibroblasts show diminished DNA damage foci but retain checkpoint capacity to oxidative stress. FEBS Lett. 580:6669–6673 10.1016/j.febslet.2006.11.023 [DOI] [PubMed] [Google Scholar]
- Chen Z., Trotman L.C., Shaffer D., Lin H.K., Dotan Z.A., Niki M., Koutcher J.A., Scher H.I., Ludwig T., Gerald W., et al. 2005. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 436:725–730 10.1038/nature03918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffinier C., Chang S.Y., Nobumori C., Tu Y., Farber E.A., Toth J.I., Fong L.G., Young S.G. 2010. Abnormal development of the cerebral cortex and cerebellum in the setting of lamin B2 deficiency. Proc. Natl. Acad. Sci. USA. 107:5076–5081 10.1073/pnas.0908790107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado M., Gil J., Efeyan A., Guerra C., Schuhmacher A.J., Barradas M., Benguría A., Zaballos A., Flores J.M., Barbacid M., et al. 2005. Tumour biology: senescence in premalignant tumours. Nature. 436:642 10.1038/436642a [DOI] [PubMed] [Google Scholar]
- Cosme-Blanco W., Shen M.F., Lazar A.J., Pathak S., Lozano G., Multani A.S., Chang S. 2007. Telomere dysfunction suppresses spontaneous tumorigenesis in vivo by initiating p53-dependent cellular senescence. EMBO Rep. 8:497–503 10.1038/sj.embor.7400937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang W., Steffen K.K., Perry R., Dorsey J.A., Johnson F.B., Shilatifard A., Kaeberlein M., Kennedy B.K., Berger S.L. 2009. Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature. 459:802–807 10.1038/nature08085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Cecco M., Jeyapalan J., Zhao X., Tamamori-Adachi M., Sedivy J.M. 2011. Nuclear protein accumulation in cellular senescence and organismal aging revealed with a novel single-cell resolution fluorescence microscopy assay. Aging (Albany NY). 3:955–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Cecco M., Criscione S.W., Peckham E.J., Hillenmeyer S., Hamm E.A., Manivannan J., Peterson A.L., Kreiling J.A., Neretti N., Sedivy J.M. 2013. Genomes of replicatively senescent cells undergo global epigenetic changes leading to gene silencing and activation of transposable elements. Aging Cell. 12:247–256 10.1111/acel.12047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vos W.H., Houben F., Kamps M., Malhas A., Verheyen F., Cox J., Manders E.M., Verstraeten V.L., van Steensel M.A., Marcelis C.L., et al. 2011. Repetitive disruptions of the nuclear envelope invoke temporary loss of cellular compartmentalization in laminopathies. Hum. Mol. Genet. 20:4175–4186 10.1093/hmg/ddr344 [DOI] [PubMed] [Google Scholar]
- Demidenko Z.N., Zubova S.G., Bukreeva E.I., Pospelov V.A., Pospelova T.V., Blagosklonny M.V. 2009. Rapamycin decelerates cellular senescence. Cell Cycle. 8:1888–1895 10.4161/cc.8.12.8606 [DOI] [PubMed] [Google Scholar]
- Di Micco R., Sulli G., Dobreva M., Liontos M., Botrugno O.A., Gargiulo G., dal Zuffo R., Matti V., d’Ario G., Montani E., et al. 2011. Interplay between oncogene-induced DNA damage response and heterochromatin in senescence and cancer. Nat. Cell Biol. 13:292–302 10.1038/ncb2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan E.M., Muratore-Schroeder T.L., Cook R.G., Garcia B.A., Shabanowitz J., Hunt D.F., Allis C.D. 2008. Cathepsin L proteolytically processes histone H3 during mouse embryonic stem cell differentiation. Cell. 135:284–294 10.1016/j.cell.2008.09.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldser D.M., Greider C.W. 2007. Short telomeres limit tumor progression in vivo by inducing senescence. Cancer Cell. 11:461–469 10.1016/j.ccr.2007.02.