Abstract
The antibody-lectin sandwich arrays (ALSA) is a powerful new tool for glycoproteomics research. ALSA enables precise measurements of the glycosylation states of multiple proteins captured directly from biological samples. The platform can be used in a high-throughput mode with low sample consumption, making it well suited to biomarker research exploring glycan alterations on specific proteins. This article provides detailed descriptions of the use of ALSA, with a particular focus on biomarker research. The preparation and selection of antibodies and lectins, the preparation and use of the arrays and samples, and special considerations for using the platform for biomarker research are covered.
Keywords: antibody arrays, antibody-lectin sandwich arrays, glycan profiling, lectin detection, serum biomarkers
1. Introduction
This chapter covers the use of antibody-lectin sandwich arrays (ALSA) to probe glycosylation levels on multiple proteins captured directly from biological samples, with a particular focus of the use of ALSA for biomarker studies (1, 2). The use of altered glycoforms of specific proteins as biomarkers has great potential (3), due to the fact that altered glycosylation can be more specifically associated with disease as compared to changes in protein abundance. The key to harnessing that information for biomarker studies is the ability to sensitively and reproducibly detect changes in glycosylation on specific proteins in biological samples. Furthermore, biomarker studies benefit from high-throughput sample processing and low consumption of clinical samples. Conventional glycobiology methods based on separations or mass spectrometry, although providing invaluable information on structure, do not score well on these points, since throughput can be low, sample requirements high, with no ability to precisely measure changes between samples.
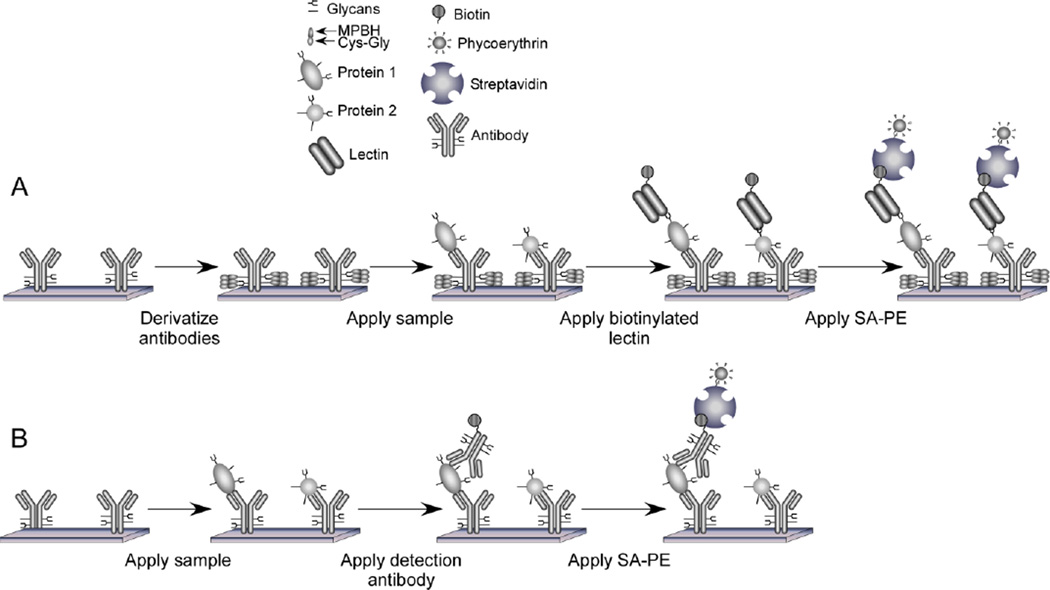
A graphical overview of the method is given in Figure 1. A biological sample, such as serum, is incubated on the surface of a microarray of immobilized antibodies, and proteins bind to the antibodies according to their specificities. The levels of specific glycan structures on the captured proteins are probed using lectins (proteins with glycan-binding activity) or antibodies targeting glycan epitopes. Different types of lectins and glycan-binding antibodies can be used to probe various glycan structures. An important first step in this procedure is a method to chemically derivatize the glycans on the immobilized antibodies. This step alters the glycans so that they are no longer recognized by the lectins or glycan-binding antibodies, ensuring that only the glycans on the captured proteins are probed.
Figure 1.
Glycan and protein detection on antibody arrays. A) Glycan detection. The drawing depicts antibodies immobilized on a planar surface. The glycans on the antibodies are derivatized to prevent lectin binding; a sample is incubated on the antibody array; proteins are captured by the antibodies; biotinylated lectins bind to the glycans on the captured proteins; and the level of bound lectin is determined by scanning for fluorescence from streptavidin-B-phycoerythrin. B) Protein detection. This approach provides measurements of the levels of the core proteins detected in (A). Antibody derivatization is not required, and individual proteins are detected using specific antibodies.
