ABSTRACT
Treatment with streptomycin enhances the growth of human commensal Escherichia coli isolates in the mouse intestine, suggesting that the resident microbial community (microbiota) can inhibit the growth of invading microbes, a phenomenon known as “colonization resistance.” However, the precise mechanisms by which streptomycin treatment lowers colonization resistance remain obscure. Here we show that streptomycin treatment rendered mice more susceptible to the development of chemically induced colitis, raising the possibility that the antibiotic might lower colonization resistance by changing mucosal immune responses rather than by preventing microbe-microbe interactions. Investigation of the underlying mechanism revealed a mild inflammatory infiltrate in the cecal mucosa of streptomycin-treated mice, which was accompanied by elevated expression of Nos2, the gene that encodes inducible nitric oxide synthase. In turn, this inflammatory response enhanced the luminal growth of E. coli by nitrate respiration in a Nos2-dependent fashion. These data identify low-level intestinal inflammation as one of the factors responsible for the loss of resistance to E. coli colonization after streptomycin treatment.
IMPORTANCE
Our intestine is host to a complex microbial community that confers benefits by educating the immune system and providing niche protection. Perturbation of intestinal communities by streptomycin treatment lowers “colonization resistance” through unknown mechanisms. Here we show that streptomycin increases the inflammatory tone of the intestinal mucosa, thereby making the bowel more susceptible to dextran sulfate sodium treatment and boosting the Nos2-dependent growth of commensal Escherichia coli by nitrate respiration. These data point to the generation of alternative electron acceptors as a by-product of the inflammatory host response as an important factor responsible for lowering resistance to colonization by facultative anaerobic bacteria such as E. coli.
Introduction
Human isolates of Escherichia coli are generally poor colonizers of the murine gastrointestinal tract, but robust growth in the large intestine is observed in streptomycin-treated mice (1, 2). The streptomycin-treated mouse model has been used widely to study whether the elaboration of smooth lipopolysaccharide (3, 4), attachment mediated by fimbriae (5–8), motility (9), and the utilization of certain nutrients and trace elements (10–17) are properties important for the in vivo growth of commensal E. coli. It also provides a model to study carbon utilization (14, 16, 18–20) and the role of Shiga-like toxin (21–26) during infection with enterohemorrhagic E. coli. However, the mechanism by which streptomycin treatment promotes colonization with E. coli remains elusive.
Marjorie Bohnhoff and coworkers first introduced the concept that microbial communities in the intestine (the microbiota) can inhibit the growth of invading pathogens by conferring “colonization resistance” after noticing that the intestinal tracts of mice pretreated with streptomycin become highly susceptible to Salmonella enterica serovar Enteritidis infection (27, 28). Pretreatment of mice with streptomycin enhances the luminal growth of S. enterica serovar Typhimurium (S. Typhimurium) in the murine cecum by lowering colonization resistance (29). In addition, the antibiotic exacerbates the severity of S. Typhimurium-induced colitis (30). The enhanced severity of colitis in streptomycin-treated mice infected with S. Typhimurium has been ascribed to a decreased “immune resistance,” a term coined recently to describe specific host-commensal interactions associated with colonization resistance that confer protection against mucosal damage (31). Streptomycin treatment is proposed to reduce colonization resistance by lowering growth competition between the microbiota and incoming S. Typhimurium (32). Interestingly, conditions that lower the resistance to colonization by commensal E. coli also lower immune resistance to S. Typhimurium-induced colitis (33), but the identities of factors that contribute to these phenomena are obscure.
Here we describe the identification and characterization of one of the mechanisms responsible for loss of resistance to E. coli colonization after streptomycin treatment.
RESULTS
Streptomycin treatment lowers immune resistance to infectious and noninfectious colitis.
Pretreatment of mice with streptomycin (a single dose of 20 mg/animal) enhances the susceptibility of mice to the development S. Typhimurium-induced colitis (30) (Fig. 1A), presumably because the antibiotic lowers growth competition between the microbiota and the incoming pathogen (32). Should streptomycin enhance inflammation because it lowers microbe-microbe interactions that limit pathogen growth, the antibiotic would not be expected to increase the susceptibility of mice to the development of colitis caused by exposure to chemicals. To test this prediction, mice received a subpathological concentration of dextran sulfate sodium (DSS) in their drinking water (1%) for 8 days. Four days after being switched to DSS-containing drinking water, some mice were treated with streptomycin (a single dose of 20 mg/animal) and their ceca were collected 96 h later. Treatment of mice with streptomycin and/or 1% DSS did not result in significant weight loss (see Table S1 in the supplemental material). Drinking water containing a subpathological concentration of DSS did not induce marked gut inflammation in mice that were not treated with streptomycin (Fig. 2A), which was consistent with a previous report of a study that used this dose to identify conditions that exaggerate susceptibility to DSS (34). Surprisingly, streptomycin-treated mice that received a subpathological concentration of DSS developed mild inflammation, as indicated by a significantly (P < 0.01) increased histopathology score in the cecum (P < 0.05) (Fig. 2A). Thus, in addition to the known enhancement of the susceptibility to S. Typhimurium-induced colitis (30), these data suggest that streptomycin treatment also lowers immune resistance to DSS-induced mucosal damage. Collectively, these data suggest that streptomycin treatment makes the bowel generally more irritable. Importantly, while it is conceivable that inhibition of pathogen growth by microbe-microbe interactions, such as competition for nutrients or metabolic exclusion, explains the enhanced susceptibility of streptomycin-treated mice to the development of S. Typhimurium-induced colitis, these potential mechanisms did not provide a compelling explanation for the increased susceptibility of streptomycin-treated mice to DSS-induced colitis.
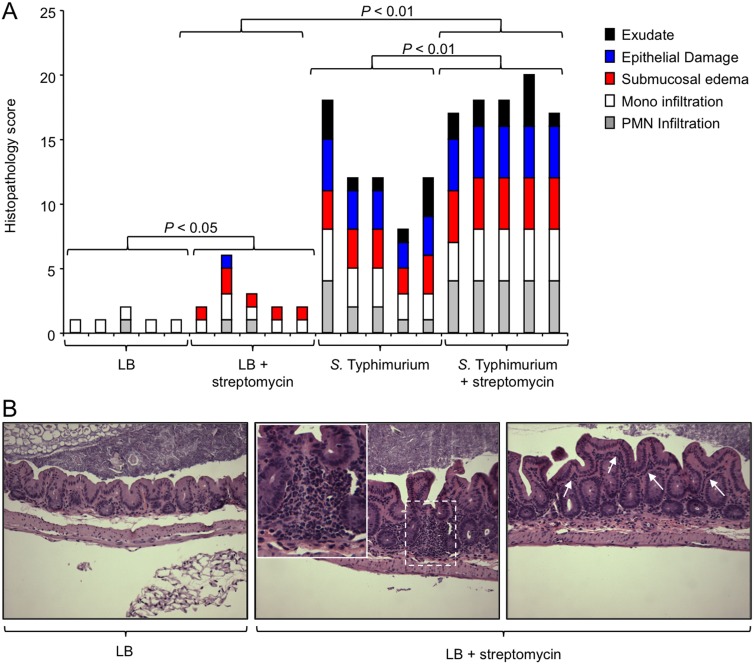
FIG 1 .
