Abstract
Transcript function is determined by sequence elements arranged on an individual RNA molecule. Variation in transcripts can affect mRNA stability, localization, and translation2, or give rise to truncated proteins with differing subcellular localizations3 and functions4. Given the existence of overlapping, variable transcript isoforms, determining the functional impact of the transcriptome requires identification of full-length transcripts, rather than just the genomic regions that are transcribed5,6. Here, by jointly determining both transcript ends for millions of RNA molecules (TIF-Seq), we reveal an extensive layer of isoform diversity previously hidden among overlapping RNA molecules. Variation in transcript boundaries appears to be the rule rather than the exception, even within a single population of yeast cells. Over 26 major transcript isoforms per protein-coding gene were expressed in yeast. Hundreds of short coding RNAs and truncated versions of proteins are concomitantly encoded by alternative transcript isoforms, increasing protein diversity. In addition, ~70% of genes express alternative isoforms that vary in posttranscriptional regulatory elements, and tandem genes frequently produce overlapping or even bicistronic transcripts. This extensive transcript diversity is generated by a relatively simple eukaryotic genome with limited splicing, and within a genetically homogeneous population of cells. Our findings have implications for genome compaction, evolution, and phenotypic diversity between single cells. They also suggest that isoform diversity as well as RNA abundance should be considered when assessing the functional repertoire of genomes.
Transcript isoform variation and its functional relevance have been studied in detail for several single genes. For example, pluripotent cells express a dominant, truncated version of p53 that inhibits the function of the full protein, thereby promoting cell proliferation4. However, the genome-wide characterization of isoform variation has been limited. Identifying either 5′ or 3′ transcript boundaries individually7-9 cannot determine the respective co-occurrence of start and end sites, which is essential for ascertaining the functional potential of a transcript. Thus, most studies have attributed variations at either transcript end to changes in the full-length messages. This interpretation is inaccurate in general, due to transcripts that could arise from neighbouring genes, short abortive transcripts, bicistronic messages, and transcripts with differing lengths that overlap a gene. Thus, an important dimension of transcriptome complexity has remained largely unexplored. Here, we characterized the heterogeneity of transcript isoforms in S. cerevisiae by jointly sequencing the 5′ and 3′ ends of each RNA molecule using an approach we term Transcript IsoForm sequencing (TIF-Seq).
To capture S. cerevisiae transcript isoforms, capped and polyadenylated RNAs were converted into full-length cDNA molecules that were subjected to intramolecular ligation, fragmentation, and capture of the 5′-3′ junctions via a biotin tag (Fig. 1a, Supplementary Fig. S1 and Table S1-S2). The start and end sites of individual RNA molecules were then identified at single-nucleotide resolution by paired-end sequencing of the tagged fragments. We applied TIF-Seq to wild-type yeast grown in two conditions (with glucose (YPD) or galactose (YPGal) as the carbon source). We identified the exact 5′ cap and 3′ polyadenylation sites of more than 19 million individual RNA molecules (Fig. 1b-c). These transcripts are arranged in a remarkably complex, overlapping pattern across the genome (Fig. 1d). In addition to genes with variations in their untranslated regions (UTRs) (e.g., CBK1), we discerned overlapping tandem genes (e.g., GIM3-YCK2) and bicistronic transcripts (e.g., PGA1-IGO1) (Fig. 1d). A comparison of our data with separate 5′ and 3′ end maps illustrates that the former cannot distinguish mono-from bicistronic transcripts, and the latter cannot distinguish 3′ UTR variation from short, overlapping 3′ end transcripts (e.g., YNL155W and the antisense transcript of YCK2 in Fig. 1d).
Figure 1. Genome-wide measurement of transcript isoform diversity using TIF-Seq.
a, The TIF-Seq method consists of RNA oligo capping, generation of full-length cDNA, circularization, and paired-end sequencing. b-c, TIF boundaries agree overall with previous determinations of transcript 5′ starts (b) and 3′ ends (c) derived from tiling array annotations5. As expected, TIF-Seq of non-capped mRNAs does not produce many 5′ reads at the annotated transcript start sites (b). d, Complex landscape of the yeast transcriptome in glucose, showing strand-specific RNA-Seq26 in comparison to TIF-Seq 5′ start and 3′ end profiles, as well as TIF-Seq coverage in logarithmic scale (dark red/blue upper tracks). Individual TIFs are represented by red or blue lines (Watson(+) or Crick(−) strand, respectively), each line designating one TIF. Nucleosome positions (green track, darkness indicates significance27), expression measured by tiling arrays (blue heatmap; darkness indicates expression level), and genome annotation5 are shown in the centre: annotated ORFs (red and blue boxes for Watson and Crick strands, respectively), their UTRs (black lines), SUTs (yellow boxes), and CUTs (purple boxes). Coordinates are indicated in base pairs. SUT, stable unannotated transcript; CUT, cryptic unstable transcript.