Some of the advantages of ALSA for biomarker studies stem the use of affinity reagents—molecules that can be used to detect particular targets through specific binding interactions. Affinity reagents enable reproducible and sensitive detection in the presence of highly complex biological backgrounds such as from blood serum. The ability to directly detect analytes in biological samples reduces the time and variability of assays, due to the reduced number of experimental steps. The use of lectins—carbohydrate-binding proteins—as reagents to detect glycan levels has been explored in many different settings (4).
Other advantages of ALSA stem from the use of the microarray platform (5). The usefulness of the microarray platform is in its multiplexing capability, enabling the acquisition of many data points in parallel, and its miniaturization, resulting in very small consumption of reagents and samples. These qualities are valuable for biomarker research because multiple candidate biomarkers can be evaluated in parallel with low consumption of precious clinical samples.
This chapter covers the procedures and important considerations for using this technology for biomarker studies. We do not cover the fabrication of antibody arrays. Several robotic microarrayers are available for producing arrays, each with particular performance features that might influence parameters such as the composition of the print solution or the substrate onto which the antibodies are printed. Previous methods chapters give some practical instruction and considerations for printing antibody microarrays and the handling and preparation of antibodies (6–9). Here we cover the selection and preparation of antibodies and lectins; the derivatization of antibody arrays to prevent lectin binding; sample incubation and detection; and high-throughput processing methods.
2. Materials
2.1. Reagents
NaIO4 (Pierce Biotechnology, Rockford, IL)
4-(4-N-Maleimidophenyl) butyric acid hydrazide hydrochloride (MPBH) (Pierce Biotechnology, Rockford, IL)
Cysteine-Glycine (CysGly) dipeptide (Sigma-Aldrich, St. Louis, MO)
Streptavidin-B-Phycoerythrin (Invitrogen, Carlsbad, CA)
Protease Inhibitors (Complete Tablet, Roche Applied Science, Indianapolis, IN).
Biotinylated lectins (Vector Labs, Burlingame, CA, and other suppliers)
Mouse, goat, sheep, and rabbit IgG antibodies, and chicken IgY antibodies (Jackson ImmunoResearch Labs, West Grove, PA)
Tween-20 (Sigma-Aldrich, St. Louis, MO)
Brij-35 (Sigma-Aldrich, St. Louis, MO)
NHS-biotin (Pierce Biotechnology, Rockford, IL)
2.2 Solutions
1X Coupling Buffer (0.02 M sodium acetate, pH 5.5)
Coupling Buffer + 0.1% Tween-20
Phosphate buffered saline (PBS), pH 7.4 (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, 1.4 mM KH2PO4)
PBST0.1: PBS + 0.1% Tween-20
PBST0.5: PBS + 0.5% Tween-20
PBST0.1 + 1 mM CysGly (prepare immediately before use)
Coupling Buffer + 200 mM NaIO4 (prepare immediately before use)
Coupling Buffer + 1 mM MPBH + 1mM CysGly (prepare immediately before use)
PBST0.5 + 1% bovine serum albumin (BSA)
PBST0.1 + 0.1% (BSA) + 1 µg/ml Streptavidin-B-Phycoerythrin
2.3 Hardware and instruments
Dialysis chambers (Slide-A-Lyzer Mini Dialysis Units, Pierce Biotechnology, 69550-69574)
Microscope slide staining chambers with slide racks (Shandon Lipshaw, cat. No. 121)
Microscope slide boxes (several versions available)
Wafer handling tweezers (Techni-Tool, Worcester, PA, cat. No. 758TW178, style 4WF)
Slide Imprinter, for printing wax partitions on slides (The Gel Company, San Francisco, CA)
Clinical centrifuge with flat swinging buckets for holding slide racks (Beckman Coulter, Fullerton, CA, among others)
Microarray scanner (several models available)
Ultracentrifuge (several models available)
3. Methods
The methods are divided into the following sections: 1) Selecting and preparing antibodies and lectins; 2) Derivatizing antibody arrays; 3) Sample incubation and detection; and 4) High-throughput sample processing.
3.1. Selecting the capture antibodies and the detection lectins
3.1.1. Selecting and preparing the capture antibodies
Antibody performance can vary widely with respect to affinity, non-specific binding, and information content in the relevant application. The microarray platform is ideal for making side-by-side comparisons of antibody performance. The affinities and specificities of multiple antibodies can be compared by running antibody array experiments (using the methods described below) on solutions containing various concentrations of the targeted proteins in a complex background, such as fetal-bovine serum. The signal at each antibody with respect to protein concentration provides information on performance. A linear response combined with low signal when the protein is absent indicate acceptable performance, while high background and lack of consistent responses indicate poor performance. Affinity and specificity alone do not predict the ultimate value of particular antibodies in the actual applications; some antibodies may provide more valuable information because they target distinct isoforms of proteins that have varying associations with particular disease conditions. Therefore it is useful to include multiple functional antibodies against each candidate biomarker.