Streptomycin treatment is associated with a modest increase in the severity of histopathological changes. Mice received streptomycin or remained untreated. One day later, mice were inoculated with sterile medium (LB) or infected with S. Typhimurium. Three days after infection, their ceca were collected for analysis. (A) A veterinary pathologist performed blinded scoring of hematoxylin-eosin-stained sections of the cecal mucosa for histopathological changes. Each bar represents the combined histopathology score of one individual animal. The statistical significances of differences between groups are indicated above the brackets. (B) Representative images of sections from mice receiving LB (left) or LB plus streptomycin (middle and right). Arrows indicate mild diffuse inflammatory infiltrates in the lamina propria (right). A focal inflammatory infiltrate in the cecal mucosa is shown in the dashed square, and a larger magnification of this region is presented in the inset. Mono, monocyte; PMN, polymorphonuclear neutrophil.
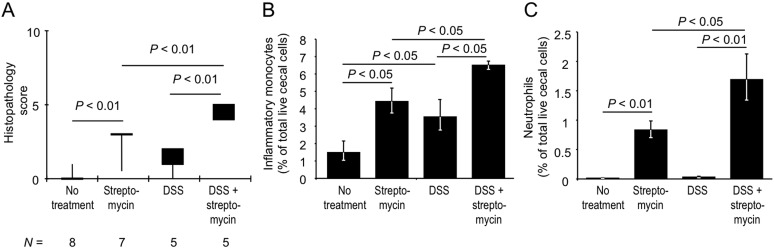
FIG 2 .
Streptomycin induces colitis in mice treated with a subpathological concentration of DSS. Mice were housed normally or received drinking water supplemented with 1% DSS. Four days after the beginning of DSS treatment, some mice received a single dose of streptomycin and their ceca were collected 4 days later for analysis. (A) A veterinary pathologist performed blinded scoring of hematoxylin-eosin-stained sections of the cecal mucosa for histopathological changes. Boxes in whisker plots represent the second and third quartiles of combined histopathology scores, while lines indicate the first and fourth quartiles. The number of animals (N) in each group is indicated at the bottom. (B and C) Analysis of cecal cell suspensions from at least four animals per group was performed by flow cytometry. Bars represent the geometric means ± the standard errors of the numbers of inflammatory monocytes (B) and neutrophils (C) expressed as percentages of the total number of live cecal cells (A to C). The statistical significances of differences between groups are indicated above the bars.
Streptomycin treatment increases the inflammatory tone of the cecal mucosa.
Interestingly, we noted that sections of the cecal mucosa collected from mice treated with streptomycin, followed by inoculation with sterile medium (LB broth), exhibited mild inflammatory changes, unlike those from mice inoculated with sterile medium alone (P < 0.05) (Fig. 1A). These inflammatory changes included mild diffuse inflammatory infiltrates in the lamina propria and occasional focal infiltrates in the mucosa (Fig. 1B). To further investigate possible inflammatory changes induced by streptomycin treatment, we next analyzed the composition of cells present in the cecal mucosa by using flow cytometry.
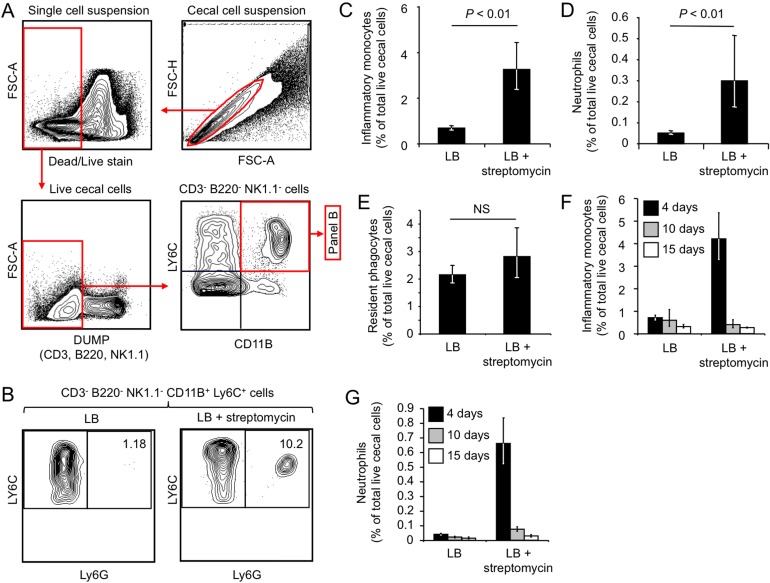
Mice were treated with streptomycin (a single dose of 20 mg/animal) and sterile medium (LB broth) or with sterile medium alone, and a single-cell suspension was generated from the cecum 96 h later. After doublet elimination and exclusion of dead cells (dead/live aqua staining), live cecal cells were gated for a population that was negative for the T cell marker CD3 (cluster of differentiation 3), the B cell marker B220, and the NK/NKT cell marker NK1.1. Inflammatory phagocytes present in the CD3− B220− NK1.1− population were identified as cells expressing CD11B, a chain of CR3 (complement receptor 3), and Ly6C (lymphocyte antigen 6 complex, locus C), a marker expressed by neutrophils and inflammatory monocytes but absent from tissue-resident macrophages and tissue-resident dendritic cells (Fig. 3A). Finally, CD3− B220− NK1.1− CD11B+ Ly6C+ phagocytes were further differentiated into neutrophils and monocytes on the basis of the expression of the neutrophil marker Ly6G (Fig. 3B). The Ly6G/Ly6C markers are superior to Gr-1 (granulocyte differentiation antigen 1) for the identification of neutrophils/monocytes, and previous work suggests that the mouse macrophage marker F4/80 is not required for the identification of these subsets (35).
FIG 3 .
Streptomycin treatment elicits infiltrates of inflammatory monocytes and neutrophils in the cecal mucosa. (A) Gating strategy for analysis of cecal cell suspensions. After doublet elimination (top right), live cells were gated (top left) and CD3+ B220+ NK1.1+ cells were eliminated by using a dump channel (bottom left). CD3− B220− NK1.1− cells were then analyzed for CD11B and Ly6C expression (bottom right). (B) CD3− B220− NK1.1− CD11B+ Ly6C+ phagocytes were separated into Ly6G− cells (inflammatory monocytes) and Ly6G+ cells (neutrophils) (C to G). Bars represent the geometric means ± the standard errors of the numbers of inflammatory monocytes (C and F), neutrophils (D and G), and resident phagocytes (E) expressed as percentages of the total number of live cecal cells (C to E). The statistical significance of the difference between groups is indicated at the top of each graph. NS, not statistically significantly different. For panels A to E, the ceca of groups of mice (n = 9) were collected 4 days after streptomycin treatment, and for panels F and G, the ceca of groups of mice (n = 3) were collected 4 (black bars), 10 (gray bars), or 15 (white bars) days after streptomycin treatment.
Remarkably, streptomycin treatment significantly (P < 0.01) increased the fraction of live cecal cells that expressed markers of inflammatory monocytes (i.e., CD3− B220− NK1.1− CD11B+ Ly6C+ Ly6G− cells) (Fig. 3C). Furthermore, streptomycin treatment was associated with a significant (P < 0.01) increase in the fraction of neutrophils (i.e., CD3− B220− NK1.1− CD11B+ Ly6C+ Ly6G+ cells) present within the live cecal cell suspension (Fig. 3B and D). In contrast, the number of resident phagocytes (i.e., CD3− B220− NK1.1− CD11B+ Ly6C− Ly6G− cells) present in the live cecal cell population remained unchanged after streptomycin treatment (Fig. 3E). We next repeated the experiment to monitor inflammatory changes at later time points after streptomycin treatment. The results revealed that the infiltrates of inflammatory monocytes (Fig. 3F) and neutrophils (Fig. 3G) were relatively short-lived, because the fraction of both cell types in live cecal cell suspensions returned to background levels within 15 days after streptomycin treatment.
