ABSTRACT
Paramyxoviruses include major pathogens with significant global health and economic impact. This large family of enveloped RNA viruses infects cells by employing two surface glycoproteins that tightly cooperate to fuse their lipid envelopes with the target cell plasma membrane, an attachment and a fusion (F) protein. Membrane fusion is believed to depend on receptor-induced conformational changes within the attachment protein that lead to the activation and subsequent refolding of F. While structural and mechanistic studies have considerably advanced our insight into paramyxovirus cell adhesion and the structural basis of F refolding, how precisely the attachment protein links receptor engagement to F triggering remained poorly understood. Recent reports based on work with several paramyxovirus family members have transformed our understanding of the triggering mechanism of the membrane fusion machinery. Here, we review these recent findings, which (i) offer a broader mechanistic understanding of the paramyxovirus cell entry system, (ii) illuminate key similarities and differences between entry strategies of different paramyxovirus family members, and (iii) suggest new strategies for the development of novel therapeutics.
PARAMYXOVIRUS MEMBRANE FUSION MACHINERIES: MOLECULAR TRIGGERS OF DISEASE
Paramyxoviruses are enveloped, nonsegmented, negative-stranded RNA viruses causing important human and animal diseases with major clinical and economic impact. For instance, measles virus (MeV), respiratory syncytial virus (RSV), mumps virus (MuV), parainfluenza virus 5 (PIV5), human parainfluenza viruses 1 to 4 (hPIV1 to -4), and the recently identified human metapneumovirus (hMPV) may induce significant morbidity and mortality in humans. Newcastle disease virus (NDV), avian metapneumovirus (AMPV), and canine distemper virus (CDV) have a serious impact on animal health. In addition, emerging paramyxoviruses include highly pathogenic zoonotic agents (e.g., members of the genus Henipavirus, Nipah virus [NiV] and Hendra virus [HeV]) (1, 2).
Fusion of the viral envelope with cellular membranes is essential for all enveloped viruses to enter target cells and initiate disease. However, in contrast to other major human pathogens, such as influenza virus, HIV, or Ebola virus, the paramyxovirus entry machinery is composed of two separately encoded envelope glycoproteins (3). In tight cooperation, the attachment and fusion proteins of the Paramyxovirinae mediate membrane merging at neutral pH (3). Nascent F is first synthesized as inactive precursors (F0), which initially fold into a trimeric metastable prefusion conformation. Subsequently, for most of the paramyxovirus F proteins, proteolytic maturation into two disulfide-linked subunits (F1 and F2) occurs in the Golgi apparatus. Prefusion F trimers are thought to undergo large-scale structural rearrangements upon attachment protein-mediated activation that lead to the merger of the viral envelope with cellular membranes for fusion pore formation (3–5).
An extensive body of evidence supports the notion that the tetramer represents the physiological oligomer of paramyxovirus attachment proteins (3, 6–10). Each monomer contains a short luminal tail, a single membrane-spanning domain, and a large ectodomain. The latter consists of a stalk region supporting a globular head domain. X-ray structures of various paramyxoviral head domains consistently revealed a common six-bladed beta-propeller fold, reminiscent of sialidases (9–17). Members of the subfamily Paramyxovirinae that bind to sialic acid-containing surface molecules (the genera Rubulavirus, Avulavirus, and Respirovirus) indeed carry hemagglutinin-neuraminidase (HN) attachment proteins (3, 18), which perform both hemagglutination (of sialic acid-bearing erythrocytes) and neuraminidase (releasing particles from the cell) functions. In contrast, viruses belonging to the genera Morbillivirus and Henipavirus carry attachment proteins that lack the ancestral neuraminidase activity; these pathogens evolved to infect cells through specific interactions with proteinaceous receptors (19–23). Of these, the attachment proteins of morbilliviruses still show hemagglutination activity (H attachment proteins), whereas henipavirus attachment glycoproteins lack both neuraminidase and hemagglutination activity (G attachment protein) (3).
PARAMYXOVIRUS CELL SURFACE RECEPTORS
The identification of SLAM and nectin-4 as MeV and CDV entry receptors provided groundbreaking new insights into our overall understanding of the pathogenesis induced by MeV and, by extension, members of the genus Morbillivirus as a whole (21, 23, 24). Indeed, extensive in vitro and in vivo studies indicate that MeV and CDV invade the host by initially infecting SLAM-positive alveolar macrophages and/or airway epithelium-associated dendritic cells (25–28). Following viral amplification in local lymphoid tissues, viremia and systemic dissemination of the virus in epithelia of different organs ensue. Notably, infected lymphocytes and dendritic cells located within the airways presumably allow cell-to-cell transfer of both morbilliviruses to nectin-4-expressing epithelial cells. Finally, further viral spread throughout the airway epithelium leads to release of infectious particles into the lumen of the respiratory tract and viral spread (21, 29, 30). Interestingly, the regulator of complement activation CD46 was first identified as a host cell entry receptor for MeV (20). However, a large body of evidence supports the notion that only attenuated MeV strains are able to productively interact with CD46, an ability likely acquired during multiple passages of MeV in tissue culture cells.
The molecules ephrinB2 and ephrinB3 were recently discovered and identified as entry receptors for henipaviruses (19, 22). These receptors play key roles in cell-cell signaling by interacting with surface-exposed molecules expressed on opposing cells (i.e., neuronal development and angiogenesis). Both transmembrane proteins are expressed in endothelial cells and in neurons, a tissue distribution which is consistent with the natural tropism of henipaviruses. Because ephrinB3 is highly expressed in the brain stem (31), this might contribute to virus-mediated brain dysfunctions and, ultimately, fatal outcomes.
In contrast to morbilliviruses and henipaviruses, which bind to proteinaceous receptors, rubula-, avula-, and respiroviruses recognize sialic acid receptors, which are widely expressed on the surfaces of target cells (3, 18). Although sialic acid binding is required for cell entry and pathogenicity, the tissue specificity of these paramyxovirus family members is not equally closely controlled by the attachment protein-receptor interaction.
Without doubt, receptor binding by Paramyxoviridae attachment proteins leading to F activation and membrane fusion constitutes a first and essential step of infection by this large group of viruses irrespective of whether viremia, respiratory illnesses, skin lesions, or chronic neurological infections will result.
STRUCTURAL INSIGHT INTO PARAMYXOVIRUS ATTACHMENT PROTEINS
Sialidase-like head domain.
Receptor binding by attachment proteins is thought to result in triggering of irreversible large-scale conformational changes of the F trimer (3, 7, 32–35). These structural rearrangements are believed to bring the cellular and viral membranes into close proximity and are thus considered to be directly coupled to membrane fusion and pore formation. In the last decade, groundbreaking structural and functional studies advanced our understanding of the molecular requirements for productive binding of paramyxoviruses to host cells.