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feser J., Truong D., Das C., Carson J.J., Kieft J., Harkness T., Tyler J.K. 2010. Elevated histone expression promotes life span extension. Mol. Cell. 39:724–735 10.1016/j.molcel.2010.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraga M.F., Ballestar E., Paz M.F., Ropero S., Setien F., Ballestar M.L., Heine-Suñer D., Cigudosa J.C., Urioste M., Benitez J., et al. 2005. Epigenetic differences arise during the lifetime of monozygotic twins. Proc. Natl. Acad. Sci. USA. 102:10604–10609 10.1073/pnas.0500398102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freund A., Laberge R.M., Demaria M., Campisi J. 2012. Lamin B1 loss is a senescence-associated biomarker. Mol. Biol. Cell. 23:2066–2075 10.1091/mbc.E11-10-0884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funayama R., Saito M., Tanobe H., Ishikawa F. 2006. Loss of linker histone H1 in cellular senescence. J. Cell Biol. 175:869–880 10.1083/jcb.200604005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glatz K., Hartmann C., Antic M., Kutzner H. 2010. Frequent mitotic activity in banal melanocytic nevi uncovered by immunohistochemical analysis. Am. J. Dermatopathol. 32:643–649 10.1097/DAD.0b013e3181d7ce6f [DOI] [PubMed] [Google Scholar]
- Goldman R.D., Shumaker D.K., Erdos M.R., Eriksson M., Goldman A.E., Gordon L.B., Gruenbaum Y., Khuon S., Mendez M., Varga R., Collins F.S. 2004. Accumulation of mutant lamin A causes progressive changes in nuclear architecture in Hutchinson-Gilford progeria syndrome. Proc. Natl. Acad. Sci. USA. 101:8963–8968 10.1073/pnas.0402943101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray-Schopfer V.C., Cheong S.C., Chong H., Chow J., Moss T., Abdel-Malek Z.A., Marais R., Wynford-Thomas D., Bennett D.C. 2006. Cellular senescence in naevi and immortalisation in melanoma: a role for p16? Br. J. Cancer. 95:496–505 10.1038/sj.bjc.6603283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray-Schopfer V., Wellbrock C., Marais R. 2007. Melanoma biology and new targeted therapy. Nature. 445:851–857 10.1038/nature05661 [DOI] [PubMed] [Google Scholar]
- Greer E.L., Maures T.J., Hauswirth A.G., Green E.M., Leeman D.S., Maro G.S., Han S., Banko M.R., Gozani O., Brunet A. 2010. Members of the H3K4 trimethylation complex regulate lifespan in a germline-dependent manner in C. elegans. Nature. 466:383–387 10.1038/nature09195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayflick L., Moorhead P.S. 1961. The serial cultivation of human diploid cell strains. Exp. Cell Res. 25:585–621 10.1016/0014-4827(61)90192-6 [DOI] [PubMed] [Google Scholar]
- Houben F., De Vos W.H., Krapels I.P., Coorens M., Kierkels G.J., Kamps M.A., Verstraeten V.L., Marcelis C.L., van den Wijngaard A., Ramaekers F.C., Broers J.L. 2013. Cytoplasmic localization of PML particles in laminopathies. Histochem. Cell Biol. 139:119–134 10.1007/s00418-012-1005-5 [DOI] [PubMed] [Google Scholar]
- Howard B.H. 1996. Replicative senescence: considerations relating to the stability of heterochromatin domains. Exp. Gerontol. 31:281–293 10.1016/0531-5565(95)00022-4 [DOI] [PubMed] [Google Scholar]
- Hunt C.R., Pandita R.K., Laszlo A., Higashikubo R., Agarwal M., Kitamura T., Gupta A., Rief N., Horikoshi N., Baskaran R., et al. 2007. Hyperthermia activates a subset of ataxia-telangiectasia mutated effectors independent of DNA strand breaks and heat shock protein 70 status. Cancer Res. 67:3010–3017 10.1158/0008-5472.CAN-06-4328 [DOI] [PubMed] [Google Scholar]