Collectively, our results suggested that a single dose of streptomycin increased the inflammatory tone of the cecal mucosa temporarily, which was characterized by an infiltrate with neutrophils and inflammatory monocytes that peaked a few days after antibiotic treatment.
Streptomycin induces colitis in mice treated with a subpathological concentration of DSS.
We next analyzed phagocyte populations in mice treated with a subpathological concentration of DSS by using flow cytometry, because this method provides a higher resolution for the characterization of mild inflammatory changes than histopathology scoring does (Fig. 2A). Streptomycin-treated mice (a single dose of 20 mg/animal) receiving a subpathological concentration of DSS (1%) harbored significantly (P < 0.05) greater fractions of inflammatory monocytes (Fig. 2B) and neutrophils (Fig. 2C) in their live cecal cell population than did mice receiving streptomycin alone or mice receiving a subpathological concentration of DSS alone. These results further corroborated our observation that streptomycin treatment lowers immune resistance to DSS-induced mucosal damage (Fig. 2A).
Streptomycin-treated mice exhibit a marked increase in mucosal Nos2 expression.
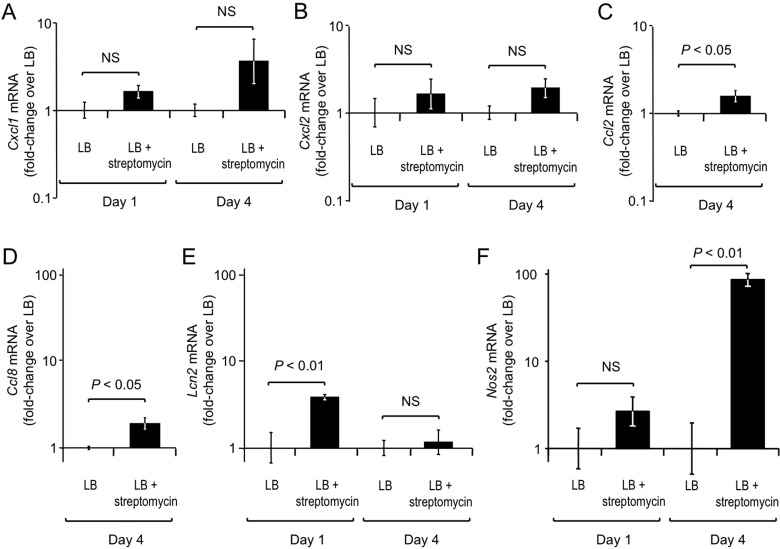
To start investigating whether there is a mechanistic link between streptomycin-induced inflammation and reduced resistance to E. coli colonization, we determined whether streptomycin treatment (a single dose of 20 mg/animal) altered the expression of inflammatory markers in the cecal mucosa by quantitative real-time PCR (Fig. 4). Streptomycin treatment resulted in a modest increase in mRNA levels of Kc (also known as Cxcl1) (Fig. 4A), which encodes the neutrophil chemoattractant KC (keratinocyte-derived cytokine), and Mip2 (also known as Cxcl2) (Fig. 4B), which encodes the neutrophil chemoattractant MIP2 (macrophage inflammatory protein 2), but these differences did not reach statistical significance. There was a small but significant (P < 0.05) increase in mRNA levels of Mcp1 (also known as Ccl2) (Fig. 4C), which encodes MCP-1 (monocyte chemoattractant protein 1), and Mcp2 (also known as Ccl8) (Fig. 4D), which encodes MCP-2, after streptomycin treatment. The small increases in the expression of genes that encode chemoattractants that were observed after streptomycin treatment were consistent with the mild infiltrate of neutrophils (Fig. 3B and D) and inflammatory monocytes (Fig. 3C) detected in live cecal cell suspensions.
FIG 4 .
Streptomycin treatment markedly enhances mucosal Nos2 mRNA levels. Groups of mice (n = 4) were mock treated (LB) or streptomycin treated (LB plus streptomycin), and RNA was extracted from their ceca 1 or 4 days after inoculation. Transcript levels of Cxcl1 (A), Cxcl2 (B), Ccl2 (C), Ccl8 (D), Lcn2 (E), and Nos2 (F) were determined by quantitative real-time PCR. Bars represent the geometric means ± the standard errors of n-fold changes in mRNA levels of the streptomycin-treated group versus the mock-treated group at each time point. The statistical significance of differences between groups is indicated at the top of each graph. NS, not statistically significantly different.
Next, we investigated the expression of other genes whose mRNA levels are markedly increased during intestinal inflammation (36, 37), including Lcn2, which encodes the antimicrobial protein lipocalin-2, and Nos2, which encodes inducible nitric oxide (NO) synthase (iNOS). Consistent with the modest increase in mRNA levels observed for other markers of inflammation, we observed a small (3.8-fold) but significant (P < 0.01) increase in the expression of Lcn2 in the cecal mucosa at 1 day after streptomycin treatment (Fig. 4E). Remarkably, levels of Nos2 mRNA were, on average, 85.6-fold elevated at 96 h after streptomycin treatment (P < 0.01) (Fig. 4F). This marked increase in the abundance of Nos2 mRNA was of particular interest, because iNOS has recently been implicated in enhanced nitrate production in the intestinal lumen, thereby enhancing the growth of E. coli through anaerobic nitrate respiration (38). Thus, analysis of the expression of genes that encode inflammatory markers suggested a possible mechanistic link between the streptomycin-induced increase in Nos2 mRNA levels (Fig. 4F) and the reduced resistance of mice to E. coli colonization.
Streptomycin treatment enhances the growth of E. coli by nitrate respiration.
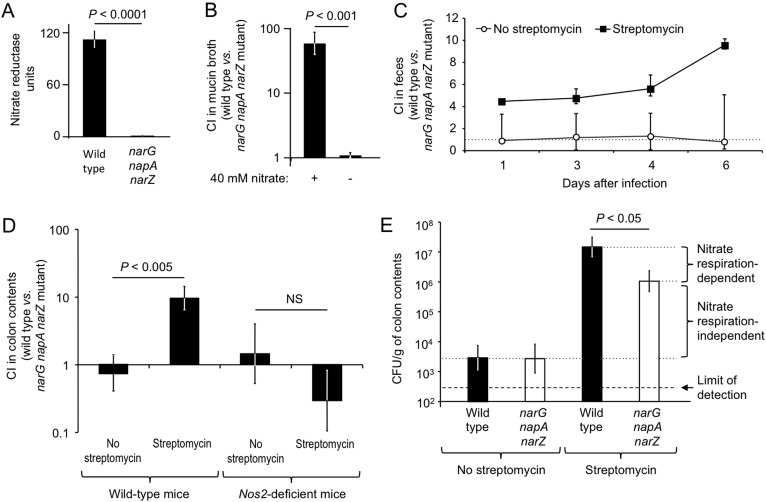
To test the hypothesis that a streptomycin-induced increase in Nos2 mRNA levels fuels the growth of E. coli in the murine large intestine by nitrate respiration, we used the commensal E. coli strain Nissle 1917. E. coli possesses three nitrate reductases encoded by the narGHJI, narZYWV, and napFDAGHBC operons (39). Inactivation of the narG, napA, and narZ genes in E. coli Nissle 1917 resulted in loss of nitrate reductase activity (Fig. 5A). To mimic the growth conditions in the lumen of the large bowel, mucin broth (i.e., hog mucin dissolved in a buffered solution) was inoculated with an equal mixture of streptomycin-resistant E. coli Nissle 1917 (CAL225) and a nitrate respiration-deficient narG napA narZ triple mutant (CAL222) and incubated overnight anaerobically. The two strains were recovered in equal numbers after anaerobic growth in mucin broth, suggesting that inactivation of the narG, napA, and narZ genes did not impair the growth of E. coli in the absence of nitrate. However, the nitrate respiration-deficient narG napA narZ triple mutant was outcompeted by the wild-type strain during competitive anaerobic growth in mucin broth supplemented with nitrate (P < 0.001) (Fig. 5B). These and previous data (17) suggest that nitrate respiration confers a fitness advantage upon E. coli under growth conditions that mimic those encountered in the large intestine.