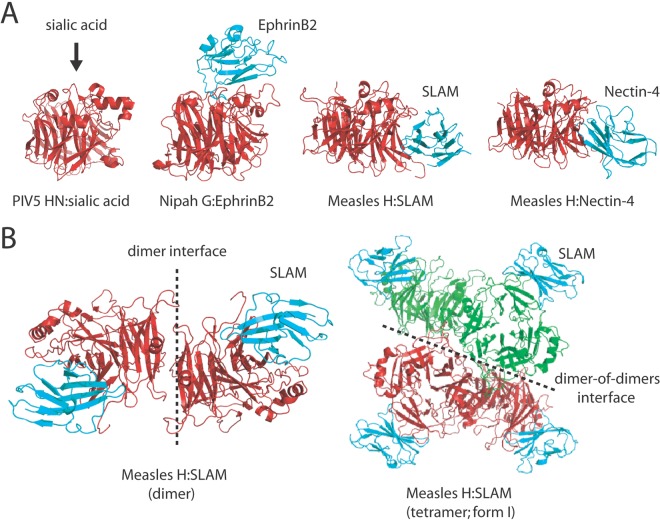
X-ray structures of receptor-bound and free hPIV3 and PIV5 HN head domains located the receptor binding site (RBS) near the top of the conserved six-bladed beta-propeller structures of the attachment protein head domains (Fig. 1A, left) (10, 15). Strikingly, however, only very minor conformational differences were observed between receptor-bound and free monomeric head structures. While a receptor-complexed structure is not available for NDV HN, the structural analysis of this protein revealed two different dimeric conformations (35). Importantly, again, no significant structural changes within individual head monomers were observed when these distinct dimer assemblies were compared.
FIG 1 .
(A) Side views of PIV5 HN (PDB code, 1Z4Z), NiV G bound to ephrinB2 (PDB code, 3D12), MeV H monomer bound to SLAM (PDB code, 3ALZ) and MeV H monomer bound to nectin-4 (PDB code, 4GJT). Beta-propeller structures of paramyxovirus attachment protein head domains are shown in red, whereas the proteinaceous receptors are shown in cyan. (B) Top views of the MeV H dimer (left) and tetramer bound to SLAM (right) (PDB code, 3ALZ). In the MeV H tetramer, one dimer of dimers is shown in red, and the other is in green.
For the henipaviruses, again, crystal structures of liganded and unliganded HeV and NiV G revealed slight modifications at the binding interface, but no major conformational changes of the overall six-bladed beta-propeller structures were found (34, 36). As described for the PIV5, hPIV3, and NDV HN attachment proteins, the RBSs of henipavirus G proteins are confined to pockets located near the top of the beta-propeller, although henipavirus receptors are proteinaceous by nature (Fig. 1A, left and middle) (34, 36).
In the case of MeV, crystal structures of the attachment protein (H) resolved in the presence (16) or absence of CD46 (12, 13) and, more recently, SLAM and nectin-4 (14, 37) provided intriguing insights into the mechanism of morbillivirus cell entry. While H crystallized as a monomer with nectin-4, in the presence of CD46 a dimeric structure was resolved. Remarkably, two distinct H tetramer conformations were found when chimeric polypeptides consisting of H head domains linked to SLAM receptor moieties were subjected to crystallization. Consistent with other X-ray structures, the monomeric MeV H head conformations remain mostly unaltered, independent of whether they are in a dimeric or tetrameric setting and/or free or complexed with receptor. In contrast to other paramyxovirus attachment proteins, however, the two MeV H head monomers were found to be tilted almost 90° relative to each other when engaged in dimers (Fig. 1B, left) (13). Furthermore, consistent with previously resolved tetrameric structures of hPIV3, PIV5, and NDV HN, MeV H tetramers appeared to be able to assemble either into a largely planar (Fig. 1B, right, and Fig. 2A, left) or more staggered spatial organization (Fig. 2B, right) (14).
FIG 2 .
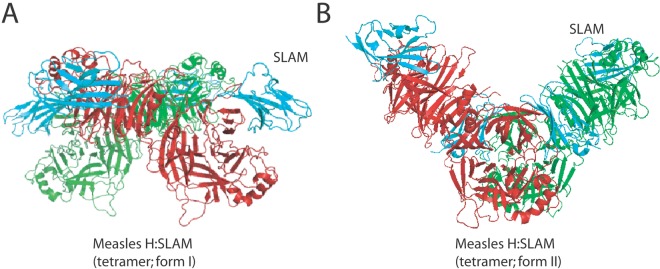
Side views of two forms of MeV-H tetramer complexed with SLAM (cyan). One MeV-H dimer is shown in red, and the other is in green. The tetramers assume two distinct conformations (A [PDB code, 3ALZ] and B [PDB code, 3ALX]).
A recently released structure of the NDV HN head and partial stalk domain introduced a new tetrameric conformation of the attachment protein complex (17). Remarkably, in this structure, both the head dimers are not in contact with each other, but the lower head of each dimer is engaged in short-range interaction with the stalk, which is present in a four-helical-bundle (4HB) configuration (Fig. 3A) (17). Since the connectors linking the upper 4HB stalk to the heads were not revealed in the X-ray structure, spatial flexibility of this domain was inferred, and changes in the head dimer/stalk assembly were proposed to contribute to F activation (see below).
FIG 3 .
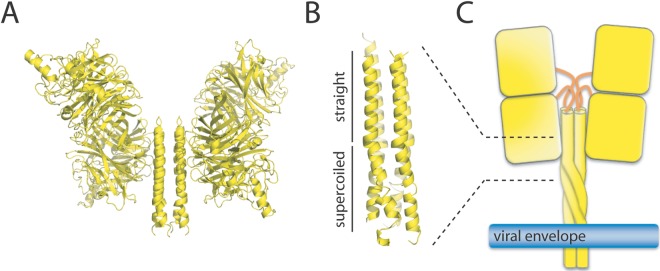
(A) Structure of the soluble NDV HN tetramer ectodomain in the heads-down conformation (PDB code, 3T1E). In this form, the two lower heads of each dimer interact with the stalk (in the 4HB conformation). The four connectors linking the top of the stalk to the four different heads are not present in the X-ray structures. (B) Structure of the soluble PIV5 HN stalk domain, consisting of an upper “straight” region and a lower “supercoiled” region (PDB code, 3TSI). (C) Putative pre-receptor-binding conformation of membrane-embedded full-length PIV5/NDV HN tetramers.
Concerning the MeV H RBS, structural and functional information identifies a common lateral face of each head monomer to mediate binding to the multiple receptors: CD46, SLAM, and the recently identified nectin-4 (Fig. 1A, right) (14, 16, 28, 38). Furthermore, within each MeV H head dimer, both RBSs are positioned (i) distal from the dimer interface and (ii) opposite each other (Fig. 1B, left). In the context of the two available tetrameric MeV H/SLAM crystal structures, the more planar head arrangement positions the RBS of each monomer distal from the dimer-of-dimers interface and near the rim of the head assembly (Fig. 1B, right), while in the more staggered alternative, only the RBSs of the two upper head monomers are readily accessible for receptor binding; the RBSs of the lower two head monomers are sandwiched between the dimers (Fig. 2B).
Stalk domain.
Early studies revealed that paramyxovirus-induced fusion activity requires coexpression of the F protein with its homotypic HN/H/G protein in the same cell (39–41). Subsequently, many studies have implicated the attachment protein stalk region in short-range interaction with the F protein and thus indicated that it contributes to the control of the overall fusion process (4, 42–47).