- Ichijima Y., Sakasai R., Okita N., Asahina K., Mizutani S., Teraoka H. 2005. Phosphorylation of histone H2AX at M phase in human cells without DNA damage response. Biochem. Biophys. Res. Commun. 336:807–812 10.1016/j.bbrc.2005.08.164 [DOI] [PubMed] [Google Scholar]
- Iglesias-Bartolome R., Patel V., Cotrim A., Leelahavanichkul K., Molinolo A.A., Mitchell J.B., Gutkind J.S. 2012. mTOR inhibition prevents epithelial stem cell senescence and protects from radiation-induced mucositis. Cell Stem Cell. 11:401–414 10.1016/j.stem.2012.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S., Kitano H. 1998. Heterochromatin islands and their dynamic reorganization: a hypothesis for three distinctive features of cellular aging. Exp. Gerontol. 33:555–570 10.1016/S0531-5565(98)00037-0 [DOI] [PubMed] [Google Scholar]
- Ishidoh K., Saido T.C., Kawashima S., Hirose M., Watanabe S., Sato N., Kominami E. 1998. Multiple processing of procathepsin L to cathepsin L in vivo. Biochem. Biophys. Res. Commun. 252:202–207 10.1006/bbrc.1998.9613 [DOI] [PubMed] [Google Scholar]
- Kanfi Y., Naiman S., Amir G., Peshti V., Zinman G., Nahum L., Bar-Joseph Z., Cohen H.Y. 2012. The sirtuin SIRT6 regulates lifespan in male mice. Nature. 483:218–221 10.1038/nature10815 [DOI] [PubMed] [Google Scholar]
- Kang T.W., Yevsa T., Woller N., Hoenicke L., Wuestefeld T., Dauch D., Hohmeyer A., Gereke M., Rudalska R., Potapova A., et al. 2011. Senescence surveillance of pre-malignant hepatocytes limits liver cancer development. Nature. 479:547–551 10.1038/nature10599 [DOI] [PubMed] [Google Scholar]
- Kennedy A.L., Morton J.P., Manoharan I., Nelson D.M., Jamieson N.B., Pawlikowski J.S., McBryan T., Doyle B., McKay C., Oien K.A., et al. 2011. Activation of the PIK3CA/AKT pathway suppresses senescence induced by an activated RAS oncogene to promote tumorigenesis. Mol. Cell. 42:36–49 10.1016/j.molcel.2011.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D.J., Abeliovich H., Agostinis P., Agrawal D.K., Aliev G., Askew D.S., Baba M., Baehrecke E.H., Bahr B.A., Ballabio A., et al. 2008. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 4:151–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreiling J.A., Tamamori-Adachi M., Sexton A.N., Jeyapalan J.C., Munoz-Najar U., Peterson A.L., Manivannan J., Rogers E.S., Pchelintsev N.A., Adams P.D., Sedivy J.M. 2011. Age-associated increase in heterochromatic marks in murine and primate tissues. Aging Cell. 10:292–304 10.1111/j.1474-9726.2010.00666.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizhanovsky V., Yon M., Dickins R.A., Hearn S., Simon J., Miething C., Yee H., Zender L., Lowe S.W. 2008. Senescence of activated stellate cells limits liver fibrosis. Cell. 134:657–667 10.1016/j.cell.2008.06.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krtolica A., Parrinello S., Lockett S., Desprez P.Y., Campisi J. 2001. Senescent fibroblasts promote epithelial cell growth and tumorigenesis: a link between cancer and aging. Proc. Natl. Acad. Sci. USA. 98:12072–12077 10.1073/pnas.211053698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuilman T., Michaloglou C., Mooi W.J., Peeper D.S. 2010. The essence of senescence. Genes Dev. 24:2463–2479 10.1101/gad.1971610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamark T., Perander M., Outzen H., Kristiansen K., Øvervatn A., Michaelsen E., Bjørkøy G., Johansen T. 2003. Interaction codes within the family of mammalian Phox and Bem1p domain-containing proteins. J. Biol. Chem. 278:34568–34581 10.1074/jbc.M303221200 [DOI] [PubMed] [Google Scholar]