FIG 5 .
Streptomycin boosts the Nos2-dependent growth of E. coli through nitrate respiration. (A) Nitrate reductase activities of E. coli Nissle 1917 (wild type) and an isogenic nitrate respiration-deficient mutant (narG napA narZ mutant). (B) Competitive anaerobic growth of the wild type and narG napA narZ mutant in mucin broth in the presence (+) or absence (−) of 40 mM nitrate. (C) Competitive indices (CI) of the wild type and the narG napA narZ mutant recovered over time from feces of streptomycin-pretreated mice (black squares) or untreated mice (open circles). (D) Competitive indices (CI) of the wild type and the narG napA narZ mutant recovered from the colon contents of wild-type mice (C57BL/6) or Nos2-deficient mice 6 days after infection (B to D). Data represent the geometric means ± the standard errors of competitive indices (CI) of the wild type and the narG napA narZ mutant. (E) Absolute numbers of wild-type (black bars) and narG napA narZ mutant (white bars) E. coli CFU recovered from colon contents. The magnitudes of nitrate respiration-dependent and nitrate respiration-independent contributions to colonization resistance are indicated on the right. A dashed line indicates the limit of detection. (A, B, D, and E) The statistical significance of differences between groups is indicated at the top of each graph. NS, not statistically significantly different.
We next investigated whether streptomycin treatment confers a growth advantage upon E. coli in the mouse large intestine. Groups of mice were inoculated with streptomycin (a single dose of 20 mg/animal) or left untreated and were inoculated 1 day later with an equal mixture of streptomycin-resistant E. coli Nissle 1917 (CAL225) and an isogenic nitrate respiration-deficient narG napA narZ triple mutant (CAL222). In the absence of streptomycin treatment, the wild type and the narG napA narZ mutant colonized the mouse intestine poorly and were recovered in equal numbers from the feces (Fig. 5C) and colon contents (Fig. 5D). In contrast, the wild-type strain was recovered in significantly (P < 0.005) higher numbers than the narG napA narZ mutant from streptomycin-treated mice 6 days after infection (Fig. 5D). These data suggested that nitrate respiration conferred a fitness advantage in the large intestines of streptomycin-treated mice but not during growth in the intestines of conventional mice. Furthermore, the overall numbers of E. coli bacteria recovered from the colon contents of streptomycin-treated mice were significantly (P < 0.01) higher than those recovered from untreated control mice (Fig. 5E), illustrating the colonization resistance-lowering effect of streptomycin. Nitrate respiration accounted for only part of this difference, pointing to the existence of additional mechanisms that contribute to the loss of colonization resistance after treatment with streptomycin.
Finally, we wanted to test whether there is a causal link between the streptomycin-induced increase in Nos2 mRNA levels (Fig. 4F) and the streptomycin-induced growth of E. coli by nitrate respiration. To this end, we inoculated groups of Nos2-deficient mice with streptomycin or left them untreated and inoculated them 1 day later with an equal mixture of streptomycin-resistant E. coli Nissle 1917 (CAL225) and an isogenic narG napA narZ mutant (CAL222). Remarkably, the wild type and the narG napA narZ mutant were recovered in equal numbers from the colon contents of Nos2-deficient mice, regardless of streptomycin treatment (Fig. 5D). These data suggested that the fitness advantage conferred by nitrate respiration in streptomycin-treated mice was Nos2 dependent, thereby providing a causal link between streptomycin-induced inflammation and the enhanced growth of (i.e., loss of resistance to colonization by) E. coli.
DISCUSSION
It is known that conditions of intestinal inflammation can lead to a microbial imbalance (dysbiosis) in the intestine that is characterized by a marked decrease in the representation of obligate anaerobic bacteria (i.e., members of the classes Bacteroidia and Clostridia) and an increased relative abundance of facultative anaerobic bacteria, which are commonly members of the family Enterobacteriaceae (reviewed in reference 40). In other words, intestinal inflammation is a known mechanism of reducing resistance to colonization by Enterobacteriaceae. However, since no overt inflammatory changes are detected as a consequence of treating mice with streptomycin (30, 41, 42), elimination of microbe-microbe interactions is commonly considered a more likely explanation for the associated loss of resistance to E. coli colonization. Specifically, streptomycin treatment is assumed to reduce the colonization resistance of mice because the antibiotic kills microbes that prevent colonization by incoming human E. coli isolates by metabolic exclusion or competition for nutrients, thereby freeing an otherwise occupied niche (reviewed in reference 31). This hypothesis suggests that colonization resistance is defined as the presence of a factor, namely, the presence of certain microbes that successfully compete with E. coli. In contrast, results presented here support the alternative view that colonization resistance is defined, at least in part, as the absence of a factor, namely, the absence of intestinal inflammation.
Our data suggest that streptomycin reduces colonization resistance, at least in part, by generating a mild inflammatory response that supports the growth of E. coli through nitrate respiration, thereby generating a new niche rather than clearing an existing niche of competitors. Inflammation induces the expression of iNOS (43), an enzyme that generates NO (44). NO can react with superoxide radicals produced during inflammation to yield peroxynitrite (ONOO−) (45), which can be converted to nitrate (NO3−) (46). Nitrate respiration enhances the growth of commensal E. coli in mice with DSS-induced colitis. In contrast, iNOS-deficient mice with DSS-induced colitis or normal mice that lack intestinal inflammation do not support the growth of E. coli by nitrate respiration (40). These data suggest that a by-product of intestinal inflammation is the generation of alternative electron acceptors, such as nitrate, that support the growth of E. coli and other members of the family Enterobacteriaceae by anaerobic respiration (38, 47, 48). Anaerobic respiration enables facultatively anaerobic bacteria to utilize nonfermentable carbon sources, thereby sidestepping the competition with obligate anaerobic bacteria that rely on the fermentation of carbohydrates and amino acids for growth (49). Through this mechanism, anaerobic respiration provides E. coli with a fitness advantage during growth in the inflamed intestine (38). Conversely, the absence of this fitness advantage in the healthy gut is manifested as resistance to E. coli colonization.
The unexpected identification of intestinal inflammation as one of the mechanisms by which streptomycin treatment reduces colonization resistance puts the spotlight on a previously unknown consequence of using this antibiotic. That is, how does streptomycin treatment induce the mild inflammatory changes in the cecal mucosa that ultimately drive bacterial growth by nitrate respiration? An increased inflammatory tone of the intestinal mucosa following antibiotic therapy is not without precedent, as mice exhibit increased macrophage and NK cell infiltration in the intestinal mucosa after oral metronidazole treatment (42). However, the elucidation of underlying mechanisms is complicated because factors that confer immune resistance to intestinal inflammation are not identical to those that enhance resistance to colonization by Enterobacteriaceae. For example, the abundance within the microbial community of bacteria belonging to the family Porphyromonadaceae (phylum Bacteroidetes) correlates with protection against S. Typhimurium-induced colitis (i.e., with increased immune resistance) but does not affect the growth of the pathogen in the intestinal lumen (i.e., it does not alter colonization resistance) (50). Our finding that streptomycin lowers immune resistance to DSS-induced colitis is of interest because the onset of irritable bowel syndrome, a condition characterized by low-level intestinal inflammation and diarrhea, often follows surgery or repeated courses of antibiotics (reviewed in reference 51). Thus, the streptomycin-treated mouse might be a model of human irritable bowel syndrome rather than a model of the healthy human gut.
