Abstract
Protein phosphorylation is a major form of posttranslational modification critical to cell signaling that also occurs in mitochondrial proteome. Yet, only very limited studies have been performed to characterize mitochondrial-targeted protein kinases or phosphatases. Recently, we identified a novel member of PP2C family (PP2Cm) that is a resident mitochondrial protein phosphatase which plays an important role in normal development and cell survival. In this chapter, we will describe the methods applied in the identification of PP2Cm as a resident mitochondrial protein phosphatase based on sequence analysis and biochemical characterization. We will also provide experimental protocols used to establish the intracellular localization of PP2Cm, to achieve loss and gain function of PP2Cm in cultured cells and intact tissue, and to assess the impact of PP2Cm deficiency on cell death, mitochondria oxidative phosphorylation and permeability transition pore opening.
1. Introduction
Protein phosphorylation/dephosphorylation is regulated by the interplay of protein kinases and phosphatases and plays a pivotal role in cell signaling important for a wide spectrum of cell functions. There are at least 518 protein kinases (human kinome) identified in the human genome and many of them have been extensively investigated (Caenepeel et al., 2004). However, our knowledge on protein phosphatases is still relatively limited. Tyrosine phosphatase has the largest number of family members whereas dual-specific phosphatases and Ser/Thr protein phosphatases represent minorities in human phosphatome. Ser/Thr protein phosphatases are further divided into four sub-families based upon sequence and structural features, namely, PP1, PP2A, PP2B (calcineurin), and PP2C (Cohen, 1994). Different from other phosphatase sub-families, the PP2C family members are monomeric with two characteristic structural domains: a common N-terminal catalytic domain and a C-terminal region with substantial structure and sequence variations among different isoforms. PP2Cs belong to Mg2+- or Mn2+-dependent protein phosphatases (PPM) family and are highly conserved in both eukaryotes and some prokaryotes (Cohen, 1994). Recent findings have established PP2C isoforms as novel players in cell signaling regulation with exquisite specificity and potency (Lu and Wang, 2008; Stern et al., 2007). Thus, PP2C family members are interesting players in cell signaling that merit detailed investigations.
The mitochondrial network is an essential organelle for cellular metabolism and survival, it is also a convergent and integrative site for cell signaling pathways (Horbinski and Chu, 2005; Ravagnan et al., 2002; Newmeyer and Ferguson-Miller, 2003). Wide-spread and dynamic phosphorylation/dephosphorylation has been reported in mitochondrial proteome (Hopper et al., 2006). A number of Ser/Thr protein kinases, including PKA, PKB/AKT, PKC, and JNK, are also located in mitochondria (Alto et al., 2002; Huang et al., 1999; Nantel et al., 1999; Wang et al., 2003; Wiltshire et al., 2002). These kinases have been implicated in metabolic or apoptotic regulations by targeting matrix, inner, outer membrane components ranging from the ATP synthase to VDAC and BAD/Bax (Baines et al., 2003; Bijur and Jope, 2003; Brichese et al., 2004; Chen et al., 2001; Cohen et al., 2000; Deng et al., 2000; Kang et al., 2003; Scacco et al., 2000; Schroeter et al., 2003; Technikova-Dobrova et al., 2001). Since protein phosphorylation is a dynamic process involving a balancing act of kinases and phosphatases (Shenolikar, 1994), mitochondrial protein phosphorylation will also most likely be regulated by mitochondrial-targeted protein phosphatases. Protein phosphatase 1 (PP1) and PP2A are localized to mitochondria (Brichese et al., 2004; Dagda et al., 2003; Ruvolo et al., 2002; Tamura et al., 2004), although their specific contribution to mitochondrial function remains unclear. Another novel tyrosine protein phosphatase, PTPMT1, has been reported to be located specifically in the mitochondrial matrix and to play an important role in both ATP production and insulin secretion (Pagliarini et al., 2005). In short, protein phosphorylation and dephosphorylation in mitochondria are an important mechanism of cell signaling to regulate metabolic and apoptotic activities.
Recently, we identified a novel Ser/Thr protein phosphatase, named PP2Cm, that is targeted exclusively to the mitochondrial matrix (Lu et al., 2007). In this chapter, we will describe the bioinformatics and biochemical methods used in the identification of PP2Cm as a resident mitochondrial protein phosphatase. We will also provide experimental protocols utilized to establish the mitochondrial localization of PP2Cm, to achieve loss and gain function of PP2Cm in cultured cells, and to assess the impact of PP2Cm on cell death. Finally, we will describe the experimental approaches employed to establish gain- and loss-of function of PP2Cm in intact mouse liver as an in vivo model system to gain insights into its function in regulating mitochondrial oxidative phosphorylation and the mitochondrial permeability transition pore.
2. Identification of Protein Phosphatases in Mitochondria
2.1. Databases used for identification of mitochondria targeting sequence in PP2C family members
PP2Cs are Mg2+- or Mn2+-dependent protein phosphatases with a number of isoforms encoded by different homologous genes (Cohen, 1994; Stern et al., 2007). In mammals, at least 17 PP2C family members have been identified. They include PP2Cα, PP2Cβ, PP2Cγ, PP2Cε, PP2Cη, PP2Cm, TA-PP2C, ILKAP, NERRP, Wip1/PPM1D, POPX1, POPX2, PHLPP1, PHLPP2, PDP1, PDP2, and PPM1H (Lu and Wang, 2008). Among them, human PP2Cm (also named PP2Cκ, PPM1K, NM 152542) was listed in Genbank as a putative protein with no known function prior to our publication. Based on sequence alignment, PP2Cm is conserved among human, mouse and zebrafish, but no homolog was identified in Drosophila genome. PP2Cm contains a highly conserved catalytic domain in its C-terminal portion as commonly seen in other PP2C family members (Lu et al., 2007). However, the N-terminus of PP2Cm contains a putative mitochondrial targeting sequence as predicted using two different programs: Mitoprot (http://ihg.gsf.de/ihg/mitoprot.html) that gave a probability of 0.9738 for mitochondrial targeting and iPSORT (http://hc.ims.u-tokyo.ac.jp/iPSORT/), see Table 14.1 for the data output.