- Laszlo A., Fleischer I. 2009. The heat-induced gamma-H2AX response does not play a role in hyperthermic cell killing. Int. J. Hyperthermia. 25:199–209 10.1080/02656730802631775 [DOI] [PubMed] [Google Scholar]
- Lee B.Y., Han J.A., Im J.S., Morrone A., Johung K., Goodwin E.C., Kleijer W.J., DiMaio D., Hwang E.S. 2006. Senescence-associated beta-galactosidase is lysosomal beta-galactosidase. Aging Cell. 5:187–195 10.1111/j.1474-9726.2006.00199.x [DOI] [PubMed] [Google Scholar]
- Maures T.J., Greer E.L., Hauswirth A.G., Brunet A. 2011. The H3K27 demethylase UTX-1 regulates C. elegans lifespan in a germline-independent, insulin-dependent manner. Aging Cell. 10:980–990 10.1111/j.1474-9726.2011.00738.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus K.J., Hendzel M.J. 2005. ATM-dependent DNA damage-independent mitotic phosphorylation of H2AX in normally growing mammalian cells. Mol. Biol. Cell. 16:5013–5025 10.1091/mbc.E05-01-0065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaloglou C., Vredeveld L.C., Soengas M.S., Denoyelle C., Kuilman T., van der Horst C.M., Majoor D.M., Shay J.W., Mooi W.J., Peeper D.S. 2005. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature. 436:720–724 10.1038/nature03890 [DOI] [PubMed] [Google Scholar]
- Michishita E., McCord R.A., Berber E., Kioi M., Padilla-Nash H., Damian M., Cheung P., Kusumoto R., Kawahara T.L., Barrett J.C., et al. 2008. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 452:492–496 10.1038/nature06736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita M., Nũnez S., Heard E., Narita M., Lin A.W., Hearn S.A., Spector D.L., Hannon G.J., Lowe S.W. 2003. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 113:703–716 10.1016/S0092-8674(03)00401-X [DOI] [PubMed] [Google Scholar]
- Narita M., Narita M., Krizhanovsky V., Nuñez S., Chicas A., Hearn S.A., Myers M.P., Lowe S.W. 2006. A novel role for high-mobility group a proteins in cellular senescence and heterochromatin formation. Cell. 126:503–514 10.1016/j.cell.2006.05.052 [DOI] [PubMed] [Google Scholar]
- Narita M., Young A.R., Arakawa S., Samarajiwa S.A., Nakashima T., Yoshida S., Hong S., Berry L.S., Reichelt S., Ferreira M., et al. 2011. Spatial coupling of mTOR and autophagy augments secretory phenotypes. Science. 332:966–970 10.1126/science.1205407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Sullivan R.J., Kubicek S., Schreiber S.L., Karlseder J. 2010. Reduced histone biosynthesis and chromatin changes arising from a damage signal at telomeres. Nat. Struct. Mol. Biol. 17:1218–1225 10.1038/nsmb.1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omholt K., Karsberg S., Platz A., Kanter L., Ringborg U., Hansson J. 2002. Screening of N-ras codon 61 mutations in paired primary and metastatic cutaneous melanomas: mutations occur early and persist throughout tumor progression. Clin. Cancer Res. 8:3468–3474 [PubMed] [Google Scholar]
- Pankiv S., Clausen T.H., Lamark T., Brech A., Bruun J.A., Outzen H., Øvervatn A., Bjørkøy G., Johansen T. 2007. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282:24131–24145 10.1074/jbc.M702824200 [DOI] [PubMed] [Google Scholar]
- Passos J.F., Nelson G., Wang C., Richter T., Simillion C., Proctor C.J., Miwa S., Olijslagers S., Hallinan J., Wipat A., et al. 2010. Feedback between p21 and reactive oxygen production is necessary for cell senescence. Mol. Syst. Biol. 6:347 10.1038/msb.2010.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleg S., Sananbenesi F., Zovoilis A., Burkhardt S., Bahari-Javan S., Agis-Balboa R.C., Cota P., Wittnam J.L., Gogol-Doering A., Opitz L., et al. 2010. Altered histone acetylation is associated with age-dependent memory impairment in mice. Science. 328:753–756 10.1126/science.1186088 [DOI] [PubMed] [Google Scholar]