Interestingly, transfer of a normal complex microbiota can restore the resistance of mice to colonization by S. Typhimurium (52). Consistent with a possible role for microbiota-induced mucosal responses in mediating colonization resistance, intestinal bacteria and their metabolic by-products are known to directly impact the repertoire and activity of intestinal immune cells (53–57). However, previous metagenomic studies did not succeed in establishing a correlation between reduced colonization resistance induced by antibiotic treatment and a decreased abundance of a specific bacterial species or family within the microbial community (32, 41, 50, 52). As a result, the identification of microbiota-host interactions underlying colonization resistance remains an important challenge for future research.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
S. Typhimurium strain IR715 (58) is a fully virulent, nalidixic acid-resistant derivative of wild-type isolate ATCC 14028 (American Type Culture Collection). Bacteria were cultured overnight aerobically at 37°C in Luria-Bertani (LB) broth or on LB plates (15 g/liter agar) with the appropriate antibiotics at the following concentrations: nalidixic acid, 0.05 mg/ml; carbenicillin, 0.1 mg/ml; kanamycin, 0.1 mg/ml.
Plasmid construction.
E. coli strains used for mouse infections contained a streptomycin resistance marker to facilitate recovery. To this end, a streptomycin resistance cassette (Ω) was isolated by digesting plasmid pHP45Ω (59) with the restriction enzyme BamHI (New England Biolabs). The Ω cassette was then inserted into the BamHI site of pWSK29 or pWSK129 (60) via ligation to yield plasmid pCAL62 or pCAL61, respectively. Plasmid pCAL62 was transformed into wild-type E. coli Nissle 1917 (61) to create CAL225, and plasmid pCAL61 was transformed into the isogenic napA narZ narG mutant (SW930) (38) to yield CAL222.
Competitive-growth assays.
For competition assays, bacteria were grown in mucin medium containing 0.25% type II porcine mucin (Sigma-Aldrich), 40 mM morpholinepropanesulfonic acid buffer, trace elements (62), and magnesium sulfate (265 mg/liter) dissolved in sterile water. Either sterile water as a negative control or 40 mM sodium nitrate (Sigma-Aldrich) was added to the medium immediately prior to inoculation with a 1:1 ratio of CAL225 and CAL222 at a total concentration of 105 CFU/ml. Bacteria were allowed to grow anaerobically for 16 h at 37°C (Bactron I anaerobic chamber; Sheldon Manufacturing, Cornelius, OR), and then the ratio of the two strains was determined by spreading serial 10-fold dilutions on LB agar plates containing the appropriate antibiotics. In vitro competition assays were performed in triplicate with cultures inoculated from different colonies. The competitive index was calculated by dividing the number of bacteria carrying the wild-type allele by the number of bacteria with the respective mutant allele and corrected by the ratio of these strains in the inoculum.
Nitrate reductase activity assay.
Overnight cultures of E. coli strains were diluted 1:50 in fresh LB broth supplemented with 40 mM sodium nitrate. Cultures were statically incubated for 3 h at 37°C, and the relative nitrate reductase activity was measured as described previously (63). Briefly, the nitrate reductase assay measures the reduction of nitrate to nitrite with methyl viologen as the electron donor. Nitrate is added to the reaction medium containing lysed bacterial cells, and nitrite (from nitrate reductase activity) is measured on the basis of its formation of a colored azo compound, which is quantified with a spectrophotometer. Assays were performed in triplicate with different colonies.
Animal experiments.
All animal experiments were approved by the Institutional Animal Care and Use Committee at the University of California, Davis, and performed according to Association for Assessment and Accreditation of Laboratory Animal Care guidelines. Female 10- to 12-week-old C57BL/6 mice (The Jackson Laboratory) or Nos2-deficient mice (stock no. 2609; The Jackson Laboratory) were used for mouse experiments. In brief, mice were inoculated intragastrically with streptomycin (0.1 ml of a 200-mg/ml solution in distilled water) as described previously (30). For infected groups, 24 h later, mice were inoculated intragastrically either with sterile LB broth or with bacteria (0.1 ml containing approximately 1 × 109 CFU/ml). At the indicated time points after infection, mice were euthanized and samples of their ceca were collected for the isolation of mRNA, for histopathological analysis, or for collection of cells. For bacteriologic analysis, cecal contents, colon contents, or fecal pellets were collected at the indicated time points and homogenized and serial 10-fold dilutions were spread on agar plates containing the appropriate antibiotics. For low-level DSS experiments, mice were given 1% DSS salt (catalogue no. 160110; MP Biomedicals) in their drinking water continuously for 8 days as described previously (34). DSS was replaced with fresh DSS solution every 2 to 3 days during this time. Mice were then inoculated with streptomycin at day 4 of DSS treatment as described above. Mouse body weights were determined daily.
Quantitative real-time PCR.
Transcript levels of murine genes in RNA isolated from the cecal mucosa were determined as described previously (36). For quantitative analysis of mRNA levels, 1 µg of RNA from each sample was reverse transcribed in a 50-µl volume (TaqMan reverse transcription reagent; Applied Biosystems), and 4 µl of cDNA was used for each real-time reaction. Real-time PCR was performed with Sybr green (Applied Biosystems) and a 7900HT fast real-time PCR system. The data were analyzed by a comparative cycle threshold method (Applied Biosystems). Increases in cytokine expression in infected mice were calculated relative to the average level of the respective cytokine in four control animals from the corresponding time point. A list of the genes analyzed in this study with the respective primers is provided in Table 1.
TABLE 1 .
Primers used for real-time PCR to quantify mRNA levels of murine genes
| Gene | Primer 1 | Primer 2 |
|---|---|---|
| Ccl2 | 5-ATTGGGATCATCTTGCTGGT-3′ | 5′ CCTGCTGTTCACAGTTGCC 3′ |
| Ccl8 | 5′ GAAGGGGGATCTTCAGCTTT 3′ | 5′ TCTTTGCCTGCTGCTCATAG 3′ |
| Lcn2 | 5′ ACATTTGTTCCAAGCTCCAGGGC 3′ | 5′ CATGGCGAACTGGTTGTAGTCCG 3′ |
| Cxcl1 | 5′ TGCACCCAAACCGAAGTCAT 3′ | 5′ TTGTCAGAAGCCAGCGTTCAC 3′ |
| Cxcl2 | 5′ AGTGAACTGCGCTGTCAATGC 3′ | 5′ AGGCAAACTTTTTGACCGCC 3′ |
| Nos2 | 5′ TTGGGTCTTGTTCACTCCACGG 3′ | 5′ CCTCTTTCAGGTCACTTTGGTAGG 3′ |
| Gapdh | 5′ TGTAGACCATGTAGTTGAGGTCA 3′ | 5′ AGGTCGGTGTGAACGGATTTG 3′ |
Histopathology.