The spotted capture antibodies should be as pure as possible (see Note 1). Considerations on the purification of antibodies and additives to their solutions were provided earlier (6, 8). In addition to standard purification steps, we recommend dialysis and ultracentrifugation to obtain optimal purity. Dialysis removes impurities that are smaller than the molecular weight cutoff of the dialysis membrane. This removal can be important for fluorescence detection, since some small contaminants are weakly fluorescent and contribute to background signal. Dialyze the antibody solutions for two hours against PBS at 4 °C. Ultracentrifugation removes protein aggregates. Aggregates, which can be caused by protein denaturation due to age or repeated freezing and thawing, can be a source of non-specific binding within an antibody spot. If an ultracentrifuge is available, spin the antibody solutions at 85,000 × g for one hour at 4 °C (see Note 2). After preparation and purification, we recommend aliquotting the antibody solutions so that a fresh aliquot can be used with every round of microarray production.
3.1.2. Selecting the detection lectins
Lectins are proteins that have carbohydrate-binding activities. The selection of the lectins to use as detection reagents in the ALSA assay depends on the glycan targets, the analytical performance of the lectins, and the availability of the lectins. The determination of the glycans to target is based on experimental or literature information about which glycans are overexpressed or involved in the disease of interest. Once the target glycans are determined, one may search for lectins or glycan-binding antibodies that bind those glycans. Basic information on the specificities of many lectins can be found from lectin suppliers and literature reports (10).
A tool that has facilitated detailed explorations of lectin specificities is the glycan microarray (11, 12). Glycan microarrays are composed of numerous biologically-relevant glycans. By incubating a lectin on a glycan microarray and detecting the level of lectin binding at each glycan, one may determine the preferred ligands for that lectin. The availability of glycan microarray data has been promoted by the Consortium for Functional Glycomics (CFG) (13). Participating researchers submit lectins and glycan-binding antibodies to the CFG for glycan array analysis, and the results of the experiments are made available on the CFG website (www.functionalglycomics.org).
This extensive experimental information is extremely valuable in providing detailed information about lectin specificities. With this information in hand, researchers may be better able to search for lectins with defined specificities to use as analytical reagents. The key for using glycan microarray data in this way is the ability to extract specificity information and to make it available in a searchable form. We recently developed an automated method for extracting specificity information from glycan microarray data (14) and have assembled the results from these analyses in a searchable database (http://vai-apps.tgen.org/haab/). Researchers may search for lectins that bind a user-defined glycan motif or obtain information on the binding specificities of given lectins. This tool provides access to detailed information about lectin specificity and may be useful for identifying lectins that are valuable as analytical reagents to detect specific glycans.
3.2. Sample incubation and detection
Prior to running the experiments, three experimental factors should be determined: the level of cross-reactivity between the detection lectins and the capture antibodies; the amount by which the experimental samples should be diluted; and the optimal concentration of the detection reagents.
3.2.1. Blocking cross-reactivity to the capture antibodies
Since antibodies contain glycans, one must confirm that the detection lectins do not cross-react with the glycans on the spotted capture antibodies. The amount of cross-reactivity between the detection lectins and the capture antibodies can be determined in preliminary experiments. Use the protocol given in section 3.3.3, except use PBS buffer as the sample instead of an experimental sample, and use all the detection lectins and antibodies planned for the actual experiments. Record the amount of signal at each capture antibody for each detection lectin. If low signal is observed, the targeted glycan of that detection lectin is not present, and it is not necessary to block cross-reactivity. (See Note 3) If strong signal is observed, for example greater than twice that seen when no lectin is present, it will be necessary to treat the glycans on the capture antibodies to prevent lectin binding. If a mixture of cross-reactive and non-cross-reactive antibodies is present on a single array, the entire array could be blocked for the sake of the cross-reactive antibodies, or the antibodies could be split into two arrays, one to be blocked and the other to be left untreated.
A convenient method to prevent lectin binding to the glycans on the capture antibodies is mild oxidation followed by reaction with hydrazide reagents (1). Hydrazide (N3) reacts with the aldehyde groups that are produced after oxidation using sodium periodate. We use a hydrazide reagent that is conjugated to another molecule to sterically hinder lectin binding to nearby non-oxidized glycans. We prefer the cysteine-glycine dipeptide as the conjugate since it is large enough to block access to the glycan but not so large as to interfere with antibody activity. We have shown that the oxidation step can slightly reduce antibody affinity, but that the drop in affinity is minor relative to the drop in non-specific binding of detecting lectins (1). The details of this blocking protocol were given earlier (15).