Yuan et al. (17) first documented the partial crystal structure of any paramyxovirus attachment protein stalk domain. As mentioned above, in this structure, the stalk is arranged in a tetrameric, four-helical bundle (4HB) conformation (Fig. 3A). More recently, the atomic structure of a soluble form of the PIV5 HN stalk domain was also determined, which revealed a comparable 4HB organization (48). Interestingly, this longer fragmented portion of the PIV5 HN stalk 4HB featured a partition into an upper (membrane-distal) straight and a lower (membrane-proximal) left-handed supercoiled section (Fig. 3B and C).
RECEPTOR-INDUCED CONFORMATIONAL CHANGES IN THE ATTACHMENT PROTEIN
Head movements.
Collectively, these structural studies have established a tangible framework for the understanding of paramyxovirus cell attachment and fusion triggering. However, static structural information can only partially reflect the highly dynamic protein refolding processes. Recent functional and biochemical analyses have probed the physiological relevance of individual structures and provided mechanistic context. The exact sequence and nature of the structural rearrangements taking place in paramyxovirus attachment proteins that lead to F refolding begin only now to emerge and demand further in-depth investigation.
Based on the available structural information, it has emerged as an overarching concept that F triggering is initiated by receptor-induced changes on the attachment protein oligomer rather than the monomer level. Mahon and colleagues have pioneered a disulfide linking strategy for the NDV HN monomer-monomer interface (49), which has revealed that engineered disulfide bonds between two head monomers do not prevent membrane fusion. Rather, fusion activity was enhanced, arguing against a mechanism in which receptor binding affects the monomer-monomer interface within the head dimers.
In agreement with the findings by Mahon and colleagues, the recently solved MeV H-SLAM cocrystal revealed two different H tetrameric organizations. Hashiguchi and colleagues (14) proposed that the more planar conformation may represent a pre-F triggering form of the H tetramer, whereas the more staggered structure corresponds to a post-F triggering state. Transition between conformations was predicted to occur through a “sliding” movement of the H dimer of dimers, which in turn would lead to the disassembling of the tetrameric H stalk into two discrete dimeric sections. Because H stalks are involved in short-range interactions with prefusion F trimers (4, 42–47), such a drastic conformational change may release F from H, and the ensuing F-refolding cascade may initiate. In this model, the H head section would act as a “signal transducer domain,” which transmits the triggering signal received from the interaction with the receptor to the stalk domain.
In support of this assumption, Brindley and colleagues found that some mutated MeV H dimers could not interact with each other, which provided the first direct evidence that MeV H tetramers are indeed capable of assuming distinct spatial organizations in situ (50). Native-PAGE analyses of intact H tetramers detected discrete gel migration profiles, which correlated with F binding-competent and -incompetent MeV H mutants. Most importantly, heat treatment or addition of soluble SLAM to receptor-competent MeV H tetramers switched gel migration patterns from F interaction-competent to F interaction-incompetent profiles, thereby strongly supporting the physiological relevance of biochemically recorded H conformational changes for F triggering (50).
The recently resolved crystal structure of the NDV HN tetramer further supports the model suggested by Hashiguchi and coworkers, where both H head dimers may move in sync upon receptor binding to promote the fusion process. As documented above (Fig. 3A), the two NDV HN dimers are not in direct contact with each other, but one head of each dimer is proposed to engage in short-range interactions with the stalk domain (17). This back-folded organization may preclude productive lateral interaction with prefusion F trimers. Consequently, Yuan and colleagues speculated that upon receptor binding, the two dimers move into a “heads-up” position, which uncovers the F interaction sites in the HN stalk region and facilitates HN/F interactions (51). Consistent with this model, HN-type paramyxovirus attachment proteins are thought not to interact with F trimers in the secretory system of the host cell and typically do not coprecipitate with F (52).
The recent report documenting that the PIV5 HN tetrameric stalk expressed in the absence of its four globular head domains (headless PIV5 HN) inferred a refined model. Indeed, headless PIV5 HN could significantly enhance fusion activity when cotransfected with the homotypic PIV5 F protein (51). Bose and colleagues propose a model of F activation in which, in addition to the heads-down/up movements presented above, the HN heads regulate fusion triggering by controlling F docking to the HN stalk (51). In this view, the HN head section serves as “inhibitory domain” that prevents premature F interaction with the stalk. Once HN reaches the cell surface in the prereceptor heads-down configuration, interaction with the receptor switches the tetramer to the heads-up state (51). This mechanism then allows productive F/HN stalk interaction, which ultimately results in F triggering. Consistent with this model, HN and F do not interact intracellularly with each other (52), thereby preventing premature intracellular F triggering in the absence of receptor.
Although this model has been suggested by Bose and colleagues to be a potential universal model for paramyxovirus membrane fusion triggering, it is unclear whether headless attachment protein stalks of other Paramyxovirinae family members can also trigger their homotypic F proteins. In the case of the morbillivirus fusion machinery, available experimental evidence is not immediately compatible with the model. In strong contrast to the HN-type proteins, the F binding sites in MeV H are always accessible, since MeV H and F oligomerize intracellularly and readily coprecipitate (53). While this could reflect the idea that the F-contacting and -activating domains within MeV H stalks may be spatially distinct (outlined in more detail below), generating H variants with elongated stalks revealed that insertion of segments above the F-contacting region does not abolish membrane fusion activity (54). Structural and functional mapping of both H stalk microdomains indicated that they are membrane proximal, relative to the stalk extensions (54), suggesting that back-folding of the MeV H head domain to the stalk, should it ever occur, does not serve to prevent contact of prefusion F with H. Possibly, the unusual inherent ability of PIV5 F trimers to achieve membrane fusion in the absence of attachment proteins (55) (which may be dependent on PIV5 F binding to a coreceptor) may set the stage for accelerated F activation by headless PIV5 HN (51).
At present, it is unclear whether the different configurations of the morbillivirus H tetramers likewise include major head movements resembling the heads-down and heads-up configurations of the HN proteins. Further structural, biochemical, and mechanistic studies will be required to probe the distinct roles of head domains of different paramyxovirus attachment protein types in F activation.
Stalk movements.
The latest model described by Bose and colleagues for PIV5 fusion-triggering suggests that HN head acts as a “regulator,” whereas the stalk acts as an “activator” (51). While receptor-induced conformational changes within the HN stalk domain have yet to be demonstrated experimentally, Bose and colleagues speculated that headless HN could spontaneously assume a receptor-bound or “trigger-competent” conformation that may destabilize F trimers upon transient HN–F association (51). Alternatively, one cannot exclude the possibility that HN stalks may preserve a pre-receptor-binding state until the protein reaches the cell surface. In this case, either random interaction of F with headless HN may directly result in F activation (without any conformational changes within the stalk) or the stalk may undergo spontaneous conformational changes that, in turn, drive F refolding.