Tissue samples were fixed in formalin, processed according to standard procedures for paraffin embedding as described previously (36), sectioned at 5 µm, and stained with hematoxylin and eosin. A veterinary pathologist scored inflammatory changes by blind sample analysis. Neutrophil and monocyte counts were determined by high-magnification (×400) microscopy, and numbers of cells in 10 microscopic fields were averaged for each animal.
Isolation of intestinal lymphocytes.
The isolation of intestinal lymphocytes for experiments has been described previously (64). To isolate cecal lymphocytes for measurements of in vivo cellular analysis, control and experimental mouse groups were sacrificed at the designated time points. The cecum and proximal colon were collected, and fat and connective tissue were removed. Intestinal sections were cut longitudinally from the proximal colon to the tip of the cecum. The cecal content was removed by gentle scraping with the flat edge of scissors. The sections were subsequently washed in cold 1× Hanks balanced salt solution (catalogue no. 14185; Gibco) containing 0.015 M HEPES (catalogue no. 15630; Gibco) a total of six times to remove mucus and remaining fecal matter. To isolate lymphocytes, the tissue was added to 10 ml of prewarmed (37°C) 1× RPMI (Sigma catalogue no. R1145) containing 10% fetal bovine serum, 1% penicillin-streptomycin (catalogue no. 15240-062; Gibco), 0.015 M HEPES, and 0.010 M glutamine. Additionally, collagenase digestive units (furylacryloyl-leucine-glycyl-propyl-alanine) (Liberase, catalogue no. 05401127001; Roche) at 2.9 U/ml and 300 U of DNase (Roche catalogue no. 04716728001) were added to the medium. The tissue was then processed with a GentleMACS tissue disruptor according to a protocol provided by the manufacturer (Miltenyi).
Flow cytometry.
The analysis of surface marker expression in intestinal lymphocytes (C57BL/6; The Jackson Laboratory) has been described previously (64). In brief, a total of 4 × 106 intestinal cells were resuspended in 2 ml Dulbecco’s phosphate-buffered saline (PBS) without calcium and magnesium and stained with aqua live/dead cell discriminator (no. L34597; Invitrogen) in accordance with the manufacturer’s protocol. Cells were then rinsed and resuspended in 50 µl fluorescence-activated cell sorting (FACS) buffer (PBS containing 1% bovine serum albumin and 1 mM EDTA). Four microliters of the anti-CD16/32 blocking antibody (clone 93; eBioscience) was added to each sample. Cells were then stained for 20 min in the dark at 4°C with optimized concentrations of anti-CD3–phycoerythrin (PE) (clone 17A2; eBioscience), anti-B220–PE (clone RA3-6B2; eBioscience), anti-NK1.1–PE (eBioscience clone PK136), anti-Ly6C–Pacific Blue (clone HK1.4; eBioscience), anti-Ly6G–PerCP Cy5.5 (clone 1A8; BD Pharmingen), and anti-CD11b–allophycocyanin Cy7 (clone M1/70; BioLegend) antibodies. Cells were washed twice with FACS buffer and subsequently fixed in 4% paraformaldehyde for 1 h. Cells were then washed twice and resuspended in FACS buffer and analyzed with an LSR II flow cytometer (Becton Dickinson, San Jose, CA). The data were analyzed by using FlowJo software (TreeStar, Inc., Ashland, OR). Gates were set on singlets and then on live cells. Subsequent gates were based on fluorescence minus one and unstained controls.
Statistical analysis.
To determine the statistical significance of differences between treatment groups in the animal experiments, an unpaired Student t test was used. A P value of less than 0.05 was considered to be significant.
SUPPLEMENTAL MATERIAL
Body weights of mice after treatment with 1% DSS and/or streptomycin.
ACKNOWLEDGMENTS
We acknowledge support by Public Health Service grants AI088122 and AI107393 to A.J.B. C.A.L. was supported by Public Health Service grant AI060555. D.D.K. was supported by Public Health Service grant 8-T32-OD11147-25. We acknowledge NIH facilities infrastructure grant 1-C06-RR12088-01.
We have no conflicts of interest to declare.
Footnotes
Citation Spees AM, Wangdi T, Lopez CA, Kingsbury DD, Xavier MN, Winter SE, Tsolis RM, Bäumler AJ. 2013. Streptomycin-induced inflammation enhances Escherichia coli gut colonization through nitrate respiration. mBio 4(4):e00430-13. doi:10.1128/mBio.00430-13.
REFERENCES
- 1. Saito K. 1961. Studies on the habitation of pathogenic Escherichia coli in the intestinal tract of mice. I. Comparative experiments on the habitation of each type of resistant pathogenic Escherichia coli under an administration of streptomycin. Paediatr. Jpn. 65:385–393 [PubMed] [Google Scholar]
- 2. Saito K. 1961. Studies on the habitation of pathogenic Escherichia coli in the intestinal tract of mice. II. Experimental inoculation of type 055 Escherichia coli after long-term administration of streptomycin. Paediatr. Jpn. 65:394–399 [PubMed] [Google Scholar]
- 3. Myhal ML, Cohen PS, Laux DC. 1983. Altered colonizing ability for mouse large intestine of a surface mutant of a human faecal isolate of Escherichia coli. J. Gen. Microbiol. 129:1549–1558 [DOI] [PubMed] [Google Scholar]
- 4. Møller AK, Leatham MP, Conway T, Nuijten PJ, de Haan LA, Krogfelt KA, Cohen PS. 2003. An Escherichia coli MG1655 lipopolysaccharide deep-rough core mutant grows and survives in mouse cecal mucus but fails to colonize the mouse large intestine. Infect. Immun. 71:2142–2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wadolkowski EA, Laux DC, Cohen PS. 1988. Colonization of the streptomycin-treated mouse large intestine by a human fecal Escherichia coli strain: role of adhesion to mucosal receptors. Infect. Immun. 56:1036–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wadolkowski EA, Laux DC, Cohen PS. 1988. Colonization of the streptomycin-treated mouse large intestine by a human fecal Escherichia coli strain: role of growth in mucus. Infect. Immun. 56:1030–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. McCormick BA, Franklin DP, Laux DC, Cohen PS. 1989. Type 1 pili are not necessary for colonization of the streptomycin-treated mouse large intestine by type 1-piliated Escherichia coli F-18 and E. coli K-12. Infect. Immun. 57:3022–3029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cohen PS, Kjelleberg S, Laux DC, Conway PL. 1990. Escherichia coli F-18 makes a streptomycin-treated mouse large intestine colonization factor when grown in nutrient broth containing glucose. Infect. Immun. 58:1471–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. McCormick BA, Laux DC, Cohen PS. 1990. Neither motility nor chemotaxis plays a role in the ability of Escherichia coli F-18 to colonize the streptomycin-treated mouse large intestine. Infect. Immun. 58:2957–2961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Stojiljković I, Cobeljić M, Trgovcević Z, Salaj-Smic E. 1991. The ability of rifampin-resistant Escherichia coli to colonize the mouse intestine is enhanced by the presence of a plasmid-encoded aerobactin-iron(III) uptake system. FEMS Microbiol. Lett. 69:89–93 [DOI] [PubMed] [Google Scholar]
- 11. Stojiljkovic I, Cobeljic M, Hantke K. 1993. Escherichia coli K-12 ferrous iron uptake mutants are impaired in their ability to colonize the mouse intestine. FEMS Microbiol. Lett. 108:111–115 [DOI] [PubMed] [Google Scholar]