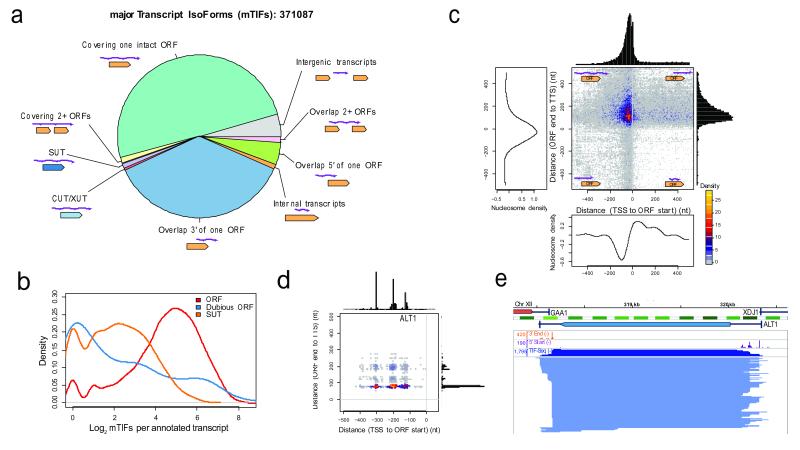
Altogether, in a genome containing only ~6000 ORFs10, we detected over 1.88 million unique transcript isoforms (TIFs) (or 776,874 supported by at least two sequencing reads, Supplementary Data S1) that are defined by a unique combination of 5′ and 3′ end sites at single-nucleotide resolution. To enable analysis of major differences, we clustered the transcripts with each of their 5′ and 3′ end sites co-occurring within 5 nucleotides, and selected the highest expressed TIF per cluster as the representative mTIF (major Transcript Isoform, see Methods), yielding 371,087 mTIFs genome-wide (Supplementary Fig. S2 and Data S2). This total corresponds to about half of all TIFs supported by two or more sequencing reads, demonstrating that there are both minor and substantial variations in transcript boundaries. Our further analysis uses TIFs and mTIFs supported by at least two sequencing reads. These numbers represent conservative estimates for transcript diversity, as our detection of isoforms is limited by sequencing depth, RNA abundance, and length (Supplementary Information). We verified the accuracy of our transcript isoform mapping with extensive controls and independent confirmations (Supplementary Information, Figs. S3-S8 and Table S3).
Our dataset reveals the extent to which different classes of transcripts are affected by isoform variation (Fig. 2a and Supplementary Fig. S9). We detected a median of 26 mTIFs (48 TIFs) that cover the coding region per verified or uncharacterized ORF in glucose and galactose (Fig. 2b-e). These mTIFs display a median positional variation of 75 nucleotides (26 for the 5′ start and 36 for the 3′ end, when considered independently) (Supplementary Fig. S10). Notably, this diversity is not dominated by a few highly abundant isoforms: a median of 10 mTIFs (or 29 TIFs) per gene is required to explain 80% of the mRNA population (Fig. S11). Isoform heterogeneity is also found in non-coding genes, including an average of 7 mTIFs per stable unannotated transcript (SUT5) (Fig. 2b and Supplementary Fig. S12). In addition, we detected thousands of multicistronic mTIFs that cover two or more ORFs (Fig. 2a). Although the number of TIFs is likely higher than what we observed, we estimate a maximum of ~100 mTIFs (or 500 TIFs) per gene (Supplementary Fig. S13). Altogether, 5211 ORFs were covered by at least one mTIF, including 86% of verified or uncharacterized ORFs and 223 dubious ORFs10 (Supplementary Data S3).
Figure 2. Extensive isoform diversity revealed among overlapping RNA populations, both at the genomic and single-gene level.
a, Categories of mTIFs identified in glucose and galactose. XUT, XRN1-sensitive unstable transcript. b, Log2-scale distribution of clustered mTIFs per annotated transcript that cover characterized or uncharacterized ORFs (ORFs), dubious ORFs, or overlap more than 80% of stable unannotated transcripts (SUTs)5. c, Transcript end distance to ORF stop codon (y-axis) vs. transcript start distance to ORF start codon (x-axis) genome-wide, revealing that most mTIFs cover the entire ORF. Decreased nucleosome density28 coincides with peaks in transcript start and end site distributions. d, Boundaries of TIFs covering ALT1 relative to ORF boundaries (as in c). e, Structure of TIFs overlapping ALT1 in glucose. 5′ start, 3′ end, and TIF-Seq coverage in natural scale. Nucleosome and genome annotations as in Fig. 1d.