3.3.2. Determining the sample dilution factor and the detection reagent concentration
Prior to running the experimental samples, it is critical to determine the proper dilution level of the sample and the optimal concentration of the detection reagent (the lectin or antibody). It is important that the concentrations of the targeted analytes are in the linear response range of the assay. The linear range of the assay depends upon the antibodies and lectins used and therefore must be determined independently for each new assay. In order to match the analyte concentrations in the experimental samples to the linear ranges of the assays, the samples must be diluted by an appropriate amount. High-concentration analytes require high dilution, and low-concentration analytes require little or no dilution.
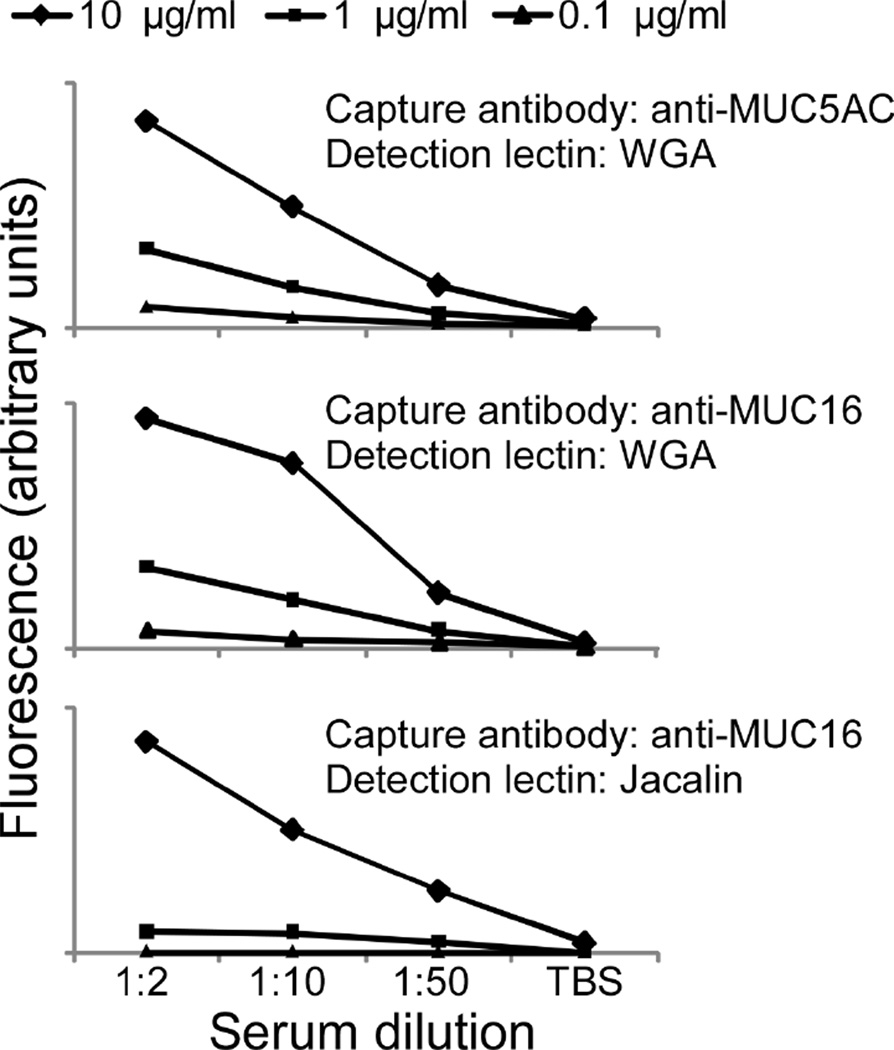
Pools of experimental samples are useful for this determination, since they contain each analyte at its average concentration over the sample set. The sample pools should be serially diluted and analyzed on antibody arrays using the detection reagents of interest. We recommend testing each detection reagent at several concentrations, since the detection reagent concentration can affect the linear range of the assay (see Note 4). The signal from each assay (each lectin-antibody combination) should be plotted with respect to dilution factor for each detection reagent concentration (Figure 2). Record the dilution factors that produce signal in the middle of the linear range and the detection reagent concentrations at which linearity is achieved. These conditions should be selected for future experiments. A challenge for array experiments is to find conditions that are optimal for multiple assays. If analytes will be targeted that have greatly divergent concentrations, it may be necessary to use separate arrays to respectively detect the high- and low-abundance analytes.