- 12. Sweeney NJ, Laux DC, Cohen PS. 1996. Escherichia coli F-18 and E. coli K-12 eda mutants do not colonize the streptomycin-treated mouse large intestine. Infect. Immun. 64:3504–3511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sweeney NJ, Klemm P, McCormick BA, Moller-Nielsen E, Utley M, Schembri MA, Laux DC, Cohen PS. 1996. The Escherichia coli K-12 gntP gene allows E. coli F-18 to occupy a distinct nutritional niche in the streptomycin-treated mouse large intestine. Infect. Immun. 64:3497–3503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Miranda RL, Conway T, Leatham MP, Chang DE, Norris WE, Allen JH, Stevenson SJ, Laux DC, Cohen PS. 2004. Glycolytic and gluconeogenic growth of Escherichia coli O157:H7 (EDL933) and E. coli K-12 (MG1655) in the mouse intestine. Infect. Immun. 72:1666–1676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jones SA, Chowdhury FZ, Fabich AJ, Anderson A, Schreiner DM, House AL, Autieri SM, Leatham MP, Lins JJ, Jorgensen M, Cohen PS, Conway T. 2007. Respiration of Escherichia coli in the mouse intestine. Infect. Immun. 75:4891–4899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fabich AJ, Jones SA, Chowdhury FZ, Cernosek A, Anderson A, Smalley D, McHargue JW, Hightower GA, Smith JT, Autieri SM, Leatham MP, Lins JJ, Allen RL, Laux DC, Cohen PS, Conway T. 2008. Comparison of carbon nutrition for pathogenic and commensal Escherichia coli strains in the mouse intestine. Infect. Immun. 76:1143–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jones SA, Gibson T, Maltby RC, Chowdhury FZ, Stewart V, Cohen PS, Conway T. 2011. Anaerobic respiration of Escherichia coli in the mouse intestine. Infect. Immun. 79:4218–4226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jones SA, Jorgensen M, Chowdhury FZ, Rodgers R, Hartline J, Leatham MP, Struve C, Krogfelt KA, Cohen PS, Conway T. 2008. Glycogen and maltose utilization by Escherichia coli O157:H7 in the mouse intestine. Infect. Immun. 76:2531–2540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Snider TA, Fabich AJ, Conway T, Clinkenbeard KD. 2009. E. coli O157:H7 catabolism of intestinal mucin-derived carbohydrates and colonization. Vet. Microbiol. 136:150–154. [DOI] [PubMed]
- 20. Maltby R, Leatham-Jensen MP, Gibson T, Cohen PS, Conway T. 2013. Nutritional basis for colonization resistance by human commensal Escherichia coli strains HS and Nissle 1917 against E. coli O157:H7 in the mouse intestine. PLoS One 8:e53957. 10.1371/journal.pone.0053957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lindgren SW, Melton AR, O’Brien AD. 1993. Virulence of enterohemorrhagic Escherichia coli O91:H21 clinical isolates in an orally infected mouse model. Infect. Immun. 61:3832–3842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sheoran AS, Chapman S, Singh P, Donohue-Rolfe A, Tzipori S. 2003. Stx2-specific human monoclonal antibodies protect mice against lethal infection with Escherichia coli expressing Stx2 variants. Infect. Immun. 71:3125–3130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shimizu K, Asahara T, Nomoto K, Tanaka R, Hamabata T, Ozawa A, Takeda Y. 2003. Development of a lethal Shiga toxin-producing Escherichia coli-infection mouse model using multiple mitomycin C treatment. Microb. Pathog. 35:1–9 [DOI] [PubMed] [Google Scholar]
- 24. Calderon Toledo C, Rogers TJ, Svensson M, Tati R, Fischer H, Svanborg C, Karpman D. 2008. Shiga toxin-mediated disease in MyD88-deficient mice infected with Escherichia coli O157:H7. Am. J. Pathol. 173:1428–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Roxas JL, Koutsouris A, Bellmeyer A, Tesfay S, Royan S, Falzari K, Harris A, Cheng H, Rhee KJ, Hecht G. 2010. Enterohemorrhagic E. coli alters murine intestinal epithelial tight junction protein expression and barrier function in a Shiga toxin independent manner. Lab. Invest. J. Tech. Methods Pathol. 90:1152–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang XH, He KW, Zhang SX, Lu WC, Zhao PD, Luan XT, Ye Q, Wen LB, Li B, Guo RL, Wang XM, Lu LX, Zhou JM, Yu ZY, Mao AH. 2011. Subcutaneous and intranasal immunization with Stx2B-Tir-Stx1B-Zot reduces colonization and shedding of Escherichia coli O157:H7 in mice. Vaccine 29:3923–3929 [DOI] [PubMed] [Google Scholar]
- 27. Bohnhoff M, Drake BL, Miller CP. 1954. Effect of streptomycin on susceptibility of intestinal tract to experimental salmonella infection. Proc. Soc. Exp. Biol. Med. 86:132–137 [DOI] [PubMed] [Google Scholar]
- 28. Bohnhoff M, Miller CP. 1962. Enhanced susceptibility to salmonella infection in streptomycin-treated mice. J. Infect. Dis. 111:117–127 [DOI] [PubMed] [Google Scholar]
- 29. Que JU, Hentges DJ. 1985. Effect of streptomycin administration on colonization resistance to Salmonella typhimurium in mice. Infect. Immun. 48:169–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Barthel M, Hapfelmeier S, Quintanilla-Martínez L, Kremer M, Rohde M, Hogardt M, Pfeffer K, Rüssmann H, Hardt WD. 2003. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect. Immun. 71:2839–2858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lawley TD, Walker AW. 2013. Intestinal colonization resistance. Immunology 138:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Stecher B, Robbiani R, Walker AW, Westendorf AM, Barthel M, Kremer M, Chaffron S, Macpherson AJ, Buer J, Parkhill J, Dougan G, von Mering C, Hardt WD. 2007. Salmonella enterica serovar Typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 5:2177–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Stecher B, Chaffron S, Kappeli R, Hapfelmeier S, Freedrich S, Weber TC, Kirundi J, Suar M, McCoy KD, von Mering C, Macpherson AJ, Hardt WD. 2010. Like will to like: abundances of closely related species can predict susceptibility to intestinal colonization by pathogenic and commensal bacteria. PLoS Pathog. 6:e1000711. 10.1371/journal.ppat.1000711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Brandl K, Sun L, Neppl C, Siggs OM, Le Gall SM, Tomisato W, Li X, Du X, Maennel DN, Blobel CP, Beutler B. 2010. MyD88 signaling in nonhematopoietic cells protects mice against induced colitis by regulating specific EGF receptor ligands. Proc. Natl. Acad. Sci. U. S. A. 107:19967–19972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rose S, Misharin A, Perlman H. 2012. A novel Ly6C/Ly6G-based strategy to analyze the mouse splenic myeloid compartment. Cytometry A J. Int. Soc. Anal. Cytol. 81:343–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Godinez I, Haneda T, Raffatellu M, George MD, Paixão TA, Rolán HG, Santos RL, Dandekar S, Tsolis RM, Bäumler AJ. 2008. T cells help to amplify inflammatory responses induced by Salmonella enterica serotype Typhimurium in the intestinal mucosa. Infect. Immun. 76:2008–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Raffatellu M, George MD, Akiyama Y, Hornsby MJ, Nuccio SP, Paixao TA, Butler BP, Chu H, Santos RL, Berger T, Mak TW, Tsolis RM, Bevins CL, Solnick JV, Dandekar S, Bäumler AJ. 2009. Lipocalin-2 resistance confers an advantage to Salmonella enterica serotype Typhimurium for growth and survival in the inflamed intestine. Cell Host Microbe 5:476–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Winter SE, Winter MG, Xavier MN, Thiennimitr P, Poon V, Keestra AM, Laughlin RC, Gomez G, Wu J, Lawhon SD, Popova IE, Parikh SJ, Adams LG, Tsolis RM, Stewart VJ, Bäumler AJ. 2013. Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science 339:708–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Blattner FR, Plunkett G, Bloch CA, Perna NT, Burland V, Riley M, Colladovides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis NW, Kirkpatrick HA, Goeden MA, Rose DJ, Mau B, Shao Y. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1543–1562 [DOI] [PubMed] [Google Scholar]