- Pollock P.M., Harper U.L., Hansen K.S., Yudt L.M., Stark M., Robbins C.M., Moses T.Y., Hostetter G., Wagner U., Kakareka J., et al. 2003. High frequency of BRAF mutations in nevi. Nat. Genet. 33:19–20 10.1038/ng1054 [DOI] [PubMed] [Google Scholar]
- Pospelova T.V., Demidenko Z.N., Bukreeva E.I., Pospelov V.A., Gudkov A.V., Blagosklonny M.V. 2009. Pseudo-DNA damage response in senescent cells. Cell Cycle. 8:4112–4118 10.4161/cc.8.24.10215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhoy S.M., Kolker S.E., Murry T.C. 2011. Mitotic activity within dermal melanocytes of benign melanocytic nevi: a study of 100 cases with clinical follow-up. Am. J. Dermatopathol. 33:167–172 10.1097/DAD.0b013e3181f3dba3 [DOI] [PubMed] [Google Scholar]
- Sagiv A., Biran A., Yon M., Simon J., Lowe S.W., Krizhanovsky V. 2012. Granule exocytosis mediates immune surveillance of senescent cells. Oncogene. 10.1038/onc.2012.206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H., Kirmizis A., Nelson C., Bartke T., Saksouk N., Cote J., Kouzarides T. 2009. Histone H3 tail clipping regulates gene expression. Nat. Struct. Mol. Biol. 16:17–22 10.1038/nsmb.1534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaffidi P., Misteli T. 2005. Reversal of the cellular phenotype in the premature aging disease Hutchinson-Gilford progeria syndrome. Nat. Med. 11:440–445 10.1038/nm1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano M., Lin A.W., McCurrach M.E., Beach D., Lowe S.W. 1997. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 88:593–602 10.1016/S0092-8674(00)81902-9 [DOI] [PubMed] [Google Scholar]
- Shimi T., Pfleghaar K., Kojima S., Pack C.G., Solovei I., Goldman A.E., Adam S.A., Shumaker D.K., Kinjo M., Cremer T., Goldman R.D. 2008. The A- and B-type nuclear lamin networks: microdomains involved in chromatin organization and transcription. Genes Dev. 22:3409–3421 10.1101/gad.1735208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimi T., Butin-Israeli V., Adam S.A., Hamanaka R.B., Goldman A.E., Lucas C.A., Shumaker D.K., Kosak S.T., Chandel N.S., Goldman R.D. 2011. The role of nuclear lamin B1 in cell proliferation and senescence. Genes Dev. 25:2579–2593 10.1101/gad.179515.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebold A.P., Banerjee R., Tie F., Kiss D.L., Moskowitz J., Harte P.J. 2010. Polycomb repressive complex 2 and trithorax modulate Drosophila longevity and stress resistance. Proc. Natl. Acad. Sci. USA. 107:169–174 10.1073/pnas.0907739107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolle J., Soyer H.P., Juettner F.M., Hoedl S., Kerl H. 1989a. Karyometry of melanocytic lesions: quantitative assessment of the ‘maturation to the depth’. Anal. Cell. Pathol. 1:133–138 [PubMed] [Google Scholar]
- Smolle J., Soyer H.P., Kerl H. 1989b. Proliferative activity of cutaneous melanocytic tumors defined by Ki-67 monoclonal antibody. A quantitative immunohistochemical study. Am. J. Dermatopathol. 11:301–307 10.1097/00000372-198908000-00002 [DOI] [PubMed] [Google Scholar]