Table 14.1.
Prediction of mitochondria targeting for PP2Cm
| MitoProt II (http://ihg.gsf.de/ihg/mintoprot.html): | |
|---|---|
| Net charge of query sequence | :−4 |
| Analyzed region | : 29 |
| Number of basic residues in targeting sequence | : 5 |
| Number of acidic residues in targeting sequence | : 0 |
| Cleavage site | : 30 |
| Cleaved sequence | : MSTAALITLVRSGGNQVRRVLLSSRLLQ |
| Probability of export to mitochondria | : 0.9738 |
| iPSORT Prediction (http://he.ims.u-tokyo.ac.jp/IPSORT/): | ||||
|---|---|---|---|---|
| Values used for reasoning | ||||
| Node | Answer | View | Substring | Value(s) |
| 1. Signal peptide? | No | Average hydropathy (KYTJ820101) | [6,20] | −0.453333 (≥0.953? No) |
| 2. Mitochondrial? | Yes | Average net charge (KLEP 840101) | [1,30] | 0.133333(≥0.083? Yes) |
| Indexing:All | ||||
| Pattern:221121122(ins/del≤3) | [1,30] | MSTAALITLVRSGGN QV-R-RRVL LSSRLLQD 222222122212000 22-1-1122 22212220 221121122 | ||
2.2. Molecular cloning of mitochondria PP2Cm
Full-length human, mouse, and zebrafish PP2Cm cDNA are amplified by PCR from a human EST clone (BG713950), a mouse heart cDNA library and an embryonic cDNA library using primers listed in Table 14.2. The full-length ORF of human and mouse PP2Cm are cloned into pShuttle-CMV vectors (Stratagene) with either GFP or 3XFLAG tag at the C-terminus for adenovirus construction. Human PP2Cm truncation mutant lacking the mitochondrial targeting signal deletion mutant are created by PCR and subcloned into pShuttle-CMV vector tagged with 3XFLAG at the C terminus. Human PP2Cm wild-type and point mutants H129A, R236G, and D298A are generated by site directed mutagenesis using PCR and cloned into pGEX vector (Amersham) for generation of recombinant proteins. All adenoviruses are concentrated with a cesium chloride gradient, and further purified on PD10 gel filtration column (Amersham), and stored at −80 °C in PBS plus 10% glycerol before usage as described earlier (Lu et al., 2006).
Table 14.2.
Primers and template information for cloning PP2Cm
| Gene | Primer sequences for PCR | Template |
|---|---|---|
| Human PP2Cm: | Sense: 5′-TAAAAGATCTGCCACCATGTCAACAGCTGCCTTA-3′ Anti-sense: 5′-TAAACTCGAGTCAGGCCCATCGTCCACTGGAGGC-3′ |
Human EST clone (BG713950) |
| Mouse PP2Cm: | Sense: 5′-TAAAGCGGCCGCCACCATGTTATCAGCGGGCTT-3′ Anti-sense: 5′-TAAACTCGAGTCAGGCCCATCTCCCACTGGA-3′ |
Mouse heart cDNA library |
3. Characterization of Mitochondrial Localization of PP2Cm in Mammalian Cells
3.1. Detecting mitochondrial localization of PP2Cm by immunofluorescent microscopy
Neonatal rat ventricular myocytes or COS1 cells are cultured on 12 mm coverslips coated with laminin. Forty eight hours after adenovirus transfection, cells are incubated with 200 nM Mito-Tracker Red (Molecular Probes) for 45 min at 37 °C. For detection of Flag-tagged PP2Cm, cells are washed twice with PBS, fixed with 10% formalin for 10 min, permeabilized with 0.2% Triton X-100 for 10 min, and blocked in PBS with 3% BSA and 5% donkey serum for 1 h. After incubation with anti-FLAG M2 antibody (1:5000) for 2 h, cells are washed fours times with PBS and then incubated with Alexa488 conjugated Donkey anti-Mouse IgG (Molecular Probes) for 2 h. For GFP-tagged PP2Cm, cells are washed and fixed only without further processing. Coverslips are extensively rinsed with PBS and mounted onto glass slides with Anti-Fade regents (Molecular Probes). Images are captured using a laser scanning confocal microscope (Olympus Fluoview) equipped with an Argon 488 laser for Alexa 488 or GFP signals, a HeNe Green 543 laser for MitoTracker Red signal, respectively. Different fluorescent signals from the same images are recorded separately as digital image files and analyzed using MetaMorph program (Universal Imaging Corp) to generate merged images (Fig. 14.1A). Co-localization of the two signals can be quantified if necessary based on protein proximity index (PPI) calculated using a custom made software program as described elsewhere (Lu et al., 2006).
Figure 14.1.
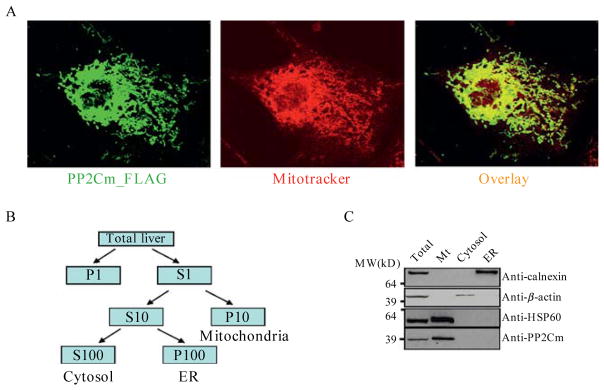
Intracellular localization of PP2Cm in mitochondria. (A) Immunofluorescent images of PP2Cm-Flag and Mitotracker, as well as their merged image in COS1 cells expressing PP2Cm-Flag fusion gene. (B) Fractionation scheme for cytosol, ER and mitochondria from mouse liver. S, supernatant; P, pellet; 1, 10, 100 represent 1000g, 10,000g, and 100,000g centrifugations, respectively. (C) Immunoblot of cellular fractions using specific antibodies as shown. Mt, mitochondria. Reproduced from (Lu et al., 2007) with permission from CSHL Press.