- 40. Winter SE, Lopez CA, Bäumler AJ. 2013. The dynamics of gut-associated microbial communities during inflammation. EMBO Rep. 14:319–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sekirov I, Tam NM, Jogova M, Robertson ML, Li Y, Lupp C, Finlay BB. 2008. Antibiotic-induced perturbations of the intestinal microbiota alter host susceptibility to enteric infection. Infect. Immun. 76:4726–4736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wlodarska M, Willing B, Keeney KM, Menendez A, Bergstrom KS, Gill N, Russell SL, Vallance BA, Finlay BB. 2011. Antibiotic treatment alters the colonic mucus layer and predisposes the host to exacerbated Citrobacter rodentium-induced colitis. Infect. Immun. 79:1536–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Salzman A, Denenberg AG, Ueta I, O’Connor M, Linn SC, Szabó C. 1996. Induction and activity of nitric oxide synthase in cultured human intestinal epithelial monolayers. Am. J. Physiol. 270:G565–G573 [DOI] [PubMed] [Google Scholar]
- 44. Palmer RM, Ashton DS, Moncada S. 1988. Vascular endothelial cells synthesize nitric oxide from l-arginine. Nature 333:664–666 [DOI] [PubMed] [Google Scholar]
- 45. De Groote MA, Granger D, Xu Y, Campbell G, Prince R, Fang FC. 1995. Genetic and redox determinants of nitric oxide cytotoxicity in a Salmonella typhimurium model. Proc. Natl. Acad. Sci. U. S. A. 92:6399–6403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Szabó C, Ischiropoulos H, Radi R. 2007. Peroxynitrite: biochemistry, pathophysiology and development of therapeutics. Nat. Rev. Drug Discov. 6:662–680 [DOI] [PubMed] [Google Scholar]
- 47. Lopez CA, Winter SE, Rivera-Chavez F, Xavier MN, Poon V, Nuccio SP, Tsolis RM, Bäumler AJ. 2012. Phage-mediated acquisition of a type III secreted effector protein boosts growth of salmonella by nitrate respiration. mBio 3:e00143-12. 10.1128/mBio.00143-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Winter SE, Thiennimitr P, Winter MG, Butler BP, Huseby DL, Crawford RW, Russell JM, Bevins CL, Adams LG, Tsolis RM, Roth JR, Bäumler AJ. 2010. Gut inflammation provides a respiratory electron acceptor for salmonella. Nature 467:426–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Thiennimitr P, Winter SE, Winter MG, Xavier MN, Tolstikov V, Huseby DL, Sterzenbach T, Tsolis RM, Roth JR, Bäumler AJ. 2011. Intestinal inflammation allows salmonella to use ethanolamine to compete with the microbiota. Proc. Natl. Acad. Sci. U. S. A. 108:17480–17485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ferreira RB, Gill N, Willing BP, Antunes LC, Russell SL, Croxen MA, Finlay BB. 2011. The intestinal microbiota plays a role in salmonella-induced colitis independent of pathogen colonization. PLoS One 6:e20338. 10.1371/journal.pone.0020338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Spiller R, Campbell E. 2006. Post-infectious irritable bowel syndrome. Curr. Opin. Gastroenterol. 22:13–17 [DOI] [PubMed] [Google Scholar]
- 52. Endt K, Stecher B, Chaffron S, Slack E, Tchitchek N, Benecke A, Van Maele L, Sirard JC, Mueller AJ, Heikenwalder M, Macpherson AJ, Strugnell R, von Mering C, Hardt WD. 2010. The microbiota mediates pathogen clearance from the gut lumen after non-typhoidal salmonella diarrhea. PLoS Pathog. 6:e1001097. 10.1371/journal.ppat.1001097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Atarashi K, Nishimura J, Shima T, Umesaki Y, Yamamoto M, Onoue M, Yagita H, Ishii N, Evans R, Honda K, Takeda K. 2008. ATP drives lamina propria T(H)17 cell differentiation. Nature 455:808–812 [DOI] [PubMed] [Google Scholar]
- 54. Satoh-Takayama N, Vosshenrich CA, Lesjean-Pottier S, Sawa S, Lochner M, Rattis F, Mention JJ, Thiam K, Cerf-Bensussan N, Mandelboim O, Eberl G, Di Santo JP. 2008. Microbial flora drives interleukin 22 production in intestinal NKp46+ cells that provide innate mucosal immune defense. Immunity 29:958–970 [DOI] [PubMed] [Google Scholar]
- 55. Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, Tanoue T, Imaoka A, Itoh K, Takeda K, Umesaki Y, Honda K, Littman DR. 2009. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139:485–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gaboriau-Routhiau V, Rakotobe S, Lécuyer E, Mulder I, Lan A, Bridonneau C, Rochet V, Pisi A, De Paepe M, Brandi G, Eberl G, Snel J, Kelly D, Cerf-Bensussan N. 2009. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity 31:677–689 [DOI] [PubMed] [Google Scholar]
- 57. Salzman NH, Hung K, Haribhai D, Chu H, Karlsson-Sjöberg J, Amir E, Teggatz P, Barman M, Hayward M, Eastwood D, Stoel M, Zhou Y, Sodergren E, Weinstock GM, Bevins CL, Williams CB, Bos NA. 2010. Enteric defensins are essential regulators of intestinal microbial ecology. Nat. Immunol. 11:76–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Stojiljkovic I, Bäumler AJ, Heffron F. 1995. Ethanolamine utilization in Salmonella typhimurium: nucleotide sequence, protein expression and mutational analysis of the cchA cchB eutE eutJ eutG eutH gene cluster. J. Bacteriol. 177:1357–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Park SR, Kim MK, Kim JO, Cho SJ, Cho YU, Yun HD. 2000. Cloning and sequencing of cel5Z gene from Erwinia chrysanthemi PY35. Mol. Cells 10:269–274 [PubMed] [Google Scholar]
- 60. Wang RF, Kushner SR. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195–199 [PubMed] [Google Scholar]
- 61. Cress BF, Linhardt RJ, Koffas MA. 2013. Draft genome sequence of Escherichia coli Strain Nissle 1917 (serovar O6:K5:H1). Genome Announc. 1:e0004713. 10.1128/genomeA.00047-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Price-Carter M, Tingey J, Bobik TA, Roth JR. 2001. The alternative electron acceptor tetrathionate supports B12-dependent anaerobic growth of Salmonella enterica serovar Typhimurium on ethanolamine or 1,2-propanediol. J. Bacteriol. 183:2463–2475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Stewart V, Parales J., Jr. 1988. Identification and expression of genes narL and narX of the nar (nitrate reductase) locus in Escherichia coli K-12. J. Bacteriol. 170:1589–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Keestra AM, Godinez I, Xavier MN, Winter MG, Winter SE, Tsolis RM, Bäumler AJ. 2011. Early, MyD88-dependent induction of interleukin-17A expression during Salmonella colitis. Infect. Immun. 79:3131–3140 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Body weights of mice after treatment with 1% DSS and/or streptomycin.