If F binding residues are always accessible in the morbillivirus H attachment protein stalk and H-F hetero-oligomers are preassembled, what may trigger F refolding upon receptor binding? New insight into the biochemical nature of morbillivirus F triggering comes from a recent study that biochemically confirmed the existence of distinct MeV H configurations resembling pre- and post-receptor-binding stages (50). Of these, only the former was capable of physically interacting with the F trimer, suggesting that receptor binding shifts the H stalk into an F-binding-incompetent configuration. Further support for this model comes from the observation that engineered disulfide bonds within the central section of the morbillivirus H stalk, the region previously identified as being engaged in short-range interaction with prefusion F, reversibly arrest membrane fusion activity (50, 56, 57). In contrast, locking membrane-proximal and -distal H-stalk regions through covalent disulfide bonds does not impede membrane fusion triggering (56, 57). Combined, these findings define a new framework for our understanding of receptor-induced conformational changes in morbillivirus H proteins that lead to F triggering: (i) H tetramers form preassembled fusion complexes with F trimers prior to receptor binding, (ii) H complexes assume distinct pre-receptor/pre-F-triggering and post-receptor/post-F-triggering conformations, (iii) the overall H-tetramer integrity is preserved throughout the fusion process (tetrameric H stalks do not disassemble), and (iv) structural flexibility within the central H stalk section is required for receptor-induced rearrangement of the F contact zone, resulting in F triggering.
Overall, the recent structural and mechanistic reports are in agreement with the notion that paramyxovirus attachment protein dimeric head domains move in sync upon receptor binding, which, in turn, translates into conformational changes in the membrane-proximal stalk region. Whether receptor-induced head movement leads to a spontaneous stalk refolding process or to a signal transduction that actively triggers the stalk’s structural rearrangements of the central section remains to be clarified.
MOLECULAR DETERMINANTS LINKING RECEPTOR BINDING TO F TRIGGERING
Discrete subdomains in MeV H.
Based on a set of new and innovative functional and biochemical assays, recent studies revealed key insights into effective MeV H interaction with the receptor and the ensuing conformational changes leading to F triggering. Specifically, three physically and functionally discrete regions within H monomers were identified: (i) a receptor binding domain mapping to one side of the head domain (14, 16, 28, 58), (ii) an F-interactive domain mapping to the upper central stalk section (44, 54), and (iii) an F-triggering domain, mapping to the lower central stalk section (6, 54, 59). Using H transcomplementation and homodimer disulfide bond engineering (which enabled the design of specific homodimer/heterotetramer assemblies), functional and nonfunctional cross talk along the H dimer-dimer interface was demonstrated (50). While receptor binding and F triggering could be efficiently complemented, H dimers lacking F interaction were complementation defective, even when combined with wild-type H dimers. These results are consistent with the notion that receptor binding of only one dimer within the tetramer is sufficient to initiate the conformational changes leading to F triggering (50).
Sustained H/HN interaction with the receptor.
We recently identified conformation-sensitive monoclonal antibodies directed against morbillivirus F proteins that discriminate between F trimers in prefusion and triggered/postfusion conformation (60). Investigation of the conformational state of membrane-bound MeV F before and after receptor binding using these antibodies revealed that the soluble receptor is sufficient to trigger F refolding (50). Intriguingly however, if triggered by the soluble receptor, F refolding does not lead to opening of a fusion pore and content mixing. In contrast, exposure to cells expressing membrane-anchored SLAM leads to productive fusion under otherwise identical conditions (50).
These data prompt us to speculate that sustained receptor binding by H may define a fusion-competent microenvironment that, possibly, supports induction of local lipid curvature in opposing donor and target membranes and/or allows continued H-F interaction beyond the initial F triggering step (61). Recent data obtained by Porotto and coworkers with NDV are in line with our conclusions and further support the notion that sustained receptor interaction by the attachment protein is a universal mechanism required by paramyxoviruses to enter target cells (61).
Interaction of NDV HN with F beyond the F activation step.
According to the current knowledge of paramyxovirus-induced membrane fusion, the F protein is triggered by the attachment protein and undergoes a series of structural rearrangements from a prefusion to a postfusion F state (passing through a putative “prehairpin” structural intermediate). Because the F protein would progress through these different structures independently, these spontaneous conformational changes are referred to as the “spring-loaded” mechanism. Recent results obtained by Porotto and colleagues with NDV HN and F drastically challenged this model. Indeed, using erythrocytes and NDV HN/F-expressing cells as a model system to study fusion activity, they obtained provocative data suggesting that HN may be required to promote fusion beyond the F activation step (even after the F protein has reached the prehairpin state) (61).
If correct, this finding alters all current models of the paramyxovirus fusion machinery and would impact the design of therapeutic strategies. However, at present, we lack biochemical evidence for the existence of a novel HN/F complex that forms after F refolding has been initiated. Clearly, further investigation is required to clarify this putative new NDV triggering mechanism.
ENERGETICS IN PARAMYXOVIRUS FUSION TRIGGERING
Previous and recent models for F activation.
An extensive body of evidence suggests that the fusion proteins of HN-containing paramyxoviruses associate with HN only as a consequence of receptor engagement by the attachment protein. In addition, from a study using conformation-sensitive anti-F monoclonal antibodies, it was reported that the PIV5 F protein can maintain the prefusion state in the absence of the attachment protein (62). These findings led to the proposal that the HN/F interaction may lead to active destabilization of intrinsically stable prefusion F trimers, often referred to as the association or “provocateur” model (63). Quite unexpectedly, this model was also recently challenged by Porotto and colleagues, who claimed provocatively not only that hPIV3 HN may interact with F prior to receptor engagement (64) but also that this HN interaction is required to stabilize F before receptor binding (65). This proposal is very reminiscent of a “clamp” relationship between attachment and F protein oligomers that was long featured for H/G attachment protein-containing paramyxoviruses (outlined in more detail below). However, in the hPIV3 F study, F trimers expressed alone were still able to induce fusion at levels about 20% of those observed after HN/F coexpression, and importantly, fusion activity drastically increased when hPIV3 F was expressed in the presence of an irrelevant receptor binding protein (influenza HA) that tethered only effector and target plasma membranes. This implies that hPIV3 F is capable of maintaining a metastable prefusion conformation in the absence of HN (65). Further work is necessary to fully appreciate the new data set in the context of the extensive body of previous studies.
As we discuss above, multiple studies have demonstrated that attachment and F glycoproteins of H/G-carrying paramyxoviruses interact intracellularly with each other (53). This made it conceivable that the attachment protein may serve as a molecular scaffold that prevents premature refolding of intrinsically unstable prefusion F proteins. Upon receptor interaction of the attachment protein, prefusion F could then be released from preassembled H/G-F complexes and undergo spontaneous refolding. This hypothesis is typically referred to as the “clamp” model of paramyxovirus glycoprotein interaction. Since HN binds sialic acid-containing molecules and H/G interact with proteinaceous receptors, it was thought that different attachment protein-dependent fusion triggering mechanisms may reflect the distinct molecular nature of the different receptors (66–69).