3.2. Determination of mitochondrial targeting of PP2Cm via fractionation
Liver is a rich source of mitochondria with relatively easy enrichment protocol. In our study, mouse liver is removed and homogenized in STE buffer (250 mM sucrose, 5 mM Tris, 1 mM EGTA, pH 7.4) using a Teflon-glass Dounce homogenizer. One to three mouse livers are broken by three to five passes with the homogenizer. Unbroken cells and cellular debris are removed by centrifugation at 1000g for 3 min. The crude mitochondrial fraction is obtained in the pellet by centrifugation at 10,000g for 10 min. The cytosolic soluble and ER enriched membrane fractions in the resulting supernatant are further separated by centrifugation at 100,000g for 1 h (see Fig. 14.1B). The specificity of each fraction is confirmed by immunoblot using specific marker proteins, including Calnexin for ER, β-actin for cytosol and Hsp60 for mitochondria. Based on immunoblot analysis of each subcellular component using anti-PP2Cm antibody, PP2Cm is detected only in mitochondrial fraction (see Fig. 14.1C), further supporting it is a mitochondrial resident protein.
3.3. Localization of mitochondrial proteins in different mitochondrial compartments
Mitochondrial proteins can be divided into four compartments, that is, outer membrane (OM), inner membrane (IM), intermembrane space (IMS), and matrix. The proteins in each compartment of mitochondria are engaged in different functions. Outer membrane and IMS proteins are critical to classic apoptotic activities; matrix and inner membrane proteins are mostly involved in respiratory chain reaction and oxidative phosphorylation; and both inner/outer membrane proteins are implicated in mitochondria permeability transition pore opening important to cell death regulation. Therefore, the sub-mitochondrial localization of mitochondrial proteins has major implications with respect to their functions. To date, it is not possible to predict with high confidence to which compartment a mitochondrial protein is located and whether a mitochondrial protein is soluble in its matured form or remains membrane anchored. These questions therefore must be addressed experimentally according to a scheme employing carbonate extraction and osmotic shock assays.
3.4. Solubility assay of PP2Cm in mitochondria
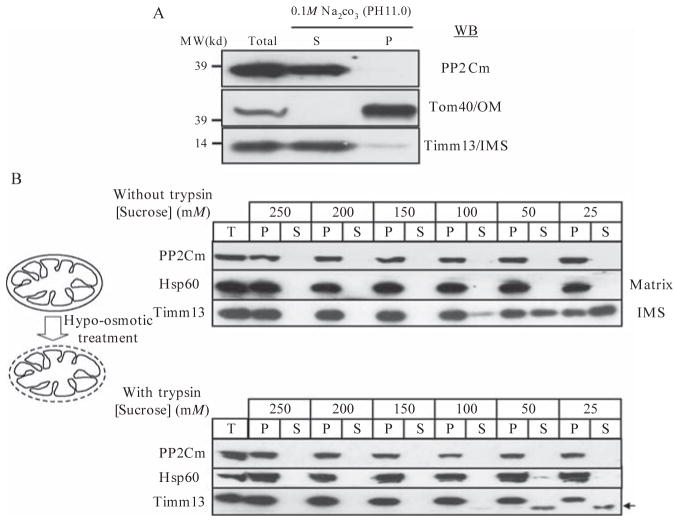
To establish first whether the mitochondrial-targeted PP2Cm is soluble or membrane associated, we perform carbonate extraction (Fujiki et al., 1982) by re-suspending mitochondria pellet in 0.1 M Na2CO3 at pH 11.0 and incubating on ice for 20 min followed by 100,000g for 10 min. The soluble proteins partition to the supernatant and the integral membrane proteins partition in pellet; fractionation is analyzed by immunoblotting using an integral membrane protein Tom40 as a positive control for the membrane fraction and an IMS soluble protein Timm13 as a positive control for the soluble fraction (Fig. 14.2A).
Figure 14.2.
Determination of PP2Cm as a soluble protein located in mitochondrial matrix. (A) Immunoblot for PP2Cm,TOM40, and Timm13 following carbonate extraction using 0.1 M Na2CO3 (pH 11.0). S, supernatant; P, pellet from centrifugation after Na2CO3 treatment. (B) Immunoblotting for PP2Cm, Hsp60 (matrix protein marker) and Timm13 (IMS marker) following mitoplasting assay using sucrose concentrations as indicated to produce hypotonic conditions.T, total; P, pellet; and S, supernatant following centrifugation. Reproduced from (Lu et al., 2007) with permission from CSHL Press.
3.5. Osmotic shock assay
In the osmotic shock assay, mitochondria are diluted into a hypotonic buffer that causes the inner membrane to swell and ruptures the outer membrane (Koehler et al., 1998). Thus, the soluble IMS contents are released and “mitoplasts,” which contain the intact inner membrane, matrix, and adherent, ruptured OM fragments, can be recovered by centrifugation. Purified mitochondria are re-suspended in serial hypotonic STE buffers containing 5.0 mM Tris, 1.0 mM EGTA, pH 7.4, plus 250, 200, 150, 100, 50, or 25 mM of sucrose. After 30 min incubation on ice followed by centrifugation at 16,000g for 10 min, the pellets, which contain the mitoplasts, and supernatants are then analyzed by regular SDS–PAGE and immunoblotting. To further demonstrate mitochondria outer membrane rupture, 50 ug/mL soybean trypsin (Sigma) is added to hypotonic STE buffer during the incubation and the mitochondrial preparation is treated with 20 ug/mL trypsin inhibitor for an additional 10 min before the centrifugation. Matrix proteins (such as Hsp60) remain in the pellets under all hypotonic conditions and resistant to trypsin digestion. On the other hand, soluble IMS proteins (such as Timm13) begin to be detected in the supernatant with increasing strength of osmotic shock (lower sucrose concentration) and become sensitive to trypsin digestion (Fig. 14.2B). Based on these assays, PP2Cm is established as a soluble protein in the mitochondrial matrix.