Most TIFs begin as expected: downstream of annotated transcription preinitiation complex (PIC) sites11 and within the +1 nucleosome (Fig. 2c, Supplementary Discussion and Figs. S14-17). Notably, some interdependence between transcript start and end sites was observed in 382 protein-coding genes (FDR<10%, Supplementary Fig. S18, Data S4 and Discussion), supporting the existence of interactions between promoters and terminators12.
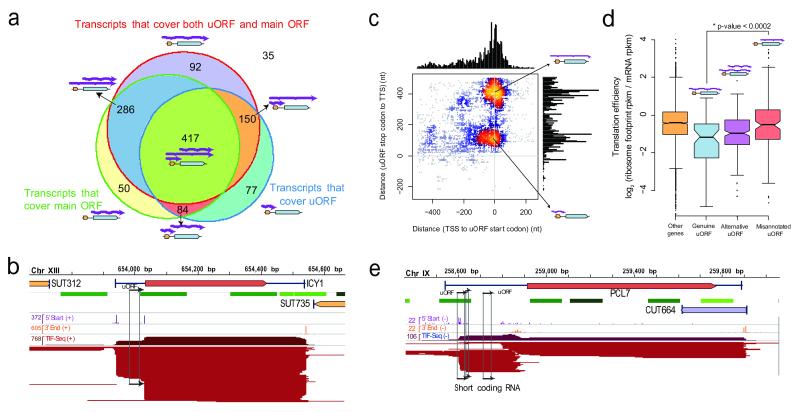
While it is unclear how much of the isoform variation is functional, we discovered several cases where phenotypic consequences would be expected. Firstly, we observed considerable variation in post-transcriptional regulatory elements. Most genes (66.9% in glucose, 71.9% across both conditions) with putative RNA-binding protein (RBP) sites13 express mTIFs with different combinations of binding sites (Supplementary Fig. S19). Furthermore, RBP sites are enriched in regions that vary between isoforms (P < 2.2 × 10−16, Supplementary Information). Secondly, we observed significant variation in upstream ORFs (uORFs), short coding regions in the 5′ UTR that modulate translation efficiency of the downstream gene14. The standard understanding is that uORFs are transcribed along with the downstream gene, as both elements must be on the same RNA to interact. Yet over half of the genes with annotated uORFs15 (703, 59% in Fig. 3a) expressed alternative mTIFs both with and without the uORF (e.g., ICY1 in Fig. 3a-b, Supplementary Data S5). This previously undetected occurrence, in addition to the variation in RNA binding sites, exemplifies transcriptional control of post-transcriptional regulatory potential: the precise isoform transcribed dictates the regulation that can be imposed on the gene.
Figure 3. Transcript isoforms with varying regulatory elements and independent short coding RNAs.
a, Number of genes whose mTIFs overlap with previously annotated upstream ORFs (uORFs) and their associated (main) ORFs15. b, ICY1 transcripts in glucose display alternative presence of uORFs (marked with arrows). c, Genome-wide plot of uORF-containing mTIFs: transcript end distance to uORF stop codon (y-axis) vs. transcript start distance to uORF start codon (x-axis). Small coding RNAs previously misannotated as uORFs represent a separate population of short overlapping RNAs. d, Genes with mTIFs that always contain uORFs display lower translation efficiency16 than those for which the uORF is independently transcribed. Genes with alternative presence of uORFs (e.g., ICY1) display intermediate translation efficiency. Significance was computed using the Wilcoxon rank sum test with continuity correction. e, Example of an scRNA that was previously misannotated as a uORF in the PCL7 locus (glucose data shown).
Notably, our dataset reveals that uORFs are not only translational regulators, but can also have an independent identity. Isoforms containing only the uORF were detected for 48% (567) of genes with known uORFs (Fig. 3c and Supplementary Fig. S20). Using ribosome profiling data16, we found that genes containing genuine uORFs (where the mTIFs always span both the uORF and the main ORF) are significantly less translated than those where the uORFs are in fact independent, misannotated transcripts (P < 2 × 10−4) (Fig. 3d). This is consistent with the expected absence of translational repression by uORFs in the latter case. In addition to re-annotating uORFs, we detected the first downstream ORFs (dORFs), defined as short coding sequences within TIFs that also cover the upstream coding gene (e.g., COX19, Supplementary Fig. S7 and Data S6).