Recent new insight into the contribution of morbillivirus and henipavirus attachment proteins to maintaining the F trimer in a prefusion conformation has altered this view. Using the conformation-sensitive pairs of anti-F monoclonal antibodies (MAbs) to probe the conformation of morbillivirus F expressed in the absence of H, it became apparent that the F trimer is inherently stable enough to maintain a prefusion conformation in the absence of the attachment protein (50, 60). Chan and colleagues obtained similar conclusions with Nipah and Hendra virus F proteins (70). Indeed, when membrane-bound henipavirus F was expressed in the absence of G, a strong reactivity with a MAb was retained, and this MAb was found to specifically bind to the prefusion F conformation (70). Thus, intracellular assembly of H with F, and likely G and F, is not required to stabilize F trimers in the prefusion state. With regard to the energetic basis for paramyxovirus membrane fusion, we propose that all paramyxovirus F proteins may employ a common mechanism for triggering, which does not rely on interaction with the attachment protein to stabilize the prefusion conformation. Rather, conformational changes of the attachment protein complex are required to alter the stalk/F interface, destabilizing prefusion F. Thus, assuming a fundamentally conserved role of the attachment protein stalk domain in F triggering within the Paramyxovirinae subfamily, we believe that stalk rearrangements affecting the interface with associated prefusion F represent the central, universally conserved step of fusion activation.
Thermodynamic control of the triggering of paramyxovirus envelope glycoproteins.
Using PIV5 F as a model system, Connolly and colleagues reported that heat can be used as surrogate for attachment protein-dependent F triggering and that the F conformation induced by heat treatment resembles that assumed when prefusion F is triggered physiologically (55, 62, 63). Fully consistent with this notion, our recent results obtained with the morbilliviruses and the data reported by Chan et al. for the henipaviruses demonstrated that brief heat shock is likewise sufficient to trigger structural rearrangements in F proteins of these two paramyxovirus genera. We also found that the antibody reactivity profile of heat-treated morbillivirus F complexes is indistinguishable from that of physiologically triggered postfusion F trimers (60).
Recent cellular and functional data furthermore indicate that thermodynamic control of paramyxovirus membrane fusion is not limited to the F protein but is equally wielded by the attachment protein complex. Nipah virus G undergoes conformational changes (monitored by a conformation-sensitive MAb) efficiently at 37°C but not at 4°C (71). Bose and colleagues showed that fusion mediated by coexpression of headless PIV5 HN with F is similar at 33°C and 37°C, while in the presence of full-length HN, limited fusion activity was observed at 33°C compared to 37°C (51).
Combined, these results are consistent with the proposal that paramyxovirus attachment proteins also feature a metastable pre-receptor-binding/pre-F-triggering state, which is governed by an inherent energy barrier that regulates the initiation of the conformational changes. The energy required for fusion could be provided by attachment protein engagement to membrane-bound receptors expressed on target cells.
Refined models for membrane fusion triggering.
Taken together, the above-mentioned findings let us propose a mechanism used by paramyxovirus attachment proteins that link receptor binding to F triggering and membrane fusion.
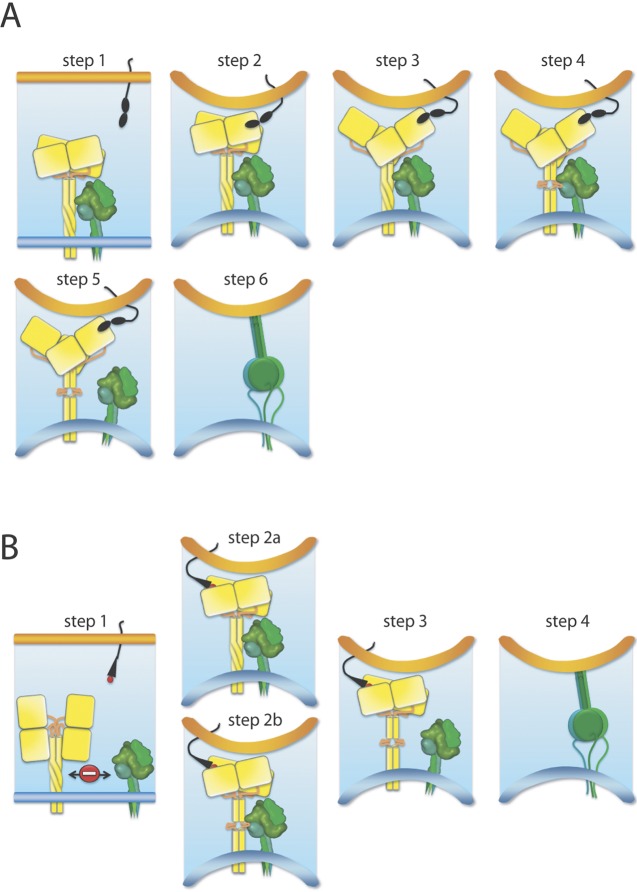
Preassembled H/G tetramer-F trimer hetero-oligomer complexes are exposed on the plasma membrane or on the viral envelope and engage in receptor binding (Fig. 4A). In the case of HN-carrying viruses, HN tetramers and F trimers associate as a consequence of HN-receptor contact and upward movement of the head domains (Fig. 4B). Reorganization of the tetrameric head assembly then translates into conformational changes in the membrane-proximal stalk section, which alters the stalk/F interface and may represent a universal trigger for F refolding. For morbillivirus H proteins, the available data support partial unwinding of the central stalk section, but the overall physical integrity of the tetramer is retained throughout the fusion triggering process (50, 56, 57). However, receptor-induced conformational changes within the HN stalk remain to be demonstrated. It is possible that simply uncovering the HN stalk through the receptor-mediated heads-up movement functionally replaces the morbillivirus stalk reorganization in F triggering. It will be interesting to see whether HN triggering-defective mutations can be identified that resemble those reported for MeV H (54, 59) which allow sustained interaction of HN/F after receptor binding but do not proceed to membrane fusion. Mechanistically, these stalk rearrangements may lower the relative F activation energy barrier (60, 63), which commits prefusion F trimers to the series of irreversible conformational changes into the postfusion conformation.
FIG 4 .
Models of receptor-induced conformational changes of the paramyxovirus attachment protein tetramer that are considered to result in F triggering. (A) Proposed model for morbilliviruses and henipaviruses. Prior to receptor binding, H/G tetrameric proteins associate with F trimers (step 1). The attachment protein then binds to its cognate cell surface receptor, which, in turn, leads to the tethering of the two opposing lipid bilayers (viral envelope and host cell plasma membrane), creating a fusion-competent microenvironment (step 2). As a result of receptor binding by the H/G head domains, the tetrameric heads undergo a “sliding” movement (step 3). Consequently, the central section of the tetrameric stalk unwinds/unfolds (step 4). Since the H/G stalk section that undergoes conformational changes is in short-range contact with F, the preassembled hetero-oligomeric H/G-F fusion complexes dissociate (step 5), and the destabilized or liberated F trimers subsequently undergo irreversible structural rearrangements (represented by F reaching the prehairpin structural intermediates; step 6). F conformational changes progress to the postfusion state, which ultimately leads to fusion pore formation (not shown). (B) Proposed model for rubulaviruses, avulaviruses, and respiroviruses. Prior to receptor binding, HN tetramers assume a four-heads-down conformation that (i) prevents functional hetero-oligomeric assembly with F trimers and (ii) stabilizes the stalk domain in a pre-receptor-binding state (step 1). Upon receptor binding, the four HN heads move up, which in turn enables F trimers to contact HN (step 2a). The HN stalk domain then refolds into the post-receptor-binding state (trigger-competent stalk domain) (step 3). Alternatively, receptor-induced movements may coincide with immediate stalks’ rearrangements and may be followed by F binding (step 2b). Common to both models, this post-receptor-binding trigger-competent central stalk section then destabilizes F trimers (step 3). As in panel A, these F trimers undergo irreversible structural rearrangements that lead to fusion pore formation (step 4). Prefusion F trimers are based on a high-resolution structural model that was morphed into a lower-resolution image using the Sculptor package. For the sake of clarity, only one receptor unit is represented in the cartoon. The host plasma membrane is in orange, and the viral envelope is in blue.