- Soutoglou E., Misteli T. 2008. Activation of the cellular DNA damage response in the absence of DNA lesions. Science. 320:1507–1510 10.1126/science.1159051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suram A., Kaplunov J., Patel P.L., Ruan H., Cerutti A., Boccardi V., Fumagalli M., Di Micco R., Mirani N., Gurung R.L., et al. 2012. Oncogene-induced telomere dysfunction enforces cellular senescence in human cancer precursor lesions. EMBO J. 31:2839–2851 10.1038/emboj.2012.132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran S.L., Haferkamp S., Scurr L.L., Gowrishankar K., Becker T.M., Desilva C., Thompson J.F., Scolyer R.A., Kefford R.F., Rizos H. 2012. Absence of distinguishing senescence traits in human melanocytic nevi. J. Invest. Dermatol. 132:2226–2234 10.1038/jid.2012.126 [DOI] [PubMed] [Google Scholar]
- Vargas J.D., Hatch E.M., Anderson D.J., Hetzer M.W. 2012. Transient nuclear envelope rupturing during interphase in human cancer cells. Nucleus. 3:88–100 10.4161/nucl.18954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergnes L., Péterfy M., Bergo M.O., Young S.G., Reue K. 2004. Lamin B1 is required for mouse development and nuclear integrity. Proc. Natl. Acad. Sci. USA. 101:10428–10433 10.1073/pnas.0401424101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeponteau B. 1997. The heterochromatin loss model of aging. Exp. Gerontol. 32:383–394 10.1016/S0531-5565(96)00155-6 [DOI] [PubMed] [Google Scholar]
- Wang J., Geesman G.J., Hostikka S.L., Atallah M., Blackwell B., Lee E., Cook P.J., Pasaniuc B., Shariat G., Halperin E., et al. 2011. Inhibition of activated pericentromeric SINE/Alu repeat transcription in senescent human adult stem cells reinstates self-renewal. Cell Cycle. 10:3016–3030 10.4161/cc.10.17.17543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood J.G., Hillenmeyer S., Lawrence C., Chang C., Hosier S., Lightfoot W., Mukherjee E., Jiang N., Schorl C., Brodsky A.S., et al. 2010. Chromatin remodeling in the aging genome of Drosophila. Aging Cell. 9:971–978 10.1111/j.1474-9726.2010.00624.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W., Zender L., Miething C., Dickins R.A., Hernando E., Krizhanovsky V., Cordon-Cardo C., Lowe S.W. 2007. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 445:656–660 10.1038/nature05529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S.H., Chang S.Y., Yin L., Tu Y., Hu Y., Yoshinaga Y., de Jong P.J., Fong L.G., Young S.G. 2011. An absence of both lamin B1 and lamin B2 in keratinocytes has no effect on cell proliferation or the development of skin and hair. Hum. Mol. Genet. 20:3537–3544 10.1093/hmg/ddr266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young A.R., Narita M., Ferreira M., Kirschner K., Sadaie M., Darot J.F., Tavaré S., Arakawa S., Shimizu S., Watt F.M., Narita M. 2009. Autophagy mediates the mitotic senescence transition. Genes Dev. 23:798–803 10.1101/gad.519709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Poustovoitov M.V., Ye X., Santos H.A., Chen W., Daganzo S.M., Erzberger J.P., Serebriiskii I.G., Canutescu A.A., Dunbrack R.L., et al. 2005. Formation of MacroH2A-containing senescence-associated heterochromatin foci and senescence driven by ASF1a and HIRA. Dev. Cell. 8:19–30 10.1016/j.devcel.2004.10.019 [DOI] [PubMed] [Google Scholar]
- Zhang R., Chen W., Adams P.D. 2007. Molecular dissection of formation of senescence-associated heterochromatin foci. Mol. Cell. Biol. 27:2343–2358 10.1128/MCB.02019-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler-Birling C., Helmrich A., Tora L., Torres-Padilla M.E. 2009. Distribution of p53 binding protein 1 (53BP1) and phosphorylated H2A.X during mouse preimplantation development in the absence of DNA damage. Int. J. Dev. Biol. 53:1003–1011 10.1387/ijdb.082707cz [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.