We confirmed the existence of several short transcripts previously misannotated as uORFs by northern blot (e.g., PCL7, Fig. 3e and Supplementary Fig. S8). The fact that these transcripts have a canonical mRNA structure (5′ capped and polyadenylated), are bound by ribosomes15, and are evolutionarily conserved (Supplementary Fig. S21) suggests that they are new short coding RNAs (scRNAs, Supplementary Data S5). Short peptides can perform crucial functions, as has recently been described in cellular differentiation17. The capacity of TIF-Seq to detect potentially peptide-encoding scRNAs opens new avenues for studying their function and regulation.
We also analyzed the impact of transcript variation on protein diversity. Previous studies have identified alternative transcript isoforms that skip the first start codon, leading to loss of N-terminal signal peptides and to alternative protein localization (e.g., SUC23, whose protein product can be either cytosolic or secreted, and VAS118). We identified 153 additional genes in which at least half of the TIFs with coding potential skipped the first start codon (Fig. 4a and Supplementary Data S7). The translation of these truncated isoforms is supported by recent ribosome profiling data16 (Fig. 4b and Supplementary Fig. S22), which along with recent proteomics data19 suggest that N-terminal truncation via alternative start codon usage is a common phenomenon. This phenomenon can be transcriptionally regulated: we detected 9 genes with significantly differential truncation between glucose and galactose (P<10−3, FDR<0.1, Fig. 4c and Supplementary Table S4). These findings suggest a more common production of truncated 5′ transcripts than previously appreciated, which can directly lead to increased protein diversity even without post-transcriptional regulation.
Figure 4. Alternative transcript isoforms increase coding diversity.
a, Differential isoform regulation between glucose and galactose produces alternative truncated proteins with differential cellular localization, as shown here for SUC23. This regulation is due to subtle variations in TSS selection (5′ start track in purple) that result in alternative inclusion of the first AUG. b, Genes producing truncated transcripts that skip the first AUG (80% of these TIFs start between the first and second AUG) are effectively translated and display the expected codon usage pattern and ribosomal protection (green) in ribosome profiling data16, starting at but not before the second in-frame methionine codon. c, Proportion of N-terminal truncated TIFs, (i.e., using the second methionine as start codon) in glucose and galactose. d, Internal polyadenylation events that introduce novel stop codons encode truncated ORFs and potentially alternative protein isoforms, as shown here for GAL10.
Our dataset also provides evidence for the production of C-terminal protein truncation via alternative polyadenylation sites. We identified 33 genes enriched for internal polyadenylation that introduces early stop codons20 (FDR<10%, Supplementary Discussion, Fig. S23, Table S5 and Data S8). Among them is GAL10 (P < 1.2 × 10−9, Fig. 4d), which encodes a bifunctional enzyme in S. cerevisiae with two enzymatic domains that are encoded by two separate genes in other organisms21. In galactose media, such early stop codons result in additional transcripts encoding proteins with only one of these domains (Fig. 4d). Our evidence of protein truncation via alternative isoform usage represents a plausible means for organisms such as yeast, in which alternative splicing is uncommon, to increase protein diversity by selective domain truncation.
Our genome-wide map of transcript boundaries enabled us to measure the extent of transcriptional compaction on each strand of the genome. Most tandem TIFs are separated by approximately 150 bp (Supplementary Fig. S24). However, chained arrangements between adjacent TIFs are common, where the end of one TIF coincides with the start of the TIF for the downstream gene. In fact, of 2747 tandem ORF pairs in the genome, 27% (743) express overlapping mTIFs (e.g., GIM3-YCK2, Fig. 1d and Supplementary Data S9) and 10.2% (279) produce bicistronic transcripts (Supplementary Data S10). Most overlapping transcripts stop within the first 100-200 bp of the downstream gene (Supplementary Discussion and Fig. S24), suggesting that upstream elongating and downstream initiating RNA polymerases are distinguished within this window22. This common overlap facilitates crosstalk between transcriptional units, wherein the expression of a gene can depend not only on its own promoter, but also on the expression of its neighbours23.
Our dataset reveals the extent of transcript isoform diversity in the yeast genome at unprecedented resolution. Since most yeast genes have <1 mRNA molecule per cell24, the sheer number of isoforms detected here, even within a single environmental condition, suggests that every cell in a clonal population has a unique transcriptome in terms of RNA abundance, sequence, and thus regulatory potential. Such cell-to-cell heterogeneity may confer evolutionary advantages, enabling more rapid adaptation of the species to unforeseen environmental challenges. The variation in transcript isoforms has functional consequences via its impact on post-transcriptional regulatory potential, as well as on protein length and localization. In addition, we discovered hundreds of short coding RNAs whose function can now be investigated. Further applications of the TIF-Seq method, or of alternative paired-end strategies such as RNA-PET25, to additional environmental conditions, genetic backgrounds, and organisms will deepen our understanding of transcriptional complexity. In multicellular organisms, the combination of transcript boundary variation and alternative splicing is expected to amplify the diversity of transcript isoforms generated from a single genomic sequence, thereby expanding the functional repertoire of the genome.