PERSPECTIVES
Membrane fusion machinery: a possible target for the development of antivirals.
Triggering the fusion machinery is an essential step in initiating paramyxovirus infection. The recent structural and functional advances open novel pathways toward the development of novel antivirals aiming at inhibiting the fusion machinery. One can envision designing molecules specifically targeting (i) the receptor-binding sites to impede receptor interaction, (ii) microdomains within the head and/or stalk domains to inhibit conformational changes that lead to F triggering, (iii) microdomains in prefusion F stabilizing the metastable state, (iv) microdomains exposed in one or more F structural intermediates to prevent completion of the F refolding cascade, or (v) putative but yet-to-be-determined host cell molecules that contribute to fusion pore formation and/or expansion. In conceptual support of the attachment protein stalk domain as a potential drug target, we found that a MAb recognizing a linear epitope in the CDV H stalk domain acts as a potent inhibitor of membrane fusion (our unpublished data).
ACKNOWLEDGMENTS
This work was supported, in part, by the Swiss National Science Foundation (reference no. 310030_132887; to P.P.) and Public Health Service grants AI083402 and AI071002 from the NIH/NIAID (to R.K.P.).
Footnotes
Citation Plattet P, Plemper RK. 2013. Envelope protein dynamics in paramyxovirus entry. mBio 4(4):e00413-13. doi:10.1128/mBio.00413-13.
REFERENCES
- 1. Chua KB, Bellini WJ, Rota PA, Harcourt BH, Tamin A, Lam SK, Ksiazek TG, Rollin PE, Zaki SR, Shieh W, Goldsmith CS, Gubler DJ, Roehrig JT, Eaton B, Gould AR, Olson J, Field H, Daniels P, Ling AE, Peters CJ, Anderson LJ, Mahy BW. 2000. Nipah virus: a recently emergent deadly paramyxovirus. Science 288:1432–1435 [DOI] [PubMed] [Google Scholar]
- 2. Murray K, Selleck P, Hooper P, Hyatt A, Gould A, Gleeson L, Westbury H, Hiley L, Selvey L, Rodwell B, Ketterer P. 1995. A morbillivirus that caused fatal disease in horses and humans. Science 268:94–97 [DOI] [PubMed] [Google Scholar]
- 3. Lamb RA, Parks GD. 2007. Paramyxoviridae: the viruses and their replication, p 1449–1496 In Knipe DM, Howley PM, Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 4. Plemper RK. 2011. Cell entry of enveloped viruses. Curr. Opin. Virol. 1:92–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yin HS, Wen X, Paterson RG, Lamb RA, Jardetzky TS. 2006. Structure of the parainfluenza virus 5 F protein in its metastable, prefusion conformation. Nature 439:38–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brindley MA, Plemper RK. 2010. Blue native PAGE and biomolecular complementation reveal a tetrameric or higher-order oligomer organization of the physiological measles virus attachment protein H. J. Virol. 84:12174–12184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Crennell S, Takimoto T, Portner A, Taylor G. 2000. Crystal structure of the multifunctional paramyxovirus hemagglutinin-neuraminidase. Nat. Struct. Biol. 7:1068–1074 [DOI] [PubMed] [Google Scholar]
- 8. Maar D, Harmon B, Chu D, Schulz B, Aguilar HC, Lee B, Negrete OA. 2012. Cysteines in the stalk of the Nipah virus G glycoprotein are located in a distinct subdomain critical for fusion activation. J. Virol. 86:6632–6642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Takimoto T, Taylor GL, Crennell SJ, Scroggs RA, Portner A. 2000. Crystallization of Newcastle disease virus hemagglutinin-neuraminidase glycoprotein. Virology 270:208–214 [DOI] [PubMed] [Google Scholar]
- 10. Yuan P, Thompson TB, Wurzburg BA, Paterson RG, Lamb RA, Jardetzky TS. 2005. Structural studies of the parainfluenza virus 5 hemagglutinin-neuraminidase tetramer in complex with its receptor, sialyllactose. Structure 13:803–815 [DOI] [PubMed] [Google Scholar]
- 11. Bowden TA, Crispin M, Harvey DJ, Aricescu AR, Grimes JM, Jones EY, Stuart DI. 2008. Crystal structure and carbohydrate analysis of Nipah virus attachment glycoprotein: a template for antiviral and vaccine design. J. Virol. 82:11628–11636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Colf LA, Juo ZS, Garcia KC. 2007. Structure of the measles virus hemagglutinin. Nat. Struct. Mol. Biol. 14:1227–1228 [DOI] [PubMed] [Google Scholar]
- 13. Hashiguchi T, Kajikawa M, Maita N, Takeda M, Kuroki K, Sasaki K, Kohda D, Yanagi Y, Maenaka K. 2007. Crystal structure of measles virus hemagglutinin provides insight into effective vaccines. Proc. Natl. Acad. Sci. U. S. A. 104:19535–19540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hashiguchi T, Ose T, Kubota M, Maita N, Kamishikiryo J, Maenaka K, Yanagi Y. 2011. Structure of the measles virus hemagglutinin bound to its cellular receptor SLAM. Nat. Struct. Mol. Biol. 18:135–141 [DOI] [PubMed] [Google Scholar]
- 15. Lawrence MC, Borg NA, Streltsov VA, Pilling PA, Epa VC, Varghese JN, McKimm-Breschkin JL, Colman PM. 2004. Structure of the haemagglutinin-neuraminidase from human parainfluenza virus type III. J. Mol. Biol. 335:1343–1357 [DOI] [PubMed] [Google Scholar]
- 16. Santiago C, Celma ML, Stehle T, Casasnovas JM. 2010. Structure of the measles virus hemagglutinin bound to the CD46 receptor. Nat. Struct. Mol. Biol. 17:124–129 [DOI] [PubMed] [Google Scholar]
- 17. Yuan P, Swanson KA, Leser GP, Paterson RG, Lamb RA, Jardetzky TS. 2011. Structure of the Newcastle disease virus hemagglutinin-neuraminidase (HN) ectodomain reveals a four-helix bundle stalk. Proc. Natl. Acad. Sci. U. S. A. 108:14920–14925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Villar E, Barroso IM. 2006. Role of sialic acid-containing molecules in paramyxovirus entry into the host cell: a minireview. Glycoconj. J. 23:5–17 [DOI] [PubMed] [Google Scholar]
- 19. Bonaparte MI, Dimitrov AS, Bossart KN, Crameri G, Mungall BA, Bishop KA, Choudhry V, Dimitrov DS, Wang LF, Eaton BT, Broder CC. 2005. Ephrin-B2 ligand is a functional receptor for Hendra virus and Nipah virus. Proc. Natl. Acad. Sci. U. S. A. 102:10652–10657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dörig RE, Marcil A, Chopra A, Richardson CD. 1993. The human CD46 molecule is a receptor for measles virus (Edmonston strain). Cell 75:295–305 [DOI] [PubMed] [Google Scholar]