Figure 2.
Dilution curves to determine optimal experimental parameters. A pool of serum samples was diluted 2-fold, 10-fold, and 50-fold, incubated on antibody arrays, and detected with either Wheat-germ agglutinin (WGA) or the Jacalin lectin at three different lectin concentrations each. Each set of curves shows the signal at a given capture antibody with respect to serum dilution. Detection of MUC16 using WGA shows saturation at the 1:2 serum dilution using the highest lectin concentration, but the other assays are still in the linear range at the 1:2 dilution and highest lectin concentration.
3.3.3. Sample preparation
Once the optimal dilution factors are determined, the samples can be prepared for use. Biological samples should be stored in frozen aliquots so that a fresh aliquot can be used for each experiment (see Notes 5 and 6). After thawing fresh aliquots of sample, dilute the samples using the buffers and additives given in the table below, to the final concentrations of each. We recommend preparing a stock solution of each at the indicated concentrations.
| Final concentration | Stock concentration | |
|---|---|---|
| Sample buffer | 1X: 0.1% Tween-20 and 0.1% Brij-35 in 1X PBS | 10X: 1% Tween-20 and 1% Brij-35 in 10X PBS |
| IgG/IgY cocktail | 1X: 100µg/ml each of mouse, goat, and sheep IgG and chicken IgY, 200µg/ml of rabbit IgG. | 4X: 400µg/ml each of mouse, goat, and sheep IgG and chicken IgY, 800µg/ml of rabbit IgG. |
| Protease inhibitor cocktail | 1X | 10X: 1 tablet into 1 ml PBS |
The detergents are important for reducing adsorption of serum proteins to the array surface, and the IgG/IgY cocktail function to prevent non-specific binding of various components of the serum samples to the capture antibodies. The protease inhibitor cocktail is necessary because certain serum proteases can be activated upon dilution of the sample.
3.3.4. Sample incubation and detection
The next step is to incubate the samples on the arrays and to detect the levels of proteins or glycans captured at each antibody.
If the spotted antibodies will be chemically derivatized to block the glycans, perform the procedure provided earlier (1, 15) immediately before the steps described below. After derivatization, wash the microarray slides in three baths of PBST0.5 for three minutes each with gentle rocking. If not derivatizing the spotted antibodies, begin with the next step.
Incubate the slides in a blocking solution of PBST0.5 + 1% BSA for one hour with gentle rocking.
Wash the slides in three baths of PBST0.5 for three minutes each.
Spin the slides at 1000 × g for one minute to dry the slides. Use a clinical centrifuge with swinging buckets and a slide rack to hold the slides vertically. The coated surface of the slide should be facing out, allowing the liquid to easily spin off the slide.
Place the slides horizontally, coated side up, in a slide box containing a moist paper towel. The paper towel will humidify the chamber and prevent evaporation during the sample incubation.
Apply the serum solutions to their designated arrays, cover the slide box, and incubate at room temperature for 1–2 hours with gentle shaking. The volume applied to each array depends on the size of the array. Using the 48-array/slide design shown in Figure 3C, about 7 µl thoroughly covers each array.
Prepare the glycan-binding detection reagents. Biotinylated lectins should be prepared in PBST0.1 with 0.1% BSA at the previously-determined optimal concentration. Biotinylated antibodies (see Note 7) should be prepared at 1 µg/ml in the same buffer as the lectins.
Wash the slides in three baths of PBST0.1 for three minutes each with gentle rocking.
Spin the slides at 1000 × g for one minute to dry the slides.
Apply the biotinylated lectin or antibody solutions to each array, and incubate the slides in a humidified slide box at room temperature for one hour with gentle shaking.
Prepare streptavidin-B-phycoerythrin in PBST0.1 + 0.1% BSA at a concentration of 1 µg/ml.
Wash the slides in three baths of PBST0.1 for three minutes each.
Spin the slides at 1000 × g for one minute to dry the slides.
Apply streptavidin-B-phycoerythrin to each array, and incubate the slides in a humidified box at room temperature for one hour with gentle shaking.
Wash the slides in three baths of PBST0.1 for three minutes each.
Spin the slides at 1000 × g for one minute to dry the slides.
Scan the slides using a microarray scanner with appropriate resolution (10 µm or better) and emission and excitation settings. If the slides will not be scanned immediately, store them vacuum-sealed with desiccant in the refrigerator. Most fluorescent reagents can be stored under these conditions for months without appreciable loss of signal.