Methods
Biological samples
S. cerevisiae strain SLS045 (MATa/α GAL2/GAL2, S288c background) was grown to mid-log phase (OD600~1) using either YPD (1% yeast extract, 2% peptone, 2% glucose) or YPGal (1% yeast extract, 2% peptone, 2% galactose). Total RNA was isolated by the standard hot phenol method and contaminant DNA was removed by DNase I treatment. Sequenced samples are shown in Supplementary Table S1.
TIF-Seq method
For the construction of the TIF-Seq libraries, we used 60 μg of DNA-free total RNA as input. As an internal control, we added capped and polyadenylated in vitro transcripts (ATCC 87482, 87483 and 87484). The 5′ end of the non-capped RNA molecules was desphosphorylated by treatment with 6 units of Shrimp Alkaline Phosphatase (SAP, Fermentas) for 30 min at 37°C in the presence of RNase inhibitor (RNasin +, Promega). The RNA was then purified by a double phenol extraction and ethanol precipitation. CAP was removed by a 1-hour incubation at 37°C with 5 units of Tobacco Acid Pyrophosphatase (TAP, Epicentre) in the presence of RNase inhibitor. The sample was phenol:chloroform purified and ethanol precipitated. Finally, for oligo ligation to the 5′ end of the formerly capped molecules, the treated RNA sample was incubated overnight at 16°C with 20 units of T4 RNA ligase 1 (NEB) in the presence of 10 mM DNA/RNA “5oligo cap” oligo (Supplementary Table S2), 10% DMSO and RNase inhibitor. The RNA was column-purified (RNEasy, Qiagen) and its integrity was checked (Bioanalyzer, Agilent).
To control for the presence of chimeras, each ligated RNA sample was divided into two aliquots and independently processed to generate two fractions of full-length cDNA (FlcDNA) with different terminal barcoding (see Supplementary information for details). Specifically, each fraction (11.2μL) was mixed with 1 μL RT priming oligo, 1μM either 3cDNANotI_A or 3cDNANotI_B (Supplementary Table S2), and 1 μL 10mM dNTPs. The sample was incubated at 65°C for 5 minutes and transferred to ice. 4μL of 5x First-Strand buffer (Invitrogen), 2 μL DTT 0.1M and 0.5 μL RNase inhibitor was added to each sample, which was incubated at 42°C for 2 min to minimize possible mispriming. 2μL of Superscript III reverse transcriptase (200U/μL, Invitrogen) and high temperature (55°C) incubation was then used for the retrotranscription to minimize the effects of RNA secondary structure. The reaction was incubated for 20 min at 42°C, 40 min at 50°C, and 20 min at 55°C with a final 15 min enzyme inactivation at 70°C. RNA removal was performed by adding 0.5 μL RNase cocktail (Ambion) and 2.5 units of RNase H (NEB) to each sample and incubating at 37°C for 30 min. Samples were purified using Agencourt Ampure XP beads (Beckman Coulter Genomics) according to the manufacturer’s instructions and eluted in 19 μL EB (10 mM TrisHCl pH 8.0). 19 μL of resulting FlcDNA samples were PCR-amplified using 20 μL 2x HF Phusion MasterMix (Finnzymes), 0.5 μL biotinylated oligo 5μM (either 5Bio_A or 5Bio_B) and 0.5 μL oligo 5μM (either 3Amp_A or 3Amp_B) (Supplementary Table S2). The following thermocycler program was used: 30 s for initial denaturation at 98°C, 10 cycles (20 s of denaturation at 98°C, 30 s of annealing at 50°C (+1°C/cycle) and 5 min elongation at 72°C (+10 s/cycle)) and a final elongation of 5 min at 72°C. Samples were purified using Ampure XP beads. The two independently barcoded aliquots were ultimately pooled together.
To generate cohesive ends, FlcDNA samples were digested for 1h at 37°C with 100 units of NotI (NEB) and heat-inactivated for 20 min at 65°C. The samples were Ampure XP purified and DNA yield was quantified. Between 300 and 600 ng of digested FlcDNA was circularized for 16 hours at 16°C by intramolecular ligation with 20 μL of T4 DNA ligase (2000 units/μL, NEB) in 600μL final volume. Non-circularized molecules were degraded by incubating the samples for 20 min at 37°C with 20 units each of Exonuclease III (NEB) and Exonuclease I (NEB). Enzymes were inactivated by adding 12μL of 0.5 M EDTA and incubating the samples at 70°C for 30 min. Circularized FlcDNAs were then phenol:chloroform purified and EtOH precipitated.