- 21. Mühlebach MD, Mateo M, Sinn PL, Prüfer S, Uhlig KM, Leonard VH, Navaratnarajah CK, Frenzke M, Wong XX, Sawatsky B, Ramachandran S, McCray PB, Jr, Cichutek K, von Messling V, Lopez M, Cattaneo R. 2011. Adherens junction protein nectin-4 is the epithelial receptor for measles virus. Nature 480:530–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Negrete OA, Levroney EL, Aguilar HC, Bertolotti-Ciarlet A, Nazarian R, Tajyar S, Lee B. 2005. EphrinB2 is the entry receptor for Nipah virus, an emergent deadly paramyxovirus. Nature 436:401–405 [DOI] [PubMed] [Google Scholar]
- 23. Tatsuo H, Ono N, Tanaka K, Yanagi Y. 2000. Slam (CDw150) is a cellular receptor for measles virus. Nature 406:893–897 [DOI] [PubMed] [Google Scholar]
- 24. Noyce RS, Bondre DG, Ha MN, Lin LT, Sisson G, Tsao MS, Richardson CD. 2011. Tumor cell marker PVRL4 (nectin 4) is an epithelial cell receptor for measles virus. PLoS Pathog. 7:e1002240. 10.1371/journal.ppat.1002240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. de Vries RD, McQuaid S, van Amerongen G, Yuksel S, Verburgh RJ, Osterhaus AD, Duprex WP, de Swart RL. 2012. Measles immune suppression: lessons from the macaque model. PLoS Pathog. 8:e1002885. 10.1371/journal.ppat.1002885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lemon K, de Vries RD, Mesman AW, McQuaid S, van Amerongen G, Yuksel S, Ludlow M, Rennick LJ, Kuiken T, Rima BK, Geijtenbeek TB, Osterhaus AD, Duprex WP, de Swart RL. 2011. Early target cells of measles virus after aerosol infection of non-human primates. PLoS Pathog. 7:e1001263. 10.1371/journal.ppat.1001263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Leonard VH, Hodge G, Reyes-Del Valle J, McChesney MB, Cattaneo R. 2010. Measles virus selectively blind to signaling lymphocytic activation molecule (SLAM; CD150) is attenuated and induces strong adaptive immune responses in rhesus monkeys. J. Virol. 84:3413–3420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Leonard VH, Sinn PL, Hodge G, Miest T, Devaux P, Oezguen N, Braun W, McCray PB, Jr, McChesney MB, Cattaneo R. 2008. Measles virus blind to its epithelial cell receptor remains virulent in rhesus monkeys but cannot cross the airway epithelium and is not shed. J. Clin. Invest. 118:2448–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kato SI, Nagata K, Takeuchi K. 2012. Cell tropism and pathogenesis of measles virus in monkeys. Front. Microbiol. 3:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Noyce RS, Richardson CD. 2012. Nectin 4 is the epithelial cell receptor for measles virus. Trends Microbiol. 20:429–439 [DOI] [PubMed] [Google Scholar]
- 31. Negrete OA, Chu D, Aguilar HC, Lee B. 2007. Single amino acid changes in the Nipah and Hendra virus attachment glycoproteins distinguish ephrinB2 from ephrinB3 usage. J. Virol. 81:10804–10814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lamb RA. 1993. Paramyxovirus fusion: a hypothesis for changes. Virology 197:1–11 [DOI] [PubMed] [Google Scholar]
- 33. Navaratnarajah CK, Oezguen N, Rupp L, Kay L, Leonard VH, Braun W, Cattaneo R. 2011. The heads of the measles virus attachment protein move to transmit the fusion-triggering signal. Nat. Struct. Mol. Biol. 18:128–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xu K, Rajashankar KR, Chan YP, Himanen JP, Broder CC, Nikolov DB. 2008. Host cell recognition by the henipaviruses: crystal structures of the Nipah G attachment glycoprotein and its complex with ephrin-B3. Proc. Natl. Acad. Sci. U. S. A. 105:9953–9958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zaitsev V, von Izstein D, Groves D, Kiefel M, Takimoto T, Portner A, Taylor G. 2004. Second sialic acid binding site in Newcastle disease virus hemagglutinin-neuraminidase: implications for fusion. J. Virol. 78:3733–3741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bowden TA, Aricescu AR, Gilbert RJ, Grimes JM, Jones EY, Stuart DI. 2008. Structural basis of Nipah and Hendra virus attachment to their cell-surface receptor ephrin-B2. Nat. Struct. Mol. Biol. 15:567–572 [DOI] [PubMed] [Google Scholar]
- 37. Zhang X, Lu G, Qi J, Li Y, He Y, Xu X, Shi J, Zhang CW, Yan J, Gao GF. 2013. Structure of measles virus hemagglutinin bound to its epithelial receptor nectin-4. Nat. Struct. Mol. Biol. 20:67–72 [DOI] [PubMed] [Google Scholar]
- 38. Tahara M, Takeda M, Shirogane Y, Hashiguchi T, Ohno S, Yanagi Y. 2008. Measles virus infects both polarized epithelial and immune cells by using distinctive receptor-binding sites on its hemagglutinin. J. Virol. 82:4630–4637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Heminway BR, Yu Y, Galinski MS. 1994. Paramyxovirus mediated cell fusion requires co-expression of both the fusion and hemagglutinin-neuraminidase glycoproteins. Virus Res. 31:1–16 [DOI] [PubMed] [Google Scholar]
- 40. Hu XL, Ray R, Compans RW. 1992. Functional interactions between the fusion protein and hemagglutinin-neuraminidase of human parainfluenza viruses. J. Virol. 66:1528–1534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Morrison T, McQuain C, McGinnes L. 1991. Complementation between avirulent Newcastle disease virus and a fusion protein gene expressed from a retrovirus vector: requirements for membrane fusion. J. Virol. 65:813–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Deng R, Mirza AM, Mahon PJ, Iorio RM. 1997. Functional chimeric HN glycoproteins derived from Newcastle disease virus and human parainfluenza virus-3. Arch. Virol. Suppl. 13:115–130 [DOI] [PubMed] [Google Scholar]
- 43. Deng R, Wang Z, Mirza AM, Iorio RM. 1995. Localization of a domain on the paramyxovirus attachment protein required for the promotion of cellular fusion by its homologous fusion protein spike. Virology 209:457–469 [DOI] [PubMed] [Google Scholar]
- 44. Lee JK, Prussia A, Paal T, White LK, Snyder JP, Plemper RK. 2008. Functional interaction between paramyxovirus fusion and attachment proteins. J. Biol. Chem. 283:16561–16572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tanabayashi K, Compans RW. 1996. Functional interaction of paramyxovirus glycoproteins: identification of a domain in Sendai virus HN which promotes cell fusion. J. Virol. 70:6112–6118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tsurudome M, Kawano M, Yuasa T, Tabata N, Nishio M, Komada H, Ito Y. 1995. Identification of regions on the hemagglutinin-neuraminidase protein of human parainfluenza virus type 2 important for promoting cell fusion. Virology 213:190–203 [DOI] [PubMed] [Google Scholar]
- 47. Wang Z, Mirza AM, Li J, Mahon PJ, Iorio RM. 2004. An oligosaccharide at the C-terminus of the F-specific domain in the stalk of the human parainfluenza virus 3 hemagglutinin-neuraminidase modulates fusion. Virus Res. 99:177–185 [DOI] [PubMed] [Google Scholar]