4. Functional Characterization of PP2Cm in Cultured Cells
4.1. Characterization of recombinant PP2Cm phosphatase activity
Protein phospatase 2C family has no known specific inhibitors and the specificities of each isoforms toward their targets are unclear based on earlier studies. Therefore, there is no method available to directly measure the specific activity of the endogenous PP2Cm in tissues or cells. However, quantitative enzymatic activity can be measured in vitro using recombinant proteins on generic phosphor–protein or phosphor–peptide substrates.
4.1.1. Radioactive assay
Ser/Thr protein phosphatase activity is determined by using myelin basic protein (MBP) pre-phosphorylated by the catalytic subunit of cyclic AMP-dependent protein kinase (PKA). To prepare phosphorylated MBP as substrate, 500 μg of MBP (M1891, Sigma) is incubated with 25 units of PKA (P2645, Sigma) in kinase buffer (50 mM Tris–HCl (pH 7.5), 10 mM MgCl2, 1 mM DTT, and 3.70 MBq [γ-32P]ATP) at 30 °C overnight. The phosphorylated MBP was then purified by Quick Spin protein desalting column (Cat# 100973, Roche) and further concentrated by Micron Millipore filter unit (Cat#42404, Milllipore). Phosphatase assays are performed using 50–500 ng purified GST fusion protein and 1.0–10.0 μg of 32P-labeled MBP substrate in 50 mM Tris–HCl (pH 7.0), 0.1 mM EDTA, 5 mM DTT, and 0.01% Brij35. The reactions are done at 30 °C and terminated by addition of 100 μl of 20% (w/v) trichloroacetic acid at specific time points. Precipitated proteins are removed by centrifugation at 16,000g for 30 min. Radioactivity of free phosphates (32Pi) released in the supernatants and total protein substrate (32P-MBP) is measured using a scintillation counter and the percent of phosphate release (free phosphate versus total MBP signals) is used to calculate protein phosphatase activities. The dependence of PP2Cm on MnCl2 was determined by adding different concentrations of MnCl2 to the reaction buffer (between 0 and 20 mM). It is important to recognize that MBP is not a natural substrate of PP2Cm and the in vitro assay condition is far from optimal comparing with its native environment in mitochondrial matrix. Therefore, the enzyme activity of PP2Cm measured in vitro may not represent its optimal physiological activity against its endogenous substrates.
4.1.2. Colorimetric assay
PP2Cm assay can also be conducted in a more high-throughput manner using a colorimetric assay in 96-well plates with a synthetic phosphopeptide instead of 32P-labeled MBP as substrate. Specifically, 50 μl total reaction solution is prepared containing 0.1–0.5 μg of GST-PP2Cm protein and 1 μg of phosphopeptide substrate (H-Arg-Arg-Ala-pThr-Val-Ala-OH, Cat # P152–0001, Biomol) in phosphatase reaction buffer (50 mM Tris–HCl (pH 7.0), 0.1 mM EDTA, 5 mM DTT, and 0.01% Brij35). After incubation at 30 °C for a specific period of time (determined experimentally within linear reaction curve), the reaction is terminated by adding 100 μl of Biomol Green (Cat #AK111–250, Biomol) and further incubation at 30 °C for additional 30 min. The free phosphate is measured based on absorbance at 620 nm. As for any colorimetric assays, a standard curve using specific concentrations of phosphate (0–10 nM, KI-102, Biomol) should be established to ensure the absorbance values are within the linear detection range. It is also important to make sure the recombinant proteins are prepared in phosphate-free buffer (Tris based) and eliminate contamination of other free phosphate throughout the assay. Since there is no known commercial source of small molecule inhibitors with sufficient specificity and potency against PP2C family of protein phosphatases, this assay can be useful for high-throughput screening of novel compounds as PP2C inhibitors or agonists.
4.2. Generation of phosphatase-dead PP2Cm mutants
Loss of function study can be very powerful to reveal novel gene functions. A loss of function can be achieved by generating a phosphatase-dead mutant or knocking down endogenous PP2Cm expression using siRNA strategy. Based on sequence alignment and earlier studies on PP2Cα/β isoforms, H129, R236, and D298 of PP2Cm are predicted to be essential residues necessary for its phosphatase activity. Site-directed mutagenesis is carried out to replace alanine in the three sites and the resulting H129A, R236A, and D298A recombinant proteins are tested for enzymatic activity in vitro as described above (Lu et al., 2007). Indeed, the mutant PP2Cm proteins show little phosphatase activity comparing to wild-type PP2Cm, suggesting further that PP2Cm is a bona fide protein phosphatase, although the endogenous substrate remains to be revealed.
4.3. Knocking down PP2Cm with siRNA expressing vector
PP2Cm protein and mRNA are detected at high levels in adult mouse brain and heart, and its expression in heart is diminished in hypertrophic or failing hearts induced by mechanical stress (Lu et al., 2007). To establish the physiological function of endogenous PP2Cm and to better understand the functional significance of PP2Cm downregulation in response to stress, a loss of function approach is employed to knockdown PP2Cm expression in cultured cardiac myocytes. Due to the low efficiency of transfection, recombinant adenovirus vectors are used to achieve efficient gene transfer. First, synthetic 64-nt oligonucleotide hairpin pairs targeting mouse and rat PP2Cm (same sequence) are annealed and ligated into a modified version of pSUPER vector (Oligoengine), pShuttle-pSUPER, which is constructed by subcloning the H1-RNA expression cassette into pShuttle (Adeasy, stratagene) (kind gift from Dr. J. Han). As a control, recombinant adenoviral shRNA vector targeting firefly luciferase is also constructed. The targeting sequences are listed as follows:
PP2Cm shRNA1, 5′-TCTGGGATAACCGCATTGA-3′; PP2Cm shRNA2, 5′-GAAGCTGACCACTGACCAT-3′; PP2Cm shRNA3, 5′-GAAGCTGACCACTGACCAT-3′; Luciferase shRNA, 5′-CTGACGCGGAATACTTCGA-3′.