Purified, circularized FlcDNAs were resuspended in 130 μL EB and sonicated with a Covaris S220 (4 min, 20% Duty Cycle, Intensity 5, 200 cycles/burst). The fragmented DNA was purified with Ampure XP beads and eluted with 20 μL EB. Biotin-containing fragments were captured by incubating the samples for 30 min at room temperature with 20 μL of Streptavidin-conjugated Dynabeads M-280 (Invitrogen) and washed according to the manufacturer’s instructions.
Addition of forked barcoded adapters to the captured molecules was performed using the standard Illumina DNA-Seq library generation protocol with some small modifications. Specifically, purifications using Ampure beads were replaced with separation on magnetic dynabeads and NEBNext Master Mixes (NEB) were used. A 20 cycle-PCR enrichment was performed using Phusion polymerase (Finnzymes). 300 bp libraries were isolated using e-Gel 2% SizeSelect (Invitrogen) and sequenced by HiSeq 2000 (Illumina) and paired-end sequencing of 105 bp reads.
TIF-Seq method for long mRNA molecules
We used the same method as described above, but introduced an additional size selection step. Specifically, after the initial PCR amplification, the FlcDNA samples were size-selected on a 1.5% agarose gel, and fragments over 2kb were purified using QIAquick Gel Extraction Kit (Qiagen). The recovered FlcDNA samples were reamplified using 10 cycles of PCR before the NotI digestion.
TIF-Seq method for non-capped mRNA molecules (mono- and triphosphorylated mRNAs)
We used the same method as described above, but modified steps prior to the ssRNA ligation. RNA was dephosphorylated using Shrimp Alkaline Phosphatase as described earlier, but instead of proceeding to treatment with Tobacco Acid Pyrophosphatase, RNA was rephosphorylated for 1 h at 37°C using T4 Polynucleotide Kinase (NEB).
RNA circularization and targeted sequencing
60 μg of DNA-free RNA samples were SAP-and TAP-treated as described above to obtain full-length RNA molecules with 5′ phosphate ends. RNA circularization was performed in the presence of RNAse inhibitor, 10%DMSO, T4 RNA ligase buffer, and 50 units of T4 RNA ligase 1 (ssRNA ligase, NEB) for 16h at 16°C. The circularized RNA was purified with RNAeasy columns (Qiagen) and subjected to random hexamer retrotranscription. The resulting cDNAs were used as a template for standard PCR amplification with divergent oligos (Supplementary Table S2). The PCR products were cloned using the TOPO TA cloning system (Invitrogen). Individual clones were bidirectionally sequenced with Sanger sequencing to determine both 5′ and 3′ ends. Only clone sequences spanning a poly(A)-tail were taken into consideration.
Northern blot
The DIG Starter Northern kit (Roche) was used according to the manufacturer’s instructions. Strand-specific RNA-DIG probes were generated by in vitro transcription.
Sequencing read processing and alignment
Sequencing reads were de-multiplexed and barcode sequences were removed. The presence of internal chimera control barcodes was assessed by the Needleman-Wunsch global alignment method provided by the R Biostrings package from Bioconductor (http://www.bioconductor.org/). Samples were classified into 4 groups (putative intermolecular (A-B and A-B) or intramolecular events (A-A and B-B)). Only high-confidence reads with both chimera control barcodes and a poly(A)-tail were considered for further analysis.
Pairs of 5′ sequences and 3′ sequences were trimmed and then aligned to the reference genome using novoalign V2.07.10 (http://www.novocraft.com) using default parameters separately. The S288c S. cerevisiae genome (SGD R64, http://www.yeastgenome.org), along with the sequences of the in vitro transcripts that were included as spike-in controls, were used as reference sequences. Only sequences where both ends mapped to the reference were further analysed. Intermolecular pairs (A-B and B-A) were discarded. To exclude any other possible intermolecular cDNA species, only TIFs with both ends mapping to the same chromosome with a length ranging from 40 to 5000 bp were considered for further analysis.