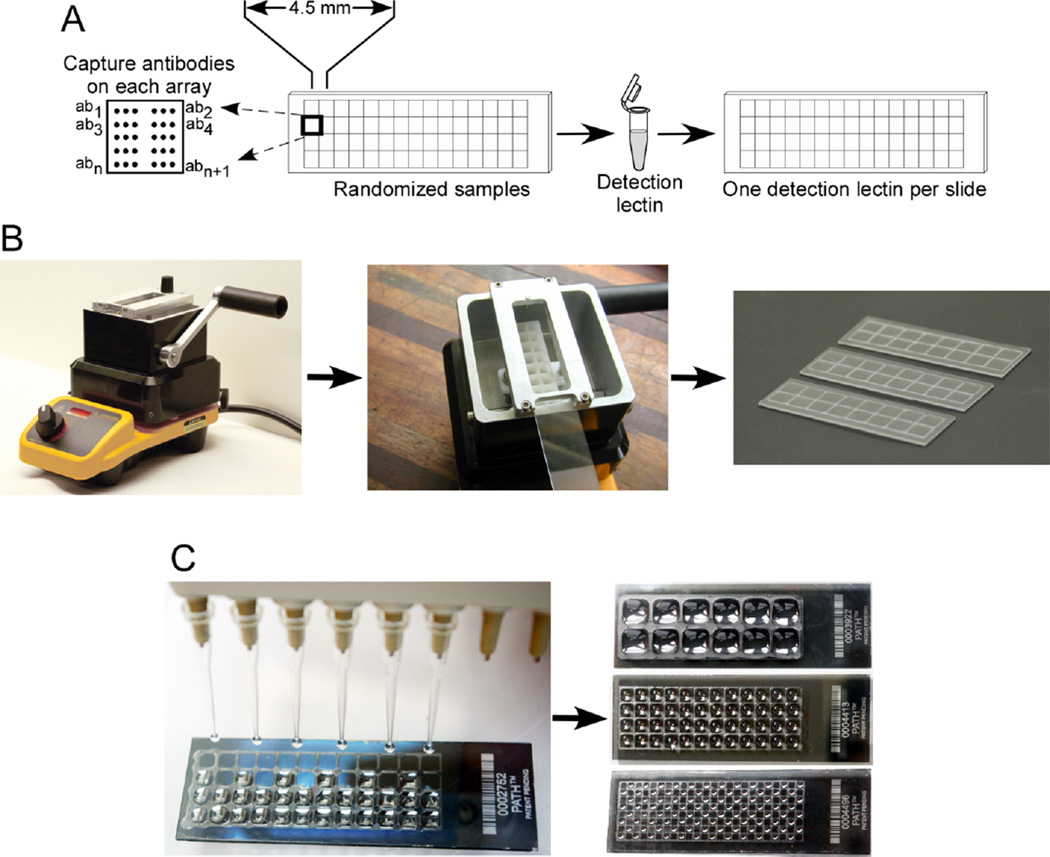
Figure 3.
High-throughput processing of antibody arrays. A) Multiple arrays can be printed on single slides using 4.5 mm spacing between arrays. A convenient approach is to incubate randomized samples on one slide followed by detection using a single lectin. B) Partitioning the arrays. A SlideImprinter device (The GelCompany, San Francisco, CA) is used to imprint wax patterns onto microscope slides. A bath of wax is melted on a hotplate, and a slide is inserted upside-down into a holder above the bath. Pulling the level forward elevates a stamp out of the bath to contact the slide, leaving a pattern of wax on the slide. C) Multi-channel liquid handling can reduce loading times and variability between the arrays. Various designs of stamps can be used, for example that partition 12, 48, or 192 arrays (right), with smaller array sizes holding proportionately smaller liquid volumes. The slide printed with 192 arrays contains samples in alternating arrays.
3.4. Using antibody arrays in biomarker experiments
3.4.1. High-throughput processing of antibody arrays
The ALSA platform can have greatly increased value when coupled with the ability to process the arrays in high-throughput. Certain research projects call for the analysis of many samples or conditions. For example, in biomarker research, measurements from dozens to hundreds of subjects can be required to properly assess the performance of a candidate marker. Samples may need to be run multiple times to characterize reproducibility, or multiple conditions may need to be tested to identify their effects on performance. Furthermore, experimental optimization can require the systematic and repeated testing of many different conditions. In these cases, the ability to efficiently and rapidly process many samples is valuable.