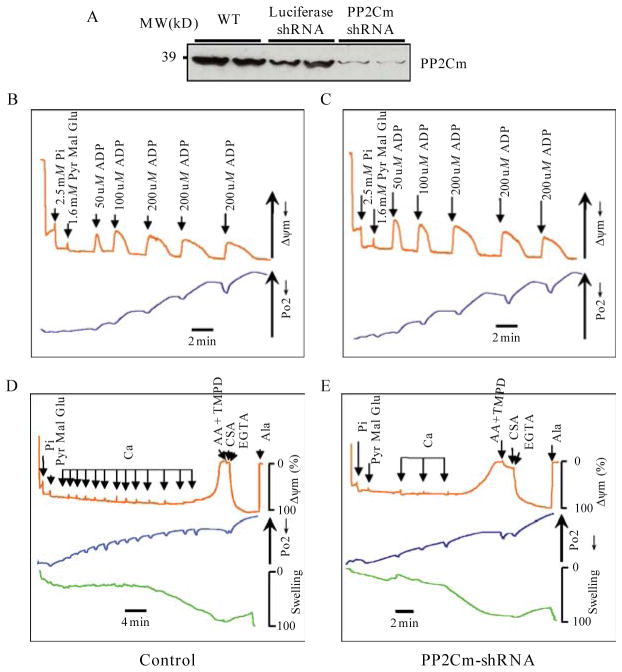
The recombinant adenoviruses expressing PP2Cm and Luciferase shRNAs are generated according to an established protocol (Lu et al., 2006). The efficacy of shRNA mediated knockdown is confirmed by immunoblotting using a PP2Cm specific antibody developed in our lab (Fig. 14.3A).
Figure 14.3.
PP2Cm deficiency on mitochondrial function. (A) Immunoblot of PP2Cm showing efficient shRNA mediated knockdown in mouse liver following adv-shRNA injection. (B) and (C) Innermembrane potential (ψΔm) and oxygen consumption simultaneously recorded in response toADP pulses in Control and PP2Cm deficient mitochondria. (D) and (E) Innermembrane potential (ψΔm), oxygen consumption and mitochondrial swelling simultaneously recorded in response to 2 μM Ca2+ stimulations. Reproduced from (Lu et al., 2007) with permission from CSHL Press.
4.4. Adenovirus-mediated gene deliver in mouse liver
To establish the physiological function for PP2Cm, a genetic-targeted mouse model would be ideal. However, in the absence of a readily available PP2Cm−/− mouse model, adenovirus mediated gene transfer can be used to achieve efficient gain and loss of function studies in targeted tissue, including liver (Wang et al., 1996). As mentioned earlier, liver is the rich source of mitochondria and thus ideally suited to adenovirus-mediated gene knockdown. For PP2Cm, the Adv-PP2Cm–shRNA vectors are generated as described earlier. Twenty to twenty-five microliters of viral solution (approximately 5 × 1011 particles/ml) are injected via tail vein for each adult mouse (2 months old, C57BL/5) to achieve an average dose of 5 × 109 particle/g body weight. In addition to PP2Cm–shRNA vectors, an adv-luciferase shRNA vector is required as a negative control. Five to seven days post-viral infection, liver tissues are excised for mitochondria isolation as described above. It is well established that systemic adenovirus administration will preferentially target liver hepatocytes (Wang et al., 1996). However, one of the drawbacks of this approach is the transient nature of the target gene expression (with peak expression within a week) and viral associated inflammatory response which may complicate the data interpretation. Finally, the efficacy of knockdown for PP2Cm is confirmed by immunoblotting as PP2Cm protein is significantly reduced following shRNA expression (Fig. 14.3A).
5. Functional Characterization of PP2Cm in Cell Death and Mitochondrial Regulation
5.1. PP2Cm deficiency on cell viability and mitochondria inner membrane potential using JC-1 fluorescent assay
Neonatal myocytes are cultured on collagen coated glass cover slips in serum free media for 1 day following isolation when they are infected with Adv-PP2Cm–shRNA or control vectors. Under normal conditions, the neonatal myocytes would survive without appreciable level of cell death for more than 5 days. However, PP2Cm–shRNA infected myocytes have a much higher death rate. To directly demonstrate if the cell death is caused by mitochondria permeability transition pore opening, we use JC-1 fluorescent activity assay to measure mitochondrial inner membrane potential (ΔΨm) in living myocytes (Reers et al., 1995). JC-1 is a cationic fluorescent dye (5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazolylcarbocyanine iodide, Molecular Probes) and has a unique ability to accumulate in polarized mitochondria when added to the medium, resulting in the formation of mitochondrial specific red fluorescent aggregates (absorbance/emission maxima of 585/590 nm). In apoptotic cells, mitochondrial inner membrane potential ΔΨ collapsed and JC-1 fails to accumulate in mitochondria and remains in the green fluorescent monomeric form (absorbance/emission maxima of 510/527 nm). Therefore, the relative intensity of red versus green fluorescence serves as an indirect measurement of DY polarity (Reers et al., 1995). To perform JC-1 assay, cells are stained with 2 μg/mL of JC-1 (Molecular Probes) in growth medium at 37 °C for 20 min following by a double wash with growth media. The fluorescent signals are viewed using a laser scanning confocal microscope (Olympus Fluoview). The red fluorescence JC-1 aggregates in healthy polarized mitochondria is detected at 585–640 nm, and the green fluorescence of JC-1 monomers in apoptotic mitochondria is detected at 510–530 nm. We use a 488 Argon Laser to excite green fluorescent signal and a 543 HeNe laser light to excite red fluorescence. The red and green fluorescence emissions from each view are recorded digitally under the same setting of exposure and light intensity without digital manipulation. Their respective pixel intensities were quantified by MetaMorph (Universal Imaging Corp). Finally, the ratio of total red (aggregated JC-1 from polarized intact mitochondria) versus total green (monomers from cells with collapsed mitochondria ΔΨm) signals from the same image was calculated as an indirect value of mitochondria membrane potential. Our results from this experiment suggests that loss of PP2Cm leads to cell death associated with loss of ΔΨm. Thus, PP2Cm is a critical molecule for mitochondrial permeability transition pore regulation and cell survival.