TIF clustering and mTIF definition
We clustered the transcripts with 5′ and 3′ end sites co-occurring within 5 bp (Supplementary Fig. S2a). Specifically, we defined TSS and TTS clusters separately. Each cluster was defined by both a window and a mTSS/TTS (the most abundant within that window). Clusters of TSS/TTSs were assigned iteratively in decreasing order of expression. In this process, each TSS/TTS site was compared to previously defined clusters, and the site was: 1) defined as a new mTSS/TTS with a 5bp window (+-2 bp up/down stream) if the window did not overlap with previous clusters; 2) defined as a new mTSS/TTS with a smaller window (< 5bp) to avoid overlap with previously defined clusters, if the TSS/TTS was >=5bp away from the closest previously defined mTSS/TTS but <=2bp from the closest cluster; 3) merged with a previously defined cluster if the TSS/TTS was <5 bp away from the closest mTSS/TTS, in which case the original cluster window was extended to include the newly assigned TSS/TTS (thus the maximum window size of one cluster would be 9 bp); if the TSS/TTS overlapped with 2 previously defined mTSS/TTS in <5 bp, it was merged with the cluster with the closer (or higher expressed) mTSS/TTS. After this assignment process, only clusters defined by mTSS/TTSs with at least 3 supporting reads were considered. mTIFs were defined as connections between mTSS and mTTS supported by at least 2 reads connecting the associated clusters. All TIFs that shared a given TSS/TTS cluster were assigned to the corresponding mTIF cluster.
TIF annotation
The TIFs were aligned to genome annotation features and classified as: (1) ORFs, if they covered an intact ORF coding region; (2) bicistronic TIFs, if they covered 2 or more ORFs; (3) SUTs, if the common region between the TIF and SUT included more than 80% of the length of both the TIF and the SUT; (4) CUTs/XUTs, same as SUTs; (5) Overlapping 2 ORFs, if they overlapped two ORFs but did not entirely covered either; (6) Overlapping 5′ of one ORF, if they overlapped only the 5′ of one ORF; (7) Overlapping 3′ of one ORF, if they overlapped only the 3′ of one ORF; (8) Intergenic TIF, if they did not overlap with any annotated ORFs or overlapped with less than 80% of annotated SUTs, CUTs or XUTs. Annotated transcripts (ORF-Ts, SUTs and CUTs) with TSSs and TTSs are from our previous study that used tiling arrays5.
Comparing TSSs and TTSs to nucleosome data
Nucleosome raw data are derived from the study by Kaplan et al.28 Normalized nucleosome occupancy values for the regions flanking the TSSs or TTSs (+/− 500bp) were extracted and the median values in each position were calculated and plotted. TSSs or TTSs for mTIFs were used.
Supplementary Material
Acknowledgments
We thank R. Aiyar for considerable help in editing and refining the manuscript. We thank W. Huber, C. Zhu, A.I. Järvelin, S. Clauder-Münster, J. Zaugg, S. Adjalley, G. Lin and the members of the Steinmetz lab for helpful discussions and critical comments on the manuscript. We thank V.N. Gladyshev and C. Pineau for sharing published data. This study was technically supported by the EMBL Genomics Core Facility. This study was financially supported by the National Institutes of Health (to L.M.S). V.P. was supported by an EMBO fellowship.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author contributions. W.W., V.P. and L.M.S. conceived the project. V.P. developed the TIF-Seq method and performed experiments. W.W. and V.P. performed the analysis. V.P., W.W., and L.M.S wrote the manuscript.
The data reported in this paper have been deposited in GEO under accession number GSE39128 and are also accessible at http://steinmetzlab.embl.de/TIFSeq.
The authors declare no competing financial interests.
References
- 1.Djebali S, et al. Landscape of transcription in human cells. 1Nature. 2012;489:101–108. doi: 10.1038/nature11233. doi:10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Di Giammartino DC, Nishida K, Manley JL. Mechanisms and consequences of alternative polyadenylation. Mol Cell. 2011;43:853–866. doi: 10.1016/j.molcel.2011.08.017. doi:10.1016/j.molcel.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carlson M, Botstein D. Two differentially regulated mRNAs with different 5′ ends encode secreted with intracellular forms of yeast invertase. Cell. 1982;28:145–154. doi: 10.1016/0092-8674(82)90384-1. [DOI] [PubMed] [Google Scholar]