The approach presented here is practical and can be implemented with limited investment in new equipment. Begin by printing many replicate microarrays on single microscope slides (Figure 3A). The number of replicate arrays depends on the size of each array. Antibody microarray experiments are often designed to test specific candidate molecules, which may be just a few or up to a few dozen, and sometimes the size of the array is limited by the availability or cost of the antibodies. A useful array size is a 12 × 12 grid of 144 spots, which could contain 48 different elements each spotted in triplicate. At a typical spacing of 250 µm between spots, each array has a width of 2.75 mm. At this width, a convenient spacing between the arrays is 4.5 mm, the spacing of a 384-well microtiter plate. This spacing is compatibility with multichannel pipettes for parallel loading of the arrays or automation using liquid-handling robots. A 1 × 3 inch microscope slide can fit a grid of 4 × 15 arrays, or 60 arrays, at a spacing of 4.5 mm between the arrays. We have found this the optimal arrangement for balancing the ability to print many spots on each array and the ability to print many arrays on each slide.
The next step is to segregate the arrays so that samples do not spill over between the arrays. Segregation can be achieved either by placing an appropriate gasket on top of the slide, or by imprinting hydrophobic borders on the slide in between the arrays. Hydrophobic borders can be imprinted on microscope slides using a stamping device that imprints wax patterns (SlideImprinter, The Gel Company, San Francisco, CA) (Figure 3B). The device lifts of stamp out of a melted wax bath to contact the surface of a microscope slide, thus depositing the pattern of wax. Any design of border can be imprinted, depending on the stamp (Figure 3C). The thin borders remain on the slide throughout the experiment and the scanning. Gaskets also work well but are not as suitable for small arrays and in our experience are not as convenient, as they require extra steps to assemble and remove pressure-based or adhesive seals to ensure the lack of leaking between arrays. An advantage of small arrays segregated by hydrophobic borders is the small sample volume required for each array; the design using arrays spaced at 4.5 mm requires only 6 µl of solution, which is a significant advantage for biomarker work using clinical samples.
3.4.2. Experimental considerations for biomarker research
For biomarker research, a primary consideration is to minimize experimental variability between samples, especially between the case and control samples, and between experiment sets run on different days. A basic rule is to treat all samples identically, and especially to eliminate experimental differences between the case and control samples. The best way to eliminate differences between the case and control samples is to blind the researchers to the sample identities and to randomize the samples. If the samples are not blinded, they should be randomized and coded with identifiers that do not indicate their disease status.
It is useful to use a single detection lectin or antibody for all the arrays on a microscope slide, with case and control samples randomized on that slide (Figure 3A). If more samples will be analyzed than will fit on one microscope slide, the samples can be randomized over multiple slides, ideally run in the same experimental set to minimize variability. Automated liquid handling and pipetting (Figure 3C), if available, can facilitate high-throughput studies and further reduce variability between samples.
For a high-throughput profiling experiment design, it is useful to include a dilution curve within an experimental unit. For a 48-pad microarray slide, a typical design is to use four arrays for pooled samples diluted at three dilutions plus a TBS buffer sample. The dilutions should span the dilution used for the samples. The curve serves as a quality control measure to ensure proper functioning of the assay and the detection of the analytes in the linear response range.
Footnotes
A useful method to evaluate the purity, concentration, and structural integrity of antibodies is one-dimensional SDS-PAGE in both denaturing and non-denaturing modes. Gels can be stained with Coomassie Blue to reveal protein content. A pure, intact antibody should show a single band at 155 kD in the non-denaturing gel and single bands at 50 kD (the heavy chain) and 25 kD (the light chain) in denaturing gels. Extra bands could indicate contamination or degradation of the antibody.
An ultracentrifuge that accepts low-volume tubes (25–100 µl) is useful because high volumes of antibody might not be available. When removing the antibody solution from the tube with a pipette tip, avoid touching the sides or bottom, so that precipitated material is not also removed.
To properly assess lection performance, it is desirable to have proper positive and negative controls spotted on the arrays. Such controls might be proteins that are known to either contain or not contain the glycan targeted by each lectin. Commercially-available and inexpensive control glycoproteins can be tested in preliminary experiments for their reactivity to the lectins of interest.
Typical optimal concentrations of detection reagents range from 0.1 to 2 µg/ml. Several factors affect performance, including affinity, avidity, non-specific binding, and self-interference effects, so the optimal range must be determined experimentally for each reagent.
Repeated freezing and thawing can be damaging to proteins and therefore should be minimized. The damage is variable between proteins, but proteomics studies have shown in general that up to three thaws is acceptable for most proteins (16). The proper aliquotting of biological samples enables the use of a fresh aliquot for each experiment with no more than three thaws for each aliquot. The volume of each aliquot should be similar to that used in each experiment to reduce waste of sample.
The thawing process should be carried out slowly to minimize damage. Remove samples from −80 °C storage and place on ice, allowing the samples to gradually thaw over the course of an hour.