5.2. PP2Cm deficiency on mitochondria oxidative phosphorylation and respiration
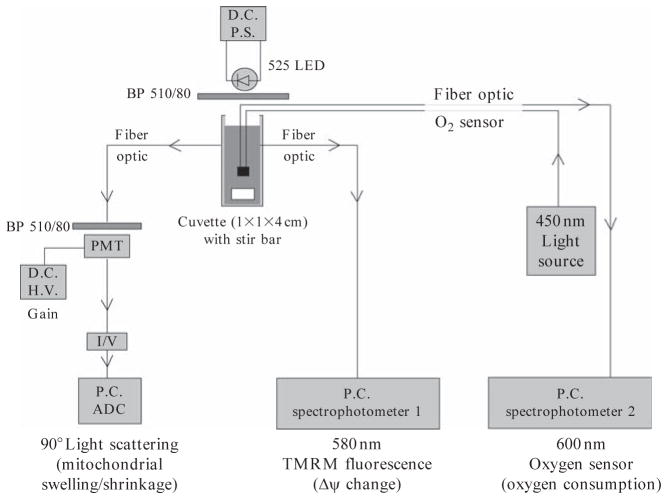
After achieving an efficient knockdown of endogenous PP2Cm (Fig. 14.3A), mitochondria are isolated from control and PP2Cm deficient liver as described in Section 3.2.To measure oxidative phosphorylation (Korge et al., 2005), isolated mitochondria (0.25 mg/mL) are added into 100 KCl buffer containing 10mM HEPES pH7.4 and 0.2% BSA in a closed, continuously stirred cuvette at room temperature. Mitochondrial membrane potential ΔΨm, matrix volume, and oxygen consumption are simultaneously recorded. For mitochondria ΔΨm, an optic fiber spectrofluorimeter (Ocean Optics) is used to detect tetramethylrhodamine methyl ester (TMRM) fluorescence at 580 nm. For mitochondrial matrix volume, 90° light scattering is recorded at 520 nm. For oxygen consumption, pO2 in the buffer is measured via a fiber–optic oxygen sensor (Fig. 14.4). Mitochondria are energized using 2.5 mM Pi and complex I substrates (1.6 mM of pyruvate, malate, and glutamate). After being stabilized for approximately 5 min, ADP is added to cuvette at 50, 100, 200 μM doses with sufficient time interval to allow complete recovery of ΔΨm. The integrity of mitochondrial respiratory activity is determined based on the time and magnitude of ΔΨm recovery associated with oxygen consumption. If there is a defect observed in experimental samples related to controls, different complex substrates can be used to detect the specific defects in respiration chain. Our studies suggest that PP2Cm deficiency does not affect mitochondria oxidative phosphorylation (Fig. 14.3B and C).
Figure 14.4.
Configuration of mitochondrial assay system. Mitochondria preparation is suspended in stirred cuvette. Excitation source is light emitting diode (LED) and BP 510/80 Fluorescence filter. Mitochondrial swelling isdetected by 90 degree scattered light at photomultipler tube (PMT).TMRM fluorescence is detected by Spectrophotometer1, and oxygen is detected using a fiber–optic sensor inside of the mitochondrial solution by Spectrophotometer 2. Signals are logged by analog to digital converter (ADC) in a PC.
5.3. PP2Cm deficiency on mitochondria permeability transition pore regulation (Weiss et al., 2003)
In the same preparation of isolated mitochondria described above, mitochondria (0.25 mg/mL) are added in 150 mM KCl buffer containing 0.2% BSA. Mitochondria are also energized with 2.5 mM Pi and 1.6 mM complex I substrates (pyruvate, malate, and glutamate). Mitochondrial membrane potential ΔΨm, matrix volume, and oxygen consumption are simultaneously recorded. Ca2+ pulses (2.0 μM) are added successively with a sufficient time delay (usually 2–3 min) to allow full recovery of ΔΨm between pulses. After a sufficient number of Ca2+ has been added to trigger the permeability transition, as indicated by depolarization of inner membrane potential (ΔΨm approaching 0) with simultaneous matrix swelling, we add the complex IV substrates ascorbic acid (2 mM) and N,N,N′, N′-tetramethyl-p-phenylenediamine (TMPD, 0.2 mM), together with the mitochondrial permeability transition pore inhibitor Cyclosporin A (CsA, 1.5 μM) and EGTA (1 mM). These treatments should recover the mitochondrial membrane potential if the earlier loss of Ψm is truly mediated by the opening of permeability transition pore. Finally, to quantify the extent of permeability transition pore opening, 10 μg alamethicin (Ala) is added to induce maximal mitochondrial swelling and Ψm dissipation. In our experiments, the number of Ca2+ pulses (2 μM) required for 95% mitochondrial matrix swelling relative to alamethicin was greater in control than in PP2Cm knockdown mitochondrial preparations (Fig. 14.3D and E). This result shows PP2Cm deficient mitochondria are much more sensitive to Ca2+ induced permeability transition pore opening.
Our studies in isolated mitochondria suggest that PP2Cm deficiency sensitizes mitochondria permeability transition pore opening in response to Ca2+ overload and can predispose cells to apoptosis. To further investigate this findings, we carried out direct apoptosis assays on PP2Cm deficient liver tissue using TUNEL immunostaining methods. This is a well-established procedure based on caspase induced DNA fragmentation present in the apoptotic nuclei. In addition to counter staining with DAPI to illustrate total number of nuclei, a positive control is performed using DNase I pretreated slides (Lu et al., 2007).
6. Mitochondrial Phosphatase in Cell Death Regulation
Based on this set of comprehensive studies, we establish the intracellular location of a novel PP2C family member as a mitochondrial resident protein. PP2Cm is present in soluble form in mitochondria matrix. It is essential to cell survival and permeability transition pore opening, but has not effect on oxidative phosphorylation and respiratory chain activity. PP2Cm is a novel regulator in cell death regulation and plays an important role in normal development. Further efforts are needed to identify specific downstream targets of PP2Cm and the mechanisms involved in its down-regulation by stress in heart. Protein phosphorylation remains poorly characterized in the mitochondrial proteome. Our study shows the potential importance of protein phosphorylation/dephosphorylation in mitochondria matrix for cellular survival and function. Research in this area holds great promise for uncovering novel mechanisms in cell signal transduction mediated via mitochondrial regulation, toward the ultimate goal of identifying molecular targets for novel therapies to treat human diseases.