- 4.Ungewitter E, Scrable H. Delta40p53 controls the switch from pluripotency to differentiation by regulating IGF signaling in ESCs. Genes Dev. 2010;24:2408–2419. doi: 10.1101/gad.1987810. doi:10.1101/gad.1987810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu Z, et al. Bidirectional promoters generate pervasive transcription in yeast. Nature. 2009;457:1033–1037. doi: 10.1038/nature07728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nagalakshmi U, et al. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. doi:10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carninci P, et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat Genet. 2006;38:626–635. doi: 10.1038/ng1789. doi:10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- 8.Ozsolak F, et al. Comprehensive polyadenylation site maps in yeast and human reveal pervasive alternative polyadenylation. Cell. 2010;143:1018–1029. doi: 10.1016/j.cell.2010.11.020. doi:10.1016/j.cell.2010.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Z, Dietrich FS. Mapping of transcription start sites in Saccharomyces cerevisiae using 5′ SAGE. Nucleic Acids Res. 2005;33:2838–2851. doi: 10.1093/nar/gki583. doi:10.1093/nar/gki583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cherry JM, et al. Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res. 2012;40:D700–705. doi: 10.1093/nar/gkr1029. doi:10.1093/nar/gkr1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rhee HS, Pugh BF. Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature. 2012;483:295–301. doi: 10.1038/nature10799. doi:10.1038/nature10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tan-Wong SM, et al. Gene loops enhance transcriptional directionality. Science. 2012;338:671–675. doi: 10.1126/science.1224350. doi:10.1126/science.1224350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Riordan DP, Herschlag D, Brown PO. Identification of RNA recognition elements in the Saccharomyces cerevisiae transcriptome. Nucleic Acids Res. 2011;39:1501–1509. doi: 10.1093/nar/gkq920. doi:10.1093/nar/gkq920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hood HM, Neafsey DE, Galagan J, Sachs MS. Evolutionary roles of upstream open reading frames in mediating gene regulation in fungi. Annu Rev Microbiol. 2009;63:385–409. doi: 10.1146/annurev.micro.62.081307.162835. doi:10.1146/annurev.micro.62.081307.162835. [DOI] [PubMed] [Google Scholar]
- 15.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. doi:10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerashchenko MV, Lobanov AV, Gladyshev VN. Genome-wide ribosome profiling reveals complex translational regulation in response to oxidative stress. Proc Natl Acad Sci U S A. 2012;109:17394–17399. doi: 10.1073/pnas.1120799109. doi:10.1073/pnas.1120799109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kondo T, et al. Small peptides switch the transcriptional activity of Shavenbaby during Drosophila embryogenesis. Science. 2010;329:336–339. doi: 10.1126/science.1188158. doi:10.1126/science.1188158. [DOI] [PubMed] [Google Scholar]
- 18.Chatton B, Walter P, Ebel JP, Lacroute F, Fasiolo F. The yeast VAS1 gene encodes both mitochondrial and cytoplasmic valyl-tRNA synthetases. J Biol Chem. 1988;263:52–57. [PubMed] [Google Scholar]
- 19.Fournier CT, et al. Amino Termini of Many Yeast Proteins Map to Downstream Start Codons. J Proteome Res. 2012 doi: 10.1021/pr300538f. doi:10.1021/pr300538f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yao P, et al. Coding Region Polyadenylation Generates a Truncated tRNA Synthetase that Counters Translation Repression. Cell. 2012 doi: 10.1016/j.cell.2012.02.018. doi:10.1016/j.cell.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Majumdar S, Ghatak J, Mukherji S, Bhattacharjee H, Bhaduri A. UDPgalactose 4-epimerase from Saccharomyces cerevisiae. A bifunctional enzyme with aldose 1-epimerase activity. Eur J Biochem. 2004;271:753–759. doi: 10.1111/j.1432-1033.2003.03974.x. [DOI] [PubMed] [Google Scholar]
- 22.Mayer A, et al. CTD tyrosine phosphorylation impairs termination factor recruitment to RNA polymerase II. Science. 2012;336:1723–1725. doi: 10.1126/science.1219651. doi:10.1126/science.1219651. [DOI] [PubMed] [Google Scholar]
- 23.Xu Z, et al. Antisense expression increases gene expression variability and locus interdependency. Mol Syst Biol. 2011;7:468. doi: 10.1038/msb.2011.1. doi:10.1038/msb.2011.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miura F, et al. Absolute quantification of the budding yeast transcriptome by means of competitive PCR between genomic and complementary DNAs. BMC Genomics. 2008;9:574. doi: 10.1186/1471-2164-9-574. doi:10.1186/1471-2164-9-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruan X, Ruan Y. Genome wide full-length transcript analysis using 5′ and 3′ paired-end-tag next generation sequencing (RNA-PET) Methods Mol Biol. 2012;809:535–562. doi: 10.1007/978-1-61779-376-9_35. doi:10.1007/978-1-61779-376-9_35. [DOI] [PubMed] [Google Scholar]
- 26.Wilkening S, et al. An efficient method for genome-wide polyadenylation site mapping and RNA quantification. Nucleic Acids Res. 2013;41:e65. doi: 10.1093/nar/gks1249. doi:10.1093/nar/gks1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Venters BJ, Pugh BF. A canonical promoter organization of the transcription machinery and its regulators in the Saccharomyces genome. Genome Res. 2009;19:360–371. doi: 10.1101/gr.084970.108. doi:10.1101/gr.084970.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaplan N, et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458:362–366. doi: 10.1038/nature07667. doi:10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.