Lectins can be purchased biotinylated or can be biotinylated using the appropriate labeling reagents (see Materials section). We recommend modifications to the manufacturer-supplied protocols for labeling with N-hydroxysuccinimide reagents, in which we lower the pH, temperature, and time of the reaction to limit the number of amine groups that are labeled. See reference (6) for details.
References
- 1.Chen S, LaRoche T, Hamelinck D, Bergsma D, Brenner D, Simeone D, Brand RE, Haab BB. Multiplexed analysis of glycan variation on native proteins captured by antibody microarrays. Nature methods. 2007;4:437–444. doi: 10.1038/nmeth1035. [DOI] [PubMed] [Google Scholar]
- 2.Yue T, Goldstein IJ, Hollingsworth MA, Kaul K, Brand RE, Haab BB. The prevalence and nature of glycan alterations on specific proteins in pancreatic cancer patients revealed using antibody-lectin sandwich arrays. Mol Cell Proteomics. 2009;8:1697–1707. doi: 10.1074/mcp.M900135-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dube DH, Bertozzi CR. Glycans in cancer and inflammation--potential for therapeutics and diagnostics. Nat Rev Drug Discov. 2005;4:477–488. doi: 10.1038/nrd1751. [DOI] [PubMed] [Google Scholar]
- 4.Hirabayashi J. Lectin-based structural glycomics: glycoproteomics and glycan profiling. Glycoconjugate journal. 2004;21:35–40. doi: 10.1023/B:GLYC.0000043745.18988.a1. [DOI] [PubMed] [Google Scholar]
- 5.Yue T, Haab BB. Microarrays in glycoproteomics research. Clin Lab Med. 2009;29:15–29. doi: 10.1016/j.cll.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haab BB, Zhou H. Multiplexed protein analysis using spotted antibody microarrays. Methods in molecular biology (Clifton, N.J. 2004;264:33–45. doi: 10.1385/1-59259-759-9:033. [DOI] [PubMed] [Google Scholar]
- 7.Chen S, Haab BB. Antibody Microarrays for Protein and Glycan Detection. In: Van Eyk J, Dunn M, editors. Clinical Proteomics. Germany: Wiley, VCH, Weinheim; 2007. [Google Scholar]
- 8.Haab BB. Multiplexed protein analysis using antibody microarrays and label-based detection. Methods Mol Med. 2005;114:183–194. doi: 10.1385/1-59259-923-0:183. [DOI] [PubMed] [Google Scholar]
- 9.Haab BB, Lizardi PM. RCA-enhanced protein detection arrays. Methods in molecular biology (Clifton, N.J. 2006;328:15–29. doi: 10.1385/1-59745-026-X:15. [DOI] [PubMed] [Google Scholar]
- 10.Hirabayashi J. Concept, strategy and realization of lectin-based glycan profiling. Journal of biochemistry. 2008;144:139–147. doi: 10.1093/jb/mvn043. [DOI] [PubMed] [Google Scholar]
- 11.Culf AS, Cuperlovic-Culf M, Ouellette RJ. Carbohydrate microarrays: survey of fabrication techniques. Omics. 2006;10:289–310. doi: 10.1089/omi.2006.10.289. [DOI] [PubMed] [Google Scholar]
- 12.Wang D. Carbohydrate microarrays. Proteomics. 2003;3:2167–2175. doi: 10.1002/pmic.200300601. [DOI] [PubMed] [Google Scholar]
- 13.Blixt O, Head S, Mondala T, Scanlan C, Huflejt ME, Alvarez R, Bryan MC, Fazio F, Calarese D, Stevens J, Razi N, Stevens DJ, Skehel JJ, van Die I, Burton DR, Wilson IA, Cummings R, Bovin N, Wong CH, Paulson JC. Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:17033–17038. doi: 10.1073/pnas.0407902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Porter A, Yue T, Heeringa L, Day S, Suh E, Haab BB. A motif-based analysis of glycan array data to determine the specificities of glycan-binding proteins. Glycobiology. 2009 doi: 10.1093/glycob/cwp187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen S, Haab BB. Analysis of glycans on serum proteins using antibody microarrays. Methods in molecular biology (Clifton, N.J. 2009;520:39–58. doi: 10.1007/978-1-60327-811-9_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rai AJ, Gelfand CA, Haywood BC, Warunek DJ, Yi J, Schuchard MD, Mehigh RJ, Cockrill SL, Scott GB, Tammen H, Schulz-Knappe P, Speicher DW, Vitzthum F, Haab BB, Siest G, Chan DW. HUPO Plasma Proteome Project specimen collection and handling: towards the standardization of parameters for plasma proteome samples. Proteomics. 2005;5:3262–3277. doi: 10.1002/pmic.200401245. [DOI] [PubMed] [Google Scholar]