Acknowledgments
Authors wish to thank Dr John Parker for his assistant in technical help and manuscript preparation. This work was partially supported by Laubisch Foundation and grants from National Institutes of Health to PK, CMK, JNW, and YW, Predoctoral fellowship to GL and Postdoctoral fellowship to HS from AHA Western States Affiliate, and YW is an Established Investigator of AHA.
References
- Alto NM, Soderling J, Scott JD. Rab32 is an A-kinase anchoring protein and participates in mitochondrial dynamics. J Cell Biol. 2002;158:659–668. doi: 10.1083/jcb.200204081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baines CP, Song CX, Zheng YT, Wang GW, Zhang J, Wang OL, Guo Y, Bolli R, Cardwell EM, Ping P. Protein kinase C epsilon interacts with and inhibits the permeability transition pore in cardiac mitochondria. Circ Res. 2003;92:873–880. doi: 10.1161/01.RES.0000069215.36389.8D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bijur GN, Jope RS. Rapid accumulation of Akt in mitochondria following phosphatidylinositol 3-kinase activation. J Neurochem. 2003;87:1427–1435. doi: 10.1046/j.1471-4159.2003.02113.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brichese L, Cazettes G, Valette A. JNK is associated with Bcl-2 and PP1 in mitochondria: Paclitaxel induces its activation and its association with the phosphorylated form of Bcl-2. Cell Cycle. 2004;3:1312–1319. doi: 10.4161/cc.3.10.1166. [DOI] [PubMed] [Google Scholar]
- Caenepeel S, Charydczak G, Sudarsanam S, Hunter T, Manning G. The mouse kinome: Discovery and comparative genomics of all mouse protein kinases. Proc Natl Acad Sci USA. 2004;101:11707–11712. doi: 10.1073/pnas.0306880101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Hahn H, Wu G, Chen CH, Liron T, Schechtman D, Cavallaro G, Banci L, Guo Y, Bolli R, Dorn GW, 2nd, Mochly-Rosen D. Opposing cardioprotective actions and parallel hypertrophic effects of delta PKC and epsilon PKC. Proc Natl Acad Sci USA. 2001;98:11114–11119. doi: 10.1073/pnas.191369098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen PTW. Nomenclature and chromosomal localization of human protein serine/threonine phosphatase genes. Adv Protein Phosphatases. 1994;8:371–376. [Google Scholar]
- Cohen MV, Baines CP, Downey JM. Ischemic preconditioning: From adenosine receptor to KATP channel. Annu Rev Physiol. 2000;62:79–109. doi: 10.1146/annurev.physiol.62.1.79. [DOI] [PubMed] [Google Scholar]
- Dagda RK, Zaucha JA, Wadzinski BE, Strack S. A developmentally regulated, neuron-specific splice variant of the variable subunit Bbeta targets protein phosphatase 2A to mitochondria and modulates apoptosis. J Biol Chem. 2003;278:24976–24985. doi: 10.1074/jbc.M302832200. [DOI] [PubMed] [Google Scholar]
- Deng X, Ruvolo P, Carr B, May WS., Jr Survival function of ERK1/2 as IL-3-activated, staurosporine-resistant Bcl2 kinases. Proc Natl Acad Sci USA. 2000;97:1578–1583. doi: 10.1073/pnas.97.4.1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki Y, Hubbard AL, Fowler S, Lazarow PB. Isolation of intracellular membranes by means of sodium carbonate treatment: Application to endoplasmic reticulum. J Cell Biol. 1982;93:97–102. doi: 10.1083/jcb.93.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper RK, Carroll S, Aponte AM, Johnson DT, French S, Shen RF, Witzmann FA, Harris RA, Balaban RS. Mitochondrial matrix phosphoproteome: Effect of extra mitochondrial calcium. Biochemistry. 2006;45:2524–2536. doi: 10.1021/bi052475e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horbinski C, Chu CT. Kinase signaling cascades in the mitochondrion: A matter of life or death. Free Radic Biol Med. 2005;38:2–11. doi: 10.1016/j.freeradbiomed.2004.09.030. [DOI] [PubMed] [Google Scholar]
- Huang LJ, Wang L, Ma Y, Durick K, Perkins G, Deerinck TJ, Ellisman MH, Taylor SS. NH2-Terminal targeting motifs direct dual specificity A-kinase-anchoring protein 1 (D-AKAP1) to either mitochondria or endoplasmic reticulum. J Cell Biol. 1999;145:951–959. doi: 10.1083/jcb.145.5.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang BP, Urbonas A, Baddoo A, Baskin S, Malhotra A, Meggs LG. IGF-1 inhibits the mitochondrial apoptosis program in mesangial cells exposed to high glucose. Am J Physiol Renal Physiol. 2003;285:F1013–F1024. doi: 10.1152/ajprenal.00209.2003. [DOI] [PubMed] [Google Scholar]
- Koehler CM, Merchant S, Oppliger W, Schmid K, Jarosch E, Dolfini L, Junne T, Schatz G, Tokatlidis K. Tim9p, an essential partner subunit of Tim10p for the import of mitochondrial carrier proteins. EMBO J. 1998;17:6477–6486. doi: 10.1093/emboj/17.22.6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korge P, Honda HM, Weiss JN. K+-dependent regulation of matrix volume improves mitochondrial function under conditions mimicking ischemia-reperfusion. Am J Physiol Heart Circ Physiol. 2005;289:H66–77. doi: 10.1152/ajpheart.01296.2004. [DOI] [PubMed] [Google Scholar]