- 48. Bose S, Welch BD, Kors CA, Yuan P, Jardetzky TS, Lamb RA. 2011. Structure and mutagenesis of the parainfluenza virus 5 hemagglutinin-neuraminidase stalk domain reveals a four-helix bundle and the role of the stalk in fusion promotion. J. Virol. 85:12855–12866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mahon PJ, Mirza AM, Musich TA, Iorio RM. 2008. Engineered intermonomeric disulfide bonds in the globular domain of Newcastle disease virus hemagglutinin-neuraminidase protein: implications for the mechanism of fusion promotion. J. Virol. 82:10386–10396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Brindley MA, Takeda M, Plattet P, Plemper RK. 2012. Triggering the measles virus membrane fusion machinery. Proc. Natl. Acad. Sci. U. S. A. 109:E3018–E3027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bose S, Zokarkar A, Welch BD, Leser GP, Jardetzky TS, Lamb RA. 2012. Fusion activation by a headless parainfluenza virus 5 hemagglutinin-neuraminidase stalk suggests a modular mechanism for triggering. Proc. Natl. Acad. Sci. U. S. A. 109:E2625–E2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Paterson RG, Johnson ML, Lamb RA. 1997. Paramyxovirus fusion (F) protein and hemagglutinin-neuraminidase (HN) protein interactions: intracellular retention of F and HN does not affect transport of the homotypic HN or F protein. Virology 237:1–9 [DOI] [PubMed] [Google Scholar]
- 53. Plemper RK, Hammond AL, Cattaneo R. 2001. Measles virus envelope glycoproteins hetero-oligomerize in the endoplasmic reticulum. J. Biol. Chem. 276:44239–44246 [DOI] [PubMed] [Google Scholar]
- 54. Paal T, Brindley MA, St Clair C, Prussia A, Gaus D, Krumm SA, Snyder JP, Plemper RK. 2009. Probing the spatial organization of measles virus fusion complexes. J. Virol. 83:10480–10493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Paterson RG, Russell CJ, Lamb RA. 2000. Fusion protein of the paramyxovirus SV5: destabilizing and stabilizing mutants of fusion activation. Virology 270:17–30 [DOI] [PubMed] [Google Scholar]
- 56. Ader N, Brindley MA, Avila M, Origgi FC, Langedijk JP, Örvell C, Vandevelde M, Zurbriggen A, Plemper RK, Plattet P. 2012. Structural rearrangements of the central region of the morbillivirus attachment protein stalk domain trigger F protein refolding for membrane fusion. J. Biol. Chem. 287:16324–16334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Navaratnarajah CK, Negi S, Braun W, Cattaneo R. 2012. Membrane fusion triggering: three modules with different structure and function in the upper half of the measles virus attachment protein stalk. J. Biol. Chem. 287:38543–38551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Vongpunsawad S, Oezgun N, Braun W, Cattaneo R. 2004. Selectively receptor-blind measles viruses: identification of residues necessary for SLAM- or CD46-induced fusion and their localization on a new hemagglutinin structural model. J. Virol. 78:302–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Corey EA, Iorio RM. 2007. Mutations in the stalk of the measles virus hemagglutinin protein decrease fusion but do not interfere with virus-specific interaction with the homologous fusion protein. J. Virol. 81:9900–9910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ader N, Brindley M, Avila M, Örvell C, Horvat B, Hiltensperger G, Schneider-Schaulies J, Vandevelde M, Zurbriggen A, Plemper RK, Plattet P. 2013. Mechanism for active membrane fusion triggering by morbillivirus attachment protein. J. Virol. 87:314–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Porotto M, DeVito I, Palmer SG, Jurgens EM, Yee JL, Yokoyama CC, Pessi A, Moscona A. 2011. Spring-loaded model revisited: paramyxovirus fusion requires engagement of a receptor binding protein beyond initial triggering of the fusion protein. J. Virol. 85:12867–12880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Connolly SA, Leser GP, Yin HS, Jardetzky TS, Lamb RA. 2006. Refolding of a paramyxovirus F protein from prefusion to postfusion conformations observed by liposome binding and electron microscopy. Proc. Natl. Acad. Sci. U. S. A. 103:17903–17908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Connolly SA, Leser GP, Jardetzky TS, Lamb RA. 2009. Bimolecular complementation of paramyxovirus fusion and hemagglutinin-neuraminidase proteins enhances fusion: implications for the mechanism of fusion triggering. J. Virol. 83:10857–10868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Porotto M, Palmer SG, Palermo LM, Moscona A. 2012. Mechanism of fusion triggering by human parainfluenza virus type III: communication between viral glycoproteins during entry. J. Biol. Chem. 287:778–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Porotto M, Salah ZW, Gui L, DeVito I, Jurgens EM, Lu H, Yokoyama CC, Palermo LM, Lee K, Moscona A. 2012. Regulation of paramyxovirus fusion activation: the hemagglutinin-neuraminidase protein stabilizes the fusion protein in a pre-triggered state. J. Virol. 86:12838–12848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Chang A, Dutch RE. 2012. Paramyxovirus fusion and entry: multiple paths to a common end. Viruses 4:613–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Iorio RM, Mahon PJ. 2008. Paramyxoviruses: different receptors—different mechanisms of fusion. Trends Microbiol. 16:135–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lee B, Ataman ZA. 2011. Modes of paramyxovirus fusion: a henipavirus perspective. Trends Microbiol. 19:389–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Plemper RK, Brindley MA, Iorio RM. 2011. Structural and mechanistic studies of measles virus illuminate paramyxovirus entry. PLoS Pathog. 7:e1002058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Chan YP, Lu M, Dutta S, Yan L, Barr J, Flora M, Feng YR, Xu K, Nikolov DB, Wang LF, Skiniotis G, Broder CC. 2012. Biochemical, conformational, and immunogenic analysis of soluble trimeric forms of henipavirus fusion glycoproteins. J. Virol. 86:11457–11471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Aguilar HC, Ataman ZA, Aspericueta V, Fang AQ, Stroud M, Negrete OA, Kammerer RA, Lee B. 2009. A novel receptor-induced activation site in the Nipah virus attachment glycoprotein (G) involved in triggering the fusion glycoprotein (F). J. Biol. Chem. 284:1628–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]