- Lu G, Kang YJ, Han J, Herschman HR, Stefani E, Wang Y. TAB-1 modulates intracellular localization of p38 MAP kinase and downstream signaling. J Biol Chem. 2006;281:6087–6095. doi: 10.1074/jbc.M507610200. [DOI] [PubMed] [Google Scholar]
- Lu G, Ren S, Korge P, Choi J, Dong Y, Weiss J, Koehler C, Chen JN, Wang Y. A novel mitochondrial matrix serine/threonine protein phosphatase regulates the mitochondria permeability transition pore and is essential for cellular survival and development. Genes Dev. 2007;21:784–796. doi: 10.1101/gad.1499107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G, Wang Y. Functional diversity of mammalian type 2C protein phosphatase isoforms: New tales from an old family. Clin Exp Pharmacol Physiol. 2008;35:107–112. doi: 10.1111/j.1440-1681.2007.04843.x. [DOI] [PubMed] [Google Scholar]
- Nantel A, Huber M, Thomas DY. Localization of endogenous Grb10 to the mitochondria and its interaction with the mitochondrial-associated Raf-1 pool. J Biol Chem. 1999;274:35719–35724. doi: 10.1074/jbc.274.50.35719. [DOI] [PubMed] [Google Scholar]
- Newmeyer DD, Ferguson-Miller S. Mitochondria: Releasing power for life and unleashing the machineries of death. Cell. 2003;112:481–490. doi: 10.1016/s0092-8674(03)00116-8. [DOI] [PubMed] [Google Scholar]
- Pagliarini DJ, Wiley SE, Kimple ME, Dixon JR, Kelly P, Worby CA, Casey PJ, Dixon JE. Involvement of a mitochondrial phosphatase in the regulation of ATP production and insulin secretion in pancreatic beta cells. Mol Cell. 2005;19:197–207. doi: 10.1016/j.molcel.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Ravagnan L, Roumier T, Kroemer G. Mitochondria, the killer organelles and their weapons. J Cell Physiol. 2002;192:131–137. doi: 10.1002/jcp.10111. [DOI] [PubMed] [Google Scholar]
- Reers M, Smiley ST, Mottola-Hartshorn C, Chen A, Lin M, Chen LB. Mitochondrial membrane potential monitored by JC-1 dye. Methods Enzymol. 1995;260:406–417. doi: 10.1016/0076-6879(95)60154-6. [DOI] [PubMed] [Google Scholar]
- Ruvolo PP, Clark W, Mumby M, Gao F, May WS. A functional role for the B56 alpha-subunit of protein phosphatase 2A in ceramide-mediated regulation of Bcl2 phosphorylation status and function. J Biol Chem. 2002;277:22847–22852. doi: 10.1074/jbc.M201830200. [DOI] [PubMed] [Google Scholar]
- Scacco S, Vergari R, Scarpulla RC, Technikova-Dobrova Z, Sardanelli A, Lambo R, Lorusso V, Papa S. cAMP-dependent phosphorylation of the nuclear encoded 18-kDa (IP) subunit of respiratory complex I and activation of the complex in serum-starved mouse fibroblast cultures. J Biol Chem. 2000;275:17578–17582. doi: 10.1074/jbc.M001174200. [DOI] [PubMed] [Google Scholar]
- Schroeter H, Boyd CS, Ahmed R, Spencer JP, Duncan RF, Rice-Evans C, Cadenas E. c-Jun N-terminal kinase (JNK)-mediated modulation of brain mitochondria function: New target proteins for JNK signalling in mitochondrion-dependent apoptosis. Biochem J. 2003;372:359–369. doi: 10.1042/BJ20030201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenolikar S. Protein serine/threonine phosphatases—New avenues for cell regulation. Annu Rev Cell Biol. 1994;10:55–86. doi: 10.1146/annurev.cb.10.110194.000415. [DOI] [PubMed] [Google Scholar]
- Stern A, Privman E, Rasis M, Lavi S, Pupko T. Evolution of the metazoan protein phosphatase 2C superfamily. J Mol Evol. 2007;64:61–70. doi: 10.1007/s00239-006-0033-y. [DOI] [PubMed] [Google Scholar]
- Tamura Y, Simizu S, Osada H. The phosphorylation status and anti-apoptotic activity of Bcl-2 are regulated by ERK and protein phosphatase 2A on the mitochondria. FEBS Lett. 2004;569:249–255. doi: 10.1016/j.febslet.2004.06.003. [DOI] [PubMed] [Google Scholar]
- Technikova-Dobrova Z, Sardanelli AM, Speranza F, Scacco S, Signorile A, Lorusso V, Papa S. Cyclic adenosine monophosphate-dependent phosphorylation of mammalian mitochondrial proteins: Enzyme and substrate characterization and functional role. Biochemistry. 2001;40:13941–13947. doi: 10.1021/bi011066p. [DOI] [PubMed] [Google Scholar]
- Wang Y, Krushel LA, Edelman GM. Targeted DNA recombination in vivo using an adenovirus carrying the cre recombinase gene. Proc Natl Acad Sci USA. 1996;93:3932–3936. doi: 10.1073/pnas.93.9.3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WL, Yeh SF, Chang YI, Hsiao SF, Lian WN, Lin CH, Huang CY, Lin WJ. PICK1, an anchoring protein that specifically targets protein kinase C alpha to mitochondria selectively upon serum stimulation in NIH 3T3 cells. J Biol Chem. 2003;278:37705–37712. doi: 10.1074/jbc.M304619200. [DOI] [PubMed] [Google Scholar]
- Weiss JN, Korge P, Honda HM, Ping P. Role of the mitochondrial permeability transition in myocardial disease. Circ Res. 2003;93:292–301. doi: 10.1161/01.RES.0000087542.26971.D4. [DOI] [PubMed] [Google Scholar]
- Wiltshire C, Matsushita M, Tsukada S, Gillespie DA, May GH. A new c-Jun N-terminal kinase (JNK)-interacting protein, Sab (SH3BP5), associates with mitochondria. Biochem J. 2002;367:577–585. doi: 10.1042/BJ20020553. [DOI] [PMC free article] [PubMed] [Google Scholar]